Abstract
The WxL domain recently has been identified as a novel cell wall binding domain found in numerous predicted proteins within multiple Gram-positive bacterial species. However, little is known about the function of proteins containing this novel domain. Here, we identify and characterize 6 Enterococcus faecium proteins containing the WxL domain which, by reverse transcription-PCR (RT-PCR) and genomic analyses, are located in three similarly organized operons, deemed WxL loci A, B, and C. Western blotting, electron microscopy, and enzyme-linked immunosorbent assays (ELISAs) determined that genes of WxL loci A and C encode antigenic, cell surface proteins exposed at higher levels in clinical isolates than in commensal isolates. Secondary structural analyses of locus A recombinant WxL domain-containing proteins found they are rich in β-sheet structure and disordered segments. Using Biacore analyses, we discovered that recombinant WxL proteins from locus A bind human extracellular matrix proteins, specifically type I collagen and fibronectin. Proteins encoded by locus A also were found to bind to each other, suggesting a novel cell surface complex. Furthermore, bile salt survival assays and animal models using a mutant from which all three WxL loci were deleted revealed the involvement of WxL operons in bile salt stress and endocarditis pathogenesis. In summary, these studies extend our understanding of proteins containing the WxL domain and their potential impact on colonization and virulence in E. faecium and possibly other Gram-positive bacterial species.
INTRODUCTION
Enterococcus faecium is one of the “no ESKAPE” pathogens responsible for a considerable percentage of nosocomial infections; its ability to cause life-threatening infections and resistance to antibiotics cause considerable difficulties for patients and physicians (1). To date, many of the potential virulence determinants that have been described in E. faecium are MSCRAMMs (microbial surface components recognizing adhesive matrix molecules); these include adhesins or pili anchored to the cell surface by an LPxTG-like motif and contain IgG-like folds (2). Our group previously performed a bioinformatic search for novel surface proteins that might be virulence factors in the E. faecium endocarditis strain TX16 (also known as DO) (3). During this search, we identified a locus which included predicted proteins possessing the previously described WxL domain colocated with a predicted LPxTG-like protein (Fms6) (3, 4).
The WxL domain is comprised of several conserved residues along with a highly conserved YXXX(L/I/V)TWXLXXXP motif and a second WxL proximal motif in the last ∼120 to 190 C-terminal residues of a protein (4, 5). Genes encoding proteins with the WxL domain were first described in Lactobacillus plantarum as part of a novel gene cluster found in a subset of low-G+C-content Gram-positive bacterial species, one of which is E. faecium (6, 7). Bioinformatics and transcriptome data in L. plantarum predicted these loci form cell surface protein complexes involved in carbon source acquisition and stress survival adaptation (7). When studying Enterococcus faecalis, Brinster et al. used fusion constructs to demonstrate that the WxL domain conferred protein localization to the cell wall fraction (4). In Lactobacillus coryniformis, a WxL protein named Cpf (coaggregation-promoting factor) was shown to mediate coaggregation with itself and other pathogens (8). Schachtsiek et al. showed Cpf could be removed from cells by treatment with 5 M LiCl and subsequently could be reattached to the cell surface by using spent culture supernatants or recombinant Cpf, which resulted in restoration of the coaggregation. Therefore, it is highly probable that proteins containing the WxL domain are noncovalently anchored cell wall-associated proteins. This work was corroborated by Brinster et al. when E. faecalis WxL domain recombinant protein fusions were shown to bind other Gram-positive bacteria, and experiments indicated peptidoglycan as the binding ligand (4). Moreover, Brinster et al. also deleted one of the 27 WxL genes in E. faecalis V583 encoding an internalin-like protein (ElrA), which led to significant attenuation in a mouse peritonitis model and reduced bacterial dissemination to the spleen, liver, and host macrophages (9). However, it is unclear if the involvement of elrA in virulence is associated with WxL proteins in general, or if this observation was specific for the ElrA protein.
As little data exist regarding the function of these potentially important novel surface proteins, particularly their role in host-pathogen interaction, we sought to characterize the E. faecium proteins containing the WxL domain. Specifically, we aimed to characterize the genomic arrangement of loci containing WxL proteins, predict structural elements of proteins within specific WxL loci, and determine if certain proteins containing the WxL domain are involved in virulence or colonization-related functions in E. faecium.
MATERIALS AND METHODS
Strains, plasmids, and cultivation of bacteria.
All relevant characteristics of bacterial strains and plasmids used in this study can be found in Table S1 in the supplemental material. Unless otherwise stated, all Escherichia coli cells were grown in LB broth/agar, and all E. faecalis or E. faecium cells were grown in brain heart infusion (BHI) agar and broth at 37°C.
Identification, in silico genomic analysis, and structural analysis of putative WxL proteins.
The closed reference genome TX16 (endocarditis clade A isolate) was used to search for open reading frames (ORFs) encoding proteins with the WxL domain using NCBI BLAST. All ORFs generated through BLAST were searched for the two C-terminal WxL motifs (YXXX[L/I/V]TWXLXXXP) (Fig. 1) within the C-terminal 150 amino acid residues (4, 7, 10). The WxL domain was confirmed using CLUSTALW alignment with previously predicted WxL proteins (11). The accession number for TX16 (DO) is CP003583.1. The locus tags for WxL locus A of TX16 (DO) are HMPREF0351_12112 to HMPREF0351_12116, where SwpA is HMPREF0351_12116 and LwpA is HMPREF0351_12114. The locus tags for the WxL locus B of TX16 (DO) are HMPREF0351_10614 to HMPREF0351_10617, where SwpB is HMPREF0351_10617 and LwpB is HMPREF0351_10614. The locus tags for the WxL locus C of TX16 (DO) are HMPREF0351_10114 to HMPREF0351_10119, where SwpC is HMPREF0351_10114 and LwpC is HMPREF0351_10115. NCBI BLAST then was used to find the WxL loci in TX1330 (a community-associated [CA] commensal isolate of clade B) and TX82 (a hospital-associated [HA] clinical isolate of clade A that we have used extensively in experimental models) using the sequences obtained from TX16 (12–17). Confirmatory sequencing of the TX1330 and TX82 WxL loci were performed using the primers in Table S2 in the supplemental material. The gene organization surrounding the predicted WxL proteins also was analyzed (7). InterProScan determined protein family, domains, and functional sites (18). TMHMM v2.0 and TMpredServer determined possible transmembrane domains (19). Signal P v3.0 determined the presence of N-terminal signal sequences (20). SoftBerryBPROM and SoftBerryFindTerm, as well as an RNA secondary structure predictor (www.genebee.msu.su), were used to predict promoters and terminators of the loci (21). Secondary and tertiary structures were predicted using PHYRE2 (22). DisEMBL v1.5 predicted intrinsic protein disorder (23). Nucleotide sequences of WxL protein-encoding genes were found using BLAST analyses in 21 E. faecium strains available from NCBI. Pairwise analysis was performed using ClustalW alignment of the amino acid sequences. Percent identity matrices were created using Geneious software, version 6.1 (see Table S3) (24).
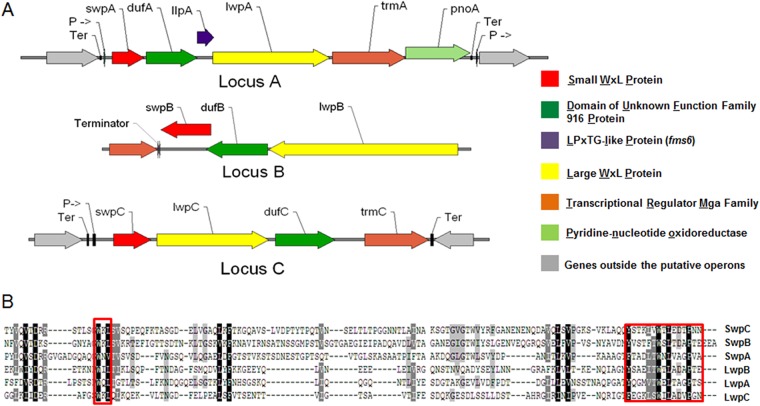
FIG 1.
WxL operons and domains of E. faecium TX82. (A) A pictorial representation of the three putative operons (loci A, B, and C) containing the 6 genes found to have the WxL domain found in E. faecium TX82 based on in silico analyses. Each arrow represents a predicted ORF named for the protein it encodes. Small WxL proteins are in red, DUF916 family proteins are in green, LPxTG-like proteins are in purple, Large WxL proteins are in yellow, transcriptional regulators are in orange, and genes outside the operon are in gray. Predicted terminators (Ter) and promoters (P->) also are labeled. (B) An alignment of the C terminus of the 6 WxL proteins found in E. faecium TX82. Highly conserved residues are in black, moderately conserved residues are in gray, and the red boxes indicate the YXXX(L/I/V)TWXLXXXP terminal motif and the second WxL proximal motif.
Colony hybridizations, RNA extraction, and RT-PCR.
Preparation of colony lysates on nylon membranes and colony hybridization under high-stringency conditions were performed as described previously (25). DNA probes for the WxL locus genes and intergenic regions were obtained with the primers listed in Table S4 and S5 in the supplemental material, respectively. Total RNA extraction from TX1330 and TX82, grown to mid-log phase in 10 ml of BHI, and reverse transcription-PCRs (RT-PCRs) were performed as described previously (3).
Construction of expression plasmids and purification of recombinant proteins.
Genomic DNA was extracted from TX1330 and TX82 using a DNeasy blood and tissue kit (Qiagen). DNA regions of SwpATX1330, SwpATX82, LwpATX1330, LwpATX82, DufATX82, SwpCTX1330, SwpCTX82, LwpCTX1330, and LwpCTX82 were amplified using the primers listed in Table S6 in the supplemental material (all recombinant proteins lack the signal peptide, LwpA and LwpC lack the WxL domain, and DufA lacks the transmembrane domains). Fragments were cloned into the expression vector pQE30 (Qiagen) and transformed into M15 (pREP4) cells (Qiagen). Recombinant proteins with N-terminal His6 tags were expressed and purified with fast-performance liquid chromatography (FPLC) using a 5-ml nickel-charged HiTrap chelating column (GE Healthcare) followed by a 5-ml anion exchange Sepharose column (GE Healthcare) (26). Fractions obtained from the anion exchange column were concentrated and dialyzed against phosphate-buffered saline (PBS) with 10 mM EDTA (at the appropriate pH for the pI of each recombinant protein). Protein concentrations were determined by absorption spectroscopy at 280 nm using calculated molar absorption coefficient values. Molecular masses were determined with matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS). Predicted molecular weights, theoretical pI, and extinction coefficients were determined using the ExPASy ProtParam tool (see Table S7 in the supplemental material) (27).
Production of polyclonal antibodies and purification of antigen-specific antibodies.
Production of antiserum was performed in Sprague-Dawley rats against individual WxL recombinant proteins from TX1330 as previously described (28). Antisera were tested for antibody titers by enzyme-linked immunosorbent assay (ELISA) (29). Antigen-specific antibodies were purified as described previously using the corresponding antigen coupled to CNBr-activated Sepharose columns (3). The antibodies were concentrated by ultrafiltration, and concentrations were determined via absorption spectroscopy.
Mutanolysin cell wall extraction and Western blot analysis.
Mutanolysin extracts of late-exponential-phase cells were prepared as described previously (29, 30). Protein concentrations were determined via bicinchoninic acid (BCA) assay according to the manufacturer's instructions (Pierce). Equal amounts of mutanolysin cell wall extracts were run on 4 to 15% gradient SDS-PAGE gels under reducing conditions, transferred to polyvinylidene difluoride (PVDF) membranes, and probed using the specific antibodies to SwpA, LwpA, SwpC, and LwpC, followed by horseradish peroxidase (HRP)-conjugated anti-rat IgG secondary antibodies. Antisera from preimmune rats were used as a control.
Immunogold electron microscopy.
Immunogold electron microscopy was performed as described previously (31). Briefly, TX82 cells were grown on BHI agar overnight. The cells were harvested from the plates by scraping and then washed with 0.1 M NaCl. For immunogold labeling, the bacterial suspension was spotted and fixed onto carbon grids, washed three times with 2% bovine serum albumin (BSA) in PBS, and blocked with 0.1% gelatin for 1 h. The samples then were incubated with the specific anti-WxL antibody or preimmune serum in a 1:100 or 1:50 dilution for 1 h, followed by washing and blocking as described above. The samples then were treated with 12-nm or 18-nm gold–anti-rat IgG (1:20 dilution) in 2% BSA in PBS for 1 h. The samples then were washed with water and stained with 1% uranyl acetate and viewed using a Jeol 1400 transmission electron microscope.
Whole-cell ELISA.
The presence of SwpA, LwpA, SwpC, and LwpC on the surface of bacterial cells was detected using affinity-purified antigen-specific antibodies against individual WxL recombinant proteins from TX1330 in a whole-cell ELISA as previously described (32). Antiserum against formalin-killed TX16 cells was used as a positive control to verify equal binding of whole cells to the microtiter plate. Statistical significance (P values) of the surface exposure of each WxL protein between clinical and community strains was calculated using an unpaired t test.
Detection of antibodies to WxL proteins in patient sera.
Detection of antibodies in patient sera was performed as described previously (33). Briefly, recombinant SwpA, LwpA, SwpC, and LwpC from TX1330 was run on 12% SDS-PAGE gels and transferred to PVDF membranes. The membranes were blocked with 2% milk, 0.1% Tween 20 and incubated with either endocarditis patient serum or healthy volunteer serum (1:2,000). After washing with PBS-Tween (PBST), antibodies were detected with goat anti-human IgG (1:5,000) conjugated to HRP. The Western blot was developed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
CD spectroscopy.
Recombinant proteins DufA, LwpA, and SwpA were dialyzed in PBS buffer (0.8 mM Na2HPO4, 0.2 mM KH2PO4, 0.3 mM KCl, 13.7 mM NaCl, pH 7.4). Circular dichroism (CD) measurements in the far-UV region (190 to 260 nm) were carried out at ambient temperature on a Jasco J-720 spectropolarimeter (Easton, MD) using a 0.5-mm cuvette. Ten scans were collected and averaged at a scan speed of 100 nm/min, with a time constant of 2 s and bandwidth of 1 nm. The spectra were background corrected with the CD signal obtained from the buffer. Analysis of the spectra was performed on the DichroWeb server (http://dichroweb.cryst.bbk.ac.uk). Secondary-structure composition for each protein is an average of the deconvolution results from SELCON, CDSSTR, and CONTINLL using reference data set 4.
SPR.
Surface plasmon resonance (SPR)-based Biacore binding experiments were performed at 25°C on a Biacore 3000 (GE Healthcare). Using an amine-coupling procedure as recommended by the manufacturer, 20 μg/ml fibronectin (Fn; Millipore), 20 μg/ml Fn-NTD (Sigma), and 10 μg/ml Fn-Hep2 (Millipore) were immobilized on a CM3 sensor chip. Rat tail collagen I (20 μg/ml; R&D Systems), collagen IV from human cell culture (Sigma), laminin (Invitrogen), and fibrinogen (Enzyme Research laboratory) were immobilized on CM5 sensor chips. DufA (20 μg/ml), SwpA (5 μg/ml), and LwpA (10 μg/ml) also were immobilized on CM5 sensor chips. The immobilization solution (10 mM sodium acetate, pH 5.5) was used for most of the proteins, except for laminin and collagen IV, where 50 mM sodium acetate, pH 4.0, was used. A reference surface was prepared with activation and deactivation treatments but no protein coupled. PBS with 0.005% Tween 20 was used as the running buffer. Soluble proteins at different concentrations were injected onto the immobilized ligand surface to obtain SPR response curves (sensorgrams). Baseline-corrected sensorgrams (with the buffer blank run further subtracted) were globally fitted to a predefined binding model using BIAevaluation software (version 4.1).
Construction of WxL deletion mutants.
Mutants were created using a PheS*-derived system as described previously (34). For deletion of WxL loci, the deletion construct fragments were amplified using the primers found in Table S8 in the supplemental material and cloned into pHOU1. The insert was confirmed by sequencing and the plasmid electroporated into E. faecalis CK111. Blue colonies were recovered on BHI gentamicin (125 μg/ml)–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (200 μg/ml) plates. Recombinant plasmids then were introduced into TX82 through filter mating with CK111 as the donor. Single-crossover integrants then were selected on BHI gentamicin-vancomycin (50 μg/ml) or BHI gentamicin-erythromycin (200 μg/ml) plates, replica plated onto BHI gentamicin-ampicillin (125 μg/ml) plates, and confirmed by sequencing. Positive colonies then were streaked onto MM9YEG plates with 10 mM 4-chloro-dl-phenylalanine (p-Chl-Phe) and incubated at 37°C. Colonies that grew on p-Chl-Phe were replica plated on BHI gentamicin to confirm the excision of the plasmid. Colonies susceptible to gentamicin were screened by PCR for the deletion and sequenced, and pulsed-field gel electrophoresis (PFGE) was performed to confirm the parental background. The ΔwxlABC triple-locus mutant was made in the following sequence: WxL locus B, then WxL locus A, and finally WxL locus C (22).
Bile salt assay.
Resistance to bile salts after 24 h was determined as described previously (35). E. faecium cells were exposed to BHI with 0.3% bile salts for 24 h. CFU were determined at t = 0 min and t = 24 h; the fraction of cells that survived was calculated by dividing the CFU after 24 h by the CFU at time zero (35). An unpaired t test was used to compare the mean survival values between strains.
Experimental endocarditis.
Aortic valve endocarditis was induced in white Sprague-Dawley rats as previously published (28). Overnight-grown wild-type (WT; TX82) and ΔwxlABC mutant cells were harvested, the concentrations were adjusted using optical density, the cells were suspended together in saline (1:1), and they were intravenously inoculated via the tail vein 24 h after catheterization. The percentage of each strain used in the inoculum was verified by plating CFU. TX82 and ΔwxlABC cells comprised 44.5% and 55.5% of the mixed inocula, respectively, based on two independent experiments (mean percent). Animals were euthanized ∼48 h after bacterial inoculation, and hearts were aseptically removed. The vegetation was excised, weighed in preweighed sterile tubes, and homogenized in 1 ml of saline, and dilutions were plated onto Enterococcosel agar plus 6 μg/ml vancomycin. After ∼48 h, all colonies that grew (up to 47 CFU/rat) were picked into microtiter plate wells containing brucella broth plus 15% glycerol, grown overnight, and used to prepare DNA lysates on Hybond N+ membranes (GE Healthcare), followed by high-stringency hybridization (25), using intragenic DNA probes for ddl (36) and lwpB (wxlB-F, GGTGGATTCGCTCACAATTTCGG; wxlB-R, GGATGATTCCGCGATTGTTGTCATTGG) in order to generate the percentages of TX82 and ΔwxlABC colonies recovered from tissue homogenate. Data were expressed as percentages of the WT and mutant per tissue homogenate and analyzed by the paired t test using GraphPad Prism v4.0. The animal procedures were carried out by following the animal welfare committee guidelines and approved protocol at the University of Texas Health Science Center at Houston.
RESULTS
Six genes encoding WxL proteins in Enterococcus faecium are located among three similarly organized operons.
In the closed reference genome of TX16, also known as DO, six genes encoding proteins with the WxL domain were found. The endocarditis isolate, TX82 (clade A), and a community fecal isolate TX1330 (clade B), strains we have used extensively, also were analyzed for the presence of the 6 genes encoding WxL proteins, and the same 6 were found (Fig. 1) (13, 37). In silico analysis determined that these are in three clusters flanked by predicted rho-independent terminators in each genome, resembling the pattern found in E. faecalis and L. plantarum clusters (Fig. 1) (7, 9). All three clusters (named WxL loci A, B, and C) encode proteins which can be divided into 3 different classes based on size, domains, and motif characteristics described previously (7): (i) a small WxL protein (SwpA-C), analogous to CscB in Lactobacillus plantarum; (ii) a large WxL protein (LwpA-C), analogous to CscC; and (iii) a DUF916 (domain of unknown function) family protein (DufA-C), analogous to CscA (7). Only WxL locus A encoded a protein with an LPxTG-like motif (Fms6), analogous to CscD (7). Locus A and locus C both are associated with Mga-like transcriptional regulators, which often are regulators of virulence genes important for colonization and immune evasion (38). Directly upstream of locus B is a LytR-family transcriptional regulator which has been shown to be involved in extracellular polysaccharide biosynthesis, fimbriation, expression of exoproteins, and cell autolysis (39).
Bioinformatic analyses indicated that each of the three gene clusters were operons. To validate this prediction, we carried out RT-PCR of the individual WxL genes (data not shown) and the intergenic regions using the strains TX1330 (data not shown) and TX82 (see Tables S4 and S5 and Fig. S1 in the supplemental material). Our results indicate that the 6 genes encoding proteins with the WxL domain are expressed under standard laboratory conditions in both hospital-associated (clade A) and community-associated commensal (clade B) strains and support the operon structure depicted in Fig. 1.
The WxL genes from all three loci are widespread among E. faecium isolates; however, the loci differ between HA and CA clades.
As introduced above, we recently reported the existence of two subpopulations of E. faecium, one containing primarily community-associated strains (CA clade; also called clade B) and one containing mostly hospital-associated strains (HA clade; also called clade A) (12, 15). These two clades differ by ∼3.5 to 4.2% of their nucleotides at the core genome level. The swpC gene (previously called wlcA, for WxL Locus C protein A) was one of the alleles used to demonstrate clade separation in our previous report (40). Thus, we analyzed the distribution and homology of the genes within the three WxL operons using 21 sequenced E. faecium genomes available from NCBI. All genes within the three loci have homologs within the 21 genomes. Pairwise analysis using all three WxL loci and deduced amino acid sequences from these strains determined that the nucleotide identity of WxL genes ranges from 85 to 100%, with as much as 16.8% amino acid difference between the WxL locus proteins (DufC) derived from HA and CA clade strains (see Table S3 in the supplemental material).
In addition to examining sequenced genomes, colony hybridizations were performed under high-stringency conditions using probes (see Table S4 in the supplemental material) for each individual WxL gene in 54 unspecified clinical infection, 10 community fecal, and 20 endocarditis isolates from our collection. While swpA, lwpA, and swpC were present in all 84 strains tested, lwpB, swpB, and lwpC were present in most, but not all, strains (90 to 98%) (data not shown). These results indicate that the WxL proteins are found in most E. faecium isolates regardless of origin, a finding that differs from those for other surface proteins, which typically are enriched in isolates of clinical origin (41).
WxL locus A and C protein structure predictions.
We continued with the analysis of WxL proteins from loci A and C in the clinical strain TX82. Locus B was not examined due to difficulties generating the complete sequence and inconsistencies found between sequenced strains. Full-length sequences of TX82 SwpA, LwpA, DufA, SwpC, LwpC, and DufC were analyzed using different protein servers. Analysis using PHYRE2 could not model SwpA, LwpA, or SwpC to a template with any reasonable confidence, while analysis using DisEMBL predicted that all three were highly disordered. Of note, MSCRAMMS also have highly disordered regions, which contributes to their ability to bind different host ECMs (42, 43). Sections of DufA and DufC both modeled to the three-dimensional model of the human transglutaminase 32 enzyme with the highest confidence level (98.8%), using 59% and 52% of the protein sequence, respectively. It is thought that bacterial proteins harboring structural domains similar to eukaryotic transglutaminases can function as proteases or cross-linking enzymes, although presently it is unclear if DufA or DufC has any enzymatic activity (44). A section of LwpA modeled to a concanavalin A (ConA)-like lectin with 100% confidence for 30% of the protein. Lectins and glucanases exhibit the common property of binding to specific complex carbohydrates, and ConA-like domains are found in bacterial proteins involved in cell recognition and adhesion (45, 46).
Cell surface expression of WxL locus A and C proteins.
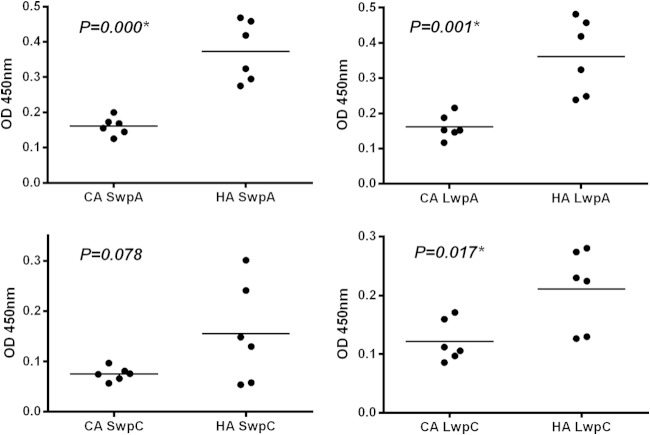
Cell wall extracts of TX82 cells generated by mutanolysin digestion were run on SDS-PAGE gels and probed with specific antibodies generated against recombinant SwpA, LwpA, SwpC, and LwpC from TX1330; each protein was found in the cell wall extracts (see Fig. S2 in the supplemental material). Six clinical and 6 community isolates then were tested by whole-cell ELISA to determine if all isolates had similar cell surface expression of WxL proteins. Although locus A and C WxL proteins were detected on the cell surface of all strains tested, HA strains gave statistically higher optical densities (approximately 2-fold higher) than CA strains for SwpA, LwpA, and LwpC (Fig. 2), even though the antibodies used had been generated against recombinant proteins from the community isolate TX1330. Results for SwpC were not statistically significant; however, the trend was similar to that for the other 3 WxL proteins. Immunogold electron microscopy (IEM) experiments also indicate that SwpC and LwpC were expressed on the surface of TX82 cells (see Fig. S3).
FIG 2.
Whole-cell ELISAs of E. faecium cells reveal differences in surface display of WxL proteins between community and hospital clade isolates. Whole-cell ELISA of native cells using 6 community-associated (CA) and 6 hospital-associated (HA) clade E. faecium strains was performed, and the surface display of WxL proteins was detected using specific antibodies to recombinant SwpA, LwpA, SwpC, or LwpC from E. faecium TX1330. Three technical replicates were performed in two independent experiments. Standard deviations were negligible, so error bars cannot be seen. Unpaired t test was used to compare surface expression levels between the community- and hospital-associated groups for each WxL protein.
Antibodies to WxL locus A and C proteins are present in serum from patients with E. faecium endocarditis.
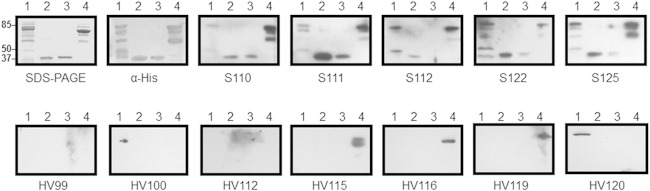
Since WxL proteins are surface expressed in vitro, we sought to determine if patients with E. faecium infections have antibodies to WxL proteins. Using Western blot analysis, we determined that all 5 endocarditis patient sera tested contained antibodies to all four WxL recombinant proteins analyzed (SwpA, LwpA, SwpC, and LwpC from TX1330) (Fig. 3). This differs from healthy volunteers, whose sera either lacked antibodies to all four WxL proteins tested (3/7) or had antibodies to only one of the four (4/7). Interestingly, the antibodies found in healthy volunteers were always to one of the large WxL proteins: three healthy volunteers had LwpC antibodies, and one had LwpA antibodies (Fig. 3).
FIG 3.
Western blot analyses using endocarditis patient sera demonstrate the in vivo immunogenicity of WxL proteins from loci A and C. Recombinant proteins from E. faecium strain TX1330 (lanes: 1, LwpA; 2, SwpA; 3, SwpC; 4, LwpC) were run on 12% SDS-PAGE gels and transferred to PVDF membranes. Each membrane was incubated with either endocarditis patient serum (S) or healthy volunteer serum (HV) (1:2,000). This was followed by goat anti-human IgG conjugated to HRP and developed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). The first gel is a Coomassie-stained gel. The second gel is the Western blot using antibody to the His tag. The numbers underneath each gel represent the patient number arbitrarily assigned.
Secondary structural analysis of recombinant WxL proteins from locus A.
We continued our study using WxL locus A from TX82, as this was the most consistent locus; that is, the genes located upstream and downstream are the same in all isolates examined, and this locus has the least diversity at the amino acid level. Recombinant protein characteristics can be found in Table S7 in the supplemental material. During the protein purification process, SwpA was obtained in two independent fractions, represented by two peaks separated by multiple fractions in both the nickel and Q column, indicating the same protein had two forms with different folding and charge. We later inferred that these were a monomer and dimer form of SwpA by comparing the size of the two independent fractions on native and reducing SDS-PAGE gels in addition to Western blot analyses. To determine the secondary structure of WxL locus A proteins from TX82, the recombinant SwpA (monomer and dimer), DufA, and LwpA were analyzed by circular dichroism (CD) spectroscopy in the far-UV region and deconvoluted using the online server Dichroweb (47). Consistent with in silico prediction described above and as shown in Fig. S4, all proteins are predominantly disordered (∼40%) and contain only 5 to 12% α-helix. Among these, DufA contains the highest percentage of α-helical structure, as indicated by two minimum bands between 205 and 225 nm, and LwpA has the highest β-sheet content, indicated by a minimum at ∼220 nm. Compared to the SwpA monomer, the dimer contains less disorder and more α-helix, as shown by a slightly stronger minimum band beyond 210 nm.
Recombinant proteins from the WxL locus A bind human type I collagen and fibronectin.
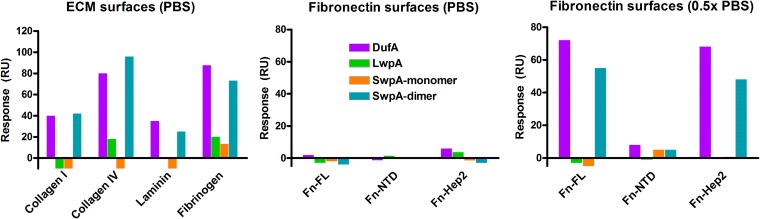
Surface plasmon resonance (SPR) was used to detect the binding of recombinant SwpA (monomer and dimer), LwpA, and DufA from TX82 to multiple immobilized human extracellular matrix proteins (ECMs) (Fig. 4). In addition to showing that all three recombinant proteins bound a number of different ECM proteins (including human collagen type I [CI], collagen type IV [CIV], fibrinogen [Fg], and fibronectin [Fn]), these preliminary analyses also showed differences in the activities between the monomer and dimer forms of SwpA and the importance of salt concentration (PBS versus 0.5× PBS) for their binding to Fn and its fragment, Hep2-40K (Fig. 4). Due to these observations, we continued our analyses with only the dimer form of SwpA and used 0.5× PBS (low salt) for Fn interaction analysis. The interactions between TX82 WxL locus A recombinant proteins and human extracellular proteins were further characterized using Biacore analyses, which revealed a range of KD values of 0.005 μM (Fn) to 1.34 μM (Fg) for DufA and 0.07 μM (CI) to 0.87 μM (Fg) for SwpA (dimer) (Table 1; also see Fig. S5 in the supplemental material). Thus, WxL locus A proteins exhibit the highest affinity for Fn and CI, while both showed the weakest affinity (although still detectable) for Fg. These KD values are comparable to (and, in some cases, stronger than) those for other cases of ECM binding in proteins of the same or similar species, such as recombinant Acm of E. faecium, which binds CI with a KD of 3.8 μM, recombinant Sfb1-4 of Streptococcus pyogenes, which binds the NTD region of Fn with KD from 60 nM, and recombinant Fss1 of E. faecium, which binds Fg with a KD of 0.7 μM (32, 48, 49). Interestingly, to date very few fibronectin binding proteins have been reported in E. faecium, and the binding kinetics of these proteins have not been determined. The finding that DufA and SwpA binding to CI and CIV are similar in affinity and SPR profile suggests that DufA and SwpA recognize a similar triple-helical collagenous structure in CI and CIV (Table 1; also see Fig. S5 in the supplemental material). LwpA was not found to interact with either immobilized CI or Fn; however, when LwpA was immobilized, it showed binding to soluble Fn (Table 1; also see Fig. S6). We further found the binding of DufA and SwpA to be specific for the C-terminal heparin-binding fragment (Hep2-40K) of Fn, with affinities of 0.075 μM and 0.60 μM, respectively. However, both soluble SwpA and DufA bound immobilized full-length fibronectin with a greater affinity than immobilized Hep2-40K fragment (Fig. 4 and Table 1; also see Fig. S5).
FIG 4.
Biacore detection of the interactions between soluble WxL and immobilized ECM proteins. SPR responses were generated from (A) binding of each WxL protein (about 100 μg/ml in PBST) to immobilized collagen I, IV, laminin, and fibrinogen and (B and C) binding of 30 μg/ml of WxL proteins to full-length fibronectin or its fragments (NTD and Hep2) under different buffer conditions (PBS or 0.5× PBS). All buffers contained 0.005% Tween 20. RU, resonance units.
TABLE 1.
Binding interactions (KD) between E. faecium WxL locus A recombinant proteins and human ECM as determined by Biacore analysis
| ECM |
KD (μM) between ECM anda: |
||
|---|---|---|---|
| DufA | LwpA | SwpA | |
| Collagen I | 0.63 | NBb | 0.07 |
| Collagen IV | 0.41 | NDc | 0.08 |
| Fibrinogen | 1.34 | ND | 0.87 |
| Fibronectin | 0.005 (1.39)d | NB (0.53) | 0.20 (1.92) |
| Fn-Hep2 | 0.075 (0.69) | NB (0.25) | 0.60 (0.51) |
The experimental data for the binding affinities (KD) of soluble WxL to immobilized ECM proteins are shown in Fig. S4 in the supplemental material.
NB, no binding.
ND, KD not determined due to low signal and lack of saturated binding.
The KD values listed in parentheses represent the reverse bindings (soluble ECM to immobilized WxL), which were performed with full-length soluble Fn and the Hep2 fragment (experimental data are shown in Fig. S5).
DufA shows self-association and association with both SwpA and LwpA.
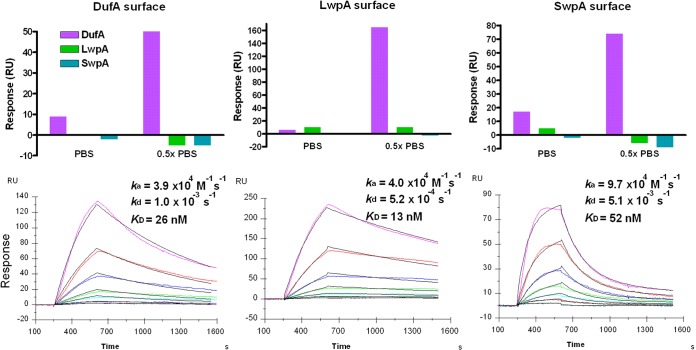
Because our hypothesis was that proteins from the same WxL operon assemble into a cell surface complex, Biacore was used to detect binding between the recombinant proteins from locus A. At first, we did not detect binding in physiological salt concentrations. Due to our previous observation that Fn binding was favorable in 0.5× PBS, we performed intrinsic tryptophan fluorescence spectroscopy, which confirmed that salt concentrations affect WxL protein conformation (see Fig. S7 in the supplemental material). Thus, we continued with 0.5× PBS, showing that only DufA recognized all WxL immobilized protein surfaces, including itself, with no detectable interaction observed between SwpA and LwpA (Fig. 5). DufA bound WxL proteins with high affinity (KD of 26 nM for DufA, 13 nM for LwpA, and 52 nm for SwpA) but with different kinetic profiles (Fig. 5). DufA binding to SwpA was faster than that to DufA and LwpA, which is indicated by a faster on-rate (ka of 9.7 × 104 compared to ∼4 × 104 M−1 s−1), while a more stable complex was seen with LwpA due to its slow off-rate (kd of 5.2 × 10−4 s−1).
FIG 5.
Biacore analysis of the interactions among WxL proteins. (A) SPR responses were obtained from a 1-min injection of each WxL protein (about 1 μM in running buffer) over the immobilized WxL surfaces in PBS or 0.5× PBS (all buffers contained 0.005% Tween 20). (B) SPR sensorgrams were generated from the injection of soluble DufA (2-fold linear dilution ranging from 1 to 64 nM) for 6 min over different WxL surfaces. Kinetic data were obtained by fitting a 1:1 (Langmuir) binding model (black lines) to the sensorgrams (colored lines, with the lower curves corresponding to lower concentrations of protein injected).
Deletion of WxL locus genes increases survival in bile salts.
In a previous study, transcriptional analysis of lactobacilli showed that WxL genes are regulated in response to bile, salt, and lactate stress (50, 51). To further investigate possible involvement of the WxL genes in bile salt stress, nonpolar WxL single-locus deletion mutants and a triple-locus deletion mutant were tested in a bile salt survival assay and compared to the wild type. It was found that all WxL mutants survived better in bile salt stress conditions than the parental strain, with the ΔwxlABC triple-locus mutant surviving the best (Fig. 6). The differences between all mutants and the parental strain TX82 were statistically significant at P < 0.0001.
FIG 6.
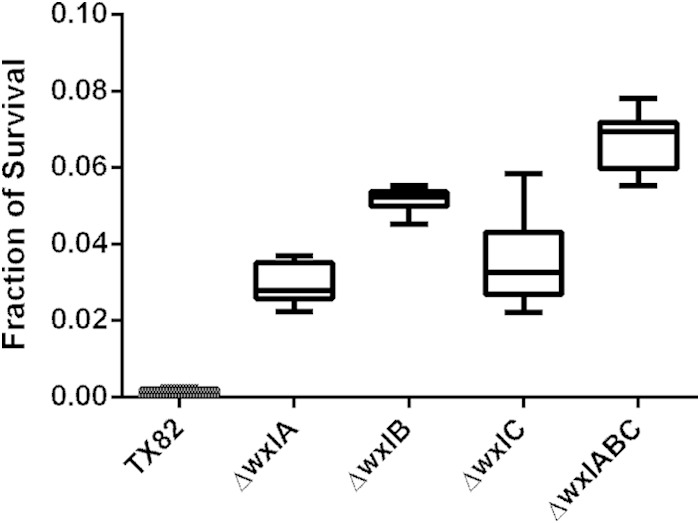
WxL mutants survive better that the parental strain after 24 h of bile salt exposure. E. faecium cells were exposed to BHI with 0.3% bile salts for 24 h. CFU were determined at t = 0 min and t = 24 h in two separate experiments, and the fractions of cells that survived after bile salt exposure for each strain were determined. The results shown represent the minimum, maximum, means, and standard deviations from 6 biological replicates, with three technical replicates each. An unpaired t test determined the difference in mean survival between all mutants and the parental strain to be statistically significant at P < 0.0001.
Deletion of all 3 WxL loci results in a decreased ability to cause infective endocarditis compared to that of the parental strain.
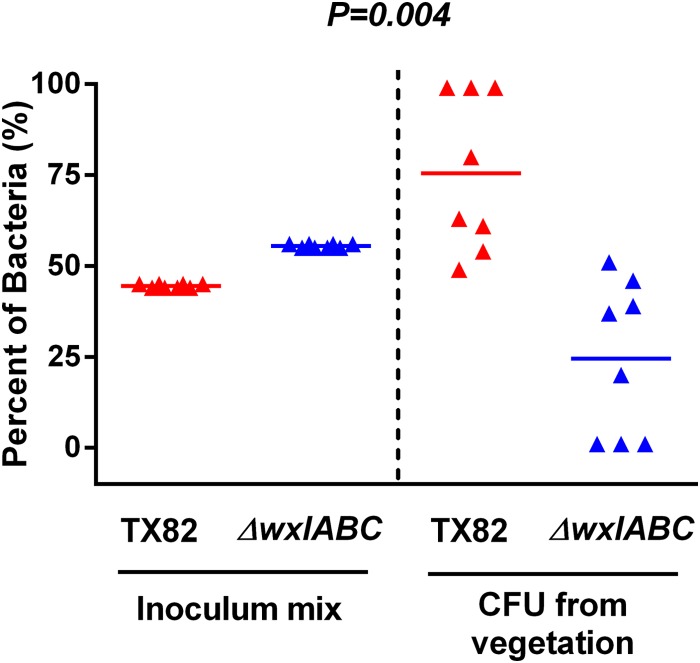
In order to further support the notion that WxL proteins are involved in virulence, we compared strain TX82 and its ΔwxlABC isogenic mutant for the ability to infect heart valves in an experimental rat endocarditis model. Our data showed that the ΔwxlABC mutant is outcompeted by the parental strain in rat endocarditis, as recovered heart vegetation contained significantly fewer ΔwxlABC colonies than wild-type TX82 colonies (P = 0.004) (Fig. 7). Although we have been unable to complement the WxL triple-locus mutant, the likelihood that extraneous mutations caused the attenuation in endocarditis is lessened by the fact that we did not see attenuation in a mouse peritonitis model or urinary tract infection model using the ΔwxlABC mutant (data not shown). In addition, we do not believe that these results are due to a growth defect, as TX82 and the ΔwxlABC mutant grow similarly in vitro. These data indicate the specificity of WxL operons for involvement in endocarditis pathogenesis, which we postulate is due to the WxL proteins' ability to bind host ECM proteins (Fig. 4 and Table 1; also see Fig. S5 and S6 in the supplemental material).
FIG 7.
Parental strain TX82 outcompetes the ΔwxlABC strain in a mixed infection assay of rat endocarditis. Percentages of TX82 and ΔwxlABC strains present in inocula (left side) and recovered from vegetation 48 h postinfection (right side) in 16 rats. Horizontal bars represent the means (P = 0.004 by paired t test) for percentages of total bacteria in the vegetation of TX82 versus those of the ΔwxlABC strain.
DISCUSSION
Proteins containing the WxL domain are a novel class of cell surface proteins that are widespread among low-G+C-content Gram-positive species. However, studies of WxL proteins have been published for only three bacterial species (4, 7, 8). These studies have been limited, focusing mostly on the role of the WxL domain in surface localization and peptidoglycan binding (4, 7, 8). Thus, further characterization and functional studies on the role of these novel surface proteins was needed.
Understanding the in vivo function of surface proteins is essential to explain the success of bacterial pathogens. Here, studies on the role of WxL surface proteins in adherence to host tissues discovered the ability of recombinant E. faecium WxL locus A proteins to bind human Fn and CI (Fig. 4 and Table 1; also see Fig. S5 and S6 in the supplemental material). Adherence of bacterial cells to various components of cardiac tissue is an important event in the pathogenesis of infective endocarditis (52). Fibronectin is an important component of the extracellular matrix of endothelium and is exposed when these tissues are injured, whereas valvular and aortic surfaces are rich in collagen (53, 54). Thus, it is likely that the ability of WxL locus proteins of E. faecium to bind fibronectin and collagen is important in colonization and infection. This is reflected in the significantly reduced presence of ΔwxlABC mutant versus wild-type TX82 colonies found in the vegetation collected from the mixed infection model of experimental endocarditis (Fig. 7).
Our data also revealed that DufA and SwpA bind a C-terminal fragment of Fn, that is, the Hep2-40K region (Fig. 4 and Table 1; also see Fig. S5 and S6 in the supplemental material), which includes Fn type III modules 12 to 14 and a variable region (55). This differs from most fibronectin-binding MSCRAMMs, such as FnBPA from Staphylococcus aureus and BBK32 from Borrelia burgdorferi, which target the N-terminal region, including NTD-30K, to which SwpA and DufA did not bind (Fig. 4 and Table 1) (56–58). To our knowledge, out of the many Fn binding proteins that have been identified, only a few bind the Hep2-40K region (59–62).
Interestingly, another function we determined for the WxL operons is an involvement in bile salt stress. Previous transcriptional analysis of Lactobacillus plantarum found that WxL genes from 2 loci were downregulated in response to bile stress (50). Our data showed that WxL locus mutants in E. faecium actually survive better than the parental strain TX82 after bile salt stress (Fig. 6). One possibility is that WxL operons allow some uptake of bile, which can be toxic in high concentrations. This suggests that during passage through the small bowel, where bile is high, these would be downregulated. In this scenario, when WxL operons are deleted, bile is no longer taken up; therefore, the mutants survive better under bile salt stress. Alternatively, perhaps the regulators associated with the WxL operons have a role in downregulating transporters that may be involved in pumping bile salt out. Loss of this repression may allow for greater survival under bile salt stress. In addition, deletion of the WxL operons might change the cell wall composition and result in some change in cell wall integrity that may be beneficial under bile salt stress. In addition to bile salt stress, we tested the WxL locus mutants' ability to form biofilms. However, we did not find any significant differences between the WxL locus mutant strains and the parental strain in their ability to form biofilm (data not shown).
Although phylogenetic and sequence analyses demonstrated genes encoding WxL proteins of E. faecium segregate into the two clade patterns found previously for this species (see Table S3 in the supplemental material), there is more variability in WxL alleles than in other E. faecium genes previously analyzed (12, 40). Moreover, even though the genes are mostly ubiquitous among the strains studied, there is a difference in surface display of the WxL proteins depending on the strain origin (Fig. 2). This is possibly a result of the numerous amino acid differences between the two types of strains, which may result in altered structures, posttranslational modification, processing at the cell surface, or regulation. In addition, the greater surface exposure seen in HA clade strains could reflect an enhanced capacity for virulence (i.e., better able to bind host ECM versus CA clade strains).
The fact that the genes within WxL loci were cotranscribed suggests a novel assembly process at the cell surface (see Fig. S1 in the supplemental material). In support of assembly of these proteins, Biacore results showed binding of DufA to itself, SwpA, and LwpA (Fig. 5). It also was found that mixed proteins formed large visible aggregates within several hours under low-salt conditions (data not shown). Interestingly, Biacore data showed that SwpA and LwpA did not bind to each other (Fig. 5), consistent with the immunogold micrographs using double labeling, which did not show colocalization of SwpC and LwpC (data not shown). Seemingly, the assembly of these proteins at the cell surface does not include a direct interaction between the large and small WxL proteins but may involve interactions with the Duf proteins. Also of interest is how LPxTG-like proteins, like the fms6 protein or others found in other species (CscD-like), play a role in cell wall surface assembly, as they are not found in all WxL operons (i.e., in E. faecium, only in WxL locus A).
Seizen et al. speculated a role for WxL clusters in carbohydrate metabolism due to the presence of concanavalin A-like lectin domains (for carbohydrate binding/degradation) and CRE (catabolite response elements) binding sites upstream or inside the gene clusters and showed WxL proteins were regulated by ccpA in lactobacilli (7). Interestingly, although we found concanavalin A-like lectin domains within LwpA, we did not find previously defined CRE sites upstream of loci or inside the gene clusters. This suggests that E. faecium has adapted an alternative role for these operons, or that CRE binding sequences for E. faecium are different from those for other species. For example, it is possible that the same domains within WxL proteins important for recognizing/sensing the environment in a nonpathogenic species (such as the concanavalin A-like lectin domains for carbohydrates) were adapted to recognize receptor domains in the host with similar structure in more pathogenic species.
As the WxL domain, which is the means by which the proteins localize to the cell surface, is the only commonality between these proteins, they are likely to have different roles. The potential variability in function is reflected in the huge range in number of WxL proteins seen between species, such as 1 in Lactobacillus coryniformis versus 27 in E. faecalis. Based on this observation, it is unlikely that, in an organism that has a high number of WxL proteins, all of them have the same function. At least in E. faecium, these proteins seem to be important for virulence-associated functions. Whether or not these attributes are applicable to other species remains to be determined, but it is probable for other pathogenic species.
In summary, these data report the identification and preliminary characterization of the 6 WxL proteins found in almost all E. faecium isolates. Sequence, phylogenetic, and structural predictions provide a foundation for a functional role in virulence as well as support the differences seen in surface expression of WxL proteins between clinical and community isolates. Further functional characterization revealed evidence of the WxL proteins' involvement in binding human extracellular matrix proteins. Moreover, genetic inactivation of enterococcal WxL proteins resulted in a decreased in vivo ability to compete in infective endocarditis. These results suggest that enterococcal proteins with the WxL domain are therapeutic targets, and additional study of WxL proteins in enterococci and other Gram-positive pathogens could elucidate novel aspects of bacterial virulence.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by NIH grant R01AI067861 (to B.E.M.) and NIH grants T32 A155449 and F31AI092891 (to J.R.G.-P.), all from the National Institute of Allergy and Infectious Diseases (NIAID).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02288-14.
REFERENCES
- 1.Rice LB. 2010. Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol 31(Suppl 1):S7–S10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 2.Hendrickx AP, Willems RJ, Bonten MJ, van Schaik W. 2009. LPxTG surface proteins of enterococci. Trends Microbiol 17:423–430. doi: 10.1016/j.tim.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Sillanpaa J, Nallapareddy SR, Prakash VP, Qin X, Hook M, Weinstock GM, Murray BE. 2008. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology 154:3199–3211. doi: 10.1099/mic.0.2008/017319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinster S, Furlan S, Serror P. 2007. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J Bacteriol 189:1244–1253. doi: 10.1128/JB.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaillou S, Champomier-Verges MC, Cornet M, Crutz-Le Coq AM, Dudez AM, Martin V, Beaufils S, Darbon-Rongere E, Bossy R, Loux V, Zagorec M. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat Biotechnol 23:1527–1533. doi: 10.1038/nbt1160. [DOI] [PubMed] [Google Scholar]
- 6.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A 100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siezen R, Boekhorst J, Muscariello L, Molenaar D, Renckens B, Kleerebezem M. 2006. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics 7:126. doi: 10.1186/1471-2164-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schachtsiek M, Hammes WP, Hertel C. 2004. Characterization of Lactobacillus coryniformis DSM 20001T surface protein Cpf mediating coaggregation with and aggregation among pathogens. Appl Environ Microbiol 70:7078–7085. doi: 10.1128/AEM.70.12.7078-7085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinster S, Posteraro B, Bierne H, Alberti A, Makhzami S, Sanguinetti M, Serror P. 2007. Enterococcal leucine-rich repeat-containing protein involved in virulence and host inflammatory response. Infect Immun 75:4463–4471. doi: 10.1128/IAI.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Gotoh O. 1995. A weighting system and algorithm for aligning many phylogenetically related sequences. Comput Appl Biosci 11:543–551. [DOI] [PubMed] [Google Scholar]
- 12.Galloway-Pena J, Roh JH, Latorre M, Qin X, Murray BE. 2012. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS One 7:e30187. doi: 10.1371/journal.pone.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nallapareddy SR, Singh KV, Murray BE. 2008. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect Immun 76:4120–4128. doi: 10.1128/IAI.00376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00318–00311. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sillanpaa J, Nallapareddy SR, Singh KV, Prakash VP, Fothergill T, Ton-That H, Murray BE. 2010. Characterization of the ebp(fm) pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 1:236–246. doi: 10.4161/viru.1.4.11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somarajan SR, Roh JH, Singh KV, Weinstock GM, Murray BE. 2014. CcpA is important for growth and virulence of Enterococcus faecium. Infect Immun 82:3580–3587. doi: 10.1128/IAI.01911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJ, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res 37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen H, Brunak S, von Heijne G. 1999. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng 12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 21.Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW. (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppage, NY. [Google Scholar]
- 22.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. 2008. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 23.Iakoucheva LM, Dunker AK. 2003. Order, disorder, and flexibility: prediction from protein sequence. Structure 11:1316–1317. doi: 10.1016/j.str.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh KV, Coque TM, Weinstock GM, Murray BE. 1998. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol Med Microbiol 21:323–331. doi: 10.1016/S0928-8244(98)00087-X. [DOI] [PubMed] [Google Scholar]
- 26.Barbu EM, Ganesh VK, Gurusiddappa S, Mackenzie RC, Foster TJ, Sudhof TC, Hook M. 2010. Beta-neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC. PLoS Pathog 6:e1000726. doi: 10.1371/journal.ppat.1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552. [DOI] [PubMed] [Google Scholar]
- 28.Singh KV, Nallapareddy SR, Sillanpaa J, Murray BE. 2010. Importance of the collagen adhesin ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog 6:e1000716. doi: 10.1371/journal.ppat.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nallapareddy SR, Qin X, Weinstock GM, Hook M, Murray BE. 2000. Enterococcus faecalis adhesin, ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun 68:5218–5224. doi: 10.1128/IAI.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, Harvey BR. 2010. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J Bacteriol 192:5489–5498. doi: 10.1128/JB.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandlik A, Das A, Ton-That H. 2008. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci U S A 105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nallapareddy SR, Weinstock GM, Murray BE. 2003. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol 47:1733–1747. doi: 10.1046/j.1365-2958.2003.03417.x. [DOI] [PubMed] [Google Scholar]
- 33.Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. 2008. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect Immun 76:4110–4119. doi: 10.1128/IAI.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panesso D, Montealegre MC, Rincon S, Mojica MF, Rice LB, Singh KV, Murray BE, Arias CA. 2011. The hylEfm gene in pHylEfm of Enterococcus faecium is not required in pathogenesis of murine peritonitis. BMC Microbiol 11:20. doi: 10.1186/1471-2180-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teng F, Nannini EC, Murray BE. 2005. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J Infect Dis 191:472–480. doi: 10.1086/427191. [DOI] [PubMed] [Google Scholar]
- 36.Dutka-Malen S, Evers S, Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol 33:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin X, Galloway-Pena JR, Sillanpaa J, Roh JH, Nallapareddy SR, Chowdhury S, Bourgogne A, Choudhury T, Muzny DM, Buhay CJ, Ding Y, Dugan-Rocha S, Liu W, Kovar C, Sodergren E, Highlander S, Petrosino JF, Worley KC, Gibbs RA, Weinstock GM, Murray BE. 2012. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol 12:135. doi: 10.1186/1471-2180-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vahling CM, McIver KS. 2006. Domains required for transcriptional activation show conservation in the mga family of virulence gene regulators. J Bacteriol 188:863–873. doi: 10.1128/JB.188.3.863-873.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolskaya AN, Galperin MY. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res 30:2453–2459. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galloway-Pena JR, Rice LB, Murray BE. 2011. Analysis of PBP5 of early U.S. isolates of Enterococcus faecium: sequence variation alone does not explain increasing ampicillin resistance over time. Antimicrob Agents Chemother 55:3272–3277. doi: 10.1128/AAC.00099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sillanpaa J, Prakash VP, Nallapareddy SR, Murray BE. 2009. Distribution of genes encoding MSCRAMMs and pili in clinical and natural populations of Enterococcus faecium. J Clin Microbiol 47:896–901. doi: 10.1128/JCM.02283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foster TJ, Geoghegan JA, Ganesh VK, Hook M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JH, Singvall J, Schwarz-Linek U, Johnson BJ, Potts JR, Hook M. 2004. BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J Biol Chem 279:41706–41714. doi: 10.1074/jbc.M401691200. [DOI] [PubMed] [Google Scholar]
- 44.Milani A, Vecchietti D, Rusmini R, Bertoni G. 2012. TgpA, a protein with a eukaryotic-like transglutaminase domain, plays a critical role in the viability of Pseudomonas aeruginosa. PLoS One 7:e50323. doi: 10.1371/journal.pone.0050323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crennell S, Garman E, Laver G, Vimr E, Taylor G. 1994. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure 2:535–544. doi: 10.1016/S0969-2126(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 46.Wedekind JE, Trame CB, Dorywalska M, Koehl P, Raschke TM, McKee M, FitzGerald D, Collier RJ, McKay DB. 2001. Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J Mol Biol 314:823–837. doi: 10.1006/jmbi.2001.5195. [DOI] [PubMed] [Google Scholar]
- 47.Whitmore L, Wallace BA. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 48.Schwarz-Linek U, Pilka ES, Pickford AR, Kim JH, Hook M, Campbell ID, Potts JR. 2004. High affinity streptococcal binding to human fibronectin requires specific recognition of sequential F1 modules. J Biol Chem 279:39017–39025. doi: 10.1074/jbc.M405083200. [DOI] [PubMed] [Google Scholar]
- 49.Sillanpaa J, Nallapareddy SR, Houston J, Ganesh VK, Bourgogne A, Singh KV, Murray BE, Hook M. 2009. A family of fibrinogen-binding MSCRAMMs from Enterococcus faecalis. Microbiology 155:2390–2400. doi: 10.1099/mic.0.027821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bron PA, Molenaar D, de Vos WM, Kleerebezem M. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J Appl Microbiol 100:728–738. doi: 10.1111/j.1365-2672.2006.02891.x. [DOI] [PubMed] [Google Scholar]
- 51.Pieterse B, Leer RJ, Schuren FH, van der Werf MJ. 2005. Unravelling the multiple effects of lactic acid stress on Lactobacillus plantarum by transcription profiling. Microbiology 151:3881–3894. doi: 10.1099/mic.0.28304-0. [DOI] [PubMed] [Google Scholar]
- 52.Kielhofner MA, Hamill RJ. 1989. Role of adherence in infective endocarditis. Tex Heart Inst J 16:239–249. [PMC free article] [PubMed] [Google Scholar]
- 53.Angrist AA, Oka M. 1963. Pathogenesis of bacterial endocarditis. JAMA 183:249–252. [DOI] [PubMed] [Google Scholar]
- 54.Hamill RJ. 1987. Role of fibronectin in infective endocarditis. Rev Infect Dis 9(Suppl 4):S360–S371. doi: 10.1093/clinids/9.Supplement_4.S360. [DOI] [PubMed] [Google Scholar]
- 55.Mostafavi-Pour Z, Askari JA, Whittard JD, Humphries MJ. 2001. Identification of a novel heparin-binding site in the alternatively spliced IIICS region of fibronectin: roles of integrins and proteoglycans in cell adhesion to fibronectin splice variants. Matrix Biol 20:63–73. doi: 10.1016/S0945-053X(00)00131-1. [DOI] [PubMed] [Google Scholar]
- 56.Foster TJ, Hook M. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol 6:484–488. doi: 10.1016/S0966-842X(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 57.Probert WS, Johnson BJ. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol Microbiol 30:1003–1015. doi: 10.1046/j.1365-2958.1998.01127.x. [DOI] [PubMed] [Google Scholar]
- 58.Schwarz-Linek U, Hook M, Potts JR. 2006. Fibronectin-binding proteins of gram-positive cocci. Microbes Infect 8:2291–2298. doi: 10.1016/j.micinf.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Dabo SM, Confer AW, Anderson BE, Gupta S. 2006. Bartonella henselae Pap31, an extracellular matrix adhesin, binds the fibronectin repeat III13 module. Infect Immun 74:2513–2521. doi: 10.1128/IAI.74.5.2513-2521.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaultney RA, Gonzalez T, Floden AM, Brissette CA. 2013. BB0347, from the Lyme disease spirochete Borrelia burgdorferi, is surface exposed and interacts with the CS1 heparin-binding domain of human fibronectin. PLoS One 8:e75643. doi: 10.1371/journal.pone.0075643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kingsley RA, Keestra AM, de Zoete MR, Baumler AJ. 2004. The ShdA adhesin binds to the cationic cradle of the fibronectin 13FnIII repeat module: evidence for molecular mimicry of heparin binding. Mol Microbiol 52:345–355. doi: 10.1111/j.1365-2958.2004.03995.x. [DOI] [PubMed] [Google Scholar]
- 62.Kuo CJ, Bell H, Hsieh CL, Ptak CP, Chang YF. 2012. Novel mycobacteria antigen 85 complex binding motif on fibronectin. J Biol Chem 287:1892–1902. doi: 10.1074/jbc.M111.298687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.