The content is available as a PDF (228.2 KB).
Figure.
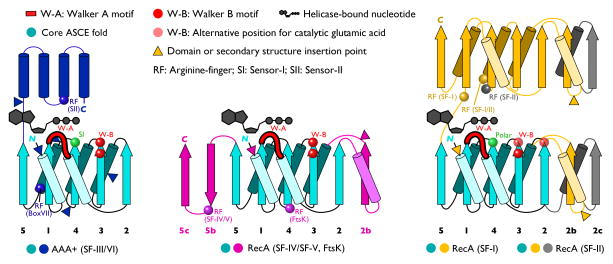
Topology diagrams of helicase and translocase superfamily members. The central RecA and AAA+ folds are built from an ancestral αβα ASCE domain (turquoise). The relative position of bound nucleotide is shown, and certain nucleoside-triphosphate binding motifs are highlighted (sensor I; sensor II; arginine-finger; Walker-A and Walker B motifs). The catalytic glutamic acid of the Walker-B motif can occupy alternative positions in some subfamilies (pale red spheres). The AAA+ sensor-I amino acid is generally a polar residue that is also found in other ASCE proteins. Triangles represent common insertion points for domains or secondary structural elements that are specific to select helicase and translocase groups (e.g. domains IB and IIB of SF-I helicases, or the pre-Sensor I β-hairpin of AAA+ proteins). The diverse placement of arginine finger elements arises from distinct tertiary and quaternary arrangements of domains or subunits in different subfamilies.
Table.
Helicase and translocase proteins constitute a broad family of nucleic acid-dependent molecular motors that consume nucleoside triphosphates (typically ATP) as fuel for directed movement. Translocases track along a DNA or RNA substrate; helicases further separate paired nucleic acid strands. The action of these motors is required for a host of essential cellular transactions, including DNA replication, recombination, and repair, the regulation of gene transcription, mRNA maturation and export, translation, and chromosome partitioning and packaging. All helicases and translocases are predicated on one of two catalytic NTP-binding domains: the RecA fold and the AAA+ fold (see Figure). These two folds are themselves predicated on an ancestral αβα domain termed an ASCE fold, which is distantly related to classic P-loop NTPase folds found in adenylate kinase and G-proteins. Within the AAA+ family of helicases and translocases, there are multiple subgroups (clades) defined by distinct insertions to the core AAA+ fold. For example, AAA+ helicases and translocases belong to the following clades: Superfamily-III (SF-III), Pre-Sensor II (PS-II) clade, HCLR (HslU, Clp, Lon, RuvB) clade, and Helix-2 (H2) insert clade. Helicases of the SF-I and SF-II groups (e.g., PcrA, eIF4A, RecQ) unwind nucleic acids or move along nucleic acid strands as monomers, but may also participate in higher-order oligomeric complexes. Helicases belonging to SF-III through SF-VII groups (e.g., DnaB, SV40 large T-antigen, Rho, MCMs) act predominantly as hexamers (although heptamers, dodecamers, tetradecamers, and even helical filaments have been observed). Many viral packaging motors belong to a diverging branch of the HerA/FtsK family of bacterial translocases and are pentamers. Although SF-IV helicases may work as 3′-5′ single-stranded DNA unwindases (or 3′-5′ RNA packaging motors), double-stranded (ds) DNA translocation activity has been reported for proteins such as DnaB and T7 gp4. The preferred substrate for SF-VI helicases (MCMs) is still under debate. Helicases of the Tip48/49 family have been described as RuvB-like and related to “classic” AAA+ ATPases such as NSF, Cdc48 (p97), and FtsH. However, the core ATP-binding subunit does not contain a β-hairpin insertion prior to the sensor I motif as is the case for RuvB, but instead has an extended β-sheet capped by an OB-fold. The AAA+ fold in the Tip48/49 eukaryotic helicases also contains a β-hairpin in place of the α-helix that connects the penultimate and final β-strands of the core fold; this insertion is unique among AAA+ ATPases. Functionally, the primary activity of RuvB is to translocate dsDNA across the RuvA tetramer to promote branch migration. Tip48/49 helicases, in contrast, display direct DNA unwinding activity. The Tip48/49 family therefore may constitute a distinct clade of AAA+ proteins. The HerA/FtsK group of bacterial translocases is an offshoot of the RecA family (particularly SF-IV helicases). These enzymes are also structural homologs of other translocases such as PilT and VirB of bacterial type IV secretory systems.
| Superfamily/Class | Protein Fold | Oligomeric state | Polarity | Function | Example Members | |
| Helicases | Superfamily I (SF-I) | RecA (tandem pair) | Monomer (dimer/multimer) | 3′-5′ (SF-IA), 5′-3′ (SF-IB) | DNA unwinding, repair and degradation | Bacterial PcrA, Rep, UvrD, RecBCD, Dda; eukaryotic Rrm3, Pif1, Dna2 |
| Superfamily II (SF-II) | RecA (tandem pair) | Monomer | 3′-5′ (SF-IIA), 5′-3′ (SF-IIB), some dsDNA translocases | RNA melting, RNA-binding protein displacement; DNA or RNA unwinding; chromatin remodeling; DNA/RNA translocation; melting and migration of Holliday junctions or branched-structures | DExD/H-box proteins (eukaryotic eIF4A, Prp2, Ski2, Vasa, Dpbs; NS3 of hepatitis C); Snf2/SWI proteins (eukaryotic Snf2, ISWI, Rad54, archaeal Hel308); bacterial RecQ, RecG, UvrB | |
| Superfamily III (SF-III) | AAA+ | Hexamer (dodecamer?) | 3′-5′ | DNA unwinding/replication | Papilloma virus E1, simian virus 40 Large T-antigen, adeno-associated virus Rep40 | |
| Superfamily IV (SF-IV) | RecA | Hexamer (other states?) | 5′-3′ (dsDNA?) | DNA unwinding/replication; ssRNA packaging | Bacterial DnaB; phage T7 gp4, T4 gp41, SPP1 G40P; pRSF1010 RepA; phage Φ 12 P4 | |
| Superfamily V (SF-V) | RecA | Hexamer | 5′-3′ | RNA translocation, RNA/DNA heteroduplex unwinding; transcription termination | Bacterial Rho | |
| Superfamily VI (SF-VI) | AAA+ (PS- II clade) | Hexamer (other states?) | 3′-5′ (dsDNA?) | DNA unwinding/replication | Eukaryotic/archaeal MCMs | |
| Superfamily VII? (SF-VII | AAA+ (new clade?) | Hexamer | 5′-3′ | Chromatin remodeling | Eukaryotic Tip48/49, Reptin/pontin | |
| Translocases | HerA/FtsK | RecA-like | Hexamer (pentamer) | dsDNA or ssDNA | Chromosome partitioning/conjugation; certain viral packaging motors | Bacterial FtsK, SpoIIIE; plasmid TrwB, TraD; podovirus Φ 29 gp16, caudovirus and herpesvirus terminase proteins |
| RuvB | AAA+ (HCLR clade) | Hexamer (dodecamer with RuvA protein) | dsDNA | Branch migration | Bacterial RuvB | |
| McrB | AAA+ (H2− insert clade) | Heptamer | dsDNA | Type IV restriction enzymes | Bacterial McrB |
Abbreviations
AAA+, ATPases associated with various cellular activities; ASCE, additional strand conserved E; dsDNA, double-stranded DNA; MCMs, minichromosomal maintenance proteins; NSF, N-ethyl maleimide sensitive factor; OB-fold, oligonucleotide/oligosaccharide binding fold; PS-II, pre-sensor II; ssRNA, single-stranded RNA.
Acknowledgments
The author’s work in this area is supported by the NIGMS (GM071747).
Recent reviews
- 1.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 2.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 3.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008 doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 5.Gallant P. Control of transcription by Pontin and Reptin. Trends Cell Biol. 2007;17:187–92. doi: 10.1016/j.tcb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Burroughs AM, Iyer LM, Aravind L. Comparative Genomics and Evolutionary Trajectories of Viral ATP Dependent DNA-Packaging Systems. Gene and Protein Evolution. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 7.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 8.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–7. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackintosh SG, Raney KD. DNA unwinding and protein displacement by superfamily 1 and superfamily 2 helicases. Nucleic Acids Res. 2006;34:4154–9. doi: 10.1093/nar/gkl501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–8. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]