Abstract
Rhodobacter sphaeroides has two different sets of flagellar genes. Under the growth conditions commonly used in the laboratory, the expression of the fla1 set is constitutive, whereas the fla2 genes are not expressed. Phylogenetic analyses have previously shown that the fla1 genes were acquired by horizontal transfer from a gammaproteobacterium and that the fla2 genes are endogenous genes of this alphaproteobacterium. In this work, we characterized a set of mutants that were selected for swimming using the Fla2 flagella in the absence of the Fla1 flagellum (Fla2+ strains). We determined that these strains have a single missense mutation in the histidine kinase domain of CckA. The expression of these mutant alleles in a Fla1− strain allowed fla2-dependent motility without selection. Motility of the Fla2+ strains is also dependent on ChpT and CtrA. The mutant versions of CckA showed an increased autophosphorylation activity in vitro. Interestingly, we found that cckA is transcriptionally repressed by the presence of organic acids, suggesting that the availability of carbon sources could be a part of the signal that turns on this flagellar set. Evidence is presented showing that reactivation of fla1 gene expression in the Fla2+ background strongly reduces the number of cells with Fla2 flagella.
INTRODUCTION
More than 40 genes are involved in the biogenesis and functioning of the bacterial flagellum. This structure has three major subcomponents, the basal body, the hook, and the filament. The basal body contains the export apparatus, an inner membrane ring (MS ring), a periplasmic ring (P ring), and, depending on the species, an outer membrane ring (L ring). The basal body also includes the flagellar motor and a rod that expands from the MS ring and crosses the L and P rings. The hook is the first extracellular structure that is assembled; it connects the basal body with the flagellar filament that is formed by thousands of flagellin subunits (reviewed in references 1–3).
In most bacterial species, the expression of the flagellar genes is highly regulated and frequently follows a hierarchical order in which the late genes are expressed until the early genes, that are higher in the hierarchy, are expressed. At the top of the hierarchy, a transcriptional regulator is responsible for the expression of the genes required to assemble the early flagellar structures that are located in the cytoplasm (i.e., export apparatus), in the cytoplasmic membrane (i.e., MS ring), and, depending on the hierarchy, in the periplasm (i.e., P ring), the outer membrane (i.e., L ring), and the extracellular milieu (i.e., hook). Within this class, additional transcription factors are expressed. These proteins are required to transcribe the late genes, such as fliC (flagellin), fliD (filament cap), and fliS (secretion of FliC) (1–4).
Different regulatory mechanisms are known to be involved in the control of these hierarchies (4). The most common scheme of regulation consists of the use of different sigma factors to transcribe the genes in different tiers of the hierarchy. For instance, in the enteric bacteria Escherichia coli and Salmonella, at the top of the hierarchy the transcriptional activator FlhD4C2 and sigma-70 promote the expression of the early genes, whereas the late promoters are dependent on σ28. In some species of Vibrio and Pseudomonas, σ54 and the activator protein FleQ promote the transcription of the genes required at the initial phases of flagellar biosynthesis, and σ28 expresses the late genes. In alphaproteobacteria, the flagellar hierarchy of Caulobacter crescentus is one of the best characterized. In this bacterium, σ70 and CtrA transcribe the genes encoding the proteins that form the export apparatus, the MS ring, and the regulators FliX and FlbD. Once the first flagellar structure is functional, FlbD becomes phosphorylated and activates Eσ54 to carry out transcription of the genes encoding the rod proteins, FlgH and FlgL (P and L rings, respectively), FlgE (the hook), and FliC (the filament). CtrA acts as a transcriptional activator when it is phosphorylated by the histidine kinase (HK) CckA and the histidine phosphotransferase ChpT (5–7). This signal transduction pathway is essential in Caulobacterales and Rhizobiales (8–11). In C. crescentus, the kinase/phosphatase activities of CckA are regulated by the histidine kinases PleC, DivJ, and DivL as well as by the response regulator DivK (12).
In contrast to C. crescentus, in many alphaproteobacteria the signaling pathway CckA/ChpT/CtrA is dispensable and the genes encoding PleC, DivJ, DivL, and DivK are not present in their genomes (13). For some species of this bacterial group, it has been reported that CtrA does not control the cell cycle, but it controls directly or indirectly the expression of the flagellar genes (14–19). In this group of bacteria, there is no evidence regarding the control of the activity of CckA, but it has been reported that the expression of cckA, chpT, and/or ctrA is reduced in quorum sensing mutants in Ruegeria sp. strain KLH11, Dinoroseobacter shibae, and Rhodobacter capsulatus (16, 17, 19, 20). However, in Ruegeria sp. KLH11, the evidence suggests that this control is indirect (16). In addition, in R. capsulatus it was shown that ctrA transcription was increased 3-fold when the cultures were grown photoheterotrophically in minimal medium compared with cultures grown in YPS-rich medium (medium containing yeast extract, peptone, and salts); in addition, a reduction in the expression of ctrA was detected in phosphate-limited cultures but not in nitrogen- or carbon-limited cultures (20).
Rhodobacter sphaeroides is an alphaproteobacterium that has two different full sets of flagellar genes (21). The fla1 set was initially characterized given that its expression is constitutive under the growth conditions commonly used in the laboratory (21–23). It has been shown that the fla1 genes are transcribed in a four-tiered hierarchy. At the top of it, the master regulator FleQ activates σ54 to promote the expression of the fleT-fliE-J operon. FleT and FleQ together activate σ54 to transcribe the genes encoding the rest of the components of the rod, the export apparatus, the L, P, and H rings, and the hook. In this class, the genes encoding σ28 and the anti-sigma σ28 factor FlgM are also expressed. After completion of the hook-basal body (HBB), FlgM is exported out of the cell, and the genes that depend on σ28 are expressed (23, 24).
Bacterial taxis is achieved through the control of flagellar rotation by the chemotactic system. Fla1-dependent taxis responds to several stimuli, some of which are not directly sensed by the chemotactic proteins. For example, it has been observed that aerotaxis is controlled in part by the two-component system RegB-RegA (also known as PrrB and PrrA) (25). These proteins form a global system that responds to the redox state of the cell to control several metabolic processes that generate energy, i.e., photosynthesis, carbon and nitrogen fixation, aerobic and anaerobic respiration, electron transport, etc. (reviewed in reference 26). Under anaerobic growth conditions, RegB is active as a kinase and phosphorylates RegA; RegA-P activates the genes encoding the apoproteins of the light-harvesting and reaction center complexes (27–29). In the presence of oxygen, RegB is inactivated and the synthesis of these complexes and proteins is halted. The absence of RegB has a negative effect on the growth rate under photoheterotrophic conditions (30), and these mutant cells do not show the negative tactic response toward high concentrations of oxygen (25).
The presence of a second set of flagellar genes (fla2) in R. sphaeroides was discovered when the genomic sequence of this bacterium was released. At that time, no other flagellar structure different from the Fla1 flagellum had been observed (31). The functionality of the fla2 genes was reported after the isolation of a mutant strain able to swim with the Fla2 flagella. This mutant, isolated in a fla1 background, showed the presence of several polar flagella, and it was proficient to swim in liquid medium, like the wild-type strain (WS8N) that assembles the Fla1 single subpolar flagellum. Neither Fla1 nor Fla2 flagella enable swarming on surfaces, and it should be stressed that the wild-type strain grown under conditions commonly used in the laboratory does not express the fla2 set (21).
Phylogenetic studies suggested that the fla1 genes were acquired by an event of horizontal transfer probably from a gammaproteobacteria, whereas the fla2 genes are the native flagellar genes of this species (21). This suggests that the fla1 genes overtook the function of the native flagellar genes or that its presence allowed R. sphaeroides to swim under growth conditions where the native genes were repressed.
So far, it is not known what is the mechanism that keeps the fla2 set transcriptionally inactive in the wild-type WS8N strain, and nothing is known about its possible regulation. In this work, we show evidence indicating that the CckA-ChpT-CtrA pathway is central to activate the fla2 genes. Our results establish that in the absence of Fla1, an activating mutation in CckA is enough to promote the expression of the fla2 genes and consequently the synthesis of the Fla2 flagella. In addition, we found that cckA is strongly repressed by a high concentration of organic acids, whereas a growth medium with a mixture of amino acids or a low concentration of succinic acid induces cckA and favors the synthesis of the Fla2 flagella. Evidence is also presented indicating that the expression of the fla1 genes reduces the synthesis of Fla2.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
Plasmids and bacterial strains used in this work are listed in Table 1. R. sphaeroides WS8N (32) was grown at 30°C in Sistrom's minimal medium (33) or in Sistrom's minimal medium without succinic acid but supplemented as indicated below. Photoheterotrophic liquid cultures were grown under continuous illumination and in completely filled screw-cap tubes. Heterotrophic liquid cultures were incubated in the dark with shaking at 200 rpm. Escherichia coli was grown in LB medium (34) at 37°C. Swimming assays were carried out with bacteria grown in liquid medium or on swimming plates containing the indicated medium and 0.22% agar. When required, the following antibiotics were added: for R. sphaeroides, kanamycin (25 μg/ml), tetracycline (1 μg/ml), and spectinomycin (50 μg/ml); for E. coli, kanamycin (50 μg/ml), spectinomycin (50 μg/ml), and ampicillin (100 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Rhodobacter sphaeroides strains | ||
| EA1 | AM1 derivative; ΔfleQ::Kanr cckAL391F ΔctrA::aadA | This work |
| EA2 | WS8N derivative; ΔctrA::aadA | This work |
| AM1 | WS8N derivative; ΔfleQ::Kanr cckAL391F | 43 |
| BV1 | WS8N derivative; ΔfleQ::Kanr cckAA387P | This work |
| BV2 | WS8N derivative; ΔfliF::aadA cckAF399C | This work |
| BV3 | AM1 derivative; ΔfleQ::Kanr cckAL391F ΔchpT::ΩSpc | This work |
| BV4 | WS8N derivative; ΔfleQ::Kanr ΔchpT::ΩSpc | This work |
| CB1 | AM1 derivative; ΔfleQ::Kanr ΔcckA::ΩSpc | This work |
| CB2 | WS8N derivative; ΔcckA::ΩSpc | This work |
| CD1 | WS8N derivative; ΔfleQ::Kanr ΔregB::ΩSpc | This work |
| CD2 | AM1 derivative; ΔfleQ::Kanr cckAL391F ΔregB::ΩSpc | This work |
| LC5 | WS8N derivative; ΔfleQ::Kanr ΔcckA::ΩSpc | This work |
| LC6 | AM1 derivative; ΔfleQ::Kanr ΔcckA::uidA-aad | This work |
| SP13 | WS8N derivative; ΔfleQ::Kanr | 22 |
| SP20 | WS8N derivative; ΔfliF::aadA | Laboratory collection |
| WS8N | Wild-type strain; spontaneous Nalr | 24 |
| Escherichia coli strains | ||
| LMG194 | Protein expression strain | Invitrogen |
| S17 | recA endA thi hsdR RP4-2-Tc::Mu::Tn7; Tpr Smr | 30 |
| TOP10 | Cloning strain | Invitrogen |
| Plasmids | ||
| pBAD/His | Expression vector of His6-tagged proteins; Apr | Invitrogen |
| pBAD/His-cCckA | pBAD/HisB expressing His6-cCckA | This work |
| pBAD/His-cCckA A387P | pBAD/HisB expressing His6-cCckA A387P | This work |
| pBAD/His-cCckA L391F | pBAD/HisB expressing His6-cCckA L391F | This work |
| pBAD/His-cCckA F399C | pBAD/HisB expressing His6-cCckA F399C | This work |
| pBBMCS53 | Transcriptional uidA fusion vector; Gmr | 34 |
| pBBMCS53_fliL2 | pBBMCS53 carrying the fliL2 promoter | This work |
| pCR2.1-TOPO | Cloning vector; Apr Kanr | Invitrogen |
| pJQ_ΔctrA::aadA | pJQ200 mp18 carrying ΔctrA::aadA | This work |
| pJQ_ΔregB::ΩSpc | pJQ200 mp18 carrying ΔregB::ΩSpc | This work |
| pJQ200_ΔcckA::ΩSpc | pJQ200 mp18 carrying ΔcckA::ΩSpc | This work |
| pJQ200_ΔchpT::ΩSpc | pJQ200 mp18 carrying ΔchpT::ΩSpc | This work |
| pJQ200mp18 | Mobilizable suicide vector for R. sphaeroides; Gmr | 27 |
| pPIRL | Vector that encodes tRNAs for rare codons; Cmr | 35 |
| pRK_cckA | pRK415 derivative expressing cckA | This work |
| pRK_cckAA387P | pRK415 derivative expressing cckAA387P | This work |
| pRK__cckAL391F | pRK415 derivative expressing cckAL391F | This work |
| pRK_cckAF399C | pRK415 derivative expressing cckAF399C | This work |
| pRK_chpT | pRK415 derivative expressing chpT | This work |
| pRK_ctrA | pRK415 derivative expressing ctrA | This work |
| pRK_regBL267 | pRK415 derivative expressing regBL267 | This work |
| pRK_regBS267 | pRK415 derivative expressing regBS267 | This work |
| pRK415 | pRK404 derivative used for expression in R. sphaeroides | 33 |
| pTOPO_cckA | pCR2.1 TOPO carrying cckA | This work |
| pTOPO_ctrAup-down | pCR2.1 TOPO carrying ctrAupdown | This work |
| pTOPO_ΔcckA::ΩSpc | pCR2.1 TOPO carrying ΔcckA::ΩSpc | This work |
| pTOPO_ΔcckA | pCR2.1 TOPO carrying ΔcckA | This work |
| pTZ_chpTup-down | pTZ19R carrying chpTupdown | This work |
| pTZ_ctrAupdown | pTZ19R BamHI− carrying ctrAupdown | This work |
| pTZ_ΔchpT::ΩSpc | pTZ19R carrying ΔchpT::ΩSpc | This work |
| pTZ_ΔctrA::aadA | pTZ19R BamHI− carrying ΔctrA::aadA | This work |
| pTZ_ΔregB | pTZ19R carrying ΔregB | This work |
| pTZ_ΔregB::ΩSpc | pTZ19R carrying ΔregB::ΩSpc | This work |
| pTZ_regBL267 | pTZ19R carrying regBL267 | This work |
| pTZ_regBS267 | pTZ19R carrying regBS267 | This work |
| pTZ19R | Cloning vector | Pharmacia |
| pTZ19R BamHI− | pTZ19R without BamHI site | Laboratory collection |
| pWM5 | Vector source of the uidA-aadA cassette | 28 |
Oligonucleotides.
The oligonucleotides used in this work are listed in Table S1 in the supplemental material.
Isolation of mutant strains.
To inactivate cckA, chpT, and ctrA, a suicide vector containing the target gene disrupted with an antibiotic resistance gene was used for allelic exchange.
To inactivate cckA, the oligonucleotides 454A and 454B were used to amplify by PCR a product of 2,460 bp containing the coding region of cckA (2,277 bp), as well as 110 and 73 bp from the downstream and upstream regions of the gene. This product was cloned into pCR2.1-TOPO plasmid, resulting in pTOPO_cckA; this plasmid was digested with EcoNI to remove most of the coding region of cckA. The digestion fragment, which included the complete vector and 792 bp from the 5′ end of cckA and 455 bp from the 3′ end, was purified and self-ligated to obtain pTOPO_ΔcckA. The allele ΔcckA in this plasmid was verified by sequencing. ΔcckA::ΩSpc was obtained by cloning the ΩSpc cassette as a 2-kb SmaI fragment into pTOPO_ΔcckA previously digested with EcoNI and end repaired with T4 DNA polymerase. The resultant plasmid was digested with XbaI and SacI, and the fragment carrying the allele ΔcckA::ΩSpc was purified and cloned in the suicide plasmid pJQ200mp18 (35). The allele ΔcckA::uidA-aadA was obtained using the same procedure as described above, but instead of cloning the ΩSpc cassette, the SmaI fragment from pWM5 (36) carrying uidA-aadA was used. This fragment allows the expression of the promotorless uidA gene creating a transcriptional fusion. The suicide plasmids carrying these alleles of cckA were introduced into R. sphaeroides by conjugation (37, 38). The double recombination events were selected as described previously (21).
Strain BV3 was obtained by cloning together two PCR products corresponding to the upstream and downstream regions of chpT in pTZ19R. The 610-bp product of the upstream region of chpT was obtained using the oligonucleotides chpTmutup1 and chpTmutup2, whereas the downstream product of 584 bp was obtained using the oligonucleotides chpTmutdown1 and chpTmutsdown2. These PCR products were joined through an EcoRV site designed in the oligonucleotides and cloned in pTZ19R. The ΩSpc cassette was cloned into pTZ_chpTup-down previously digested with EcoRV. From the resultant plasmid, pTZ_ΔchpT::ΩSpc, the DNA fragment carrying ΔchpT::ΩSpc was purified and subcloned into pJQ200 mp18.
The allele ΔctrA::aadA was obtained by cloning together in pCR2.1-TOPO two PCR products corresponding to the upstream and downstream regions of ctrA joined by a BamHI recognition site. The upstream region of ctrA (308 bp) was amplified by PCR using the oligonucleotides ctrAfor and ctrAinrev, whereas the downstream product of 294 bp was obtained using the oligonucleotides ctrAinfor and ctrArev. The upstream product encompassed 200 bp of the coding region of ctrA and 108 bp of the upstream region of the gene. The downstream product included 220 bp of the 3′ end of ctrA and 73 bp of the downstream region. These products were digested with BamHI, ligated, and cloned in pCR2.1-TOPO, resulting in pTOPO_ctrAupdown. To facilitate further manipulations, the resultant fragment (ctrAupdown) of 602 bp was subcloned into pTZ19R Bam−. The resultant plasmid, pTZ_ctrAupdown, was digested with BamHI and ligated with an internal fragment of the ΩSpc that only carries the aadA gene that confers the Spcr phenotype without the transcriptional terminator sequences existent in the omega cassette (39). The aadA fragment was obtained by PCR using the oligonucleotides aadABam1 y aadABam2. The fragment containing the allele ΔctrA::aadA was subcloned into pJQ200; the resultant plasmid was introduced into R. sphaeroides, and double-recombination events were selected.
To inactivate regB, the oligonucleotides regBupXbafw and regBdwSacIrv were used to amplify a product of 2,074 bp containing regB (1,389 bp) and its flanking regions of 300 and 385 bp upstream and downstream of regB, respectively. This product was cloned into pTZ19R, and the resultant plasmid (pTZ_regB) was subjected to inverse PCR using the oligonucleotides regBupEcoRVrv and regBdwEcoRVfw; this reaction removed 720 bp of the coding region of regB and retained 600 bp upstream and 756 bp downstream of the deletion boundary marked by an EcoRV recognition site, present in the oligonucleotides. The PCR product was purified and self-ligated to generate pTZ_ΔregB. The ΩSpc cassette was cloned in the EcoRV site of the resultant plasmid as a 2-kb SmaI fragment. The plasmid pTZ_ΔregB::ΩSpc was digested with XbaI and SacI, and the fragment carrying the allele ΔregB::ΩSpc was subcloned into pJQ200mp18 and introduced into R. sphaeroides by conjugation. The double-recombination events were selected.
Motility assays.
Motility was tested in soft-agar plates (0.22%) containing Sistrom's minimal medium or Sistrom's minimal medium without succinic acid but supplemented as indicated below. Plates were incubated aerobically (in a normal incubator) or anaerobically in a transparent polycarbonate anaerobic jar using the BD GasPak EZ anaerobe container system sachets and illuminated with incandescent bulbs (two at 75 W). After the desired time for each assay, motility was registered.
Isolation of Fla2+ strains.
To isolate additional Fla2+ mutants, we prepared soft-agar plates containing Sistrom's minimal medium with 15 mM succinic acid. These plates were inoculated with strains SP13 and SP20 and incubated for 7 days inside an anaerobic jar and under continuous illumination. In addition, soft-agar plates containing Sistrom's minimal medium with 1 mM succinic acid were inoculated with strains SP13 and SP20 and incubated aerobically for 7 days at 30°C.
β-Glucuronidase activity.
For the experiments shown in Fig. 7, strain LC6 was grown photoheterotrophically in Sistrom's minimal medium until early stationary phase. The cells were washed with medium without a carbon source, and an aliquot was used to inoculate fresh medium supplemented with a carbon source. After 14 h of photoheterotrophic growth, a 1.5-ml sample was withdrawn for analysis. For samples grown under heterotrophic conditions, aerobic cultures grown to stationary phase were diluted 10-fold and incubated at 30°C in the dark with shaking at 200 rpm until they reached an optical density at 600 nm (OD600) of 0.6; at this point, 1.5 ml from each culture was collected and concentrated 6-fold. For the experiments shown in Fig. 3, the strains were grown photoheterotrophically in Sistrom's minimal medium with 0.2% Casamino Acids until exponential phase; at this point, a sample was withdrawn for analysis. β-Glucuronidase was determined from sonicated cell extracts using 4-methyl-umbelliferyl-β-d-glucuronide as the substrate and following a previously reported protocol (40). 4-Methyl-umbelliferone (Sigma) was used as a standard. In this work, specific activities are expressed as nmol of 4-methyl-umbelliferone formed min−1 mg−1 of protein.
FIG 7.
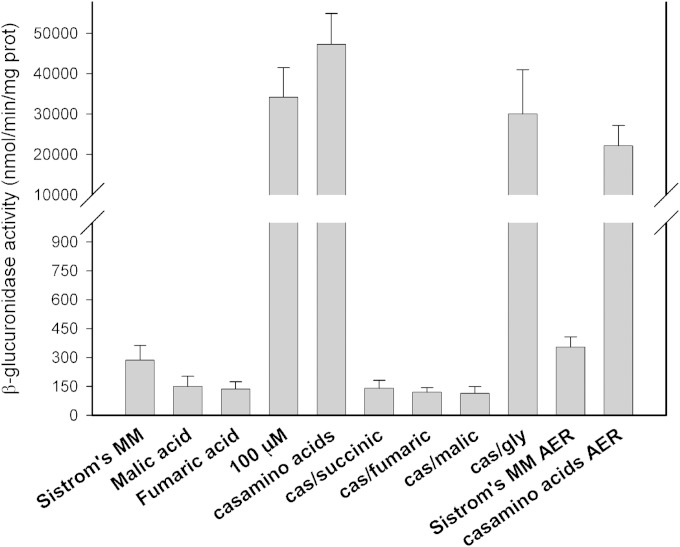
Effects of organic acids on the transcriptional activity of cckA. Shown are the β-glucuronidase activities of strain LC6 (ΔfleQ::Kanr ΔcckA::uidA-aad) grown with different carbon sources: Sistrom's minimal medium supplemented with 15 mM or 100 μM succinic acid (Sistrom's MM or 100 μM, respectively), Sistrom's minimal medium without succinic acid but supplemented with 0.2% Casamino Acids (casamino acids) or with 0.2% Casamino Acids (cas) and the indicated organic acid to a final concentration of 15 mM, or 0.6% glycerol (gly). Cultures were grown anaerobically in screw-cap tubes or aerobically (AER). Values are the means of three independent measurements. The standard deviation is also shown.
FIG 3.
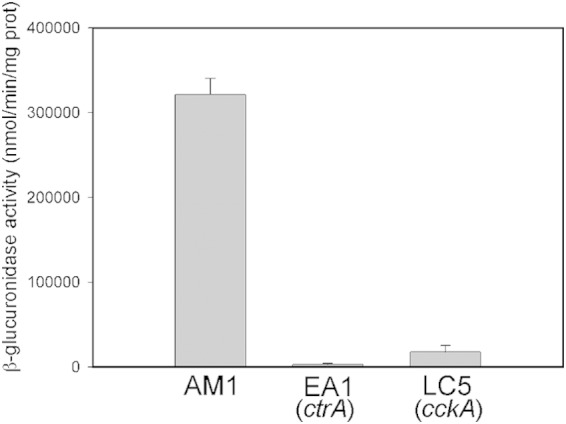
Transcriptional activity of fliL2p. β-Glucuronidase activity was determined in the following strains: AM1 (Fla2+), EA1 (ΔctrA::aadA), and LC5 (ΔcckA::ΩSpc) carrying plasmid pBBMCS53_fliL2.
Genetic and molecular biology techniques.
Standard methods were used to obtain chromosomal or plasmid DNA (34). Restriction and other DNA-modifying enzymes were acquired from Roche, New England BioLabs (NEB), or Invitrogen. For sequencing, plasmids were purified using the Illustra plasmidPrep Mini Spin kit (GE). Chromosomal or plasmid DNA was amplified with the appropriate oligonucleotides using PrimeSTAR HS DNA polymerase (TaKaRa Bio Inc.) according to the recommendations of the manufacturer. Cloning was often carried out using the TOPO TA cloning kit or Zero Blunt TOPO PCR cloning kit (Invitrogen).
Site-directed mutagenesis.
To replace the leucine codon found in regB at position 267 with serine, the plasmid pTZ_regBL267 was subject to site-directed mutagenesis using the oligonucleotides Ser267regBqchfw and Ser267regBqchrv and the QuikChange protocol (Agilent Technologies, Inc.). The change was confirmed by sequencing, and the resultant plasmid was named pTZ_regBS267. By following the same protocol and using the plasmid pBAD/His-cCckA as the substrate for site-directed mutagenesis, we obtained pBAD/His-cCckA L391F, pBAD/His-cCckA A387P, and pBAD/His-cCckA F399C. The pairs of oligonucleotides 454mutbad1 and 454mutbad2, cck A387Pfw and cck A387Prv, and cck F399Cfw and cck F399Crv were used to obtain each of these mutant versions.
Plasmids used in this work.
pRK_cckA and its allelic variants were obtained by cloning into pRK415 (41) the 2.4-kb PCR product generated with the oligonucleotides 454A and 454B; in these plasmids, cckA is expressed from the lac promoter (lacp) present in pRK415. pTZ_regBL267 was obtained by cloning into pTZ19R the 1.5-kb PCR product generated with the oligonucleotides regB1 and regB2. pRK_regBL267 and pRK_regBS267 were obtained by cloning into pRK415 the 1.5-kb XbaI-KpnI fragment obtained from pTZ_regBL267 or pTZ_regBS267; in the resultant plasmids, regB is transcribed from lacp present in pRK415. pRK_chpT was obtained by cloning in pRK415 the 774-bp PCR product generated with the oligonucleotides FwchpTXbaI and RvchpTSacI; chpT is expressed from lacp present in this plasmid. pRK_ctrA was obtained by cloning the 894-bp PCR product generated with the oligonucleotides ctrAfor and ctrArev into pRK415; in this construction, ctrA is expressed from lacp. The region of cckA encoding residues 249 to 758 was amplified by PCR using the oligonucleotides cckAHKfwpBAD and cckARR2pBAD2; the 1,558-bp product was cloned into pBAD_HisA to generate pBAD/His-cCckA. The fusion fliL2-uidA was obtained by cloning the regulatory region of fliL2 into pBBMCS53; this plasmid was designed to generate transcriptional fusions using the uidA gene present in the plasmid (42). The 335-bp regulatory region of fliL2 was obtained by PCR using the oligonucleotides fliFL3 and fliFL4.
Microscopy.
Swimming cells were observed using a Nikon E600 microscope with a 40× objective with dark-field illumination. Immunofluorescence was carried out in cells previously fixed with 3% formaldehyde for 20 min at room temperature. The cells were centrifuged at 3,000 rpm and washed in phosphate-buffered saline (PBS) to remove the paraformaldehyde. Finally, the cells were resuspended in PBS (pH 7.4) with 1% bovine serum albumin (BSA). The primary antibody, FlgE1 previously labeled with Zenon Alexa Fluor 488 or FlgE2 labeled with Zenon Alexa Fluor 546 (Invitrogen), was added at a 1:50 dilution. After incubation for 2 to 12 h with the antibody, the cells were washed twice with PBS and incubated with 3% paraformaldehyde for 20 min at room temperature. The cells were washed twice with PBS and placed on a slide covered with a thin bed of agar. Epifluorescence images were taken using a Nikon Eclipse 600 microscope equipped with a Hamamatsu Orca-ER cooled charge-coupled-device (CCD) camera. Epifluorescent images were acquired for 3 s.
Protein overexpression and purification.
Strain LMG194 was transformed with plasmid pBAD/His-cCckA, pBAD/His-cCckA L391F, pBAD/His-cCckA A387P, or pBAD/His-cCckA F399C. These strains were also transformed with the pPIRL plasmid, which increases the availability of certain tRNAs (43). Overnight cultures of these strains were diluted 1:50 and incubated at 37°C until they reached an OD600 of 0.5. At this point, 0.02% arabinose was added and incubation was allowed to proceed for 3 h. Cells were harvested and resuspended in 1/100 of the original volume in PBS (pH 7.4) with 20% glycerol and 10 mM imidazole. Lysozyme was added (1-mg/ml final concentration), and the mixture was incubated on ice for 15 min. The cell suspension was sonicated on ice using a microtip (3 mm), with three bursts of 10 s. Cell debris were removed by centrifugation (14,000 rpm for 5 min). The supernatant was mixed with nickel-nitrilotriacetic acid (Ni2+-NTA)–agarose beads (1/250 of the original culture volume) and incubated for 1 h on ice, with mixing by occasional inversion. After this time, the sample was loaded into a polypropylene column (1 ml) and washed with 10 volumes of PBS with 25 mM imidazole. The protein was eluted using PBS with 20% glycerol and 200 mM imidazole and dialyzed overnight against PBS with 20% glycerol at 4°C. The purity of the protein was determined by SDS-PAGE (44) and Coomassie blue staining.
Phosphorylation of cCckA.
A 5 μM concentration of purified His-cCckA wild type or a mutant version was incubated in HEPES buffer (pH 7.5; 33 mM HEPES, 10 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol [DTT], and 10% glycerol), the reaction was started by adding 6.6 μM [γ-32P]ATP to a final volume of 60 μl. At the desired time points, a sample of 4 μl was withdrawn and the reaction was stopped by the addition of 4× Laemmli sample buffer (44). After SDS-PAGE, radioactivity was visualized and quantified using phosphor screens and a Typhoon scanner (GE Healthcare Life Sciences).
Genome sequence of AM1 strain and analysis.
Total genomic DNA was isolated from a saturated culture of 3 ml using the GenElute bacterial genomic kit (Sigma). A library was constructed and paired-end sequenced (2 × 100 nucleotides [nt]; Illumina HiSeq 2000) using a commercial technology platform from Macrogen (Republic of Korea). Single reads were mapped against the genome of R. sphaeroides WS8N (45) using bowtie2. The .SAM file was converted to .bam and visualized with Artemis and SNVER (46–50) to identify the differences between the genome sequence and the mapped reads. The changes that were consistently present in all the reads at a particular position were confirmed by PCR followed by Sanger sequencing. For these confirmatory experiments, we tested genomic DNA from the wild-type WS8N and AM1 strains.
RESULTS
Analysis of the genome sequence of AM1.
We have previously reported the isolation of spontaneous mutant strains able to swim with the Fla2 flagella. One of these mutants was selected in a ΔflgC::Kanr background (SP18) (21), and the other was selected in a ΔfleQ::Kanr background (AM1) (51). In order to establish the mutations that led to the expression of fla2 genes, we determined the complete genome sequence of the AM1 strain. A total of 12.19 million paired reads of 100 nt were obtained and mapped against the complete genome of WS8N (45). The analysis of the mapped reads looking for single nucleotide variants (SNVs) or indels revealed the presence of the expected deletion in fleQ and 7 SNVs with an E value lower than E−150 (Table 2). To verify the identities of these positions in AM1 and also in WS8N, we amplified by PCR a fragment that included the position to be verified. The PCR products obtained from chromosomal DNA of strains WS8N and AM1 were sequenced by Sanger capillary electrophoresis. As shown in Table 2, 5 of these changes were also present in our wild-type strain (WS8N), suggesting that these mutations could have emerged in our laboratory and are not related with the Fla2+ phenotype given that our wild-type strain does not have Fla2 flagella, as occurs with other wild-type WS8N strains previously studied in other laboratories (22). In an attempt to evaluate if these mutations could reflect a frequent variation present in other strains of R. sphaeroides, we analyzed the orthologous genes in R. sphaeroides 2.4.1 and R. sphaeroides ATCC 17029. We observed that 3 out of these 5 changes were also present in R. sphaeroides 2.4.1 and ATCC 17029; therefore, it is possible that these alleles commonly emerge in different strains or that they are errors in the published sequence. The other two mutations shared by our wild-type WS8N strain and AM1 resulted in amino acids changes in a siderophore receptor and in the RegB histidine kinase. Only 2 changes are specific to AM1; one is located in cckA, and the other is located in an intercistronic region (Table 2).
TABLE 2.
Changes found in the AM1 genome
| Location in WS8Nd | WS8N (2.4.1) | AM1 | WS8Na | ATCC17029 | Amino acid in WS8N/2.4.1/WS8Na/AM1 | Product (name or domain) |
|---|---|---|---|---|---|---|
| Gene name/position | ||||||
| RSWS8N_04500 (RSP2804)/935763 | G (A) | A | A | A | D/D/D/D | Queuine tRNA-ribosyltransferase |
| RSWS8N_05655 (RSP_0088)/1195392 | G(A) | A | A | A | R/C/C/C | YciF; stress response protein |
| RSWS8N_07570 (RSP0454)/1592120 | G (G) | A | G | G | L/L/L/F | CckA |
| RSWS8N_13040 (RSP1520)/2714509 | C (C) | T | T | C | S/S/L/L | RegB |
| RSWS8N_16629 (RSP3079)/Chr II 311247 | G (G) | A | A | G | G/G/D/D | Siderophore binding protein; FatB |
| Intercistronic region position in Chr I | ||||||
| 1101564 | C (T) | T | T | T | Between RSWS8N_05200b and tRNA-Asp | |
| 3058907 | C (C) | T | C | C | 45 nt upstream of RSWS8N_14710c |
Laboratory collection since 1993.
Hypothetical acetyltransferase.
Hypothetical transcriptional regulator, Cro/CI family.
Chr, chromosome.
The change observed in the histidine kinase RegB deserved a closer inspection (Table 2) since it has been reported that RegB is involved in Fla1-dependent swimming (25). To determine the relevance of the regB allele (L267) present in our strains, we tested if the wild-type allele of regB (RegB S267) could affect swimming of the AM1 strain or if its presence would inhibit the appearance of Fla2+ strains. Under heterotrophic growth conditions, we observed a reduction of approximately 20% in the size of the swim ring of the strains lacking regB or expressing RegB S267 (strain CD2 [Fla2+ ΔregB::ΩSpc] or CD2/pRK_regBS267, respectively) compared with the swim ring of AM1 or CD2/pRK_regBL267 (see Fig. S1 in the supplemental material). However, under photoheterotrophic conditions, the strains expressing RegB S267 or L267 (AM1 [RegB L267], CD2/pRK_regBS267, and CD2/pRK_regBL267) formed similar swim rings (see Fig. S1). These results indicate that wild-type RegB (S267) and RegB L267 allow normal Fla2-dependent motility under anaerobic conditions and suggest that RegB L267 is insensitive to oxygen. In agreement with previous reports, the regB mutant CD2, grew poorly under photoheterotrophic conditions (30, 52, 53), and as a consequence, the swimming halo was barely detectable (see Fig. S1); nevertheless, from this halo it was possible to observe swimming cells under the microscope, indicating that the absence of RegB does not prevent the synthesis of Fla2 in this growth condition. In addition, the time or frequency with which Fla2+ cells emerged from a selection process using CD1/pRK_regBS267 as the parental strain is similar to that observed when the SP13 (that expresses RegB L267) strain was used. From these results, we conclude that (i) it is possible to isolate Fla2+ cells in the presence of the regBS267 allele (wild type) and that (ii) the Fla2-dependent swimming is slightly affected by the absence of RegB.
Effect of CckA and CckA L391F on the swimming motility of AM1.
As mentioned above, the only relevant change that we observed exclusively in the genome sequence of AM1 corresponded to a single substitution in cckA. This mutation replaces Leu 391 with Phe. In R. sphaeroides WS8N, CckA has been annotated as a polypeptide of 758 residues. This protein has two transmembrane (TM) regions with a short loop in the periplasmic space. After the second TM region (after residue 58), the cytoplasmic region shows a histidine kinase (HK) domain conformed by a catalytic domain (CA) that binds ATP and phosphorylates a conserved histidine located in the DHp domain. In addition to the HK domain, this protein also has a receiver domain (REC) in the C terminus. HK and REC domains are present in the so-called hybrid histidine kinases of the two-component systems (54, 55). The mutation identified in CckA is within the HK domain and is five residues away from the conserved histidine residue that is phosphorylated (H397); the presence of this change within the HK domain suggests that the properties of CckA could have been affected by this change.
Given that CckA has been reported to be involved in the control of the flagellar genes in other alphaproteobacteria (14–16), we tested if the presence of this mutant allele of cckA was enough to promote swimming with Fla2 flagella. To test this possibility, we cloned the wild type and the mutant version of cckA into pRK415. The resultant plasmids were introduced into strain LC5 (ΔcckA::ΩSpc ΔfleQ::Kanr), and swimming was evaluated. As shown in Fig. 1A, the strain carrying the plasmid with the mutant version of cckA (pRK_cckAL391F) was able to swim; in contrast, the strain that expressed the wild-type version of cckA only showed a few clusters of cells that did not move much from the inoculation point. The appearance of these clusters was dependent on the presence of the cckA gene in a multicopy plasmid, since they are not visible in the LC5 (Fig. 1) or in the SP13 (ΔfleQ::Kanr) strains (data not shown). Therefore, these results indicate that CckA L391F is responsible for the Fla2+ phenotype, whereas the expression of wild-type CckA from the plasmid promoter is not enough to promote swimming.
FIG 1.
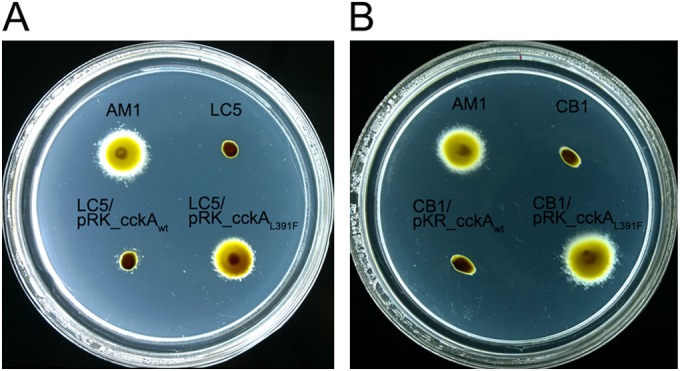
Swimming of strains expressing different cckA alleles. The swimming ability of different strains expressing different cckA alleles was tested. (A) Swimming of LC5 strain (WS8N ΔfleQ::Kanr ΔcckA::ΩSpc) expressing wild-type CckA or CckA L391F. AM1, positive control; LC5, negative control. (B) Swimming of strain CB1 (AM1 ΔfleQ::Kanr ΔcckA::ΩSpc) and complementation test with plasmids expressing CckAwt and CckA L391F. Plates were prepared with Sistrom's minimal medium containing 15 mM succinic acid and 0.22% agar. After inoculation, the plates were incubated anaerobically under continuous illumination for 60 h.
To collect further evidence supporting the role of CckA in the control of Fla2, we proceeded to inactivate cckA in the strain AM1 (resulting in strain CB1). As shown in Fig. 1B, the swimming ability of AM1 was completely abolished by the absence of cckA; the phenotype of this strain was complemented by the plasmid that expressed CckA L391F but not CckA.
Effect of mutations in CtrA and ChpT on the swimming motility of AM1.
Our results show that a single mutation in CckA is able to turn on the Fla2 system and imply that the proteins homologous to CtrA and ChpT should be involved in this phenomenon. The idea that CtrA and ChpT could also control the expression of the Fla2 genes is based on the fact that these proteins together with CckA are implicated in the expression of the flagellar genes in other bacteria (14–19). Therefore, we searched the genome of R. sphaeroides for genes encoding these proteins and found that RSWS8N_03320 (RSP_2621 in 2.4.1) encodes the response regulator CtrA, whereas RSWS8N_16219 (RSP_3825) encodes the homologue of the phosphotransfer protein ChpT. The gene encoding CtrA is located in chromosome I, whereas ChpT is encoded in chromosome II. Both genes were inactivated in WS8N (Fla1+ Fla2−) and AM1 (Fla2+ Fla1−) strains using the alleles ΔctrA::aadA and ΔchpT::ΩSpc. As shown in Fig. 2, the AM1 strain lost the ability to swim when ctrA or chpT were inactivated. In contrast, these mutant alleles did not affect the swimming motility of WS8N (see Fig. S2 in the supplemental material). Complementation using plasmids pRK_ctrA and pRK_cphT confirmed that both proteins are required for Fla2-dependent swimming (Fig. 2).
FIG 2.
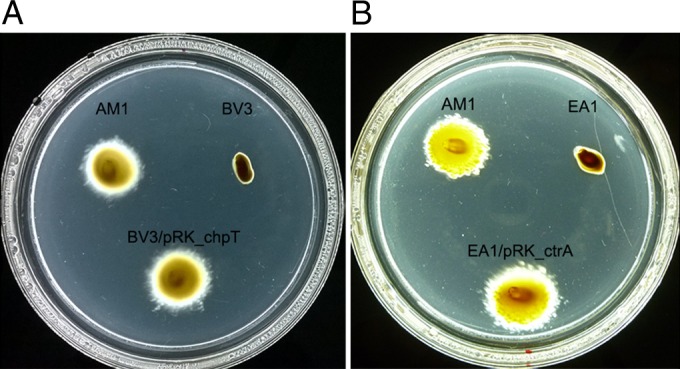
Influence of chpT and ctrA on the swimming behavior of AM1 cells. The swimming ability of chpT and ctrA mutant strains was tested. (A) Swimming of BV3 (AM1 ΔfleQ::Kanr ΔchpT::ΩSpc) strain, and complementation with plasmid pRK_chpT. (B) Swimming of EA1 (AM1 ΔfleQ::Kanr ΔctrA::aadA) and complementation with the plasmid pRK_ctrA. Plates were prepared with Sistrom's minimal medium containing 15 mM succinic acid and 0.22% agar. After inoculation the plates were incubated anaerobically under continuous illumination for 60 h.
The expression level of the fla2 genes was tested directly; for this, the reporter gene uidA, which encodes β-glucuronidase, was fused to the regulatory region of fliL2 (fliL2p) that is the first gene of a putative operon. As shown in Fig. 3, the amount of β-glucuronidase was approximately 117-fold lower in EA1 (ΔctrA::aadA) than in AM1 cells, indicating that CtrA is required, directly or indirectly, to express the fla2 genes. Unexpectedly, the expression level of fliL2p was only 20-fold lower in LC5 (ΔcckA::ΩSpc) cells compared with the level detected for AM1. This result suggests that CtrA-P accumulates in the ΔcckA strain probably due to the absence of the phosphatase activity of CckA and the phosphorylation of CtrA by a noncognate kinase or small-molecule phospho donors (56–58); alternatively, the nonphosphorylated form of CtrA could activate fliL2p in some degree. Given that in WS8N cells the expression of fliL2p is similar to that observed in EA1 cells (ΔctrA::aadA) (data not shown), we believe that the first hypothesis is correct. Nonetheless, under any of these possibilities, the low level of expression of the fla2 genes in LC5 (ΔcckA::ΩSpc) cells is not enough to promote swimming.
Isolation of additional Fla2+ strains with mutations in cckA.
To further characterize the mechanisms that allow the expression of the fla2 genes, we decided to isolate new strains showing the Fla2+ phenotype. For this, we inoculated soft-agar plates with SP13 (ΔfleQ::Kanr) or SP20 (ΔfliF::aadA) and incubated them under heterotrophic and photoheterotrophic growth conditions. After 7 days of incubation 13 Fla2+ strains were obtained, approximately the same number of mutant strains was obtained regardless of the original phenotype or growth condition. To determine if a similar mutation to that present in the AM1 strain was responsible for the Fla2+ phenotype of these strains, the DHp domain of all the cckA alleles was sequenced. We found two new mutations in two of them that generate CckA A387P and CckA F399C. The first change was identified in a strain isolated under photoheterotrophic growth conditions, the SP13 (ΔfleQ::Kanr) strain. The second mutation was identified in a strain isolated under heterotrophic growth conditions, the SP20 (ΔfliF::aadA) strain. To verify that these cckA alleles were functional, the complete cckA gene was amplified by PCR and cloned in pRK415. As expected, these mutant versions of cckA expressed from the lac promoter present in pRK415 enabled swimming of strain LC5 (ΔfleQ::Kanr ΔcckA::ΩSpc) without any selection (Fig. 4). The cckA DHp domain of the remaining Fla2+ strains did not show any change, and the full cckA gene from these strains was unable to promote swimming of LC5 (ΔfleQ::Kanr ΔcckA::ΩSpc), suggesting that cckA in these clones does not carry any mutation (data not shown). To determine if these strains had acquired a mutation in the CckA/ChpT/CtrA pathway that could explain the Fla2+ phenotype, we sequenced the ctrA and chpT genes from each strain and 75 and 74 nt upstream of the coding regions, respectively. However, no changes were detected (data not shown). These results suggest that other proteins could be involved in the control of the Fla2 system. This control could regulate the expression or activity of CckA or CtrA. Several proteins have been reported to be involved in controlling these proteins, among them, the well-conserved SciP protein, which counteracts the activity of CtrA (15, 59, 60). In C. crescentus, the level of cyclic di-GMP (c-di-GMP) regulates the stability of CtrA (61), and in other bacteria, the quorum sensing systems affect the expression of cckA and ctrA (see below).
FIG 4.
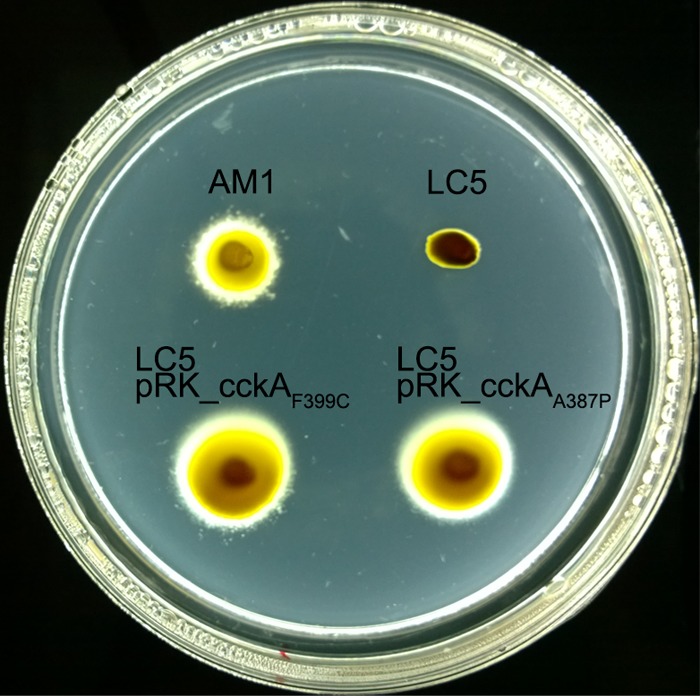
Swimming of LC5 (WS8N derivative, ΔfleQ::Kanr ΔcckA::ΩSpc) carrying plasmids pRK_cckAF399C and pRK_cckAA387P. Plates were prepared with Sistrom's minimal medium containing 15 mM succinic acid and 0.22% agar. After inoculation, the plates were incubated anaerobically under continuous illumination for 60 h.
Autokinase activity of the CckA mutants.
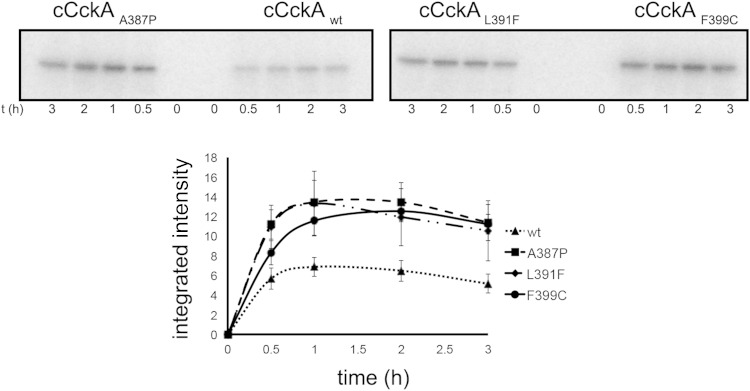
It has been reported that mutations in the DHp domain of the histidine kinases of the two-component systems usually affect the catalytic properties of the protein (62). We presume that in our case, these mutations may favor a conformation that mimics a constitutively active kinase, which would bring about activation of CtrA. To test this idea, we purified the His-tagged cytoplasmic domain of the wild-type CckA (cCckAwt) and of the three mutant versions of the protein. The phosphorylation kinetics showed an increase in the amount of phosphorylated protein for cCckA L391F, A387P, and F399C compared to the amount of phosphorylated protein accumulated for cCckAwt (Fig. 5).
FIG 5.
Autophosphorylation activity of wild-type and mutant versions of CckA. The autophosphorylation activities of the cytoplasmic fragments of different CckA versions were tested. Representative time course autoradiograms of the kinase activity of the cytoplasmic domain of wild-type CckA (cCckA) or the mutant versions are shown in the upper part. Purified proteins were incubated in the presence of [γ-32P]ATP; at the indicated time points, a sample was withdrawn for SDS-PAGE analysis and quantification. The lower part shows the densitometric quantification from three independent experiments.
Effect of the culture medium on the swimming motility of AM1.
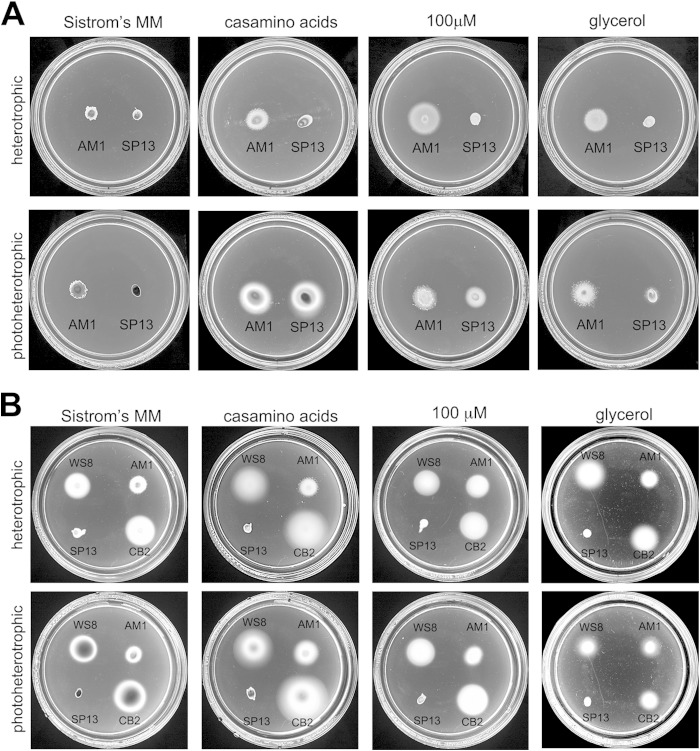
The Fla2+ mutants previously obtained were selected in Sistrom's minimal medium containing 34 mM succinic acid (21, 51). Later on, it was reported that AM1 cells displayed robust swarm halos and swimming in liquid Sistrom's medium with 100 μM succinic acid (51, 63); however, cells grow poorly at this low carbon source concentration, reaching a maximal OD600 of 0.3 in liquid medium (see Fig. S3 in the supplemental material). Therefore, we tested if other carbon sources such as glycerol and Casamino Acids could promote vigorous swimming along with better growth. Soft-agar plates containing Sistrom's medium with 15 mM or 100 μM succinic acid or Sistrom's medium without succinic acid supplemented with 0.2% Casamino Acids or 0.6% glycerol were inoculated with AM1 and SP13 (ΔfleQ::Kanr) cells and incubated under photoheterotrophic or heterotrophic conditions for 67 h. As shown in Fig. 6A, under photoheterotrophic conditions with Casamino Acids, AM1 forms a larger chemotactic ring (approximately 68%) than the one generated in Sistrom's minimal medium (15 mM succinic acid). This effect is also observed under heterotrophic conditions but to a lesser degree. The cell density reached in liquid cultures with Casamino Acids was significantly higher than in Sistrom's minimal medium with 100 μM succinic acid (see Fig. S3).
FIG 6.
Effects of different carbon sources on the swimming behavior of cells expressing either Fla1 or Fla2 flagella. The relevance of the carbon source on the swimming behavior of both flagellar types was evaluated. (A) Effect of the carbon source on swimming with the Fla2 flagellum. Strains AM1 (WS8 ΔfleQ::Kanr cckAL391F) and SP13 (WS8N ΔfleQ::Kanr) were seeded on swimming plates containing Sistrom's minimal medium with 15 mM succinic acid (Sistrom's MM), with100 μM succinic acid (100 μM), without succinic acid but supplemented with 0.2% Casamino Acids, or with 0.6% glycerol. After inoculation, the plates were incubated aerobically (upper row) or anaerobically under continuous illumination (lower row) for 60 h. (B) Effect of the carbon source on swimming with Fla1 flagella. WS8, AM1, SP13, and CB2 (AM1 ΔcckA::ΩSpc) were seeded on swarm plates as described for panel A and incubated aerobically (upper row) or anaerobically under continuous illumination (lower row) for 48 h.
Unexpectedly, in the swimming plates with a low concentration of succinic acid, with glycerol and particularly with Casamino Acids as carbon sources, we detected a swimming ring around the inoculation point of SP13 (ΔfleQ::Kanr), but only when the plates were incubated under photoheterotrophic conditions (Fig. 6A). To discard the possibility that suppressor mutants had appeared and formed this halo, cells from the periphery were reinoculated in swimming plates; again, these cells were able to swim in 100 μM succinic acid or 0.2% Casamino Acids only under photoheterotrophic conditions and not under aerobic conditions regardless of the carbon source. The swimming ring formed by these cells was similar to that presented in Fig. 6A (data not shown), further supporting the notion that these cells did not have a mutation that activates the Fla2 system. Therefore, we concluded that Fla2 flagella could be synthetized by SP13 cells without any additional mutation (besides ΔfleQ) by growing them anaerobically in either a low concentration of succinic acid or using Casamino Acids.
When AM1 and SP13 strains were grown in swimming plates containing simultaneously 0.2% Casamino Acids and a high concentration of succinic acid (15 mM), a strong reduction of the swimming ring was observed for both strains (see Fig. S4 in the supplemental material).
The fact that strain SP13 (ΔfleQ::Kanr) showed Fla2-dependent motility when grown photoheterotrophically in Casamino Acids or 100 μM succinic acid could indicate that these growth conditions could be unfavorable for Fla1-dependent swimming. Therefore, to determine if the Fla1 flagellum was active when the cells were grown in 0.2% Casamino Acids or 100 μM succinic acid, the swimming of strains WS8N and CB2 (Fla1+ ΔcckA::ΩSpc) was tested in swimming plates; the results in Fig. 6B show that swimming with Fla1 flagella is not negatively affected by these growth conditions. Given that these plates were incubated for only 48 h, the Fla2-dependent swimming of SP13 (ΔfleQ::Kanr) cells was still not detectable.
In spite of the swimming halo observed when the SP13 (ΔfleQ::Kanr) strain was grown photoheterotrophically in 0.2% Casamino Acids (Fig. 6A), in liquid medium under these growth conditions (photoheterotrophic growth with 0.2% Casamino Acids), it was very difficult to find swimming cells in the sample. This indicates that other factors are required to fully express the fla2 set. This result is in contrast with the fact that AM1 cells swim vigorously under these growth conditions (data not shown).
Transcriptional activity of cckA under different culture conditions.
To correlate the swimming behavior of AM1 under different growth conditions with the expression level of cckA, we isolated a strain carrying a transcriptional fusion of the cckA promoter with the uidA gene (ΔcckA::uidA-aadA). In this strain, the amount of β-glucuronidase, encoded by uidA, reflects the transcriptional activity of the cckA promoter. The amount of β-glucuronidase was determined in total cell extracts obtained from cultures of this strain grown under different conditions. From the results shown in Fig. 7, we observed that the amount of β-glucuronidase is maximal when the extracts were obtained from cells grown in 100 μM succinic acid (this is a very low concentration of succinic acid where growth is severely limited, see Fig. S3 in the supplemental material) or 0.2% Casamino Acids. In contrast, a pronounced reduction (100-fold) was detected when strain AM1 was grown in Sistrom's minimal medium containing 15 mM succinic acid (Fig. 7). Besides, the addition of 15 mM succinic acid to the culture medium containing Casamino Acids also reduced the amount of β-glucuronidase present in the cell extracts. These results indicate that a high concentration of succinic acid represses the expression of cckA. To further evaluate if other organic acids from the Krebs cycle negatively affect the expression of cckA, we included a 15 mM concentration of either malic or fumaric acid in the culture medium containing 0.2% Casamino Acids. As shown in Fig. 7, these compounds also diminish the expression of cckA, as was observed with succinic acid. As expected, the activity level of β-glucuronidase was also low when the strain was grown only in 15 mM fumaric or malic acid as a carbon source (Fig. 7). To test a different carbon source, we added glycerol to the culture medium with Casamino Acids. Glycerol did not affect the induction of cckA produced by the presence of Casamino Acids (Fig. 7).
Presence of Fla2 flagella in a Fla1+ Fla2+ strain.
To select for the presence of Fla2 flagella, it was necessary to eliminate the swimming motility mediated by the Fla1 flagella. However, to explore whether or not a single cell can display both types of flagella, we introduced the plasmid pRK_fleQ+ in AM1 (ΔfleQ::Kanr cckAL391F) cells. It was previously demonstrated that this plasmid restores the Fla1+ phenotype of the Fla1− strain carrying the ΔfleQ::Kanr allele (SP13) (23).
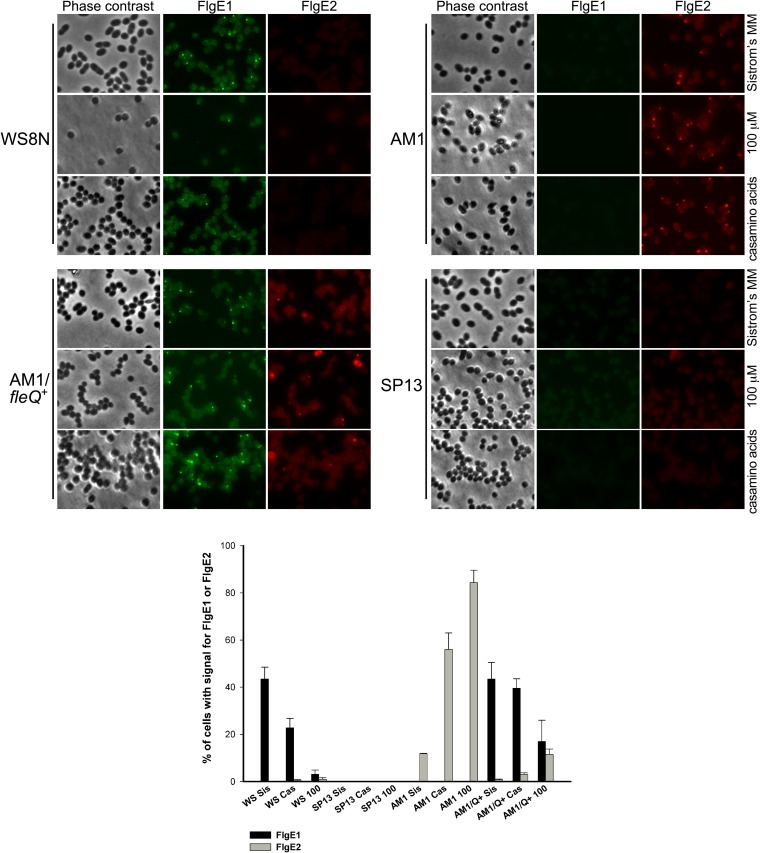
AM1/pRK_fleQ+, as well as AM1, WS8N, and SP13 (ΔfleQ::Kanr) cells, were grown photoheterotrophically in Sistrom's minimal medium until early stationary phase. The cultures were washed in medium without carbon source, and an aliquot was used to inoculate fresh medium supplemented with 100 μM succinic acid, 0.2% Casamino Acids, or 15 mM succinic acid. After 14 h of photoheterotrophic growth; cells were collected, fixed, and tested separately with anti-FlgE1 or anti-FlgE2 antibody, previously stained with Alexa Fluor 488 or 546, respectively.
Three independent experiments were carried out, and representative images obtained from one of these experiments are shown in Fig. 8, as is a quantification of a total of 3,000 cells for each strain and for each condition. From these experiments, we determined that in Sistrom's medium supplemented with 15 mM succinic acid, approximately 45% of the wild-type WS8N cells showed a single fluorescent focus corresponding to the Fla1 flagellum; whereas no signal was detected for the Fla2 flagella. In contrast, 84% of AM1 cells grown in 100 μM succinic acid showed a fluorescent focus when tested with anti-FlgE2, but no signal was detected using anti-FlgE1. As expected, SP13 cells did not show the presence of Fla1 or Fla2 when grown in 15 mM or 100 μM succinic acid or in 0.2% Casamino Acids (Fig. 8), showing that our methodology is appropriate for the detection of both types of flagella. Therefore, we tested the presence of Fla1 and Fla2 in AM1/pRK_fleQ+ cells. When this strain was grown in 15 mM succinic acid, we detected 43% of cells with a single fluorescent focus corresponding to the Fla1 flagella and 0.7% of the cells with Fla2. The same strain grown in 100 μM succinic acid showed 17% of the cells with Fla1 flagella and 11% of cells with Fla2 flagella. When this strain was grown in 0.2% Casamino Acids the same trend was observed, i.e., a dominance of Fla1 (39%) over Fla2 (3%) (Fig. 8). Interestingly, even at 100 μM succinic acid we were unable to detect a single cell showing simultaneous fluorescent signals for FlgE1 and FlgE2. This suggests a mutual exclusion between the two flagellar systems in a single cell but not in the population.
FIG 8.
In situ detection of Fla1 and Fla2 flagella. Bacterial cells obtained from the indicated culture medium were incubated in the presence of anti-FlgE1 or anti-FlgE2 labeled with Zenon Alexa Fluor 488 or Alexa Fluor 546, respectively. In the lower part, the percentage of cells showing signal for FlgE1 or FlgE2 for each condition is indicated. The graph shows the average from three independent experiments. The standard deviation is also shown.
From these experiments, we also observed that for WS8N the numbers of cells with Fla1 flagella were reduced approximately 2- and 14-fold when the strain was grown under culture conditions that promote the expression of cckA, i.e., Sistrom's minimal medium with 0.2% Casamino Acids or 100 μM succinic acid (Fig. 8). Similarly, the number of cells with Fla2 flagella decreased drastically when the AM1 strain was complemented with fleQ, further supporting the exclusion of the flagellar systems.
DISCUSSION
In this work, we show that the two-component system formed by CckA, ChpT, and CtrA regulates the expression of the fla2 genes in R. sphaeroides. The genome sequence of mutants able to swim in the absence of the Fla1 flagella revealed that a single mutation in cckA is enough to turn on the expression of fla2 under growth conditions that do not normally enable transcription of these flagellar genes (21, 22). The presence of the Fla2 flagella in strains that express these gain-of-function versions of CckA is dependent on the presence of ChpT and CtrA, indicating that the mutations in cckA do not alter the specificity of the kinase and that the signaling occurs through the known components of the pathway. Therefore, CtrA should activate the expression of the fla2 genes in R. sphaeroides, similar to the situation previously reported for other alphaproteobacteria that do not have a second flagellar gene set (14–16, 19).
Previously, it has been reported that mutations in the DHp domain of HKs such as EnvZ, NtrB, CrdS, and AgrC can affect the kinase, phosphatase activity, or both (62, 64–66). In this work, we show that CckA L391F, A387P, and F399C autophosphorylate faster than wild-type CckA; in vivo, this should result in a higher level of CtrA-P and hence the expression of fla2. It remains to be demonstrated that autophosphorylation of wild-type CckA is the only rate-limiting step in the phosphorylation of CtrA.
A transcriptional fusion of fliL2p with a reporter gene showed that in the absence of cckA, this promoter is expressed at a higher level than in the ctrA strain or in WS8N, suggesting that CtrA could be phosphorylated by a noncognate histidine kinase, as has been observed for other response regulators (56, 58), or by small phospho donors such as acetyl phosphate (57); in WS8N, the phosphatase activity of CckA seems to reduce the level of CtrA-P, given that the expression of fliL2p is as low as that observed in the ctrA strain.
Intriguingly, we did not isolate a gain-of-function ctrA mutant even though we were able to isolate three independent mutants with changes in CckA that yield a Fla2+ phenotype, as well as 11 independent mutants with the Fla2+ phenotype with changes that did not affect cckA, chpT, or ctrA. It is possible that a constitutive mutation in CtrA could result in the repression of the fla2 genes, or perhaps an unknown gene under the control of CtrA could affect cell growth negatively. On the other hand, the fact that we were able to isolate strains with a Fla2+ phenotype without affecting the CckA pathway suggest that other elements might control the expression of the fla2 genes. It will be interesting to identify these mutations and determine if they act through the CckA pathway or independently.
In addition to the CckA pathway, our results show that the redox sensor RegB could be involved in the regulation of the fla2 genes. We noticed that under aerobic conditions, the swim ring generated by AM1 is reduced by approximately 20% if regB is deleted or when the wild-type regBS267 allele is expressed in the CD2 (AM1 ΔregB::ΩSpc) strain. However, under photoheterotrophic conditions where RegB is active (26), the CD2 (AM1 ΔregB::ΩSpc) strain carrying pRK_regBS267 or pRK_regBL267 showed a similar swim ring. It has been reported that under oxidizing conditions, the kinase activity of RegB is inactivated by the formation of cysteine sulfenic acid at position 265 (67), explaining the reduction of the swarm ring when RegB S267 is expressed under aerobic conditions. Conversely, under anaerobic conditions, RegB acts as a kinase (26), and in this state, RegB S267 promotes swimming with Fla2 flagella. However, when RegB L267 is expressed, swimming is enhanced under aerobic growth conditions, suggesting that this mutation renders RegB insensitive to oxygen. In a search for possible RegA binding sites, the cognate response regulator of RegB, using its proposed recognition sequence (26), did not reveal any positive hits in the noncoding regions upstream of cckA and ctrA, indicating that control of fla2 expression by the Reg system could be indirect or that the binding site of RegA in these regions is not conserved. Alternatively, RegB could influence the aerotactic response mediated by Fla2.
In this work, we also show that cckA transcription is strongly repressed by the presence of a high concentration of organic acids in the culture medium (i.e., 15 mM succinic, malic, and fumaric acids) but is strongly induced when 0.2% Casamino Acids or a low concentration of succinic acid (100 μM) is used as a carbon source. Cell growth is compromised in the presence of 100 μM succinic acid (see Fig. S3 in the supplemental material), but the cultures reach a high OD600 in 0.2% Casamino Acids, suggesting that the induction of cckA is not dependent on the cell density of the culture. However, it cannot be discarded that the quorum sensing system of R. sphaeroides that is controlled by the proteins CerR and CerI (homologs of LuxR and LuxI, respectively) (68) could influence cckA expression, as occurs in other alphaproteobacteria (16, 20).
The repression of cckA and the consequent absence of Fla2 flagella mediated by the presence of organic acids is contrary to what has been reported for R. capsulatus, in which ctrA is expressed in a higher level when the cells are grown in minimal medium containing malic acid as a carbon source than in peptone-based rich medium (20). In agreement with this, a recent comparison of the expression profiles of orthologous genes between R. capsulatus and R. sphaeroides showed that the expression pattern of cckA is not conserved (69).
C4-dicarboxylates are a preferred carbon source for Pseudomonas aeruginosa and Sinorhizobium meliloti and cause catabolite repression of degradative pathways for other carbon sources (70–72). Catabolite repression in P. aeruginosa is mediated by the global regulator Crc, which represses the translation of genes involved in the uptake and catabolism of several nonpreferred carbon sources. In the presence of less preferred carbon sources, the two-component system CbrA/CbrB activates the transcription of one small RNA that binds Crc, relieving translational repression (73). In S. meliloti a histidine kinase is required to maintain a strong succinate-mediated catabolite repression (74), but the kinases of P. aeruginosa and S. meliloti are not similar. Therefore, it is difficult at this point to predict how succinate and other C4-dicarboxylic acids could repress the expression of cckA in R. sphaeroides.
R. sphaeroides is commonly grown in Sistrom's minimal medium that includes 34 or 17 mM succinic acid, partially explaining why the Fla2 flagella had not been detected previously in the wild-type strain. Nevertheless, relieving the repression exerted by organic acids is not enough to observe swimming with the Fla2 flagella, suggesting that swimming with Fla2 may require activation of CckA by an unknown mechanism. Given that R. sphaeroides does not carry the genes encoding DivL, DivK, or DivJ, which are known to control CckA activity in C. crescentus, it is possible that other proteins are involved in controlling CckA activity.
We observed that vigorous swimming of a fla1 strain with the wild-type cckA allele requires growth in soft-agar plates containing Casamino Acids as a carbon source and incubation under photoheterotrophic conditions; swimming was not detected when the same type of plates was incubated aerobically in the dark or when cells were grown photoheterotrophically in 0.2% Casamino Acids but in liquid medium. These results show that is possible to detect Fla2-dependent swimming on wild-type cckA when photoheterotrophic growth using Casamino Acids as a carbon source is combined with an unknown signal or stimuli generated during the growth in soft-agar plates. The most obvious explanation would imply that this hypothetical signal could be related with the nutrient gradient formed by cell growth. It remains to be determined if these conditions synergistically increase the expression of cckA or if other mechanisms could exist, such as CckA activation.
Independently of the hypothetical mechanism that may mediate CckA activation, our results indicate that transcriptional activation of cckA is an important step to enable the expression of the fla2 genes.
The control of fla2 genes in R. sphaeroides is particularly relevant, given that this bacterium could have developed additional mechanisms to control fla2 after acquiring the fla1 set. We present evidence that indicates that the expression of fla2 is somehow incompatible with the synthesis of the Fla1 flagellum. This idea is based on the observations that 84% of the cells of the AM1 strain grown in 100 μM succinic acid have Fla2 flagella and that, when this strain was complemented with fleQ+, a notable reduction in the number of cells that express Fla2 occurred, together with an important increase in the number of cells with Fla1 flagella. Individual cells with both flagella where not observed. This result suggests that in a single cell, the synthesis of Fla2 is incompatible with synthesis of Fla1. These results suggest that R. sphaeroides has a mechanism that ensures the expression of only one flagellar set per cell. Even though the presence of Fla1 in WS8N, or of Fla2 in AM1, follows an opposite trend depending on the culture conditions, a more complex regulation should exist, given that growth in the presence of Casamino Acids or 100 μM succinic acid does not hinder the synthesis of Fla1 in the AM1/pRK_fleQ+ strain, indicating that Fla1 somehow inhibits the synthesis of Fla2 flagella in individual cells.
It has been commonly observed in bacteria that have a polar flagellum and inducible lateral flagella (required for swarming) that lateral flagella are expressed only under conditions of increased viscosity (75), However, it was recently reported that under planktonic conditions Shewanella oneidensis has the polar flagellum and, sporadically, one or two lateral flagella are formed. In this case, the simultaneous expression of both types of flagella allows a more efficient spreading in swimming plates because the lateral flagella modify the swimming pattern (76). Although we were unable to detect R. sphaeroides cells with both types of flagella when grown in liquid media, it is still possible that this type of cells was present in swimming plates. If this is the case, Fla2 does not contribute to cell spreading, since we did not observe a reduction in the swimming halo of Fla1+ Fla2− cells even in plates incubated photoheterotrophically with Casamino Acids (Fig. 6B). In R. sphaeroides, the existence of two types of flagella that are mutually exclusive could be advantageous because of a possible specialization for each type of flagellum depending on the environmental conditions. In addition, since each of these flagella has its own chemotactic system, it is possible that the advantage of having two flagellar systems is related with a better browsing of attractants in the environment at a population level.
As mentioned above, R. capsulatus, which is closely related to R. sphaeroides, is motile in a medium containing malic acid as a carbon source, as well as in rich medium (20). To our knowledge, there are no reports of a growth condition under which this bacterium becomes nonmotile, indicating that the CckA pathway must be functional in all the growth conditions tested so far. In contrast, in R. sphaeroides we observed that this pathway seems to be inactive under several growth conditions. Therefore, it is expected that profound differences should exist in the control of these proteins in order to produce such a different outcome. It is possible that the acquisition of the Fla1 set reshaped the regulatory circuit that controls this signal transduction system. A similar situation has been reported regarding the multiple σ54 factors present in R. sphaeroides that show unique features (77, 78).
Supplementary Material
ACKNOWLEDGMENTS
We thank Aurora Osorio, Teresa Ballado, and Javier de la Mora for technical support. We also thank Andrés de Sandozequi Mijares for the gift of the FlgE1 protein and Georgina Hernández for valuable help with antibody production. We thank the IFC Molecular Biology Unit for sequence facilities.
A.R. was supported by a fellowship from CONACyT (239738). This work was partially supported by grants from PAPIIT (IN204614) and CONACyT (235996).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02429-14.
REFERENCES
- 1.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 3.Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol 270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 4.Smith TG, Hoover TR. 2009. Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv Appl Microbiol 67:257–295. doi: 10.1016/S0065-2164(08)01008-3. [DOI] [PubMed] [Google Scholar]
- 5.Curtis PD, Brun YV. 2010. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev 74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsang J, Hoover TR. 2014. Themes and variations: regulation of RpoN-dependent flagellar genes across diverse bacterial species. Scientifica 2014:681754. doi: 10.1155/2014/681754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. 2009. Dynamics of two phosphorelays controlling cell cycle progression in Caulobacter crescentus. J Bacteriol 191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. doi: 10.1016/S0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 9.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Heindl JE, Fuqua C. 2013. Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens. PLoS One 8:e56682. doi: 10.1371/journal.pone.0056682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. 2001. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol 183:3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsokos CG, Perchuk BS, Laub MT. 2011. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell 20:329–341. doi: 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belas R, Horikawa E, Aizawa S, Suvanasuthi R. 2009. Genetic determinants of Silicibacter sp. TM1040 motility. J Bacteriol 191:4502–4512. doi: 10.1128/JB.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS. 2012. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett 331:53–62. doi: 10.1111/j.1574-6968.2012.02553.x. [DOI] [PubMed] [Google Scholar]
- 16.Zan J, Heindl JE, Liu Y, Fuqua C, Hill RT. 2013. The CckA-ChpT-CtrA phosphorelay system is regulated by quorum sensing and controls flagellar motility in the marine sponge symbiont Ruegeria sp. KLH11. PLoS One 8:e66346. doi: 10.1371/journal.pone.0066346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Ziesche L, Frank O, Michael V, Martin M, Petersen J, Schulz S, Wagner-Dobler I, Tomasch J. 2014. The CtrA phosphorelay integrates differentiation and communication in the marine alphaproteobacterium Dinoroseobacter shibae. BMC Genomics 15:130. doi: 10.1186/1471-2164-15-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene SE, Brilli M, Biondi EG, Komeili A. 2012. Analysis of the CtrA pathway in Magnetospirillum reveals an ancestral role in motility in alphaproteobacteria. J Bacteriol 194:2973–2986. doi: 10.1128/JB.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller TR, Belas R. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ Microbiol 8:1648–1659. doi: 10.1111/j.1462-2920.2006.01071.x. [DOI] [PubMed] [Google Scholar]
- 20.Leung MM, Brimacombe CA, Beatty JT. 2013. Transcriptional regulation of the Rhodobacter capsulatus response regulator CtrA. Microbiology 159:96–106. doi: 10.1099/mic.0.062349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poggio S, Abreu-Goodger C, Fabela S, Osorio A, Dreyfus G, Vinuesa P, Camarena L. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J Bacteriol 189:3208–3216. doi: 10.1128/JB.01681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armitage JP, Macnab RM. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol 169:514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poggio S, Osorio A, Dreyfus G, Camarena L. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol Microbiol 58:969–983. doi: 10.1111/j.1365-2958.2005.04900.x. [DOI] [PubMed] [Google Scholar]
- 24.Martin AC, Gould M, Byles E, Roberts MA, Armitage JP. 2006. Two chemosensory operons of Rhodobacter sphaeroides are regulated independently by sigma 28 and sigma 54. J Bacteriol 188:7932–7940. doi: 10.1128/JB.00964-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romagnoli S, Packer HL, Armitage JP. 2002. Tactic responses to oxygen in the phototrophic bacterium Rhodobacter sphaeroides WS8N. J Bacteriol 184:5590–5598. doi: 10.1128/JB.184.20.5590-5598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsen S, Swem LR, Swem DL, Bauer CE. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev 68:263–279. doi: 10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eraso JM, Kaplan S. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol 176:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swem LR, Elsen S, Bird TH, Swem DL, Koch HG, Myllykallio H, Daldal F, Bauer CE. 2001. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J Mol Biol 309:121–138. doi: 10.1006/jmbi.2001.4652. [DOI] [PubMed] [Google Scholar]
- 29.Swem LR, Kraft BJ, Swem DL, Setterdahl AT, Masuda S, Knaff DB, Zaleski JM, Bauer CE. 2003. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J 22:4699–4708. doi: 10.1093/emboj/cdg461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eraso JM, Kaplan S. 1996. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J Bacteriol 178:7037–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackenzie C, Choudhary M, Larimer FW, Predki PF, Stilwagen S, Armitage JP, Barber RD, Donohue TJ, Hosler JP, Newman JE, Shapleigh JP, Sockett RE, Zeilstra-Ryalls J, Kaplan S. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth Res 70:19–41. doi: 10.1023/A:1013831823701. [DOI] [PubMed] [Google Scholar]
- 32.Sockett RE, Foster JCA, Armitage JP. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp 53:473–479. [Google Scholar]
- 33.Sistrom WR. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas spheroides. J Gen Microbiol 28:607–616. doi: 10.1099/00221287-28-4-607. [DOI] [PubMed] [Google Scholar]
- 34.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1987. Current protocols in molecular biology. John WiIey and Sons, New York, NY. [Google Scholar]
- 35.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 36.Metcalf WW, Wanner BL. 1993. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene 129:17–25. doi: 10.1016/0378-1119(93)90691-U. [DOI] [PubMed] [Google Scholar]
- 37.Davis J, Donohue TJ, Kaplan S. 1988. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J Bacteriol 170:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology (NY) 1:37–45. [Google Scholar]
- 39.Ballado T, Camarena L, Gonzalez-Pedrajo B, Silva-Herzog E, Dreyfus G. 2001. The hook gene (flgE) is expressed from the flgBCDEF operon in Rhodobacter sphaeroides: study of an flgE mutant. J Bacteriol 183:1680–1687. doi: 10.1128/JB.183.5.1680-1687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jefferson RA, Burgess SM, Hirsh D. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A 83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 42.Girard L, Brom S, Davalos A, Lopez O, Soberon M, Romero D. 2000. Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol Plant Microbe Interact 13:1283–1292. doi: 10.1094/MPMI.2000.13.12.1283. [DOI] [PubMed] [Google Scholar]
- 43.Bao K, Cohen SN. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev 15:1518–1527. doi: 10.1101/gad.896201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Porter SL, Wilkinson DA, Byles ED, Wadhams GH, Taylor S, Saunders NJ, Armitage JP. 2011. Genome sequence of Rhodobacter sphaeroides strain WS8N. J Bacteriol 193:4027–4028. doi: 10.1128/JB.05257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goecks J, Nekrutenko A, Taylor J, Galaxy T. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 89:19.10.1–19.10.21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. 2011. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res 39:e132. doi: 10.1093/nar/gkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.del Campo AM, Ballado T, de la Mora J, Poggio S, Camarena L, Dreyfus G. 2007. Chemotactic control of the two flagellar systems of Rhodobacter sphaeroides is mediated by different sets of CheY and FliM proteins. J Bacteriol 189:8397–8401. doi: 10.1128/JB.00730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeilstra-Ryalls J, Gomelsky M, Eraso JM, Yeliseev A, O'Gara J, Kaplan S. 1998. Control of photosystem formation in Rhodobacter sphaeroides. J Bacteriol 180:2801–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laratta WP, Choi PS, Tosques IE, Shapleigh JP. 2002. Involvement of the PrrB/PrrA two-component system in nitrite respiration in Rhodobacter sphaeroides 2.4.3: evidence for transcriptional regulation. J Bacteriol 184:3521–3529. doi: 10.1128/JB.184.13.3521-3529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 55.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu Rev Genet 41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 56.Siryaporn A, Goulian M. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol 70:494–506. doi: 10.1111/j.1365-2958.2008.06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A 89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guckes KR, Kostakioti M, Breland EJ, Gu AP, Shaffer CL, Martinez CR III, Hultgren SJ, Hadjifrangiskou M. 2013. Strong cross-system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc Natl Acad Sci U S A 110:16592–16597. doi: 10.1073/pnas.1315320110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan MH, Kozdon JB, Shen X, Shapiro L, McAdams HH. 2010. An essential transcription factor, SciP, enhances robustness of Caulobacter cell cycle regulation. Proc Natl Acad Sci U S A 107:18985–18990. doi: 10.1073/pnas.1014395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gora KG, Tsokos CG, Chen YE, Srinivasan BS, Perchuk BS, Laub MT. 2010. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell 39:455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. 2011. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell 43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willett JW, Kirby JR. 2012. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet 8:e1003084. doi: 10.1371/journal.pgen.1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martínez-del Campo A, Ballado T, Camarena L, Dreyfus G. 2011. In Rhodobacter sphaeroides, chemotactic operon 1 regulates rotation of the flagellar system 2. J Bacteriol 193:6781–6786. doi: 10.1128/JB.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pioszak AA, Ninfa AJ. 2003. Genetic and biochemical analysis of phosphatase activity of Escherichia coli NRII (NtrB) and its regulation by the PII signal transduction protein. J Bacteriol 185:1299–1315. doi: 10.1128/JB.185.4.1299-1315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geisinger E, Muir TW, Novick RP. 2009. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc Natl Acad Sci U S A 106:1216–1221. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsing W, Russo FD, Bernd KK, Silhavy TJ. 1998. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol 180:4538–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J, Cheng Z, Reddie K, Carroll K, Hammad LA, Karty JA, Bauer CE. 2013. RegB kinase activity is repressed by oxidative formation of cysteine sulfenic acid. J Biol Chem 288:4755–4762. doi: 10.1074/jbc.M112.413492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puskas A, Greenberg EP, Kaplan S, Schaefer AL. 1997. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol 179:7530–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peña-Castillo L, Mercer RG, Gurinovich A, Callister SJ, Wright AT, Westbye AB, Beatty JT, Lang AS. 2014. Gene co-expression network analysis in Rhodobacter capsulatus and application to comparative expression analysis of Rhodobacter sphaeroides. BMC Genomics 15:730. doi: 10.1186/1471-2164-15-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Collier DN, Hager PW, Phibbs PVJ. 1996. Catabolite repression control in the Pseudomonads. Res Microbiol 147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 71.Ucker DS, Signer ER. 1978. Catabolite-repression-like phenomenon in Rhizobium meliloti. J Bacteriol 136:1197–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valentini M, Lapouge K. 2013. Catabolite repression in Pseudomonas aeruginosa PAO1 regulates the uptake of C4 -dicarboxylates depending on succinate concentration. Environ Microbiol 15:1707–1716. doi: 10.1111/1462-2920.12056. [DOI] [PubMed] [Google Scholar]
- 73.Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 74.Garcia PP, Bringhurst RM, Arango Pinedo C, Gage DJ. 2010. Characterization of a two-component regulatory system that regulates succinate-mediated catabolite repression in Sinorhizobium meliloti. J Bacteriol 192:5725–5735. doi: 10.1128/JB.00629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCarter LL. 2004. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol 7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 76.Bubendorfer S, Koltai M, Rossmann F, Sourjik V, Thormann KM. 2014. Secondary bacterial flagellar system improves bacterial spreading by increasing the directional persistence of swimming. Proc Natl Acad Sci U S A 111:11485–11490. doi: 10.1073/pnas.1405820111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poggio S, Osorio A, Dreyfus G, Camarena L. 2002. The four different σ54 factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol Microbiol 46:75–85. doi: 10.1046/j.1365-2958.2002.03158.x. [DOI] [PubMed] [Google Scholar]
- 78.Poggio S, Osorio A, Dreyfus G, Camarena L. 2006. Transcriptional specificity of RpoN1 and RpoN2 involves differential recognition of the promoter sequences and specific interaction with the cognate activator proteins. J Biol Chem 281:27205–27215. doi: 10.1074/jbc.M601735200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.