Abstract
Our previously published data link P-selectin-reactive chondroitin sulfate structures on the surface of breast cancer cells to metastatic behavior of cells. We have shown that a particular sulfation pattern mediated by the expression of carbohydrate (chondroitin 4) sulfotransferase-11 (CHST11) correlates with P-selectin binding and aggressiveness of human breast cancer cell lines. The present study was performed to evaluate the prognostic value of CHST11 expression and determine whether aberrant DNA methylation controls CHST11 expression in breast cancer. Publicly available datasets were used to examine the association of CHST11 expression to aggressiveness and progression of breast cancer. Methylation status was analyzed using bisulfite genomic sequencing. 5-aza-2′-deoxycytidine (5AzadC) was used for DNA demethylation. Reduced representation bisulfite sequencing was performed in the CpG island of CHST11 with a minimum coverage of 10. Quantitative real-time RT-PCR was employed to confirm the expression profile of CHST11 in breast cancer cell lines. Flow cytometry was also used to confirm the expression of the CHST11 product, chondroitin sulfate A (CS-A). The expression of CHST11 was significantly higher in basal-like and Her2-amplified cell lines compared to luminal cell lines. CHST11 was also highly expressed in cancer tissues compared to normal tissues and the expression levels were significantly associated with tumor progression. We observed very low levels of DNA methylation in a CpG island of CHST11 in basal-like cells but very high levels in the same region in luminal cells. Treatment of MCF7 cells, a luminal cell line with very low expression of CHST11, with 5AzadC increased the expression of CHST11 and its immediate product, CS-A, in a dose-dependent manner. These results suggest that CHST11 may play a direct role in progression of breast cancer and that its expression is controlled by DNA methylation. Therefore, in addition to CHST11 mRNA levels, the methylation status of this gene also has potential as a prognostic biomarker.
Keywords: breast cancer, chondroitin sulfate, CHST11, gene expression, DNA methylation, 5-aza-2′-deoxycytidine
Introduction
Increased expression and alteration in sulfation pattern of chondroitin sulfate glycosaminoglycans (CS-GAGs) are observed in various neoplastic tissues, including pancreatic, lung, ovarian and breast (1–3). Several studies, including ours, suggest involvement of particular CS chains in tumor progression and metastasis (2,4–7). The sulfation by site-specific chondroitin sulfotransferases controls the biological function of CS-GAGs (8–11). CHST11 is a key enzyme in the biosynthesis of chondroitin sulfates (CS) and its action on chondroitin chains can lead to the production of chondroitin sulfate A (CS-A), B (CS-B) and E (CS-E) (12,13). We have shown that the expression of CHST11 is upregulated in tumor tissues of breast cancer patients compared to their normal tissues and that the expression levels of the gene in breast cancer cell lines correlate with CS-A expression, P-selectin binding and aggressive phenotype (14). However, the clinical significance of CHST11 expression is yet to be established and more studies are needed to evaluate the expression of this gene in relation to cancer progression.
We have observed that the expression of CHST11 is low to none in MCF7 cells with considerably higher expression in MDA-MB-231 (14). The expression levels correlated with P-selectin reactivity and general aggressiveness. The expression of P-selectin ligands on tumor cells plays a role in distant metastasis by facilitating tumor cell extravasation (15–18). Because of its potential role in breast cancer metastasis, determining mechanisms controlling CHST11 expression is of great interest. Aberrant DNA methylation is a mechanism that controls gene expression and is involved in tumor initiation and progression (19–23). In a gene profiling study of head and neck cancer it was suggested that DNA methylation may affect CHST11 expression in laryngeal carcinoma (24). In another study on breast cancer cell lines using genome wide methylation profiling, the authors reported that CHST11 is hypermethylated in ER-positive and hypomethylated in ER-negative cell lines (25). Therefore, variation in the expression of CHST11 in breast cancer cells may be controlled by DNA methylation.
Our current data suggest that the expression levels of CHST11 can predict progression of breast cancer and more aggressive phenotypes. Our investigation of the methylation status of CHST11 in breast cancer cells clearly demonstrates an association between lack of expression of this gene and hypermethylation of a CpG island of its DNA. Treatment of MCF7 cells with 5AzadC led to a dose-dependent increase in the expression of the gene and its product CS-A. Given the role of CS and the significance of CHST11 expression in tumor growth and metastasis, these findings have significant implications for the development of novel prognostic strategies in breast cancer.
Materials and methods
Reagents
Anti-CS-A mAb 2H6 was from Associates of Cape Cod/Seikagaku America (Falmouth, MA, USA). Fluorescence-conjugated, anti-mouse IgM and 5-aza-2′-deoxycytidine (5AzadC) were from Sigma (St. Louis, MO, USA). Primers were from Integrated DNA Technologies (IDT, Coralville, IA, USA). Real-time PCR reagents were from Applied Biosystems (Foster City, CA, USA). TRIzol reagent was from Invitrogen (Carlsbad, CA, USA), FailSafe PCR PreMix Selection kits and Enzyme Mix were from Epicentre Biotechnologies (Madison, WI, USA). Alkaline phosphatase (rAPid) was from Roche (Nutley, NJ, USA).
Analysis of Oncomine cancer gene microarray database
Publicly available Oncomine cancer microarray database (Compendia Biosciences; Ann Arbor, MI, USA; www.oncomine.com) was used to examine the expression of CHST11 in cancer tissue and determine association of CHST11 expression with breast cancer outcomes. Richardson et al (GEO accession GSE3744) (26), Finak et al (GEO accession GSE9014) (27), The Cancer Genome Atlas (TCGA, http://tcga-data.nci.nih.gov/tcga/) invasive breast carcinoma gene expression data and Gluck et al (GEO accession GSE22358) (28) dataset were used to compare CHST11 expression levels between cancer and normal tissues. Hoeflich et al (GEO accession GSE12777) (29) dataset was used to compare expression levels of CHST11 among a panel of cell lines that represent luminal, Her-2-amplified and basal-like molecular subtypes of breast cancer. Schuetz et al dataset (GEO accession GSE3893) (30) was used to compare CHST11 expression levels between matched ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC). Log-transformed, median-centered and normalized expression values (31) were extracted, analyzed and graphed accordingly.
Cell lines and tissue specimen
Cell lines MCF7, MDA-MB-231, MDA-MB-468, T47-D, and ZR-75-1 were from ATCC (Manassas, VA, USA). MDA-MB-231, MDA-MB-468, T47-D, and ZR-75-1 cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies), 50 U/ml penicillin and 50 μg/ml streptomycin. MCF7 were grown as described before (14). Cells are checked every six months to be free from Mycoplasma contamination using the MycoAlert® Mycoplasma Detection kit (Lonza Rockland Inc., Rockland, ME, USA).
De-identified paraffin-embedded specimens from 5 female breast cancer patients were provided by the Department of Pathology of the University of Arkansas for Medical Sciences (UAMS). For this study, an active human tissue use protocol approved by the UAMS Institutional Review Board was used.
Total RNA isolation and quantitative real-time RT-PCR (real-time RT-qPCR)
RNA isolation and quantitative real-time was performed as described earlier (14). Briefly, total RNA was isolated from cultured cells using TRIzol reagent (Life Technologies, Grand Island, NY, USA) following the manufacturer’s instructions. The quantity and quality of the isolated RNA was determined using an Agilent 2100 Bioanalyzer (Palo Alto, CA, USA). Total RNA (1 μg) was reverse-transcribed using random-hexamer primers with TaqMan Reverse Transcription reagents (Applied Biosystems). Reverse-transcribed RNA was amplified with SYBR Green PCR Master Mix (Applied Biosystems) plus 0.3 μM of gene-specific upstream and downstream primers during 40 cycles on an Applied Biosystems 7900 HT Fast Real-time system. Data were analyzed by absolute and relative quantification. In absolute quantification, data were expressed in relation to 18S RNA, where the standard curves were generated using pooled RNA from the samples assayed. In relative quantification, the 2−ΔΔCT method was used to assess the target transcript in a treatment group to that of untreated control using expression of an internal control (reference gene) to normalize data (32). Expression of GAPDH was used as internal control. The primer sequences are shown in Table I.
Table I.
Primers used in this study.
| Primer | Sequence |
|---|---|
| 18S forward | 5′-TTCGAACGTCTGCCCTATCAA-3′ |
| 18S reverse | 5′-ATGGTAGGCACGGCGACTA-3′ |
| CHST11 forward | 5′-TCCCTTTGGTGTGGACATCT-3′ |
| CHST11 reverse | 5′-CACGTGTCTGTCACCTGGTC-3′ |
| GAPDH forward | 5′-ACAGTCAGCCGCATCTTCTT-3′ |
| GAPDH reverse | 5′-ACGACCAAATCCGTTGACTC-3′ |
| CHST11 bisulfite outside, forward | 5′-TTTGATTATTGTAGTTTTGGAGGAAAT-3′ |
| CHST11 bisulfite reverse | 5′-CCTTACAATTAAAAAAACAAATTATTACTA-3′ |
| CHST11 bisulfite nested, forward | 5′-TTTTTGTGGTTTAGGTAAAGTTTTA-3′ |
18S, 18S ribosomal RNA; CHST11, carbohydrate (chondroitin 4) sulfotransferase 11; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
5AzadC treatment
5AzadC was dissolved in ice-cold phosphate-buffered saline, filter sterilized at 4°C and the resulting solution used to treat the MCF7 cell line. For dose–response experiments, cells were harvested 5 days after the initial treatment. Cell growth medium was refreshed every other day.
Extraction and bisulfite modification of DNA
DNA from cells was extracted as described before (33). DNA from the paraffin-embedded tissues of breast cancer patients was extracted using Ex-Wax DNA extraction kit (Chemicon International, Temecula, CA, USA), following the manufacturer’s instructions with the additional steps of extracting recovered DNA with phenol (Amresco, Solon, OH, USA) and then 1-bromo-3-chloropropane (Molecular Research Center Inc., Cincinnati, OH, USA). DNA was bisulfite modified with an Epitect kit (Qiagen, Valencia, CA, USA) using 300 ng of DNA per reaction. PCR was performed using a FailSafe PCR PreMix Selection kit and FailSafe Enzyme Mix. Each 25-μl PCR reaction included 1.0 μM of each primer, 2.5 U of the FailSafe Enzyme Mix and 12.5 μl of the FailSafe PCR PreMixes A or C. Bisulfite-modified genomic DNA was amplified by semi-nested PCR using two sets of primers for part of intron 1 that is within a CpG island spanning exon 1 of the CHST11 gene (Genbank NM_000012 and exon 1 located with NM_018413). The same amplification profile was used for both reactions of the semi-nested PCR: 1 cycle at 80°C for 1 min, 1 cycle at 94°C for 1 min; 1 cycle at 95°C for 1 min, 54°C for 1 min, 72°C for 1 min; 1 cycle at 95°C for 1 min, 53°C for 1 min, 72°C for 1 min; 1 cycle at 95°C for 1 min, 52°C for 1 min, 72°C for 1 min; 1 cycle at 95°C for 1 min, 51°C for 1 min, 72°C for 1 min; 36 cycles at 95°C for 1 min, 50°C for 1 min, 72°C for 1 min; 72°C for 5 min and cooling to 4°C.
A forward, outside primer and reverse primer were used for CHST11 for the first reaction (Table I). A second, semi-nested, PCR was then performed on 1 μl of the amplificate (in a 25-μl PCR reaction) using a forward nested primer and the reverse primer from the first reaction (346-bp PCR product, Table I). The primers were designed using MethPrimer web software (34) (http://www.urogene.org/methprimer/). The CpG island was defined using CpG Island Searcher set on the default criteria for defining CpG islands (35) (http://cpgislands.usc.edu/cpg.aspx).
Bisulfite genomic sequencing (BGS) and methylation level quantification
BGS was performed as described before (33) with the following minor modifications. We used rAPid alkaline phosphatase and PCR products were sequenced using the nested forward (upstream) CHST11 primer. We also analyzed DNA methylation in the region spanning >1.5 kb of the CHST11 CpG island with reduced representation bisulfite sequencing [RRBS (36,37)] data generated on breast cancer cell lines for the Illumina Idea Challenge (http://www.illumina.com/landing/idea) (25). RRBS selects DNA fragments ≤220 bp in the vicinity of MspI recognition sites (C.CGG) (36). We only considered CpG dinucleotides with coverage >10x. The percentage of methylation was inferred from the number of methylated reads divided by the sum of methylated and unmethylated reads. All reads were mapped to the hg18 release of the human genome (http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg18). Second generation sequencing DNA methylation results were plotted in Matlab (http://www.mathworks.com).
Statistical analysis
One-way ANOVA with Tukey’s post hoc procedure test was used to compare gene expression between subtypes of cell lines. For comparison of gene expression data generated by real-time PCR, the raw amount for each mRNA was log transformed and normalized to the control mRNA (18S) amount, and analyzed via one-way ANOVA with Tukey’s post hoc procedure. The Mann-Whitney U test was performed to compare gene expression between DCIS and IDC. For 5AzadC induced fold change in gene expression, the mRNA levels of the non-zero doses for each transcript and experimental replication were normalized to that of the zero-dose control, transformed to their base-10 logarithms and analyzed for trend with dose via one-way ANOVA. Statistical analyses were performed using Excel (Microsoft, Seattle, WA, USA) or GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA, USA). All P-values were 2-sided.
Results
CHST11 is overexpressed in basal-like and HER2-amplified cell lines and the elevated expression correlates with tumor progression
We have shown that the expression of CHST11 is elevated in tumor cells compared to normal cells of breast cancer patient tissue specimens (14). We used Oncomine database to confirm our original data. The data from several studies were analyzed to compare CHST11 expression in breast carcinoma versus normal breast tissue (Table II). The comparison showed average increases of 2–3.6-fold in CHST11 expression in breast cancer compared to normal tissue.
Table II.
CHST11 differential transcript expression in human breast carcinomas extracted from multiple studies in the Oncomine microarray database.
| Studya | Comparison (specimen number in each group) | Average fold increase | P-valueb |
|---|---|---|---|
| TCGAc | Invasive breast carcinoma (n=76) vs normal (n=61) | 2.6 | 2.51E-24 |
| Finak et al (27) | Invasive breast carcinoma (n=53) vs normal (n=6) | 3.6 | 9.43E-20 |
| Richardson et al (26) | Ductal breast carcinoma (n=40) vs normal (n=7) | 2.2 | 2.00E-4d |
| Gluck et al (28) | Invasive breast carcinoma (n=154) vs. normal (n=4) | 2 | 0.02 |
Studies are addressed by consortium name or first author’s last name.
Two-tailed P-values are shown.
TCGA, The Cancer Genome Atlas - Invasive Breast Carcinoma Gene Expression Data (http://tcga-data.nci.nih.gov/tcga/).
Student’s t-test was performed with Welch’s correction for unequal variances.
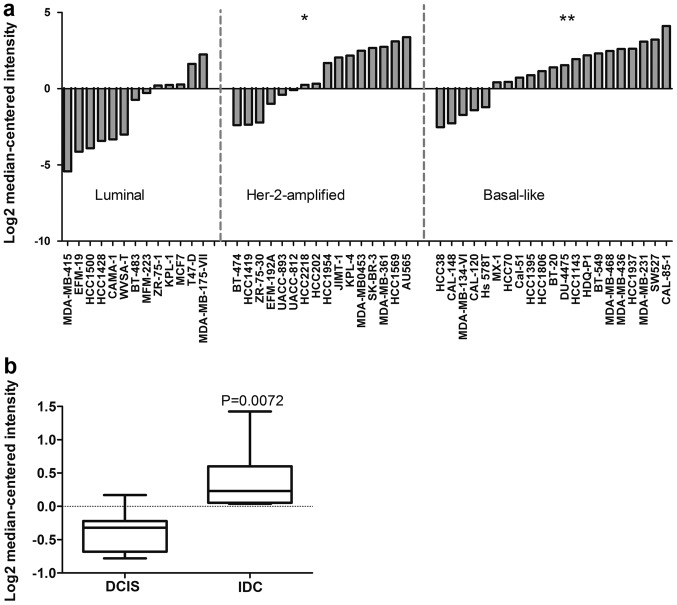
Our previously published data analyzing CHST11 expression in a set of commonly used human breast cancer cell lines suggest that CHST11 expression is low in luminal and high in basal-like cell lines (14). The data relate CHST11 expression to basal-like cancer cells. To further investigate such an association, we analyzed an Oncomine dataset that screened 50 breast cancer cell lines representative of the molecular subtypes luminal, Her2-amplified, and basal-like (29). We observed that the expression of CHST11 in basal-like cell lines was significantly higher than in luminal cell types (Fig. 1a). CHST11 was also significantly overexpressed in Her-2-amplified cell lines. Considering cells with above average expression as positive, 75% of basal-like and 56% Her-2-amplified cell lines were positive; while only 15% of luminal cell lines were positive for CHST11 expression.
Figure 1.
CHST11 expression is associated with basal-like molecular subtype and progression of breast cancer. All data are from the Oncomine database. (a) CHST11 gene expression was evaluated in a panel of breast cancer cell lines (13 luminal, 16 Her-2-amplified and 21 basal-like cell lines) (29). The log-transformed normalized expression values were analyzed by one-way ANOVA and Tukey’s post hoc comparisons. *,**Statistically significant compared to the luminal subgroup at P<0.05 and P<0.01, respectively. (b) CHST11 expression is associated with tumor progression (30). Tumor specimens (7 DCIS and 7 IDC) were isolated by LCM from each individual specimen and matched. Specimens then were compared for the expression of CHST11. The P-value shows two-sided comparison using Mann-Whitney U test.
To investigate the association of CHST11 overexpression with tumor progression, we interrogated another dataset in the Oncomine database, comparing the expression of CHST11 between ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC) (30). DCIS and IDC samples were isolated by laser capture microdissection (LCM) and matched for each individual specimen (30). We found a significant increase in the expression of CHST11 in IDC compared to DCIS samples (Fig. 1b). Notably, both groups displayed a similar distribution pattern for ER status (5 ER-positive specimens in each group) and HER2-amplification pattern (3 HER2-amplified specimens in each group) with histological grades 2 and 3. The data link the elevated expression of CHST11 to aggressive subtypes and progression of the disease.
A CpG island in the CHST11 gene sequence is hypermethylated in MCF7 and hypomethylated in MDA-MB-231 cells
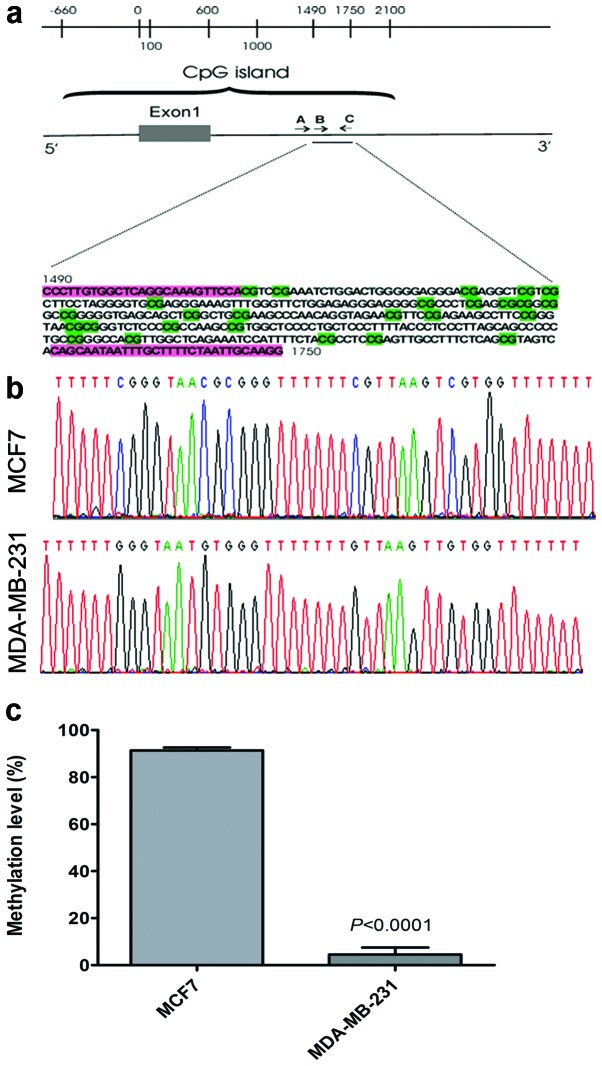
Our current and previously published data suggest that CHST11 expression plays a role in tumor progression and metastasis. Therefore understanding the mechanisms controlling CHST11 gene expression will help formulate strategies for development of biomarkers and drug targets. We demonstrated previously that CHST11 and CS-A expression was high in highly metastatic MDA-MB-231 cells, while it was significantly lower in MCF7 cells as assayed by qRT-PCR and flow cytometry (14). To examine whether DNA methylation controls variation and regulation of CHST11 in breast cancer cell lines, we examined DNA methylation of the CHST11 sequence in a section of its CpG island covering part of its promoter, its first exon, and a portion of its first intron (Fig. 2a). Using BGS and the Mquant algorithm (33) we determined methylation levels of ten CpG sites for MCF7 and MDA-MB-231 cells (Fig. 2b). This revealed very high methylation levels (91%) averaged >10 CpGs in the CHST11 sequence in MCF7 cells, and very low levels (5%) over the same 10 CpGs in MDA-MB-231 cells (Fig. 2c). These observations suggest that low expression of CHST11 in MCF7 cells is due to DNA hypermethylation.
Figure 2.
The genomic configuration of the CHST11 gene and methylation of CPG island in MCF7 and MDA-MB-231 cells. (a) Exon 1 from NCBI NM_018413.5 is shown (0 to +616) as is the CpG island (between −660 and +2,100). The horizontal arrows indicate the approximate relative positions of bisulfite genomic sequencing primers for semi-nested PCR. Primer A is the outside forward primer (Table I), primer B is the nested forward primer (position +1,490 and Table I), and primer C is the reverse primer (position +1,750 and Table I). The genomic sequence (pre-bisulfite) between the B and C primers is shown below the map and B and C primer locations are highlighted in pink. The scale is in base pairs. (b) Sections of bisulfite genomic sequencing electropherograms showing part of the CHST11 CpG island. Top, DNA of MCF-7 cells. In this top electropherogram most CpG sites have prominent cytosine (C) peaks because methylated cytosines are not changed to thymines (Ts) in the bisulfite reaction. Bottom, DNA of MDA-MB-231 cells. In this section CpG sites appear as TpG sites and are in the same sequence positions as the top CpGs. In this bottom electropherogram most CpG sites have prominent T peaks because unmethylated Cs are changed to uracils in the bisulfite reaction. (c) Quantification of methylation levels of the CpG island in MCF7 and MDA-MB-231 cells. Methylation levels were averaged >10 CpGs and 18 preparations for MCF7 and 16 preparations for MDA-MB-231 cells. Means and SEM are shown. Data were Arcsin transformed and subjected to the Mann-Whitney U test to compare means.
Treatment of MCF7 cells with 5AzadC increases the expression of CHST11 and CS-A
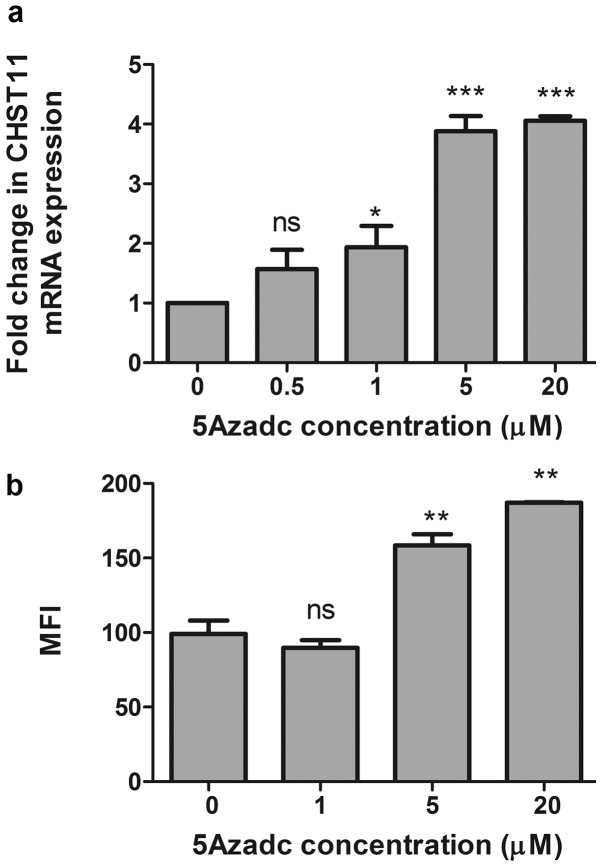
To further validate our data and confirm that low expression of CHST11 in the MCF7 cell line is due to hypermethylation status of the CHST11 CpG island, MCF7 cells were treated with 5AzadC and CHST11 gene expression was determined. Upon 5AzadC treatment, we observed a significant increase in the expression of CHST11 mRNA (Fig. 3a) that paralleled surface expression of CS-A as assayed by flow cytometry using anti-CS-A mAb 2H6 (Fig. 3b). Together, the data suggest that low expression of CHST11 in the MCF7 cells is due to promoter hypermethylation, and that DNA hypomethylation in the more aggressive mesenchymal-like MDA-MB-231 cell line is permissive for overt CHST11 expression.
Figure 3.
5AzadC treatment increases CHST11 gene expression in MCF7 cells. Gene expression was assayed by real-time RT-PCR (a) and flow cytometry (b). MCF7 cells were grown in the presence of various concentrations of 5AzadC for 5 days and then cells were harvested for RNA purification or staining with anti-CS-A mAb 2H6. (a) CHST11 mRNA levels are shown relative to mRNA levels of cells grown in medium only. GAPDH was used as the house-keeping gene to normalize mRNA-based expression data using the ΔΔCT method. Data were log transformed and subjected to one-way ANOVA with post hoc Tukey’s analysis. (b) Cells were harvested and then stained with anti-CS-A mAb 2H6. Binding was analyzed by flow cytometry. Mean fluorescent intensities (MFIs) of two independent experiments were log transformed and analyzed by ANOVA and post hoc comparison. *, **, ***Significantly different compared to control at P≤0.05, P≤0.01 and P≤0.001, respectively. NS, not significant as compared to control (0 μM 5AzadC).
Methylation of the CHST11 CpG island in other cell lines with low expression of CHST11
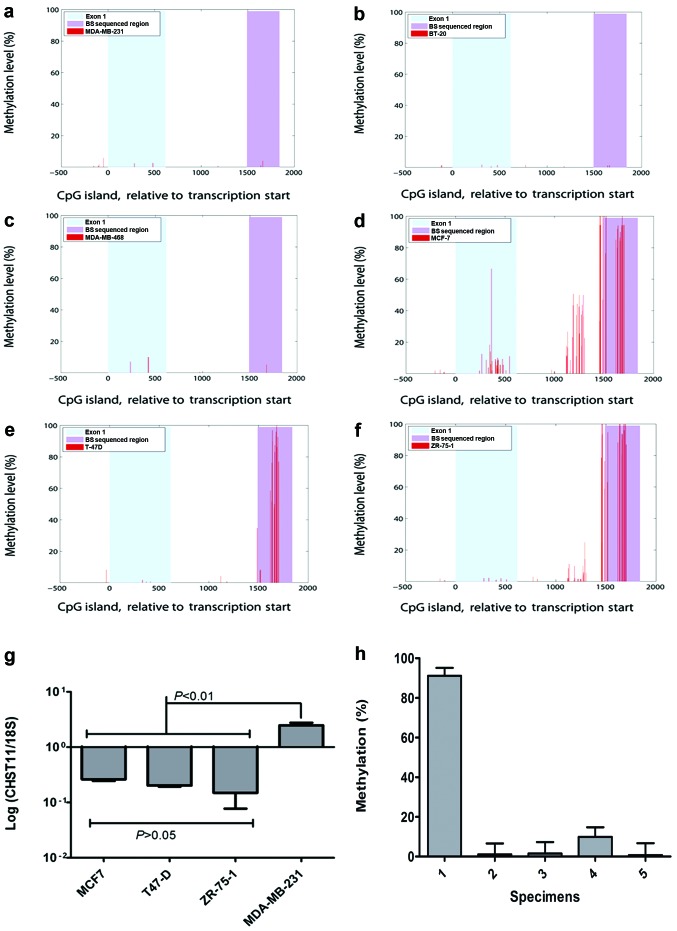
To confirm a role for methylation of the CHST11 CpG island in the expression control of CHST11, we further analyzed second generation DNA methylation sequencing data on breast cancer cell lines (Illumina Idea Challenge and ref. 25). This method provided information on DNA methylation at single nucleotide resolution across 162 out of 186 CpGs in the ~2.5 kb long CHST11 CpG island. Methylation analysis of Illumina Idea Challenge data of these cell lines indicates a close relationship between expression levels and methylation status. A very hypomethylated state was observed for MDA-MB-231 (Fig. 4a). The methylation status of two other triple-negative cell lines MDA-MB-468 and BT-20, was similar to MDA-MB-231 (Fig 4b and c). Similar to MCF7 cells, the CpG island of the CHST11 gene in the T47-D and ZR-75-1 cell lines, was hypermethylated (Fig. 4d–f). The expression of CHST11 in T47-D and ZR-75-1 cell lines was further examined by qRT-PCR (Fig. 4g) and flow cytometry (Table III), and compared with CHST11 expression levels in MCF7 and MDA-MB-231. Consistent with the methylation data, the expression of the CHST11 gene and CS-A in both of these cell lines was significantly lower than in MDA-MB-231 and comparable with that of MCF7 cells. MDA-MB-468 displayed a very hypomethylated state but the expression of CHST11 in these cells was moderate and less than what we observed for MDA-MB-231 and MDA-MET (4,14). The expression of CHST11 in tumor tissue may also be controlled by DNA methylation similar to that observed in cell lines.
Figure 4.
CHST11 expression and methylation status of its CpG island in basal-like and luminal-like breast cancer cell lines and the patient tissues. Single nucleotide resolution of DNA methylation in the CHST11 CpG island of MDA-MB-231 (a), BT-20 (b), MDA-MB-468 (c), MCF-7 (d), T-47D (e), and ZR-75-1 (f) was determined by RRBS (36). The red peaks denote Cs covered with ≥10 reads. Methylation level percentage was calculated from the number of Cs after bisulfite treatment divided by the sum of Cs (methylated sites) and Ts (unmethylated sites). There are a total of 186 CpG dinucleotides in the CpG island, of which ≥75% were accessible to RRBS with a minimum coverage of 10 (average coverage ≥40). The x-axis shows CHST11 in the coordinates relative to the transcription start site. The first exon and the genomic region we sequenced after the bisulfite treatment (BS sequenced region) are shown as indicated in the plots. (g) The expression of CHST11 was measured in MCF7, T-47D, ZR-75-1 and MDA-MB-231 cells by qRT-PCR and normalized using 18S. Data were analyzed by one-way ANOVA and post hoc analysis using data from three independent experiments. Means, standard deviations, statistically significant differences and P-values are shown. (h) Methylation levels of the CpG island in five specimens from breast cancer patients diagnosed with invasive ductal carcinoma. Methylation analysis was performed and methylation levels were quantified as described in legend to Fig. 2. Percentages were averaged >20 CpGs and ≥2 replications. Means and SEM are shown. Data were Arcsin transformed and subjected to one-way ANOVA with Tukey’s post hoc analysis to compare means. Specimen 1 is ER-positive and the other four specimens are triple negative. Specimen 1 displayed a significantly higher methylation level than the other specimens (P≤0.001).
Table III.
Summary of CHST11 gene expression and methylation status in human breast cancer cell lines tested.
| Cell line | ER status | Molecular subtype | CHST11 (qRT-PCR) | CS-A expression (2H6 mAb binding by flow cytometry) | DNA methylation status |
|---|---|---|---|---|---|
| MCF7 | Positive | Luminal | Low | Lowb | Hypermethylated |
| T47-D | Positive | Luminal | Low | Lowc | Hypermethylated |
| ZR-75-1 | Positive | Luminal | Low | Lowc | Hypermethylated |
| BT-474 | Positive | Her-2 amplified | Lowa | ND | Hypermethylated |
| MDA-MB-468 | Negative | Basal-like | Intermediate | Intermediate to highb | Hypomethylated |
| BT-20 | Negative | Basal-like | Intermediatea | ND | Hypomethylated |
| MDA-MB-231 | Negative | Basal-like | High | Highb | Hypomethylated |
| MDA-MET | Negative | Basal-like | High | Highb | Hypomethylated |
Digital gene expression with primers used to perform qRT-PCR was applied to estimate gene expression. ND, not determined.
Flow cytometry data were published before (14).
Flow cytometry was performed and no detectable binding of anti-CS-A mAb 2H6 was observed.
To determine whether the same CpG island can be methylated differently in actual breast cancer, we examined the methylation status of the CpG island of CHST11 gene in 5 de-identified clinical breast cancer specimens. We observed that in 4 triple negative (TN, i.e., negative for ER, Her2/neu and progesterone receptor) specimens the CpG island was highly hypomethylated, very similar to what was observed for ER-negative basal-like cell lines (Fig. 4h). The CpG island was hypermethylated in one ER-positive specimen tested. Therefore, methylation status of CHST11 CpG might be a useful marker to differentiate between luminal and basal-like breast cancer subtypes, however, a larger sample size is required to confirm this.
Discussion
We have previously shown that removal or blocking of CS on the surface of breast cancer cells inhibits metastasis (5,14). We have demonstrated that the expression levels of CHST11 correlate with the expression of P-selectin-reactive surface CS and aggressiveness of human breast cancer cells (14). Here, we confirm that the CHST11 gene is overexpressed in cancer tissues compared to normal breast tissues and its expression correlates with more aggressive basal-like and HER2-amplified phenotypes in cell lines. The expression analysis of CHST11 in DCIS and IDC specimens suggest a progression specific role for this gene that needs to be further validated. An association between CHST11 expression and distant metastasis in breast cancer patients has been reported (6). The data suggest that the expression of CHST11 correlates well with properties associated with prognosis and progression and should be investigated as a potential biomarker.
Our data suggest that the expression of CHST11 is controlled by DNA methylation. Aberrant DNA methylation is involved in tumor initiation and progression (22,23,38). Hypermethylation of tumor suppressor genes is a common mechanism of gene silencing observed in cancer, and similarly, DNA hypomethylation can contribute to overexpression of tumor-promoting genes. DNA hypomethylation is associated with advanced stages, metastatic phenotypes, and drug-resistant variants of breast cancer (39–41). We find that induced hypomethylation by 5Azadc led to CHST11 overexpression and increased levels of the CHST11’s immediate cell surface product CS-A. Based upon our results, it seems that a combination of hypomethylation and high expression occurs in basal-like cancer cells. Hypermethylation of CHST11 (and very low to no expression) clearly differentiates the least aggressive, luminal cells with epithelial morphology from the more aggressive cells we tested. We observed a hypomethylated CpG island in a limited number of triple negative specimens from patients. Triple-negative cancers are usually classified as basal-like while ER-positive cells are considered luminal. The data suggest that methylation analysis of this CpG island of CHST11 might be potentially used as a surrogate for detection of expression levels of this gene in clinical samples. However, more experiments are needed to evaluate the correlation of the expression of this gene with its DNA methylation levels among subtypes of breast cancer.
Our study illustrates an example of a gene, where methylation is lower and expression is higher as the cancer phenotype becomes more aggressive. In contrast, the main trend is widespread gene hypermethylation as cancers go from less aggressive to more aggressive phenotypes (42,43). Our data are consistent with some other models of metastatic and/or more aggressive cancers exemplifying tumor promoting genes like urokinase plasminogen activator (uPA), PAX3 and Ezrin that are less methylated and/or more highly expressed in the more aggressive cancer forms (40,44,45). This less frequent pattern of gene methylation between cancers with low and high aggressiveness suggests that the expression of these genes is necessary for the more aggressive phenotypes (because their change in gene specific methylation is counter to the trend of gene hypermethylation in cancer).
It needs to be pointed out that in less aggressive basal B cells that are epithelial-like (e.g., MDA-MB-468), hypomethylation is accompanied by intermediate expression of CHST11, suggesting involvement of other mechanisms in controlling the expression levels of this gene such as TGFβ1 (46,47). As we suggested before (14), in order for CHST11 activity to express the proper receptor it might need to be co-expressed with a specific proteoglycan. Future studies on well-characterized clinical specimens are needed to determine a role for the CHST11 gene alone or in combination with other genes in patient outcome. More studies are also needed to determine whether DNA methylation status can replace gene expression for prognosis.
The recognition that silencing of tumor suppressor genes through promoter hypermethylation plays a significant role in tumorigenesis (48,49) has led to the clinical use of hypomethylating agents including 5AzadC (50). However, the expression of several pro-tumor genes is induced by DNA hypomethylation (40,45). The expression of such genes, including CHST11, may be activated by the clinical use of hypomethylating agents and this may promote more aggressive forms of breast cancer. In this regard, our data, in agreement with others (51), suggest that therapeutic use of such demethylating agents may promote tumor progression and metastasis. Histone deacetylase inhibitors can also hypomethylate genes and change their expression levels in breast cancer cell lines (52) and their combination with metabolic therapies may modify their action (53). Therefore, additional studies are needed to determine if some agents or mechanisms of hypomethylation are more or less likely to promote tumor metastasis.
Our findings suggest that the expression of CHST11 correlates with aggressive phenotypes and progression of DCIS to IDC. We found out that the expression of CHST11 is modulated by DNA methylation. Therefore, DNA methylation plays an important role in the remodeling of CS in breast tumors. Our data strongly suggest that the expression of CHST11 and its role in defining metastatic potential of tumor cells should be seriously considered when demethylating agents are used for medical treatment of breast cancer patients. Moreover, the methylation and expression of the CHST11 gene have potential to be developed into novel prognostic biomarkers.
Acknowledgements
This study was supported in part by a pilot project grant to B.M.K. from the UAMS Translational Research Institute through the Clinical and Translational Science Award 1UL1RR029884 from the National Center for Research Resources.
References
- 1.Masuda H, Ozeki T, Takazono I, Tanaka Y. Composition of glycosaminoglycans in human pancreatic cancer. Biochem Med Metab Biol. 1989;41:193–200. doi: 10.1016/0885-4505(89)90026-1. [DOI] [PubMed] [Google Scholar]
- 2.Li F, Ten Dam GB, Murugan S, Yamada S, Hashiguchi T, Mizumoto S, Oguri K, Okayama M, van Kuppevelt TH, Sugahara K. Involvement of highly sulfated chondroitin sulfate in the metastasis of the Lewis lung carcinoma cells. J Biol Chem. 2008;283:34294–34304. doi: 10.1074/jbc.M806015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svensson KJ, Christianson HC, Kucharzewska P, Fagerstrom V, Lundstedt L, Borgquist S, Jirstrom K, Belting M. Chondroitin sulfate expression predicts poor outcome in breast cancer. Int J Oncol. 2011;39:1421–1428. doi: 10.3892/ijo.2011.1164. [DOI] [PubMed] [Google Scholar]
- 4.Iida J, Wilhelmson KL, Ng J, Lee P, Morrison C, Tam E, Overall CM, McCarthy JB. Cell surface chondroitin sulfate glycosaminoglycan in melanoma: role in the activation of pro-MMP-2 (pro-gelatinase A) Biochem J. 2007;403:553–563. doi: 10.1042/BJ20061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monzavi-Karbassi B, Stanley JS, Hennings L, Jousheghany F, Artaud C, Shaaf S, Kieber-Emmons T. Chondroitin sulfate glycosaminoglycans as major P-selectin ligands on metastatic breast cancer cell lines. Int J Cancer. 2007;120:1179–1191. doi: 10.1002/ijc.22424. [DOI] [PubMed] [Google Scholar]
- 6.Martinez P, Vergoten G, Colomb F, Bobowski M, Steenackers A, Carpentier M, Allain F, Delannoy P, Julien S. Over-sulfated glycosaminoglycans are alternative selectin ligands: insights into molecular interactions and possible role in breast cancer metastasis. Clin Exp Metastasis. 2013;30:919–931. doi: 10.1007/s10585-013-9592-7. [DOI] [PubMed] [Google Scholar]
- 7.Vallen MJ, van der Steen SC, van Tilborg AA, Massuger LF, van Kuppevelt TH. Sulfated sugars in the extracellular matrix orchestrate ovarian cancer development: ‘when sweet turns sour’. Gynecol Oncol. 2014 Aug 23; doi: 10.1016/j.ygyno.2014.08.023. (Epub ahead of print). pii: S0090-8258(14)01276-1. [DOI] [PubMed] [Google Scholar]
- 8.Sugahara K, Mikami T, Uyama T, Mizuguchi S, Nomura K, Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr Opin Struct Biol. 2003;13:612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Kusche-Gullberg M, Kjellen L. Sulfotransferases in glycosaminoglycan biosynthesis. Curr Opin Struct Biol. 2003;13:605–611. doi: 10.1016/j.sbi.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, Pavao MS, Tzanakakis GN, Karamanos NK. Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J. 2012;279:1177–1197. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 11.Mikami T, Kitagawa H. Biosynthesis and function of chondroitin sulfate. Biochim Biophys Acta. 18302013:4719–4733. doi: 10.1016/j.bbagen.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Mikami T, Mizumoto S, Kago N, Kitagawa H, Sugahara K. Specificities of three distinct human chondroitin/dermatan N-acetylgalactosamine 4-O-sulfotransferases demonstrated using partially desulfated dermatan sulfate as an acceptor: implication of differential roles in dermatan sulfate biosynthesis. J Biol Chem. 2003;278:36115–36127. doi: 10.1074/jbc.M306044200. [DOI] [PubMed] [Google Scholar]
- 13.Uyama T, Ishida M, Izumikawa T, Trybala E, Tufaro F, Bergstrom T, Sugahara K, Kitagawa H. Chondroitin 4-O-sulfotransferase-1 regulates E disaccharide expression of chondroitin sulfate required for herpes simplex virus infectivity. J Biol Chem. 2006;281:38668–38674. doi: 10.1074/jbc.M609320200. [DOI] [PubMed] [Google Scholar]
- 14.Cooney CA, Jousheghany F, Yao-Borengasser A, Phanavanh B, Gomes T, Kieber-Emmons AM, Siegel ER, Suva LJ, Ferrone S, Kieber-Emmons T, Monzavi-Karbassi B. Chondroitin sulfates play a major role in breast cancer metastasis: a role for CSPG4 and CHST11 gene expression in forming surface P-selectin ligands in aggressive breast cancer cells. Breast Cancer Res. 2011;13:R58. doi: 10.1186/bcr2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YJ, Borsig L, Varki NM, Varki A. P-selectin deficiency attenuates tumor growth and metastasis. Proc Natl Acad Sci USA. 1998;95:9325–9330. doi: 10.1073/pnas.95.16.9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borsig L, Wong R, Feramisco J, Nadeau DR, Varki NM, Varki A. Heparin and cancer revisited: mechanistic connections involving platelets, P-selectin, carcinoma mucins, and tumor metastasis. Proc Natl Acad Sci USA. 2001;98:3352–3357. doi: 10.1073/pnas.061615598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia J, Callewaert N, Borsig L. P-selectin mediates metastatic progression through binding to sulfatides on tumor cells. Glycobiology. 2007;17:185–196. doi: 10.1093/glycob/cwl059. [DOI] [PubMed] [Google Scholar]
- 18.Stubke K, Wicklein D, Herich L, Schumacher U, Nehmann N. Selectin-deficiency reduces the number of spontaneous metastases in a xenograft model of human breast cancer. Cancer Lett. 2012;321:89–99. doi: 10.1016/j.canlet.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, Ehrlich M. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer. 1998;77:833–838. doi: 10.1002/(SICI)1097-0215(19980911)77:6<833::AID-IJC6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 20.Szyf M, Pakneshan P, Rabbani SA. DNA methylation and breast cancer. Biochem Pharmacol. 2004;68:1187–1197. doi: 10.1016/j.bcp.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 21.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 22.Bardowell SA, Parker J, Fan C, Crandell J, Perou CM, Swift-Scanlan T. Differential methylation relative to breast cancer subtype and matched normal tissue reveals distinct patterns. Breast Cancer Res Treat. 2013;142:365–380. doi: 10.1007/s10549-013-2738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weisenberger DJ. Characterizing DNA methylation alterations from The Cancer Genome Atlas. J Clin Invest. 2014;124:17–23. doi: 10.1172/JCI69740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalathas D, Triantaphyllidou IE, Mastronikolis NS, Goumas PD, Papadas TA, Tsiropoulos G, Vynios DH. The chondroitin/dermatan sulfate synthesizing and modifying enzymes in laryngeal cancer: expressional and epigenetic studies. Head Neck Oncol. 2010;2:27. doi: 10.1186/1758-3284-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Z, Asmann YW, Kalari KR, Bot B, Eckel-Passow JE, Baker TR, Carr JM, Khrebtukova I, Luo S, Zhang L, Schroth GP, Perez EA, Thompson EA. Integrated analysis of gene expression, CpG island methylation, and gene copy number in breast cancer cells by deep sequencing. PLoS One. 2011;6:e17490. doi: 10.1371/journal.pone.0017490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 28.Gluck S, Ross JS, Royce M, McKenna EF, Jr, Perou CM, Avisar E, Wu L. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat. 2012;132:781–791. doi: 10.1007/s10549-011-1412-7. [DOI] [PubMed] [Google Scholar]
- 29.Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, Januario T, Savage H, Punnoose E, Truong T, Zhou W, Berry L, Murray L, Amler L, Belvin M, Friedman LS, Lackner MR. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 30.Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, Fehm T, Solomayer E, Riess O, Wallwiener D, Kurek R, Neubauer HJ. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66:5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Leakey TI, Zielinski J, Siegfried RN, Siegel ER, Fan CY, Cooney CA. A simple algorithm for quantifying DNA methylation levels on multiple independent CpG sites in bisulfite genomic sequencing electropherograms. Nucleic Acids Res. 2008;36:e64. doi: 10.1093/nar/gkn210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 35.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- 36.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48:226–232. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 39.Soares J, Pinto AE, Cunha CV, Andre S, Barao I, Sousa JM, Cravo M. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–118. doi: 10.1002/(SICI)1097-0142(19990101)85:1<112::AID-CNCR16>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 40.Pakneshan P, Szyf M, Farias-Eisner R, Rabbani SA. Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J Biol Chem. 2004;279:31735–31744. doi: 10.1074/jbc.M401669200. [DOI] [PubMed] [Google Scholar]
- 41.Chekhun VF, Kulik GI, Yurchenko OV, Tryndyak VP, Todor IN, Luniv LS, Tregubova NA, Pryzimirska TV, Montgomery B, Rusetskaya NV, Pogribny IP. Role of DNA hypomethylation in the development of the resistance to doxorubicin in human MCF-7 breast adenocarcinoma cells. Cancer Lett. 2006;231:87–93. doi: 10.1016/j.canlet.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 42.Andrews J, Kennette W, Pilon J, Hodgson A, Tuck AB, Chambers AF, Rodenhiser DI. Multi-platform whole-genome microarray analyses refine the epigenetic signature of breast cancer metastasis with gene expression and copy number. PLoS One. 2010;5:e8665. doi: 10.1371/journal.pone.0008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff EM, Chihara Y, Pan F, Weisenberger DJ, Siegmund KD, Sugano K, Kawashima K, Laird PW, Jones PA, Liang G. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70:8169–8178. doi: 10.1158/0008-5472.CAN-10-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurmasheva RT, Peterson CA, Parham DM, Chen B, McDonald RE, Cooney CA. Upstream CpG island methylation of the PAX3 gene in human rhabdomyosarcomas. Pediatr Blood Cancer. 2005;44:328–337. doi: 10.1002/pbc.20285. [DOI] [PubMed] [Google Scholar]
- 45.Yu Y, Zeng P, Xiong J, Liu Z, Berger SL, Merlino G. Epigenetic drugs can stimulate metastasis through enhanced expression of the pro-metastatic Ezrin gene. PLoS One. 2010;5:e12710. doi: 10.1371/journal.pone.0012710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kluppel M, Vallis KA, Wrana JL. A high-throughput induction gene trap approach defines C4ST as a target of BMP signaling. Mech Dev. 2002;118:77–89. doi: 10.1016/S0925-4773(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 47.Willis CM, Wrana JL, Kluppel M. Identification and characterization of TGFbeta-dependent and -independent cis-regulatory modules in the C4ST-1/CHST11 locus. Genet Mol Res. 2009;8:1331–1343. doi: 10.4238/vol8-4gmr673. [DOI] [PubMed] [Google Scholar]
- 48.MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–11337. doi: 10.1074/jbc.270.19.11327. [DOI] [PubMed] [Google Scholar]
- 49.Ramchandani S, MacLeod AR, Pinard M, von Hofe E, Szyf M. Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proc Natl Acad Sci USA. 1997;94:684–689. doi: 10.1073/pnas.94.2.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenaux P. Inhibitors of DNA methylation: beyond myelodysplastic syndromes. Nat Clin Pract Oncol. 2005;2(Suppl 1):S36–S44. doi: 10.1038/ncponc0351. [DOI] [PubMed] [Google Scholar]
- 51.Ateeq B, Unterberger A, Szyf M, Rabbani SA. Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia. 2008;10:266–278. doi: 10.1593/neo.07947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooney CA. Drugs and supplements that may slow aging of the epigenome. Drug Discov Today Ther Strateg. 2010;7:57–84. doi: 10.1016/j.ddstr.2011.03.001. [DOI] [Google Scholar]