Abstract
Symbiosis between intestinal microbiota and the host animal plays an important role in the homeostasis of host physiology. Since the first production of germ-free rodents in 1945, it has become increasingly clear that the intestinal immune system and the biochemical characteristics of epithelial cells differ greatly between conventional and germ-free rodents. However, questions remain about the types of microbes involved and the precise mechanism by which these microbes affect the host physiology. Here, we review experiments designed to answer these questions with the use of gnotobiotic mice. We have determined suitable biochemical and immunological markers for monitoring microbial effects in these mice. Using these markers, we have found clear differences in epithelial cell glycolipid biosynthesis and intraepithelial lymphocyte dynamics between germ-free and conventional mice. Furthermore, we have identified a key microbe that activates the mucosal immune system in the small intestine. This indigenous bacteria, called segmented filamentous bacteria, is a key symbiont in the host-microbiota interplay, including Th17 cell-inducing activity.
Keywords: segmented filamentous bacteria, intraepithelial lymphocyte, IgA, Th17 cell, fucosyltransferase, gnotobiotic mouse
1. Introduction
The human gastrointestinal tract harbors more than five hundred phylotypes of microbes based on 16S rRNA gene sequences, and approximately a hundred trillion individual microbes, which is more than the number of host cells.1,2) The contribution of this great number of gut microbes to the metabolism and physiology of the whole body cannot be ignored. Furthermore, the external organs of the body are also in close association with indigenous microbes.3) The development of methods for the cultivation of strict anaerobes in bacterial ecosystems, and the classification of these microbes, as pioneered by Mitsuoka et al.,4) have paved the way for the study of intestinal microbiota. Further advances in cultivation-independent methods for microbial analysis, including the targeting of not-yet-cultivable bacteria, have been made possible with the introduction of molecular methods based on 16S rRNA gene sequence.5,6) Moreover, it is now possible to conduct a comprehensive analysis of the microbiota by directly reading the DNA sequence of the microbes with the use of next-generation DNA sequencing. As a result, a limited number of enterotypes are postulated to be present in each individual irrespective of geographic location or sex.7)
Intestinal microbes profoundly affect the host physiology, and reciprocally, intestinal physiological conditions affect the composition and metabolism of the microbes, with two-way communication. It is assumed that the composition of the intestinal microbiota of healthy individuals is balanced with the host physiology, shaping symbiosis between host and microbes. In diseased conditions or under antibiotic usage, a different profile of microbiota to that found in healthy conditions is observed, which is associated with the occurrence and progression of diseases.8) Accordingly, if it were possible to replace the microbiota of patients with that of a healthy person, a recovery from the disease might be expected. Several clinical trials of patients suffering from Clostridium difficile infection,9,10) inflammatory bowel disease (IBD),11,12) or type II diabetes13) have been performed, and promising results have been obtained. These clinical studies strongly impact on the concept of the normal microbiota not only clinically but also microbiologically.
In animal models, many comparative studies have been performed between germ-free (GF) animals and specific pathogen–free (SPF) or conventional (CV) animals based on the concept of normal microbiota. GF rats and mice were first produced in 1945 and 1954, respectively, by Reynier and his associates of the University of Notre Dame in the United States14) and were thereafter utilized in biomedical studies. In Japan, GF guinea pigs were produced by Miyakawa in 1954, and GF rats and mice were widely utilized in the latter half of the 1960s.15) Although it was concerned whether the intestinal microbiota is indispensable for the survival of animals before appearance of GF animal, the life span of a GF rat is rather longer than that of a CV rat, partly due to the absence of infectious incidents.16)
The gastrointestinal tract, in particular the epithelial cells, is the first line of defense against the external pathogens, and is greatly influenced by environmental factors, such as microbial products. Intestinal microbes affect the proliferation and differentiation of the cryptal epithelial cells by changing the production of mucus17) or antimicrobial proteins18) by goblet cells, the production and release of biologically active peptides by enteroendocrine cells,19) or the epithelial cell kinetics.20) Intestinal metabolism by microbes deconjugates the bile acids, promoting their absorption by the ileum,21) or converts the primary bile acids into secondary ones, which are suggested to be associated with promotion of carcinogenesis.22) Short-chain fatty acids, which are major products of intestinal microbes, contribute to the energy acquisition in the colon by the host23) and are well known to affect the physiology of the gastrointestinal tract.24) Liver and brain functions are also suggested to be modified by the presence of the intestinal microbiota.25)
Currently, colonization by more than several tens of intestinal microbes isolated from indigenous microbiota is thought to be necessary to elevate the colonization resistance and IgA response in gnotobiotic mice26,27) to the levels of CV ones because normalization of the cecal size and other physiological and histological characteristics of GF animals by association with one or a few kinds of bacterial species has not been successful. The kinds of microbes responsible for the development of mucosal immune system and how the intestinal microbes affect the host physiology remains unclear. However, the use of GF animals or gnotobiotes associated with particular microbes including not-yet-cultivable bacteria is expected to resolve these questions. Because the epithelial layer is located close to the immune cells, which react to challenges from the external environment, my colleagues and I have attempted to answer these questions by examining the biochemical differences between the intestinal epithelial cells of GF and conventionalized (Cvd) mice associated with fecal microbes derived from CV mice. This review will describe differences in the intestinal glycolipids and the intestinal immune system including IELs, IgA, and Th17 cells between GF and CV mice and key intestinal microbes responsible for the development of these immunological activities.
2. Molecular markers for monitoring the effects of microbiota on the host
To evaluate the effects of microbiota on the development of the intestine, we first explored the molecular components of the epithelial cells of the small intestine because the genealogy and cell kinetics of these cells is well known compared with those of the large intestine. As the first step, GF mice and conventionalized (Cvd) mice (i.e., GF mice that were orally inoculated with a suspension of fecal microbiota from conventionally reared mice and then kept in a conventional animal room) were injected with radiolabeled precursors for the synthesis of the glycoprotein and glycolipid components of the epithelial cell surface. In this study, we used L-[6-3H]-fucose, known to be incorporated into glycoconjugates almost without conversion into other sugar nucleotides, and also D-[1-14C]-glucosamine and L-[4,5-3H]-leucine to efficiently label the carbohydrate and protein portion of the glycoproteins, respectively. The microvillar membranes (MVMs) of the small intestinal epithelial cells were then partially purified according to the methods of Fujita.28) Incorporation of radiolabeled compounds into the protein fraction of the MVMs was greater in Cvd mice than in GF mice after intraperitoneal injection with L-[6-3H]-fucose or D-[1-14C]-glucosamine, whereas L-[4,5-3H]-leucine incorporation into this fraction was almost equal in both groups.29) A similar difference between Cvd and GF mice was found for radiolabeled compounds incorporated into individual protein molecules in the MVM fraction, such as sucrase-isomaltase complex and maltase protein, which were isolated by immunoprecipitation with a polyclonal rabbit antibody against each molecule.29)
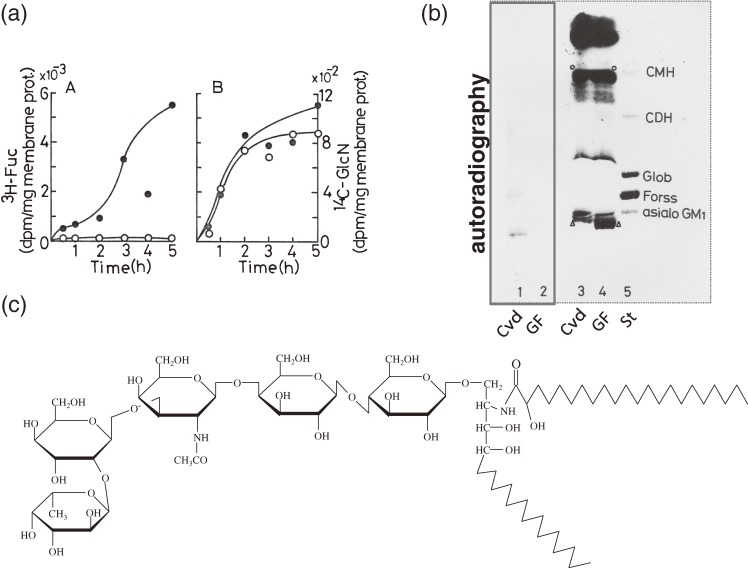
In contrast to 14C-glucosamine, 3H-fucose was scarcely incorporated into the lipid fraction of MVMs isolated from GF mice and extensively incorporated into the equivalent fraction from Cvd mice30) (Fig. 1a). Autoradiography of the thin layer chromatography (TLC) plate on which the MVM lipid fraction of Cvd mice was separated showed a radioactive band present just beneath the band corresponding to the main glycosphingolipid of mouse intestinal epithelial cells, asialo ganglioside GM1 (GA1) (Fig. 1b). These results demonstrate that the attachment of fucose moieties to lipid fraction of the MVMs of epithelial cells is drastically influenced by the association with microbiota. The chemical structure of this glycolipid was found to be α(1-2) fucosyl GA1 glycosphingolipid (FGA1) (Fig. 1c), of which the long chain base and fatty acids were C-18 phytosphingosine and 2-hydroxy fatty acids, respectively.31) Interestingly the chemical structure of the ceramide of this glycosphingolipid was different from that of glycosphingolipids isolated from other origins except kidney tissue.32) There also exists a clear difference in the sugar portion of sphingoglycolipid molecules between epithelial cells and non-epithelial tissue.33,34)
Figure 1.
Microbiota induces fucosylation of epithelial glycosphingolipid.30,31) (a) Time-course of L-[6-3H]-fucose or D-[1-14C]-glucosamine incorporation into the microvillus membrane lipid fractions of small intestinal epithelial cells in GF (○) and Cvd mice (●). (b) TLC analysis (lanes 3, 4) and autoradiography (lanes 1, 2) of the lipid fraction of both groups. St represents the glycolipid standards, ceramide monohexoside (CMH), ceramide dihexoside (CDH), Globoside (Glob), Forssman glycolipid (Forss) and asialo GM1. The open arrowheads and the open circles show the bands of the glycolipids. (c) The chemical structure of the glycolipid labeled with 3H-fucose. Panels a, b are reproduced with permission from ref. 30.
3. Activity of α(1-2) asialo GM1 fucosyltransferase as a sensitive marker for microbiota-induced development of the small intestine
We next investigated the age-dependency of FGA1 synthesis during the microbial association process by examining α(1-2) asialo GM1 fucosyltransferase (FT) activity35) in mice that were conventionalized at different ages by treatment with adult mouse feces. FT activity was determined by measuring incorporation of GDP-14C-fucose in the presence of intestinal GA1 glycolipid and Triton X-100. When GF mice were conventionalized at more than four weeks of age, FT activity was abruptly induced within a week of the microbial association. By TLC analysis of glycolipids, we confirmed conversion from GA1 into FGA1, coincident with FT activity, during the course of the conventionalization. In the case of GF mice that were conventionalized a week after birth, FT was induced in an overshoot fashion during the weaning period. In CV mice raised under conventional microbial conditions, FT increased only around the weaning period, and then returned to low levels.36) These data strongly suggest that the key microbes for FT induction expand in number around the weaning stage and are absent in the intestine before weaning.
Next, we tried to develop mice with simpler microbiota compositions than those found in CV mice. To this end, several kinds of gnotobiotic mice were prepared by treating GF mice with the following isolated strains: Escherichia coli, Proteus sp., Lactobacillus acidophilus, Lactobacillus fermentum, Lactobacillus murini, Streptococcus faecalis, Bacteroides sp., and Bifidobacterium sp. None of the monocolonized or multi-species colonized gnotobiotic mice induced substantial levels of FT activity in the small intestinal epithelial cells.37)
Spore-forming microbes that have a characteristic morphology and adhere tightly to epithelial cells have been detected historically in the small intestine of a wide range of animals, fish, and insects.38) These bacteria are idiomatically called “segmented filamentous bacteria (SFB)”. Their successful cultivation has not been reported to date. We found that monocolonization by SFB induced FT activity in the small intestine and the development of the mucosal immune system.39) Subsequently, monoassociation of Bacteroides thetaiotaomicron, a non-spore forming microbe of human origin, with GF mice was also reported to induce α(1-2) fucosylransferase.40)
4. FT induction and its physiology
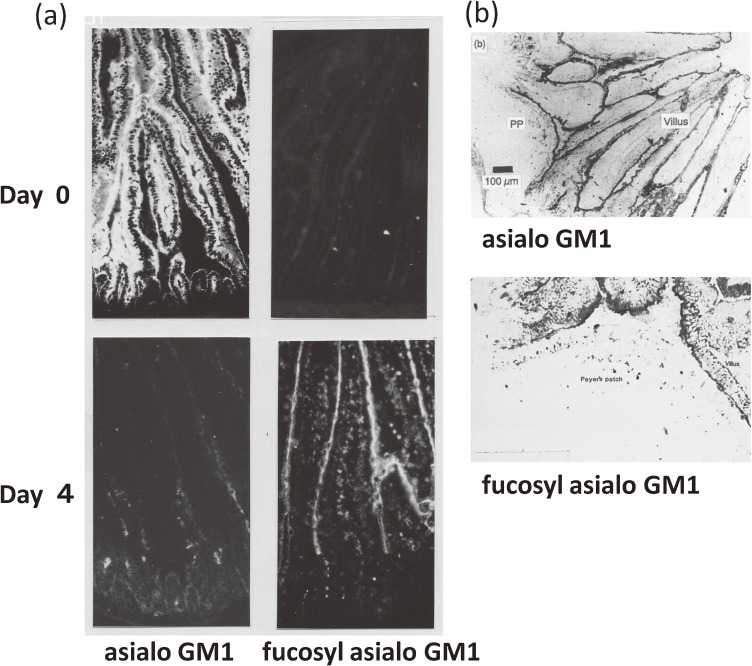
FGA1 expression and FT activity were investigated in post-mitotic development of the intestinal epithelial cells. Cryostat sections were prepared from the small intestine in the course of conventionalization of GF mice and then stained with polyclonal rabbit antibodies against GA1 or FGA1. FGA1 was much more strongly expressed in epithelial cells from the villus portion of the small intestine than in those from the crypt portion or Peyer’s patch (PP) covering follicle-associated epithelial cells, whereas GA1 was strongly expressed in the epithelial cells both from villus and crypt portions and in the follicle-associated epithelial cells in PPs41,42) (Fig. 2), and FGA1 was maximally expressed several days after conventionalization, coincident with the TLC profile mentioned above.35) In epithelial cell preparations isolated longitudinally along the villus-crypt axis, as expected from immunohistochemical stainings, the FT activity was stronger in the tip to mid villus portion compared with the crypt portion.36,42)
Figure 2.
FGA1 glycolipid expression in the villus portion of the villus-crypt axis42) but not in PP-covered epithelial cells. (a) The thin sections of the small intestine of Cvd mice on day 0 or day 4 after microbial inoculation were immunostained with rabbit anti-GA1 and anti-FGA1 antibodies followed by FITC-conjugated secondary antibody. (b) Thin sections of the PPs of Cvd mice at day 4 were immunostained with anti-GA1 or anti-FGA1 glycolipid antibody followed by peroxidase-labeled secondary antibody. Antibody binding was visualized by using the chromogen AEC (3-amino-9-ethylcarbazole). Panel a is reproduced with permission from ref. 42.
In the two differentiation process of the small intestine (i.e., post-partum and post-mitotic differentiation along the crypt-villus axis) of CV or Cvd mice, we observed that FT activity correlated with the degree of differentiation of the epithelial cells.36,42) However, FT activity was never observed in the small intestine in the absence of microbiota, except under the unusual conditions described below. Therefore, we consider that FT is another type of differentiation marker, different from the intestinal digestive enzyme such as sucrase-isomaltase complex expressed in the villus differentiated epithelial cells just in the weaning stage in both CV and GF mice.36)
In colonic tissue, FGA1 was clearly expressed in CV and Cvd mice, and even in GF mice, whereas GA1 was present at much lower levels. FT activity was present to some extent in GF mice and was elevated by conventionalization (unpublished data). We conclude that FT activity in the colon is regulated in a complex manner, either dependently or independently of the microbiota. These findings suggest that some factors required for FT activity are present in GF mice, even in the absence of microbiota.
Under GF conditions, we observed FT induction around the wounded intestinal tissue resulting from a needle puncture, or in the small intestinal epithelial cells of mice intraperitoneally injected with cycloheximide or emetine (i.e., protein synthesis inhibitors).36) Furthermore, proteinaceous substances extracted from the intestinal tissue were able to induce FT activity following intravenous injection into GF mice.36) Due to the dependency of FGA1 synthesis on MyD8843) in Cvd or CV mice but not the Severe Combined Immunodeficiency (SCID) mutation (unpublished data), innate immunity rather than acquired immunity is suggested to be involved in this event. Therefore, it is possible that substances such as cytokines derived from non-T and non-B cells such as innate lymphoid cells (ILCs) are involved in FT induction. Although intravenous administration of interferon α/β or interferon γ into GF mice did not effectively induce FT (unpublished data), many cytokines are now considered to be candidates for the role of α(1-2) fucosyltransferase inducer (Goto, Y. et al., submitted for publication). Interestingly, cycloheximide treatment and administration of the proteinaceous substances acted synergistically to induce FT,36) as observed in “super-induction” of a protein or its mRNA in cultured cells, which is previously so called because of marked enhancement of production in the presence of cycloheximide.44,45)
Analysis of the glycolipids in the intestinal epithelial cells of Fut2 gene knockout mice strongly indicates that either FT is the product of the Fut2 gene or its expression is under the control of this gene, consistent with the absence of Fut2 transcript in knockout mice.46) In humans, it was recently suggested that a nonsecretor genotype (i.e., a mutation in the FUT2 gene) is closely associated with susceptibility to IBDs such as Crohn’s disease47) or viral infection.48) Moreover, a non-secretor or low secretor genotype is reported to be associated with severe outcome in premature babies,49) while a secretor genotype is highly associated with colonization by Bifidobacterium.50) Intestinal bacteria that induce FUT2 expression, if present in humans, have the potential to stimulate colonization by Bifidobacterium and to play a protective role against viral infection.
5. Differences in dynamics and activity of intraepithelial lymphocytes between GF and Cvd mice
Because of the difficulties in discovering a specific microbe with a physiological impact on FT-mediated activation of intestinal mucosa, another evaluation system for the microbial effects was required. We considered that intraepithelial lymphocytes (IELs) might be suitable for this purpose because they reside within the intestinal epithelia and function as a first line of defense against infections. T cell receptor (TCR)αβ or TCRγδ T cells bearing CD8αα directly migrate from the thymus into the interepithelial space as “natural IELs” with self-reactivity, whereas TCRαβ T cells bearing CD8αβ or CD4 are recruited as “induced IELs” with cytotoxic activity after activation by antigens in the lamina propria, PP, or mesenteric lymph node.51) The recent observation that identical T cell clones are located in the lamina propria and interepithelial cell space and circulate in the thoracic duct lymph52) supported the differentiation pathways and functions of “induced IELs”.
We investigated the dynamics of the IEL population during the conventionalization of GF mice. The number of TCRαβ IELs bearing CD8αα or CD8αβ markedly increased to reach the maximum approximately one month after conventionalization. In contrast, the proportion of total IELs that were TCRγδ IELs or TCRαβ IELs bearing CD4 showed only a small increase or a decrease over the same time period.53) This was confirmed by our subsequent observation that the increase in BrdU-labeled TCRαβ IELs was greater than that of TCRγδ IELs during the course of the conventionalization process and in CV mice.54) Furthermore, cyclophosphamide abrogated the expansion of IELs two to three weeks after conventionalization, suggesting the involvement of a TCR-mediated pathway.55) CD8αα+ and CD8αβ+ TCRαβ IEL subsets increased equally in the course of conventionalization, whereas only CD8αβ+ TCRαβ T cell subset markedly increased in the mesenteric lymph node after conventionalization. Given that the increase in each subset of IELs was not abrogated by thymectomy, the microbiota probably affects the T cell development after selection in thymus or their migration into intraepithelial space. CD8αα+ TCRαβ “natural IELs” with self reactivity and CD8αβ+ TCRαβ “induced IELs” with cytotoxic activity are considered to bear protective functions against cancerous or injured epithelial cells and pathobiont-infected cells, respectively.51) Accordingly, conventionalization of GF mice may provide hosts with the ability to protect themselves by expansion of IELs.53)
We found that expression of MHC class II [I-A]molecules in the epithelial cells of the small intestine is induced a week after conventionalization, whereas the level of the non-classical MHC class I molecule, TLa, does not change during this process.56) This MHC class II induction was mediated by TCRγδ IELs activated by microbiota.57) In contrast, classical MHC class I molecules such as H-2K were expressed to the same extent in both GF and Cvd mice. Although the contribution of TCRγδ IELs and CD4+TCRαβ IELs to the dynamics of whole IELs population is small compared with that of CD8+ TCRαβ IELs in the course of conventionalization, TCRγδ IELs in particular are thought to play an important role in epithelial MHC class II expression.57)
We then fractionated the microbes in microbiota by using chloroform treatment, introduced them to GF mice, and monitored the dynamics of the IELs and checked the epithelial cell characteristics. Chloroform treatment of fecal suspensions is currently used to select spore-forming microbes from the microbial population.58) Treatment of GF mice with chloroform-treated fecal microbiota expanded the IEL population and caused expression of MHC class II molecules and FGA1 glycolipid in the epithelial cells;59) furthermore the IELs acquired cytolytic activity against P815 cells (derived from mouse lymphoblast-like mastocytoma).
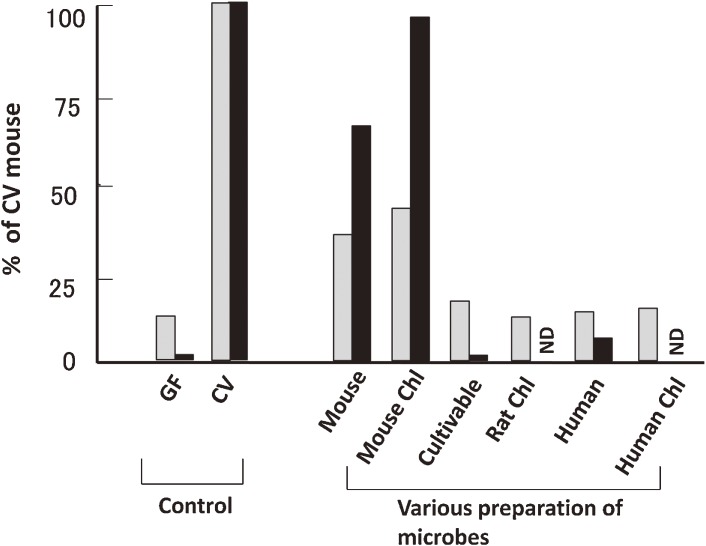
Based on these experimental results we explored the defined microbial fraction responsible for both the increase in TCRαβ IELs and the acquisition of redirected cytolytic activity against P815 cells. To this end, we compared gnotobiotic mice associated with several microbial preparations derived from various origins with or without chloroform treatment. Both the chloroform-treated and native forms of mouse fecal microbiota induced an increase in TCRαβ IELs and their cytolytic activity in mouse, but similar preparations from rat or human had no effect60) (Fig. 3). Moreover, an admixture of cultivable intestinal bacteria obtained from rodents or human at that time was also ineffective. Together, these results suggest that mouse-derived microbes that form spores are sufficient for activation of IELs in the mouse small intestine. These results strongly suggest stringent host specificity of the key microbes, consistent with observations reported later by Chung et al.61)
Figure 3.
Effect of species and chloroform-treatment of microbe preparations on induction of TCRαβ IEL recruitment and cytolytic activity (n = 2–4). The ratio of TCRαβ IELs to TCRγδ IELs (gray bars) and P815-cell specific lysis (%) in the presence of anti-TCRαβ antibody (black bars) were determined in GF mice two weeks after inoculation with microbe preparations derived from mouse, rat, or human, with or without chloroform (Chl) treatment, or an admixture of colonies grown on EG agar plate derived from mouse feces (Cultivable). GF or CV mouse feces were used as the control. All values are represented as percentage of each value of CV mice. ND: not determined. The panel is reproduced with permission from ref. 60.
6. Contribution of SFB to the development of the mucosal immune system
As mentioned above, SFB are present in the small intestine of a wide range of species, but their precise functions are still unknown, largely because they have not been cultivated to date. We decided to establish SFB-monocolonized mice to assess the immunostimulatory activity of SFB. Our main strategy for preparation of the gnotobiotic mice was to use EDTA treatment to isolate the epithelial cells of the small intestinal mucosa, a part of which were tightly bound to SFB, and then to remove all other living bacteria from these epithelial cells by repeated washing and centrifugation steps. The resultant epithelial cell preparation was treated with chloroform to exclude contamination by non-spore forming microbes. The final materials containing SFB spores were orally inoculated into GF BALB/c mice as little as possible by using a stomach tube; the minimum dose necessary to produce clearly detectable bacteria in the feces was used. One to two weeks after inoculation, a large number of SFB and a small number of Bacillus sp. colonized the GF mice. Thereafter sequential dilution of the feces of the first generation of gnotobiotic mice and administration of these feces into GF mice was repeated until the appearance of apparently SFB-monocolonized mice free from Bacillus. To check the homogeneity of the bacteria that colonized the intestine for each mouse, 16S rRNA gene sequences of approximately ten clones of randomly selected recombinants were determined by using the clone library method. The 16S rRNA gene sequence of each clone was almost identical as far as we checked. For each mouse, fluorescence-isothiocyanate in situ hybridization with a probe designed based on this 16S rRNA gene sequence showed positive staining of all the bacteria found in a smear preparation of the fecal suspension. At this point, we considered that the gnotobiotic mice were SFB-monocolonized and suitable for further analysis.39)
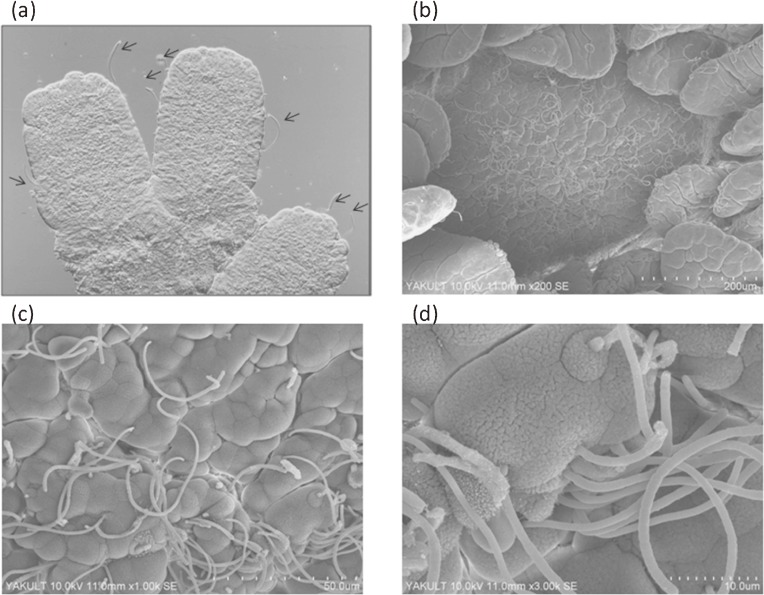
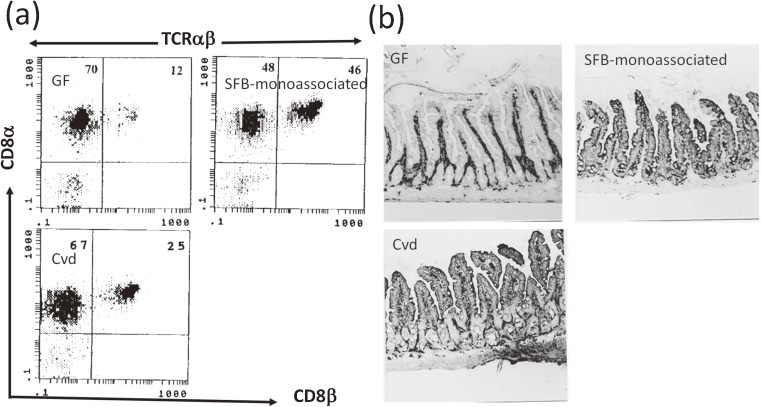
By conducting differential interference contrast and scanning electron microscopy (SEM) analyses of these SFB-monocolonized mice, we demonstrated that SFB colonized both the small and large intestines39) and tightly adhered to the MVMs of the epithelial cells of the small intestine62) (Fig. 4) and a part of the cecum, but not the colon (data not shown). In the small intestine, SFB tightly adhered to both the villus- and the PP follicle-associated epithelial cells. In Cvd and CV mice, SFB colonized the ileum at maximum density compared to other intestinal tissues.63)
Figure 4.
Tight adhesion of SFB to the small intestinal epithelial cells in SFB-monocolonized mice. Images were obtained from differential interference contrast microscopy (a, arrows) and SEM (b–d).
We then performed SFB-monocolonization of GF mice by oral inoculation of the fecal suspension of the gnotobiotic mice established above, and measured various activities of IELs and IgA production and evaluated the characteristics of the intestinal epithelial cells.
i) IELs.
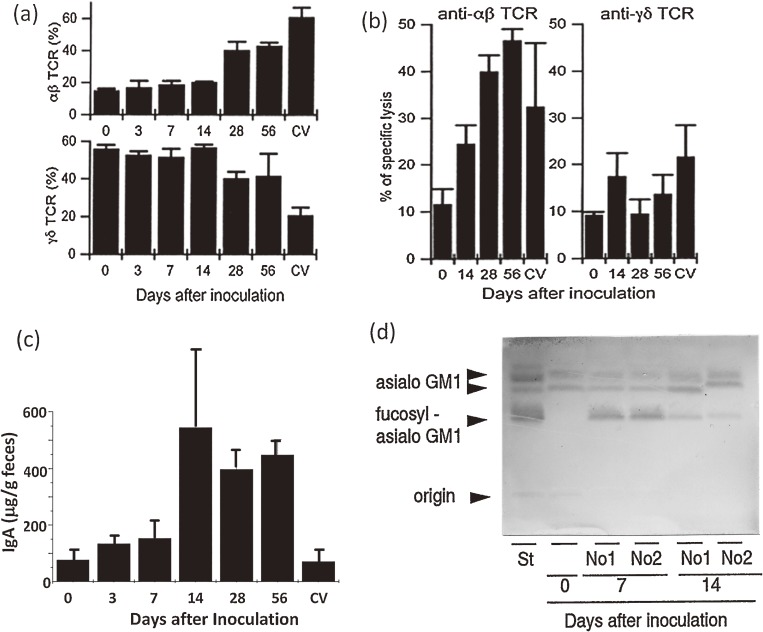
During the course of SFB monocolonization, the percentage of IEL cells in each IEL subpopulation was determined. Between two and three weeks after SFB inoculation, the percentages of TCRαβ IELs and TCRγδ IELs drastically increased and decreased, respectively, to reach a similar percentage to each other39) (Fig. 5). The percentages then remained constant until the end of the observation period (Fig. 5a). The percentage of TCRαβ IELs bearing CD8αβ heterodimer molecules in SFB-monoassociated mice was different from that in CV39) or Cvd mice64) (Fig. 6a). This is extremely interesting due to differences in the differentiation route and protective functions between “natural IELs” bearing TCRαβ and CD8αα and “induced IELs” bearing TCRαβ and CD8αβ, as described above. SFB may affect the activation process of TCRαβ CD8αβ+ T cells in the lamina propria, PPs, or mesenteric lymph node, and the subsequent recruitment process of IELs. Monoclonal antibody analysis of the Vβ usage of TCRαβ IELs and their CD8αα+ and CD8αβ+ subtypes showed almost no difference in Vβ usage, except for Vβ6 among SFB-monocolonized, Cvd, and GF mice.64) During SFB-monocolonization, only Vβ6+ TCRαβ IELs bearing CD8αβ notably increased among the Vβ repertoire examined. Redirected cytolytic activity of IELs of SFB-monocolonized mice against P815 cells was observed in the presence of anti-TCRαβ but not anti-TCRγδ monoclonal antibody, suggesting selective activation of TCRαβ IELs for cytolytic activity by SFB39) (Fig. 5b).
Figure 5.
IEL recruitment, IgA production, and epithelial fucosylation during the course of SFB monocolonization of GF mice.39) (a) Percentage of TCRαβ and TCRγδ IELs to total IELs. (b) Cytolytic activity against P815 cells in the presence of anti-TCRαβ or anti-TCRγδ antibody. (c) IgA concentration in the cecal contents. (d) TLC profile of the change from GA1 to FGA1 glycolipid. Panels a, b, and d are reproduced with permission from ref. 39.
Figure 6.
Differences in CD8 molecules of TCRαβ IELs and MHC class II expression among GF, SFB-monocolonized, and Cvd mice. (a) Flow cytometry of TCRαβ IELs with CD8αβ or CD8αα. (b) Immunohistochemistry of small intestinal tissue with anti-MHC I-Ad and MHC I-Ed antibodies. Both analyses are performed using the second generations of SFB-monocolonized and Cvd mice together with age matched GF mice. Panels a, b are reproduced with permission from ref. 64.
ii) IgA and epithelial MHC class II molecules.
IgA content in the feces was drastically elevated two weeks after SFB colonization36) (Fig. 5c), consistent with the increase in IgA-secreting cells. IgA response to SFB-monocolonization in another mouse strain (Swiss) was first reported by Klaasen et al.65) Interestingly, SFB also increased IgA-secreting cells in the colon,64) different from the IEL response described below. This finding strongly suggests that the priming for IgA synthesis occurs in PPs in the small intestine.
Immunohistochemical analysis with an admixture of antibodies against I-Ad and I-Ed MHC class II alloantigens indicated that induction of MHC class II molecules occurred at least three weeks after SFB monocolonization39) (Fig. 6b).
iii) FGA1 glycolipid and others.
FGA1 synthesis was clearly induced in the small intestinal epithelial cells within a week after SFB monocolonization39) (Fig. 5d), similar to the finding after conventionalization. FT induction by intestinal colonization of specific bacteria, SFB, was reported for the first time.39) The mitotic activity of the cryptal cells and differentiation of small intestinal epithelial cells, which were estimated by the metaphase-arrest method with vincristine and the ratio of the number of goblet cells to columnar epithelial cells, respectively, were also decreased by SFB-monocolonization39) or conventionalization of GF mice.66)
Taken together, these findings indicate that SFB-induced alterations of various characteristics of the intestinal immune system and epithelial cell surface are closely associated with physiological host responses to environmental stimuli or insults and may contribute to the maintenance of homeostasis in the digestive tract.
7. Complementary role of SFB and clostridia in the intestinal immune system
We next explored whether SFB also activate the colonic immune system. The IEL population in the large intestine, judging from the ratio of CD8+ IELs to CD4+ IELs, did not change following SFB-monocolonization of GF mice.64) Since the usual habitat of SFB is the small intestine, not the large intestine, we tried to establish in GF mice another candidate bacteria, whose habitat is the large intestine, by orally administering an admixture of 46 clostridial strains;64) these strains were previously reported to reduce cecal size to that of Cvd mice by their association with GF mice.67) In contrast to the result obtained with SFB-monocolonized mice, clostridia-colonized mice showed an increase in the number of IELs and IgA-producing cells in the colon, but not the small intestine, as well as no expression of epithelial MHC class II in the small intestine.64) IgA production in the colon was enhanced not only by clostridia but also by SFB, probably due to the priming of IgA induction in PPs of the small intestine colonized by SFB. We speculate that the effects of both types of bacteria on the mucosal functions including the immune system are complementary, with SFB affecting the small intestine and clostridia affecting the large intestine, respectively, corresponding to their primary habitats. Although the ratio of CD8αβ+TCRαβ to CD8αα+TCRαβ IELs was higher in SFB-monocolonized mice than in Cvd mice, addition of clostridia returned the ratio to normal (i.e., the level in Cvd mice).64) Thus, the mice associated with both types of bacteria show activation of the mucosal immune system along the whole intestine as observed in CV or Cvd mice.
These results led us to hypothesize that not all members of the microbiota are necessary for activation of the intestinal mucosal immune system including the epithelial functions, and that limited kinds of bacteria such as SFB and clostridia are sufficient to develop the immune system. The presence of other indigenous intestinal bacteria such as Bacteroides acidifaciens68) in addition to this bacterial combination is speculated to alter or modify the intestinal immune system beneficially or harmfully for the host. We called this minimum bacterial combination of SFB and clostridia “basal flora” or “minimal flora”.60) In the case of IBD models such as SAMP1/Yit mutant mice on the AKR background with spontaneous ileitis,69) and mice with dextran sodium sulfate-induced colitis, neither ileitis nor colitis developed in ex-GF mice that were not only associated with basal flora but also mono-associated with IBD-derived Bacteroides vulgatus.70) However, IBD-like diseases clearly occurred following addition of Bacteroides vulgatus to basal flora-associated mice (unpublished data). A similar observation was recently reported by Stephankova et al.,71) where particular microbiota members except SFB seemed to alter the immune system being developed by SFB to induce or exacerbate intestinal inflammatory responses. In this case it is very difficult to evaluate the boundary, if present, between commensal bacteria and pathogenic ones.72)
SFB and clostridia not only play a complementary role in the development of the mucosal immune system of the digestive tract, as described above, but they also contribute to the development of Th17 cells (T helper cells that produce interleukin 17) in the small intestine73,74) and Treg cells (regulatory T cells) in the colon.75,76)
8. Role of SFB as a Th17 inducer
The discovery of Treg cells and Th17 cells, and their close association with gut functions in health and disease,77–79) is big topic in the field of immunology. A large populations of Treg or Th17 cells are present in the intestinal mucosa and these cells are suggested to be involved in the occurrence and repression of infectious diseases or IBD.62,80)
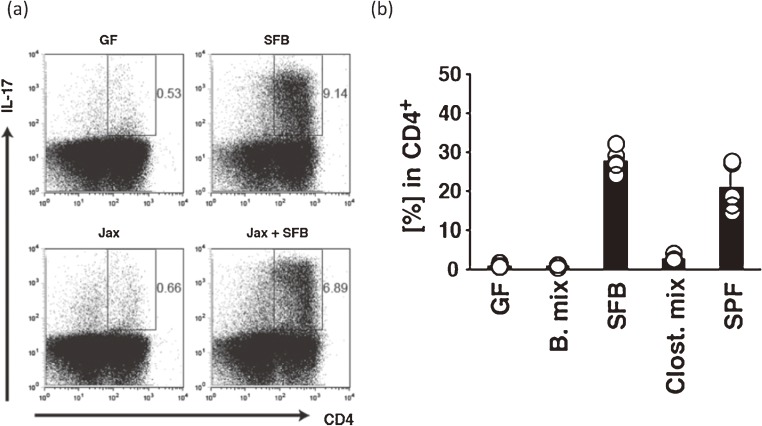
Notably, Ivanov et al. observed a close relationship between the density of Th17 cells in the lamina propria of the small intestine and the presence of particular bacteria. They found that a mouse strain C57BL/6 raised at Taconic Farms, has many more Th17 cells than the same strain raised at the Jackson Laboratory.81) This observation indicates that a limited number of characteristic bacteria, such as SFB or Lactobacillus murinus, that are present in the mice from Taconic Farms, but not the Jackson Laboratory, play a key role in Th17 cell development. Addition of SFB to Jackson Laboratory mice, which were SFB-free according to the results of a PhyloChip82) assay, clearly evoked an induction of Th17 cells73) (Fig. 7a). Furthermore, induction of Th17 cells has been confirmed in SFB-monocolonized mice but not in the mice colonized with 46 strains of clostridia73) (Fig. 7b) that were used for the study of IELs and IgA described above. Interestingly, transcripts of all isoforms of the serum amyloid A (SAA), Saa1, Saa2, and Saa3, were enormously upregulated in the epithelial cells of SFB-monocolonized mice and in Jackson Laboratory mice cohoused with Taconic Farms mice, according to the results of microarray analysis.73) Addition of SAA to in vitro cultivation of the naïve T cells with lamina propria dendritic cells (DCs) in the presence of anti-CD3 produced Th17 cell cytokines such as IL-17A, IL-17F, and RORγt. This result suggests a possibility that naïve CD4+ T cell differentiates to Th17 cell via IL-23 produced by SAA-stimulated DCs.73) More recently, analysis of Th17 cell differentiation mechanism by SFB remarkably advanced. Most Th17 cells in lamina propria of SFB-monocolonized mice recognize particular SFB peptides83) and also this SFB antigen is presented by DCs through their MHC class II expression.84)
Figure 7.
Induction of Th17 cells by SFB colonization of IQI GF mice. (a) Flow cytometry of Th17 cells in the small intestine of SFB- or Jackson Laboratory microbiota (Jax)-associated mice or the Jax mice which were additionally associated with SFB. (b) Proportion of Th17 cells (%) among CD4+ cells in the small intestine of gnotobiotic mice that were associated with SFB alone or an admixture of Bacteroides or Clostridium species. Panels a, b are reproduced with permission from ref. 73.
Physiologically, Citrobacter rodentium infection was repressed to keep the normal cryptal depth in SFB-colonized Jackson Laboratory mice compared with SFB-free Jackson Laboratory mice.73) Before the appearance of the Th17 cell concept, SFB colonization was reported to be negatively correlated with pathogenic E. coli O103 colonization in rabbits85) suggesting that it inhibits infection. More interestingly, the SFB population expands abnormally in the small intestine of mice with disruption of the activation-induced cytidine deaminase (AID) gene,86) which also greatly contributes to IgA production in the intestine.87) Thus IgA secretion is thought to play an important role in leaving intestinal microbes in their appropriate niches.
Involvement of the microbiota in autoimmunity became of great concern after this discovery of Th17 cell-inducing commensal microbe. Induction of disease by SFB-monocolonization of GF mice was attempted in several mouse models of autoimmune disease as described below. When arthritis model GF K/BxN mice, which express a TCR transgene that recognizes a self-peptide derived from glucose-6-phosphate isomerase, were inoculated with SFB, the disease score greatly increased, coincident with the appearance of Th17 cells in the lamina propria in the small intestine.88) In a mouse model of experimental autoimmune encephalomyelitis, GF mice (C57/BL6) were inoculated with an SFB isolated by us39) together with myelin oligodendrocyte glycoprotein (MOG) peptide and purtussis toxin. Disease development was observed, although it was weaker than that in SPF mice.89) In this case the Th17 cells expanded in the spinal cord rather than in the intestinal lamina propria. Furthermore, SFB affected the disease development in the type 1 diabetic model mouse following SFB-inoculation90) or following natural transmission of SFB.91) Among these autoimmune models, the characteristics of the Th17 cells, such as their anatomical site and γ-interferon production ability seem to be different. The interrelationship between autoimmune diseases and SFB is currently being clarified.
9. Host-specificity of SFB in epithelial binding and mucosal immune stimulation
As mentioned above, SFB are present in a wide range of animals.38) Because of their uncultivability, the only reported host-specific differences between SFBs relate to their colonization ability92) and 16S rRNA gene sequences.93,94) We have isolated SFB not only from BALB/c mouse (mSFB)39) but also from F344 rat (rSFB).94) Our observations in mSFB or rSFB and mouse- or rat-microbiota colonized mice, and the findings of Chung et al.,61) suggest that a strong relationship exists between the adhesion ability of SFB and their effects on the host mucosal immune system.
When mSFB and rSFB were inoculated into GF mice, both SFBs colonized almost equally both the small and large intestines. However, tight adhesion of rSFB to the mouse small intestinal epithelial cells was not detected by SEM and real-time PCR. Furthermore in ex-GF mice that were monocolonized with rSFB, IELs were not expanded, IgA scarcely increased at all, MHC class II molecules were not expressed, and Th17 cell induction in the lamina propria was not observed. As described above, the opposite was the case for mSFB-monocolonized mice. Th17 cell induction in the lamina propria was observed in mSFB-monocolonized but not rSFB-monocolonized mice. Concomitantly, when mSFB or rSFB were inoculated into GF rats (F344), only rSFB adhered tightly to the rat small intestinal epithelial cells, coincident with Th17 induction, IEL response, and IgA secretion (in preparation for submission). These findings indicate that SFB exhibit clear host-specificity between mice and rats, particularly in the small intestine. We therefore consider it likely that SFB affect the host mucosal immune system in a host-specific manner in a wide range of the animals, and that the host-specific effect of SFB contributes to the host-specific effect of the whole microbiota on the host mucosal immune system, such as the changes in Treg cells, Th17 cells, and IgA-producing cells observed in Cvd mice by Chung et al.61) In comparison with Cvd mice, the degree of the activation among various parts of the immune system in SFB-monocolonized mice is variable; for example, the number of Th17 cells is almost equal between Cvd and SFB-monocolonized mice, whereas that of IELs is much greater in Cvd mice than in SFB-monocolonized mice. In contrast, IgA content is somewhat higher in SFB-monocolonized mice than in Cvd mice, probably due to degradation by microbial IgA-protease present in Cvd mice95) but not SFB-monocolonized mice.
Importantly, host-specificity of SFB for activation of the mucosal immune system is likely to be dependent on the tight adhesion to the small intestinal epithelial cells, where polymerization of actin filaments occurs,96) as observed in signal transduction by epithelial adhesion of enteropathogenic E. coli.97) Clarification of the precise mechanisms of tight adhesion is required to elucidate how SFB activates the mucosal immune system.
10. Genomic view of differences between mSFB and rSFB
As described above, mSFB and rSFB have stringent host-specificity regarding adhesion to epithelial cells and immune stimulation. We consider that whole genome analysis might suggest a mechanism for these host-specific effects. The genome sizes of mSFB and rSFB are 1.59 Mb and 1.52 Mb, respectively63) (Table 1), which is approximately half that of clostridial genomes, and nucleotide identity between the mSFB and rSFB genomes is 86%. The larger size of the mSFB genome compared with the rSFB genome corresponds to the integration of two prophages specifically into the mSFB genome. The genome sizes of both SFBs are intermediate between those of free-living bacteria and host-dependent symbionts, suggesting that SFB may have both characters.98) Both SFB genomes encode flagellin proteins, although there is no reported SEM image of the flagellar filaments. Although the amino acid sequence of the flagellin protein FliC, which is assumed to be responsible for its binding to mouse TLR5,99,100) is almost identical between the two SFBs, rSFB was not effective for immunostimulation of mice. Furthermore, although ADP-ribosyltransferase, which may be involved in modulation of host responses, is encoded by the prophages that are integrated in the mSFB genome only, as suggested by Pamp et al.,101) rSFB was effective for immunostimulation in rats. Therefore, no promising functional gene candidate for the regulation of SFB-induced immunostimulation has yet been found, and further investigation is required. The genomic sequence of mSFB preparation isolated and determined by another group102) and that of our mSFB preparation determined by another group103) are almost identical to the sequence published by ours.63) Moreover, the DNA sequences of individual SFB microbes isolated from our SFB preparation by using a single-cell manipulator technique101) suggest that the mSFB preparation originally isolated from mice in our laboratory39) may be a mixture of SFB with slightly heterogeneous genome sequences.
Table 1.
General features of the whole genome and possible functional gene candidates of mSFB and rSFB*
| Genome size (bp) | G+C content (%) | Number of | Flagellin genes | |||||
|---|---|---|---|---|---|---|---|---|
| CDSs | rRNA operons | tRNA genes | CRISPRS | Prophages | ||||
| mSFB | 1,586,397 | 28 | 1420 | 5 | 38 | 3 | 2 | FliC1 to FliC3 |
| rSFB | 1,515,556 | 28 | 1346 | 5 | 39 | 1 | 0 | FliC1 to FliC4 |
*Modified from ref. 63.
11. Concluding remarks
While exploring the impacts of microbiota on the intestinal immune system, including epithelial cell surface changes, we discovered that enhancement of IEL recruitment and Th17 cell induction in lamina propria, and cell surface glycolipid fucosylation in the small intestine were mediated by a type of not-yet-cultivable intestinal bacteria, that is, SFB. Although the physiological roles of IELs and FGA1 glycolipid are still obscure, we consider that the mechanisms underlying these events, including Th17 induction, are likely to contain common elements because these events seem to be induced by tight adhesion of SFB to the inestinal epithelial cells.
Based on our analysis of TCR and CD8 molecules, the effect of SFB on IELs seems to differ from that of the whole intestinal microbiota in that it is limited to recruitment of a part of the IEL population that responds to the peripheral antigens, probably SFB cell components. The effects of SFB on immunological events are host-specific in that rSFB could not induce the events in mouse, and vice versa. Although the whole genome sequences of both rSFB and mSFB have been determined, the candidate genes responsible for the host-specificity have not been deduced as yet. However, it is clear that the rSFB and mSFB tightly adhere to both villus and PP-covered epithelial cells in the small intestine in a host-specific fashion (preparation for submission), suggesting that this is a starting point for the mechanism underlying the immunological events. Taken together, our results suggest that the tight adhesion of SFB to epithelial cells induces Th17 cells and simultaneously activates CD8αβ+ TCRαβ T cells in the lamina propria following recruitment in the interepithelial space. We consider it likely that increased IgA secretion is due to the enhancement of priming by SFB-adhesion in PP-covered epithelial cells.
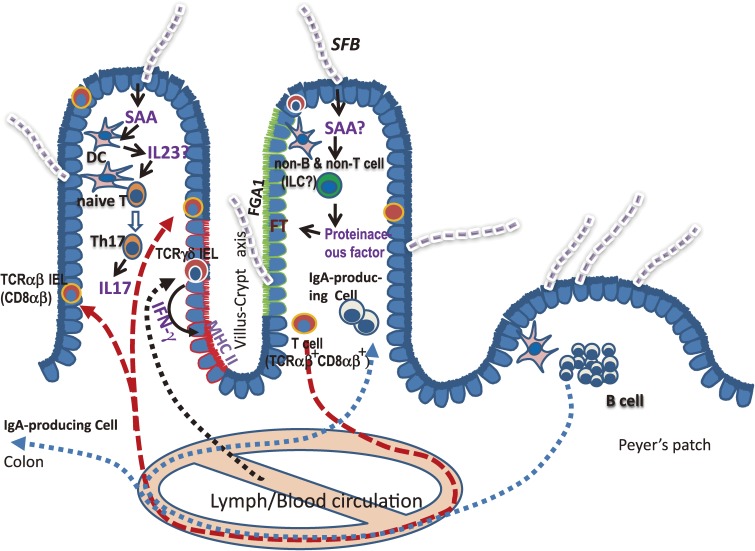
On the other hand, our observations of FT induction in SCID mice, but not MyD88 knockout mice, the involvement of a proteinaceous substance present in the small intestinal homogenate, and the selective expression of FGA1 in villus epithelial cells suggest that proteinaceous substances produced by non-T and non-B cells may mediate FT induction in villus epithelial cells. Our proposed activation scheme for the intestinal immune system, including cell surface glycosylation by SFB, is described in Fig. 8.
Figure 8.
Schematic illustration of activation of the intestinal immune system including fucosylation of epithelial glycolipid by SFB. Details and non-standard abbreviations are shown in the main text.
Finally, the issue of presence or absence of SFB or SFB equivalent microbes in human attracts much attention. The SFB population expands in the weaning stage, at least, in mice.104) Although we tried in vain to find SFB 16S rRNA gene sequence in the intestinal contents of humanized ex-germ free mice associated with microbiota of human baby feces (data not shown), there are some reports suggesting the presence of SFB in humans, according to light micrography and partial 16S rRNA gene sequences.38,105,106) Moreover, due to the presence of SFB or the morphologically similar microbes in a wide range of mammals such as rhesus monkey,38) crab-eating monkey,38,94) pig,107,108) dog,38,109,110) cat,38) cow,111) calf,112) horse,113) and rabbit,85) there is no reason to exclude human from this group at present. Although the functions of SFB are unknown at all except rodents, SFB or SFB equivalent microbes may start to colonize or transiently colonize the human small intestine around the weaning period according to the observations in the animals mentioned above. The discovery of SFB in humans and clarification of its role in human-microbiota interplay are waiting to be done in the near future.
Acknowledgements
I would like to first thank Dr. T. Honjo (Grad. Sch. Med., Kyoto Univ.) for encouraging me to write this review. Moreover, I deeply appreciate the work of my collaborators: Drs. T. Yamakawa and A. Suzuki (Fac. Med., Univ. Tokyo) for glycolipid research; Dr. K. Itoh (Grad. Sch. Agri. Life Sci., Univ. Tokyo) for clostridial gnotobiote research; Drs. K. Honda, K. Atarashi (RIKEN Inst.), Dr. K. Takeda (Grad. Sch. Med., Osaka Univ.), Drs. D. Littman and I. Ivanov (New York Univ. Sch. Med.) for immunological research; and Dr. M. Hattori (Grad. Sch. Front. Sci., Univ. Tokyo), Dr. T. Prakash (RIKEN Inst.), Dr. H. Morita and T. Nagamori (Azabu Univ.) for genome research. I also greatly thank the members of my team, Drs. S. Matsumoto, A. Imaoka, Y. Okada, H. Setoyama, R. Shima, S. Hara and M. Ando, with whom all of the work presented here was performed, and the animal care staff of our institute for production and maintenance of all experimental animals. Finally I sincerely thank the directors of our institute for their steady support and encouragement and all members of our institute for cooperation in this research.
Abbreviations
- IBD
inflammatory bowel disease
- GF
germ-free
- SPF
specific pathogen-free
- CV
conventional
- Cvd
conventionalized
- TLC
thin layer chromatography
- IEL
intraepithelial lymphocyte
- ILC
innate lymphoid cell
- MVM
microvillar membrane
- GM1
ganglioside GM1
- GA1
asialo GM1
- FGA1
fucosyl asialo GM1
- FT
α(1-2) asialo GM1 fucosyltransferase
- SFB
segmented filamentous bacteria
- PP
Peyer’s patch
- SAA
serum amyloid A
- SEM
scanning electron microscopy
Profile
Yoshinori Umesaki was born in Fukui Prefecture, Japan in 1950. He graduated from Department of Fermentation Technology, Faculty of Engineering, Osaka University in 1972 and completed a master’s degree of the same department in 1974. He started his research carrier in Yakult Central Institute for Microbiological Research, Tokyo in 1974. After a program of research student at the Department of Biochemistry, Faculty of Medicine, University of Tokyo (Professor Tamio Yamakawa) in 1980, he received Ph.D. from Department of Food Chemistry, Tohoku University (Professor Shuichi Kimura) in 1987, for his thesis “Analysis for Interactions between Intestinal Microbiota and Host”. Thereafter, he experienced visiting researcher of RIKEN Frontier Research Program for Intestinal Flora directed by Professor Tomotari Mitsuoka, 1987–1990 and Head of Basic Research Department II, 2006–2010 and Executive Manager, 2008–2010 in Yakult Central Institute. He is now in position of Senior Research Fellow of Yakult Central Institute and Visiting Professor of Azabu University. Throughout his carrier, he focused on the research for biological and medical significance of the interactions between the intestinal microbes including probiotics and host. A pioneering work of his research group using gnotobiotic mouse model largely contributed to the elucidation of the role of segmented filamentous bacteria (SFB) in the host-microbiota interplay.
References
- 1).Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. (2005) Diversity of the human intestinal microbial flora. Science 308, 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. (2006) Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Grice E.A., Segre J.A. (2011) The skin microbiome. Nat. Rev. Microbiol. 9, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Mitsuoka, T. (1978) Intestinal Bacteria and Health: An introductory Narrative. Harcourt Brace Jovanovich, Tokyo. [Google Scholar]
- 5).Mai V., Morris J.G., Jr. (2004) Colonic bacterial flora: changing understandings in the molecular age. J. Nutr. 134, 459–464. [DOI] [PubMed] [Google Scholar]
- 6).Matsuda K., Tsuji H., Asahara T., Matsumoto K., Takada T., Nomoto K. (2009) Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl. Environ. Microbiol. 75, 1961–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., Fernandes G.R., Tap J., Bruls T., Batto J.M., Bertalan M., Borruel N., Casellas F., Fernandez L., Gautier L., Hansen T., Hattori M., Hayashi T., Kleerebezem M., Kurokawa K., Leclerc M., Levenez F., Manichanh C., Nielsen H.B., Nielsen T., Pons N., Poulain J., Qin J., Sicheritz-Ponten T., Tims S., Torrents D., Ugarte E., Zoetendal E.G., Wang J., Guarner F., Pedersen O., de Vos W.M., Brunak S., Doré J., MetaHIT Consortium. Antolín M., Artiguenave F., Blottiere H.M., Almeida M., Brechot C., Cara C., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Foerstner K.U., Friss C., van de Guchte M., Guedon E., Haimet F., Huber W., van Hylckama-Vlieg J., Jamet A., Juste C., Kaci G., Knol J., Lakhdari O., Layec S., Le Roux K., Maguin E., Mérieux A., Melo Minardi R., M’rini C., Muller J., Oozeer R., Parkhill J., Renault P., Rescigno M., Sanchez N., Sunagawa S., Torrejon A., Turner K., Vandemeulebrouck G., Varela E., Winogradsky Y., Zeller G., Weissenbach J., Ehrlich S.D., Bork P. (2011) Enterotypes of the human gut microbiome. Nature 473, 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Power S.E., O’Toole P.W., Stanton C., Ross R.P., Fitzgerald G.F. (2014) Intestinal microbiota, diet and health. Br. J. Nutr. 111, 387–402. [DOI] [PubMed] [Google Scholar]
- 9).van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M., Visser C.E., Kuijper E.J., Bartelsman J.F., Tijssen J.G., Speelman P., Dijkgraaf M.G., Keller J.J. (2013) Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415. [DOI] [PubMed] [Google Scholar]
- 10).Borody T.J., Khoruts A. (2012) Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol. 9, 88–96. [DOI] [PubMed] [Google Scholar]
- 11).Bennet J.D., Brinkman M. (1989) Treatment of ulcerative colitis by implantation of normal colonic flora. Lancet 333, 164. [DOI] [PubMed] [Google Scholar]
- 12).Kunde S., Pham A., Bonczyk S., Crumb T., Duba M., Conrad H., Jr., Cloney D., Kugathasan S.J. (2013) Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. Pediatr. Gastroenterol. Nutr. 56, 597–601. [DOI] [PubMed] [Google Scholar]
- 13).Vrieze A., Van Nood E., Holleman F., Salojärvi J., Kootte R.S., Bartelsman J.F., Dallinga-Thie G.M., Ackermans M.T., Serlie M.J., Oozeer R., Derrien M., Druesne A., Van HylckamaVlieg J.E., Bloks V.W., Groen A.K., Heilig H.G., Zoetendal E.G., Stroes E.S., de Vos W.M., Hoekstra J.B., Nieuwdorp M. (2012) Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916. [DOI] [PubMed] [Google Scholar]
- 14).Pleasants J.R. (1965) History of germfree animal research at Lobund Laboratory, Biology Department, University of Notre Dame, 1928–1965. Proc. Indiana Acad. Sci. 65, 220–226. [Google Scholar]
- 15).Iijima, S. (1981) Gnotobiology research in Japan — A brief history of and a review of the present status —, In Recent Advances in Germfree Research (eds. Sasaki, S., Ozawa, A. and Hashimoto, K.). Tokai University Press, Tokyo, pp. 5–9. [Google Scholar]
- 16).Snyder D.L., Pollard M., Wostmann B.S., Luckert P. (1990) Life span, morphology, and pathology of diet-restricted germ-free and conventional Lobund-Wistar rats. J. Gerontol. 45, B52–B58. [DOI] [PubMed] [Google Scholar]
- 17).Deplancke B., Gaskins H.R. (2001) Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 73, 1131S–1141S. [DOI] [PubMed] [Google Scholar]
- 18).He W., Wang M.L., Jiang H.Q., Steppan C.M., Shin M.E., Thurnheer M.C., Cebra J.J., Lazar M.A., Wu G.D. (2003) Bacterial colonization leads to the colonic secretion of RELMbeta/FIZZ2, a novel goblet cell-specific protein. Gastroenterology 125, 1388–1397. [DOI] [PubMed] [Google Scholar]
- 19).Uribe A., Alam M., Johansson O., Midtvedt T., Theodorsson E. (1994) Microflora modulates endocrine cells in the gastrointestinal mucosa of the rat. Gastroenterology 107, 1259–1269. [DOI] [PubMed] [Google Scholar]
- 20).Khoury K.A., Floch M.H., Hersh T. (1969) Small intestinal mucosal cell proliferation and bacterial flora in the conventionalization of the germfree mouse. J. Exp. Med. 130, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Lifshitz F., Wapnir R.A., Wehman H.J., Diaz-bensussen S., Pergolizzi R. (1978) The effects of small intestinal colonization by fecal and colonic bacteria on intestinal function in rats. J. Nutr. 108, 1913–1923. [DOI] [PubMed] [Google Scholar]
- 22).Nagengast F.M., Grubben M.J., van Munster I.P. (1995) Role of bile acids in colorectal carcinogenesis. Eur. J. Cancer 31A (7–8), 1067–1070. [DOI] [PubMed] [Google Scholar]
- 23).Livesey, G. and Elia, M. (1995) Short-chain fatty acids as an energy source in the colon: metabolism and clinical applications, In Physiological and Clinical Aspects of Short-Chain Fatty Acid (eds. Cummings, J.H., Rombeau, J.L. and Sakata, T.). Cambridge University Press, Cambridge, pp. 427–481. [Google Scholar]
- 24).Topping D.L., Clifton P.M. (2001) Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81, 1031–1064. [DOI] [PubMed] [Google Scholar]
- 25).Diaz Heijtz R., Wang S., Anuar F., Qian Y., Björkholm B., Samuelsson A., Hibberd M.L., Forssberg H., Pettersson S. (2011) Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U.S.A. 108, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Welling G.W., Groen G., Tuinte J.H., Koopman J.P., Kennis H.M. (1980) Biochemical effects on germ-free mice of association with several strains of anaerobic bacteria. J. Gen. Microbiol. 117, 57–63. [DOI] [PubMed] [Google Scholar]
- 27).Koopman J.P., Kennis H.M., Mullink J.W., Prins R.A., Stadhouders A.M., De Boer H., Hectors M.P. (1984) ‘Normalization’ of germfree mice with anaerobically cultured caecal flora of ‘normal’ mice. Lab. Anim. 18, 188–194. [DOI] [PubMed] [Google Scholar]
- 28).Fujita M., Ohta H., Uezato T. (1981) Characterization of brush borders purified in iso-osmotic medium and microvillar membranes subfractionated from mouse small intestine. Biochem. J. 196, 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Umesaki Y., Tohyama K., Mutai M. (1982) Biosynthesis of microvillus membrane-associated glycoproteins of small intestinal epithelial cells in germ-free and conventionalized mice. J. Biochem. 92, 373–379. [DOI] [PubMed] [Google Scholar]
- 30).Umesaki Y., Tohyama K., Mutai M. (1981) Appearance of fucolipid after conventionalization of germ-free mice. J. Biochem. 90, 559–561. [DOI] [PubMed] [Google Scholar]
- 31).Umesaki Y., Suzuki A., Kasama T., Tohyama K., Mutai M., Yamakawa T. (1981) Presence of asialo GM1 and glucosylceramide in the intestinal mucosa of mice and induction of fucosyl asialo GM1 by conventionalization of germ-free mice. J. Biochem. 90, 1731–1738. [DOI] [PubMed] [Google Scholar]
- 32).Breimer M.E., Karlsson K.A., Samuelsson B.E. (1975) Presence of phytosphingosine combined with 2-hydroxy fatty acids in sphingomyelins of bovine kidney and intestinal mucosa. Lipids 10, 17–19. [DOI] [PubMed] [Google Scholar]
- 33).Suzuki A., Umesaki Y., Yamakawa T. (1982) Localization of asialo GM1 and Forssman antigen in the small intestine of mouse. Adv. Exp. Med. Biol. 152, 415–424. [PubMed] [Google Scholar]
- 34).Umesaki Y., Takamizawa K., Ohara M. (1989) Structural and compositional difference in the neutral glycolipids between epithelial and non-epithelial tissue of the mouse small intestine. Biochim. Biophys. Acta 1001, 157–162. [DOI] [PubMed] [Google Scholar]
- 35).Umesaki Y., Sakata T., Yajima T. (1982) Abrupt induction of GDP-fucose: asialo GM1 fucosyltransferase in the small intestine after conventionalization of germ-free mice. Biochem. Biophys. Res. Commun. 105, 439–443. [DOI] [PubMed] [Google Scholar]
- 36).Umesaki Y., Ohara M. (1989) Factors regulating the expression of the neutral glycolipids in the mouse small intestinal mucosa. Biochim. Biophys. Acta 1001, 163–168. [DOI] [PubMed] [Google Scholar]
- 37).Umesaki Y. (1989) Intestinal glycolipids and their possible role in microbial colonization of mice. Bifidobacteria Microflora 8, 13–22. [Google Scholar]
- 38).Klaasen H.L., Koopman J.P., Van den Brink M.E., Bakker M.H., Poelma F.G., Beynen A.C. (1993) Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 27, 141–150. [DOI] [PubMed] [Google Scholar]
- 39).Umesaki Y., Okada Y., Matsumoto S., Imaoka A., Setoyama H. (1995) Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 39, 555–562. [DOI] [PubMed] [Google Scholar]
- 40).Bry L., Falk P.G., Midtvedt T., Gordon J.I. (1996) A model of host-microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383. [DOI] [PubMed] [Google Scholar]
- 41).Umesaki Y., Setoyama H. (1992) Immune responses of mice to orally administered asialo GM1-specific rabbit IgG in the presence or absence of cholera toxin. Immunology 75, 386–388. [PMC free article] [PubMed] [Google Scholar]
- 42).Umesaki Y. (1984) Immunohistochemical and biochemical demonstration of the change in glycolipid composition of the intestinal epithelial cell surface in mice in relation to epithelial cell differentiation and bacterial association. J. Histochem. Cytochem. 32, 299–304. [DOI] [PubMed] [Google Scholar]
- 43).Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. (1999) Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11, 115–122. [DOI] [PubMed] [Google Scholar]
- 44).Vilcek J., Havell E.A., Kohase M. (1976) Superinduction of interferon with metabolic inhibitors: possible mechanisms and practical applications. J. Infect. Dis. 133 (Suppl), A22–A29. [DOI] [PubMed] [Google Scholar]
- 45).Osipovich O.A., Fegeding K.V., Misuno N.I., Kolesnikova T.S., Savostin I.K., Sudarikov A.B., Voitenok N.N. (1993) Differential action of cycloheximide and activation stimuli on transcription of tumor necrosis factor-alpha, IL-1 beta, IL-8, and P53 genes in human monocytes. J. Immunol. 150, 4958–4965. [PubMed] [Google Scholar]
- 46).Iwamori M., Domino S.E. (2004) Tissue-specific loss of fucosylated glycolipids in mice with targeted deletion of alpha(1,2) fucosyltransferase genes. Biochem. J. 380, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Rausch P., Rehman A., Künzel S., Häsler R., Ott S.J., Schreiber S., Rosenstiel P., Franke A., Baines J.F. (2011) Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl. Acad. Sci. U.S.A. 108, 19030–19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Shirato-Horikoshi H., Takeda N. (2007) Interaction between noroviruses and human histo-blood group antigens. Uirusu 57, 181–189(in Japanese). [DOI] [PubMed] [Google Scholar]
- 49).Morrow A.L., Meinzen-Derr J., Huang P., Schibler K.R., Cahill T., Keddache M., Kallapur S.G., Newburg D.S., Tabangin M., Warner B.B., Jiang X. (2011) Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. J. Pediatr. 158, 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Wacklin P., Mäkivuokko H., Alakulppi N., Nikkilä J., Tenkanen H., Räbinä J., Partanen J., Aranko K., Mättö J. (2011) Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE 6, e20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Cheroutre H., Lambolez F., Mucida D. (2011) The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 11, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Arstila T., Arstila T.P., Calbo S., Selz F., Malassis-Seris M., Vassalli P., Kourilsky P., Guy-Grand D. (2000) Identical T cell clones are located within the mouse gut epithelium and lamina propia and circulate in the thoracic duct lymph. J. Exp. Med. 191, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Umesaki Y., Setoyama H., Matsumoto S., Okada Y. (1993) Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology 79, 32–37. [PMC free article] [PubMed] [Google Scholar]
- 54).Imaoka A., Matsumoto S., Setoyama H., Okada Y., Umesaki Y. (1996) Proliferative recruitment of intestinal intraepithelial lymphocytes after microbial colonization of germ-free mice. Eur. J. Immunol. 26, 945–948. [DOI] [PubMed] [Google Scholar]
- 55).Kawaguchi-Miyashita M., Nanno M., Shimada S., Nagaoka N., Okada Y., Matsumoto S., Umesaki Y., Matsuoka Y., Ohwaki M. (1997) A step-wise expansion of intestinal intraepithelial T lymphocytes in association with microbial colonization is defined by sensitivity to cyclosporin A. Immunology 91, 628–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Matsumoto S., Setoyama H., Umesaki Y. (1992) Differential induction of major histocompatibility complex molecules on mouse intestine by bacterial colonization. Gastroenterology 103, 1777–1782. [DOI] [PubMed] [Google Scholar]
- 57).Matsumoto S., Nanno M., Watanabe N., Miyashita M., Amasaki H., Suzuki K., Umesaki Y. (1999) Physiological roles of gammadelta T-cell receptor intraepithelial lymphocytes in cytoproliferation and differentiation of mouse intestinal epithelial cells. Immunology 97, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Itoh K., Mitsuoka T. (1980) Production of gnotobiotic mice with normal physiological functions. I. Selection of useful bacteria from feces of conventional mice. Zeitschrift für Versuchstierkunde 22, 173–178. [PubMed] [Google Scholar]
- 59).Okada Y., Setoyama H., Matsumoto S., Imaoka A., Nanno M., Kawaguchi M., Umesaki Y. (1994) Effects of fecal microorganisms and their chloroform-resistant variants derived from mice, rats, and humans on immunological and physiological characteristics of the intestines of ex-germfree mice. Infect. Immun. 62, 5442–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Umesaki Y., Setoyama H. (2000) Structure of the intestinal flora responsible for development of the gut immune system in a rodent model. Microbes Infect. 2, 1343–1351. [DOI] [PubMed] [Google Scholar]
- 61).Chung H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B., Reading N.C., Villablanca E.J., Wang S., Mora J.R., Umesaki Y., Mathis D., Benoist C., Relman D.A., Kasper D.L. (2012) Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Atarashi K., Umesaki Y., Honda K. (2011) Microbiotal influence on T cell subset development. Semin. Immunol. 23, 146–153. [DOI] [PubMed] [Google Scholar]
- 63).Prakash T., Oshima K., Morita H., Fukuda S., Imaoka A., Kumar N., Sharma V.K., Kim S.W., Takahashi M., Saitou N., Taylor T.D., Ohno H., Umesaki Y., Hattori M. (2011) Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of Th17 cell differentiation. Cell Host Microbe 10, 273–284. [DOI] [PubMed] [Google Scholar]
- 64).Umesaki Y., Setoyama H., Matsumoto S., Imaoka A., Itoh K. (1999) Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 67, 3504–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Klaasen H.L., Van der Heijden P.J., Stok W., Poelma F.G., Koopman J.P., Van den Brink M.E., Bakker M.H., Eling W.M., Beynen A.C. (1993) Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 61, 303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Khoury K.A., Floch M.H., Hersh T. (1969) Small intestinal mucosal cell proliferation and bacterial flora in the conventionalization of the germfree mouse. J. Exp. Med. 130, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Itoh K., Mitsuoka T. (1985) Characterization of clostridia isolated from faeces of limited flora mice and their effect on caecal size when associated with germ-free mice. Lab. Anim. 19, 111–118. [DOI] [PubMed] [Google Scholar]
- 68).Momose Y., Park S.H., Miyamoto Y., Itoh K. (2011) Design of species-specific oligonucleotide probes for the detection of Bacteroides and Parabacteroides by fluorescence in situ hybridization and their application to the analysis of mouse caecal Bacteroides-Parabacteroides microbiota. J. Appl. Microbiol. 111, 176–184. [DOI] [PubMed] [Google Scholar]
- 69).Matsumoto S., Okabe Y., Setoyama H., Takayama K., Ohtsuka J., Funahashi H., Imaoka A., Okada Y., Umesaki Y. (1998) Inflammatory bowel disease-like enteritis and caecitis in a senescence accelerated mouse P1/Yit strain. Gut 43, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Ishikawa H., Akedo I., Umesaki Y., Tanaka R., Imaoka A., Otani T. (2003) Randomized controlled trial of the effect of bifidobacteria-fermented milk on ulcerative colitis. J. Am. Coll. Nutr. 22, 56–63. [DOI] [PubMed] [Google Scholar]
- 71).Stepankova R., Powrie F., Kofronova O., Kozakova H., Hudcovic T., Hrncir T., Uhlig H., Read S., Rehakova Z., Benada O., Heczko P., Strus M., Bland P., Tlaskalova-Hogenova H. (2007) Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm. Bowel Dis. 13, 1202–1211. [DOI] [PubMed] [Google Scholar]
- 72).Umesaki Y., Okada Y., Imaoka A., Setoyama H., Matsumoto S. (1997) Interactions between epithelial cells and bacteria, normal and pathogenic. Science 276, 964–965. [DOI] [PubMed] [Google Scholar]
- 73).Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., Tanoue T., Imaoka A., Itoh K., Takeda K., Umesaki Y., Honda K., Littman D.R. (2009) Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Gaboriau-Routhiau V., Rakotobe S., Lecuyer E., Mulder I., Lan A., Bridonneau C., Rochet V., Pisi A., De Paepe M., Brand G., Eberl G., Snel J., Kelly D., Cerf-Bensussan N. (2009) The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689. [DOI] [PubMed] [Google Scholar]
- 75).Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., Taniguchi T., Takeda K., Hori S., Ivanov I.I., Umesaki Y., Itoh K., Honda K. (2011) Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Nishio J., Honda K. (2012) Immunoregulation by the gut microbiota. Cell. Mol. Life Sci. 69, 3635–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. (2006) Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238. [DOI] [PubMed] [Google Scholar]
- 78).Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., Littman D.R. (2006) The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133. [DOI] [PubMed] [Google Scholar]
- 79).Littman D.R., Rudensky A.Y. (2010) Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858. [DOI] [PubMed] [Google Scholar]
- 80).Brand S. (2009) Crohn’s disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn’s disease. Gut 58, 1152–1167. [DOI] [PubMed] [Google Scholar]
- 81).Ivanov I.I., Frutos Rde L., Manel N., Yoshinaga K., Rifkin D.B., Sartor R.B., Finlay B.B., Littman D.R. (2008) Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82).Richardson P.M., Herman D.J., Tokunaga T.K., Wan J.M., Firestone M.K. (2006) Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72, 6288–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Yang Y., Torchinsky M.B., Gobert M., Xiong H., Xu M., Linehan J.L., Alonzo F., Ng C., Chen A., Lin X., Sczesnak A., Liao J.J., Torres V.J., Jenkins M.K., Lafaille J.J., Littman D.R. (2014) Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84).Goto Y., Panea C., Nakato G., Cebula A., Lee C., Diez M.G., Laufer T.M., Ignatowicz L., Ivanov I.I. (2014) Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40, 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85).Heczko U., Abe A., Finlay B.B. (2000) Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. J. Infect. Dis. 181, 1027–1033. [DOI] [PubMed] [Google Scholar]
- 86).Suzuki K., Meek B., Doi Y., Muramatsu M., Chiba T., Honjo T., Fagarasan S. (2004) Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. U.S.A. 17, 1981–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Fagarasan S., Kinoshita K., Muramatsu M., Ikuta K., Honjo T. (2001) In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature 413, 639–643. [DOI] [PubMed] [Google Scholar]
- 88).Wu H.J., Ivanov I.I., Darce J., Hattori K., Shima T., Umesaki Y., Littman D.R., Benoist C., Mathis D. (2010) Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89).Lee Y.K., Menezes J.S., Umesaki Y., Mazmanian S.K. (2011) Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl 1), 4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Yurkovetskiy L., Burrows M., Khan A.A., Graham L., Volchkov P., Becker L., Antonopoulos D., Umesaki Y., Chervonsky A.V. (2013) Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91).Kriegel M.A., Sefik E., Hill J.A., Wu H.J., Benoist C., Mathis D. (2011) Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 108, 11548–11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Tannock G.W., Miller J.R., Savage D.C. (1984) Host specificity of filamentous, segmented microorganisms adherent to the small bowel epithelium in mice and rats. Appl. Environ. Microbiol. 47, 441–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93).Snel J., Heinen P.P., Blok H.J., Carman R.J., Duncan A.J., Allen P.C., Collins M.D. (1995) Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int. J. Syst. Bacteriol. 45, 780–782. [DOI] [PubMed] [Google Scholar]
- 94).Imaoka A., Okada Y., Matsumoto S., Setoyama H., Umesaki Y. (1997) 16S Ribosomal DNA sequence divergence of segmented filamentous bacteria with special reference to inter-species and within-species variation of host animals. Syst. Appl. Microbiol. 20, 418–422. [Google Scholar]
- 95).Proctor M., Manning P.J. (1990) Production of immunoglobulin A protease by Streptococcus pneumoniae from animals. Infect. Immun. 58, 2733–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Jepson M.A., Clark M.A., Simmons N.L., Hirst B.H. (1993) Actin accumulation at sites of attachment of indigenous apathogenic segmented filamentous bacteria to mouse ileal epithelial cells. Infect. Immun. 61, 4001–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97).Rosenshine I., Donnenberg M.S., Kaper J.B., Finlay B.B. (1992) Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 11, 3551–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98).Podar M., Anderson I., Makarova K.S., Elkins J.G., Ivanova N., Wall M.A., Lykidis A., Mavromatis K., Sun H., Hudson M.E., Chen W., Deciu C., Hutchison D., Eads J.R., Anderson A., Fernandes F., Szeto E., Lapidus A., Kyrpides N.C., Saier M.H., Jr., Richardson P.M., Rachel R., Huber H., Eisen J.A., Koonin E.V., Keller M., Stetter K.O. (2008) A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol. 9, R158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99).Smith K.D., Andersen-Nissen E., Hayashi F., Strobe K., Bergman M.A., Barrett S.L., Cookson B.T., Aderem A. (2003) Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4, 1247–1253. [DOI] [PubMed] [Google Scholar]
- 100).Jacchieri S.G., Torquato R., Brentani R.R. (2003) Structural study of binding of flagellin by Toll-like receptor 5. J. Bacteriol. 185, 4243–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101).Pamp S.J., Harrington E.D., Quake S.R., Relman D.A., Blainey P.C. (2012) Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB). Genome Res. 22, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102).Kuwahara T., Ogura Y., Oshima K., Kurokawa K., Ooka T., Hirakawa H., Itoh T., Nakayama-Imaohji H., Ichimura M., Itoh K., Ishifune C., Maekawa Y., Yasutomo K., Hattori M., Hayashi T. (2011) The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 18, 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103).Sczesnak A., Segata N., Qin X., Gevers D., Petrosino J.F., Huttenhower C., Littman D.R., Ivanov I.I. (2011) The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 10, 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104).Jiang H.Q., Bos N.A., Cebra J.J. (2001) Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect. Immun. 69, 3611–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105).Yin Y., Wang Y., Zhu L., Liu W., Liao N., Jiang M., Zhu B., Yu H.D., Xiang C., Wang X. (2013) Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 7, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106).Jonsson H. (2013) Segmented filamentous bacteria in human ileostomy samples after high-fiber intake. FEMS Microbiol. Lett. 342, 24–29. [DOI] [PubMed] [Google Scholar]
- 107).Sanford S.E. (1991) Light and electron microscopic observations of a segmented filamentous bacterium attached to the mucosa of the terminal ileum of pigs. J. Vet. Diagn. Invest. 3, 328–333. [DOI] [PubMed] [Google Scholar]
- 108).Tajima K., Ohmori H., Tohno M., Ohtsu H., Tsukahara T., Aminov R. (2013) Segmented filamentous bacteria are a major group in terminal ileum of piglets. Anaerobe 23, 109–111. [DOI] [PubMed] [Google Scholar]
- 109).Hoskins J.D., Henk W.G., Abdelbaki Y.Z. (1982) Scanning electron microscopic study of the small intestine of dogs from birth to 337 days of age. Am. J. Vet. Res. 43, 1715–1720. [PubMed] [Google Scholar]
- 110).Davis C.P., Cleven D., Balish E., Yale C.E. (1977) Bacterial association in the gastrointestinal tract of beagle dogs. Appl. Environ. Microbiol. 34, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111).Ibaraki Y., Shibahara T., Kobayashi H., Ito N., Shinozuka Y., Kasuya K., Murakoshi N., Chikuba T., Kubo M. (2011) Multifocal granulomatous jejunitis associated with an argyrophilic gram-positive segmented filamentous bacterium in a Holstein cow. J. Comp. Pathol. 145, 118–121. [DOI] [PubMed] [Google Scholar]
- 112).Smith T.M. (1997) Segmented filamentous bacteria in the bovine small intestine. J. Comp. Pathol. 117, 185–190. [DOI] [PubMed] [Google Scholar]
- 113).Lowden S., Heath T. (1995) Segmented filamentous bacteria associated with lymphoid tissues in the ileum of horses. Res. Vet. Sci. 59, 272–274. [DOI] [PubMed] [Google Scholar]