Abstract
Horizontal gene transfer by conjugation plays a major role in bacterial evolution, allowing the acquisition of new traits, such as virulence and resistance to antibacterial agents. With the increased antibiotic resistance in bacterial pathogens, a better understanding of how bacteria modulate conjugation under changing environments and the genetic factors involved is needed. Despite the evolutionary advantages conjugation may confer, the process can be quite stressful for the donor cell. Here, we characterize the ability of TraR, encoded on the episomal F′ plasmid, to upregulate the σE extracytoplasmic stress pathway in Escherichia coli. TraR, a DksA homolog, modulates transcription initiation through the secondary channel of RNA polymerase. We show here that TraR activates transcription directly; however, unlike DksA, it does so without using ppGpp as a cofactor. TraR expression can stimulate the σE extracytoplasmic stress response independently of the DegS/RseA signal transduction cascade. In the absence of TraR, bacteria carrying conjugative plasmids become more susceptible to external stress. We propose that TraR increases the concentrations of periplasmic chaperones and proteases by directly activating the transcription of σE-dependent promoters; this increased protein folding capacity may prepare the bacterium to endure the periplasmic stress of sex pilus biosynthesis during mating.
INTRODUCTION
Horizontal gene transfer is a primary way for bacteria to transmit genetic material, including genes for virulence and antibiotic resistance (1). Conjugation is the most common mechanism of horizontal transfer, with the F′ plasmid being the archetypal example in Escherichia coli (2). During conjugation, the donor cell synthesizes a sex pilus apparatus to facilitate the exchange of genetic material. The pilus forms a mating bridge, which establishes proximity and formation of a pore that allows the transfer of DNA from the donor bacterium to the recipient. Regulation of the F′ pilus biosynthesis is complex; most of its structural and regulatory components are encoded by the large tra transfer operon (3, 4). The tra operon includes the pilin and regulatory genes, which together form sex pili on the cell surface (5). Synthesis and maintenance of the F′ pilus is associated with cellular and envelope sensitivity and the cpxAR and σE stress responses (6, 7). Activation of the cpxAR envelope stress response decreases F′ transfer, suggesting that F′ transfer is inhibited during cell envelope stress (8). In addition, it has been shown in E. coli and Salmonella enterica serovar Typhimurium that the transfer of F-like plasmids induces the extracytoplasmic stress pathway. An overproduction of pilus subunits leads to an increased level of unfolded proteins in the periplasm (9). Accordingly, deletion of the pilin gene from the tra operon reduces induction of the extracytoplasmic and cytoplasmic stress response genes seen in wild-type conjugating cells (6). Thus, induction of bacterial stress response pathways is likely an important cellular response during conjugation.
RpoE (σE) is the alternative sigma factor associated with extracytoplasmic and envelope stress (10). Unfolded outer membrane proteins result in activation of the σE pathway, leading to transcription of genes involved in processes such as protein folding, degradation, and envelope synthesis (11). σE activity is regulated by the membrane anti-sigma factor RseA. Under nonstress conditions, the cytoplasmic domain of RseA sequesters σE, hindering interaction of σE with RNA polymerase (RNAP) (12). Upon stress, the DegS membrane protease cleaves the periplasmic end of RseA, after which a second protease, RseP, cleaves the transmembrane domain of RseA. Finally, degradation of the cytoplasmic fragment of RseA by the ClpXP protease releases σE into the cytoplasm, allowing the activation of the envelope stress response genes (13–15). The σE response also can be activated by the action of DksA/ppGpp on RNAP (16). DksA and ppGpp are able to upregulate σE activity both directly, by modulating transcription of the σE holoenzyme at the promoter, and indirectly, by altering the competition for core RNAP between the different sigma factors (16–19).
Within the conjugative plasmid's transfer operon is TraR, a DksA-like protein that acts independently of ppGpp (20). Like DksA, TraR has been proposed to interact with the secondary channel of RNAP to modulate transcription initiation. TraR expression rescues the auxotrophy of ΔdksA and ppGpp0 mutants (20). TraR is able to downregulate rRNA genes and upregulate genes necessary for amino acid uptake and synthesis without ppGpp accumulation (20). Not surprisingly, considering its effects on transcription, induction of TraR synthesis inhibits growth (20). TraR homologs are present on a variety of conjugative plasmids and are found in phage, suggesting conservation of an important function. Nevertheless, deletion of the F′ traR does not hinder conjugation under standard laboratory conditions (21). Since DksA/ppGpp can regulate the σE pathway (19), we are interested in examining the role of TraR in the modulation of this stress response.
In this work, we show that TraR activates σE-dependent promoters in vivo and in vitro with purified components, i.e., TraR activates transcription directly. These results are consistent with the ability of TraR to activate σE-dependent promoters in vivo in the absence of proteolytic activation of the anti-sigma factor RseA and without a change in σE protein levels. Single-cell analysis via microscopy and flow cytometry shows increased σE-dependent activation in the presence of the conjugative F′ plasmid. Additionally, the presence of TraR assists cells in surviving heat and ethanol stress, a stress associated with the extracytoplasmic stress response. We propose that TraR is activated during conjugation to assist cells in dealing with the upcoming periplasmic stress associated with pilus formation by redirecting the host transcriptional machinery toward σE-dependent transcription. This work illuminates one of the functions of the TraR protein and could explain its high level of conservation across conjugative plasmids.
MATERIALS AND METHODS
Media and bacterial growth conditions.
Standard methods of E. coli genetics were performed (22). In order to better compare our assays to others in the literature, all experiments were done at 32°C, unless otherwise indicated, with either LB medium or M9 medium supplemented with FeCl2 (10 μM) and thiamine (vitamin B1) (2 μg ml−1). Media were supplemented with the following when required: sodium citrate (5 to 20 mM), glucose (0.2%), Casamino Acids (0.3%), isopropyl-β-d-thiogalactopyranoside (IPTG) (0.1 mM), ampicillin (50 μg ml−1), kanamycin (30 μg ml−1), chloramphenicol (12.5 μg ml−1), and tetracycline (3.33 μg ml−1 with sodium citrate and 10 μg ml−1 without).
Bacterial strains and plasmids.
The backgrounds, genotypes, and sources of the E. coli strains and plasmids used in this study are listed in Table S1 in the supplemental material. Unless otherwise indicated, all strains are derivatives of MG1655, and plasmids were introduced by standard transformation techniques. Mutant alleles were transferred into the desired background through either standard P1 transduction (22) or linear transformation techniques, with subsequent elimination of the drug resistance marker by FLP recombinase when necessary (23). Proper alleles were verified by antibiotic resistance selection and, when appropriate, PCR, DNA sequencing, and/or phenotypic assays. Plasmids have been described previously (20).
β-Gal activity assays.
Overnight cultures were diluted 1:100 and grown aerobically at 32°C in LB supplemented with antibiotic and IPTG. Samples (0.5 ml) were taken at appropriate OD600 intervals and assayed as previously described (22). Graphs show OD420 × (2 × 103)/time versus the OD600 to calculate β-galactosidase (β-gal) units. All graphs are representative of at least 3 independent experiments and were plotted using GraphPad Prism 5. The rate of β-gal synthesis was calculated from a linear equation using a best-fit line during the exponential growth phase.
Western blot analysis.
Protein samples were taken from liquid LB cultures at the specified OD600 by adding 0.9 ml of culture to 0.1 ml of ice-cold 50% trichloroacetic acid (TCA). Samples then were centrifuged and the pellet resuspended to a concentration of 0.250 OD600 units in SDS sample buffer. Samples then were boiled for 10 min and loaded onto 15% polyacrylamide gels. Protein levels of σE were detected using a rabbit polyclonal antibody (Carol Gross laboratory). Alexa Fluor 647–goat anti-rabbit IgG (Invitrogen) was used as a secondary antibody. Bands were visualized using a Typhoon Trio scanner (emission filter, 670BP 30 PMT 300; GE), and data were quantified. Data shown are the averages from at least 3 independent experiments.
In vitro transcription.
Multiple-round in vitro transcription assays were performed using EσE (holoenzyme containing σE) purified by standard procedures as described previously (19). C-terminally hexahistidine-tagged WT and D6N TraR were purified as described previously (20). The plasmid template, pSEB015, is a derivative of pRLG770 containing the P3 promoter region of rpoH (19) and an rrnB T1 terminator ∼140-nucleotides downstream of the promoter. Transcription reaction mixtures (25 μl) contained 50 ng DNA template, 40 mM Tris HCl, pH 8.0, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1 mg/ml bovine serum albumin (BSA), 40 mM NaCl, 500 μM ATP, 100 μM GTP, 100 μM CTP, 10 μM UTP, and 1 μCi of [α32-P]UTP. TraR (nM to μM range) was added to the transcription mix, and reactions were initiated by addition of 5 nM EσE RNAP. Reactions were carried out at 23°C, stopped after 15 min with an equal volume of formamide stop solution, and electrophoresed on 7 M urea-6% polyacrylamide gels. Transcripts were visualized by phosphorimaging and quantified using ImageQuant 5.2 software. The amount of transcription in the presence of factor was normalized to transcription in the absence of any factor. The experiments were performed independently in triplicate. Averages and standard deviations are plotted.
Fluorescence microscopy and flow cytometry.
Overnight cultures were grown at 32°C in LB plus antibiotic. Microscopy was performed using an Axio Observer Z1 inverted microscope (Zeiss) on phase contrast at ×100 magnification. Flow cytometry was performed using a BD FACSCanto II flow cytometer, measuring 10,000 events. Data were analyzed using FloJo software.
Stress assays.
Nutrient stress was examined by diluting (1:100) overnight cultures grown in LB and plating on M9/glucose with or without Casamino Acids. Plates were incubated at 32°C, and CFU/ml counts on M9/glucose plus Casamino Acids were considered to be 100% growth.
Thermotolerance was examined using a modified protocol from Strozen et al. (24). Cultures were diluted 1:1,000 and grown at 30°C in LB for 1 h. The cultures then were aliquoted into a 96-well microtiter plate (Costar) and incubated at 42°C using an Optima microplate reader (BMG Scientific). Cultures were shaken in double orbital mode with 3-cm width, and OD600 readings were taken every 20 min for 18 h.
Ethanol stress was assayed by treating overnight bacterial cultures grown at 32°C with either 10% ethanol or water for 1 h with aeration. To obtain viability counts, cells were washed twice in H2O, diluted, and plated on LB.
Statistics.
Data sets were compared using either t tests or analysis of variance (ANOVA) when appropriate. Curves shown are best-fit 3rd-order polynomials. The rate of β-gal synthesis was calculated from a linear equation using a best-fit line during exponential growth phase. All data analysis was performed using GraphPad Prism 5 software.
RESULTS
TraR is expressed when the conjugal transfer operon is active.
Previous work established that TraR is a functional homolog of DksA, inhibits transcription of the ribosomal rrnB P1 promoter, and enhances transcription of the amino acid transporter livJ (20). Building on this work, we sought to determine if TraR also is directly responsible for regulating transcription by the alternative σ factor, σE.
We have previously shown that traR from the F′ plasmid can compensate for ΔdksA auxotrophy in E. coli (20), suggesting that TraR could function at all times in the cell. However, the F′ plasmid carries a mutation in finO, a gene involved in the repression of the transfer operon, making the transfer operon somewhat leaky for transcription (25). To examine whether ΔdksA auxotrophy could be complemented by providing TraR from another conjugative system, we chose the multidrug-resistant pR1 plasmid isolated from Salmonella enterica serovar Paratyphi (26). Conjugation frequency with the wild-type pR1 plasmid is lower than that of its high-transfer mutant, pR1drd19 (27). Like the laboratory F′ plasmid, pR1drd19 has a mutation in the finOP repressor, causing it to constitutively express the transfer operon, while pR1 represses it most of the time (26, 28). Both variants of this conjugative plasmid contain pilus-encoding tra operons, and each tra operon contains a traR gene encoding a protein with the same amino acid sequence (see Fig. S1 in the supplemental material). When dksA is knocked out, the strain with the pR1 plasmid remains auxotrophic after 2 days, while the strain with the pR1drd19 plasmid becomes prototrophic to wild-type levels (Fig. 1A and B). These data support the idea that in a wild-type context (finOP+), TraR is expressed when the conjugation operon is activated.
FIG 1.
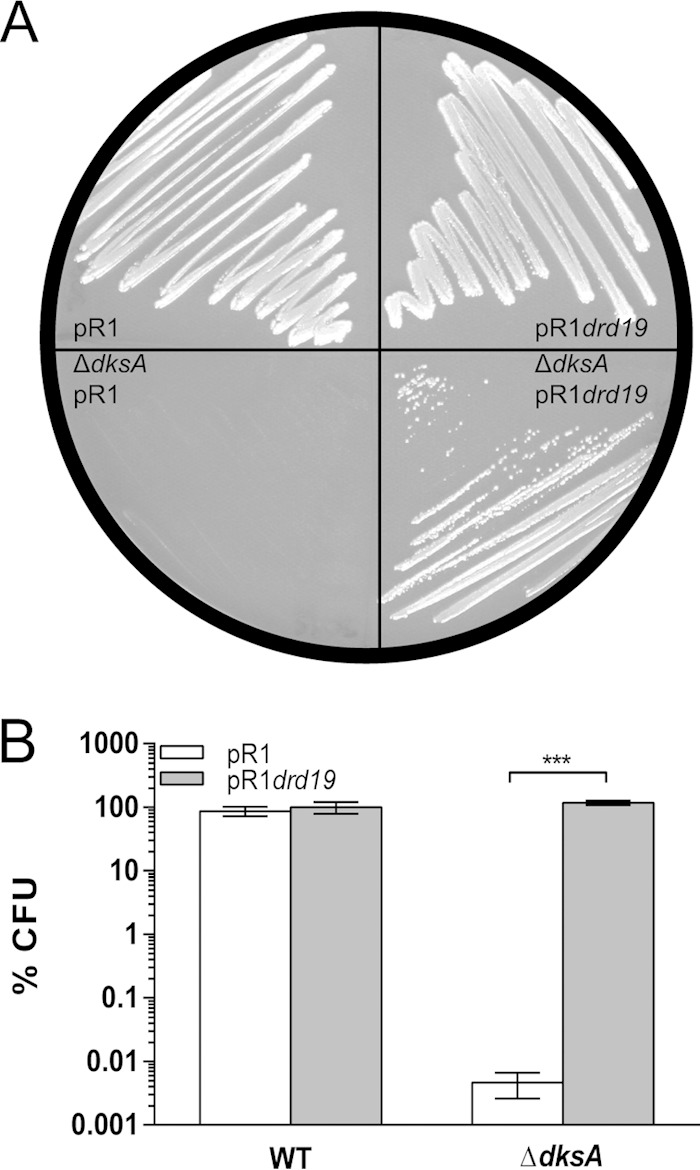
TraR is expressed during bacterial conjugation. (A) Growth of wild-type and ΔdksA mutant strains carrying either conjugative plasmid R1 or pR1drd19 (constitutive transfer) on minimal medium at 32°C after 2 days of incubation. (B) Quantification of plating efficiency on minimal medium at 32°C by E. coli wild-type and ΔdksA strains containing plasmid R1 or pR1drd19 (***, P < 0.0001 by 2-way ANOVA).
TraR induction increases σE activity.
Expression of the sex pilus during conjugation leads to extracytoplasmic stress and activation of the σE envelope stress response (9). Since TraR is expressed during conjugation and its homolog, DksA, activates the σE pathway in the presence of ppGpp, we hypothesized that TraR also could upregulate the σE pathway. To address whether TraR can activate σE, a nonconjugative, multicopy plasmid (pBA169) carrying traR under the control of an IPTG-inducible promoter was used to ensure uniform expression of TraR and prevent any confounding factors due to the F′ plasmid, such as pilus formation (Fig. 2). TraR overexpression decreases the growth rate significantly more than overexpression of DksA (20).
FIG 2.
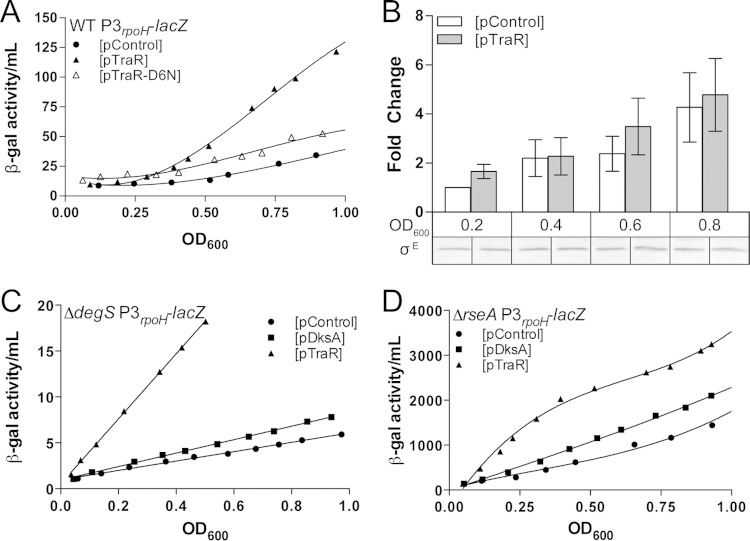
TraR induction activates σE-dependent transcription independently of the degS-rseA signal transduction cascade. Differential β-galactosidase activity of the P3rpoH-lacZ promoter fusion was measured in LB at 32°C with or without TraR induction (0.1 mM IPTG). (A, C, and D) All lines have an R2 of >0.95 and are significantly different. TraR and control plasmids are indicated by triangles and circles, respectively. The data shown are a representation of 3 independent experiments. (A) Induced expression of TraR but not TraR-D6N activates the σE-dependent P3rpoH promoter. Rates of β-galactosidase synthesis in exponential phase (from an OD600 of 0.2 to 0.8) are ∼40.17, 149.8, and 59.46 β-gal activity/OD600 unit for pControl, pTraR, and pTraR-D6N, respectively (R2 > 0.95). There is no significant difference between the slopes of pControl and pTraR-D6N. (B) Quantification of σE levels using polyclonal anti-σE antibody after induction of TraR. Wild-type culture with either pControl or pTraR was grown in early exponential phase with IPTG. Samples were taken at various OD, cell extracts from the two cultures were separated by SDS-PAGE, and σE was detected by Western blotting using a fluorescent secondary antibody. There is no statistically significant influence by either growth phase or strain (2-way ANOVA). Quantification of σE is an average from 3 independent replicates. (C) TraR expression increases the activity of P3rpoH-lacZ in a ΔdegS background, which prevents release of σE from the membrane. Rates of β-galactosidase synthesis in exponential phase (from an OD600 of 0.2 to 0.5) are 5.10 β-gal activity/OD600 unit for pControl, 7.33 for pDksA, and 35.45 for pTraR (R2 > 0.99). (D) pTraR increases the activity of P3rpoH-lacZ in a ΔrseA background, in which σE is no longer sequestered at the membrane. Rates of β-galactosidase synthesis in exponential phase (from an OD600 of 0.1 to 0.5) are 1,814 β-gal activity/OD600 unit for pControl, 2,351 for pDksA, and 3,155 for pTraR (R2 > 0.96).
Changes in σE-dependent transcription by TraR were monitored via a lacZ transcriptional fusion to the P3rpoH promoter, a promoter only sensitive to σE holoenzyme (15). β-Galactosidase activity from pControl and pTraR cultures was measured upon IPTG addition. Compared to the empty vector (pControl), TraR-expressing cells showed a 3-fold increase in σE-dependent promoter activity in exponential phase (Fig. 2A). The plasmid containing the TraR-D6N mutant, which is less active than wild-type TraR, resulted in no significant change in P3rpoH-lacZ levels (20).
To rule out the possibility that TraR activates the cytoplasmic σ32 stress response, we measured the σ32 activity of a PhtpG-lacZ transcriptional fusion. We observed a 2-fold increase in the rate of beta-galactosidase activity with TraR compared to that of the DksA or control vectors between an OD600 of 0.2 and 0.4. (see Fig. S2 in the supplemental material), but this modest induction disappears after an OD600 of 0.4. This modest and transient induction may represent an indirect effect due to the redistribution of RNAP during TraR induction, but additional experiments are required to fully understand the effect on σ32.
We also tested whether the TraR-dependent increase in the σE stress response resulted from an increase in σE protein levels. No differences in σE levels were detected by Western blotting after TraR induction relative to the plasmid control (Fig. 2B), indicating that TraR does not directly alter levels of σE protein
TraR activates the σE pathway independently of the typical DegS/RseA signal transduction pathway.
Our next step was to determine the mechanism by which TraR induces the σE stress response. Because σE regulation is predominately posttranslational, we focused on the upstream factors that sense envelope stress and subsequently trigger σE to respond. DegS is a membrane protease responsible for activating the envelope stress response by initiating the degradation of the anti-sigma factor RseA (15). Loss of DegS prevents cells from responding to periplasmic stress and, as a consequence, lowers the pool of free σE regardless of the stress status (14). To identify the mechanism by which TraR activates the σE stress response, we tested the effects of TraR induction in a ΔdegS background. (It was shown previously that the ΔdegS strain also contains a suppressor mutation[s] that accounts for its viability [15].) We reasoned that if TraR stimulates σE activity independently of DegS, there should be an increase in σE-dependent promoter activity despite little free σE being available. Upon induction of TraR in a ΔdegS mutant, we observe a 7-fold activation of the σE-dependent promoter compared to the control plasmid during logarithmic growth (Fig. 2C).
We then examined the requirement for the membrane-spanning anti-σE factor, RseA, which sequesters σE. Deletion of rseA uncouples the periplasmic stress from σE activation by releasing σE in the cytoplasm (12). If TraR increases σE activity via RseA, we would expect that TraR would have no further effect on the activation of σE-dependent transcription in an rseA deletion strain. Despite σE activity being greatly increased in a ΔrseA strain for all plasmids, we found that TraR expression further increased σE-dependent promoter activity by 5-fold (Fig. 2D), providing further evidence that TraR functions via a different mechanism than σE release by RseA degradation, perhaps by increasing the ability of σE to compete for RNAP. Taken together, these results indicate that TraR is capable of activating σE-dependent promoters separately from the DegS/RseA signal transduction cascade.
TraR directly activates transcription by EσE in vitro.
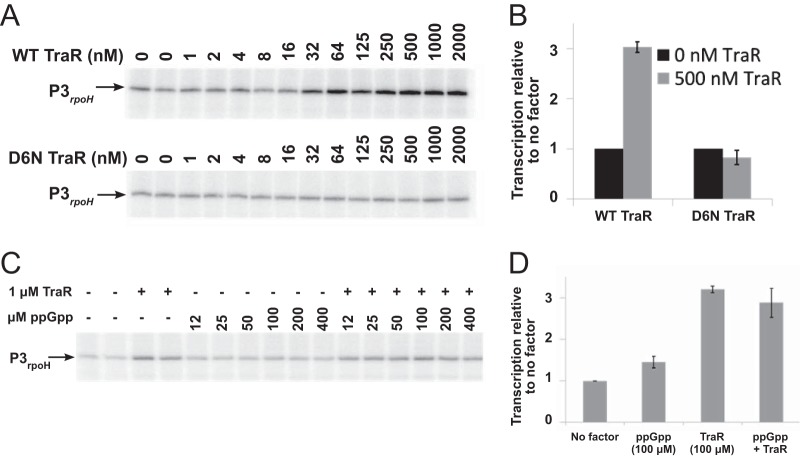
To assess whether TraR activates expression by EσE directly, we measured transcription from the P3rpoH promoter with purified components using EσE and a range of concentrations of either TraR or the mutant TraR D6N (Fig. 3A). Wild-type TraR increased transcription from P3rpoH 3-fold, while TraR D6N had no effect, consistent with the in vivo effects of TraR on P3rpoH transcription (Fig. 2A). The presence of ppGpp did not further increase activation of P3rpoH by TraR (Fig. 3C and D), consistent with the previous observation that ppGpp was not required for effects of TraR on transcription by Eσ70 (20). Although the concentration of TraR synthesized in vivo in the absence of overexpression is not known, these data strongly suggest that TraR, like DksA/ppGpp, activates transcription from σE-dependent promoters directly by changing the kinetic properties of the promoter-RNA polymerase complex. These results also are consistent with the absence of effects of TraR on σE levels and with the independence of the effects of TraR on the DegS/RseA signal transduction cascade.
FIG 3.
TraR activates transcription from P3rpoH in vitro. (A) Transcripts from a supercoiled plasmid template containing the P3rpoH promoter produced at a range of concentrations of purified wild-type TraR or mutant TraR D6N and resolved on 6% acrylamide-7 M urea gels. (B) Quantification of triplicate determinations of transcripts produced with 500 μM wild-type or D6N mutant TraR as described for panel A, normalized to transcripts produced in the absence of TraR. (C) Transcripts from P3rpoH produced in the absence of factors or in the presence of 1 μM wild-type TraR with or without the indicated concentrations of ppGpp. (D) Quantification of transcripts as described for panel C, normalized to transcription in the absence of factors (1.0).
Endogenous TraR expression from the F′ plasmid increases σE-dependent transcription.
Measurement of P2rpoE-gfp activity by fluorescence microscopy is a useful reporter of the effects of TraR on the extracytoplasmic response (i.e., on σE-dependent transcription) in an episomal context. We monitored expression of a P2rpoE-gfp fusion (29) in wild-type cells carrying either the F′ or F′ΔtraR plasmid. We observed more green fluorescent protein (GFP) fluorescence in cells containing wild-type F′ than in F′ΔtraR cells (Fig. 4A). When quantified via flow cytometry, the relative GFP level of F′-containing cells was roughly 10 times higher than that of F′ΔtraR cells (Fig. 4B). Interestingly, this activation of P2rpoE is not followed by an increase in σE level. The most likely explanation for the lack of increase in steady-state levels of σE despite activation of P2rpoE is that there is a shift from the P1 promoter to the P2 promoter of the rpoE operon, resulting in no major change in the overall transcription of the operon.
FIG 4.
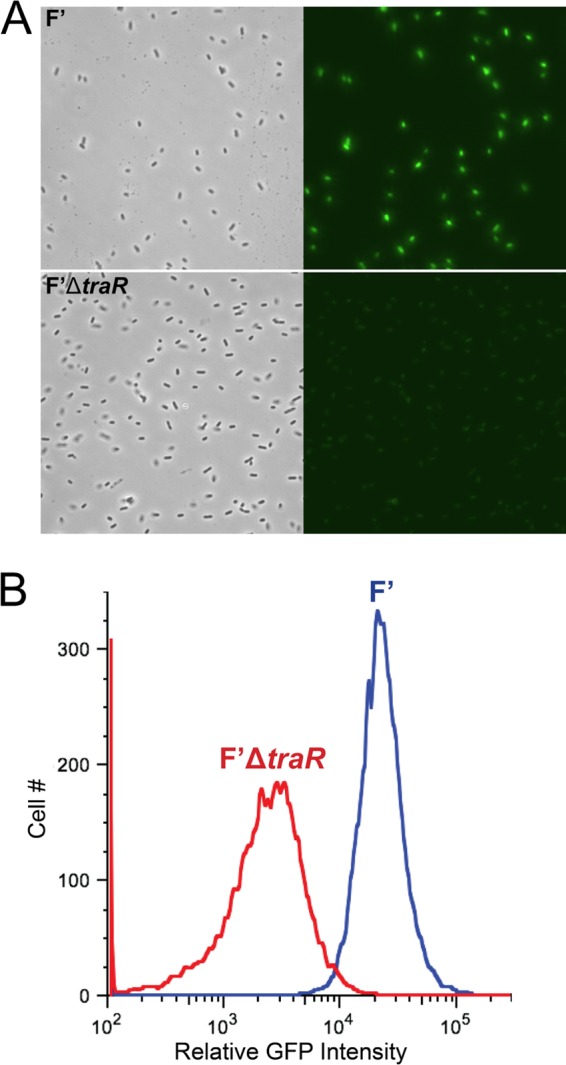
Expression of TraR from its native promoter on the F′ plasmid increases σE-dependent promoter activity. (A) Microscopy images from cultures grown at 32°C in LB medium of wild-type F′ (top) and ΔtraR mutant F′ plasmid carrying a P2rpoE-gfp fusion. The images shown are phase contrast and GFP fluorescence (from left to right). All images were taken with identical light or fluorescent exposure. (B) Quantification of σE activity by flow cytometry from cultures grown at 32°C in LB medium. σE-Dependent activity is measured with the P2rpoE-gfp fusion in cells containing wild-type F′ and F′ΔtraR′.
Therefore, a transient increase in P2rpoE activity during conjugation would not be expected to result in increased steady-state levels of σE. As a result, the observed increase in P2rpoE activity (Fig. 4) without a corresponding increase in σE levels (Fig. 2B) may be expected.
To ensure the increase in transcription from σE-dependent promoter activity resulted from TraR alone and not from other genes located on the F plasmid, we also examined GFP fluorescence of the P2rpoE-gfp fusion in cells carrying nonconjugative plasmids, either a vector control (pControl) or one carrying an IPTG-inducible TraR (pTraR). Similarly, we observed larger amounts of GFP fluorescence in cells expressing TraR (see Fig. S3 in the supplemental material). Collectively, these data also show that expression of TraR, either naturally from the F′ plasmid or by artificial induction, activates the σE envelope stress response.
Episomal TraR affects stress resistance.
To determine if the presence of TraR would confer increased stress resistance in the presence of a conjugative plasmid, we compared strains carrying an F′ plasmid with or without TraR under different stress conditions. Using temperature or ethanol stress (which both lead to σE induction), F′ and F′ΔtraR strains were compared for differences in growth and survival. Wild-type E. coli containing F′ plasmids with and without TraR were grown at 42°C. During early log phase, both strains showed identical growth, as measured by optical density, colony counts, and cell morphology. However, the strain lacking traR on the F′ plasmid virtually stopped growing at an OD600 of about 0.2 (Fig. 5A).
FIG 5.
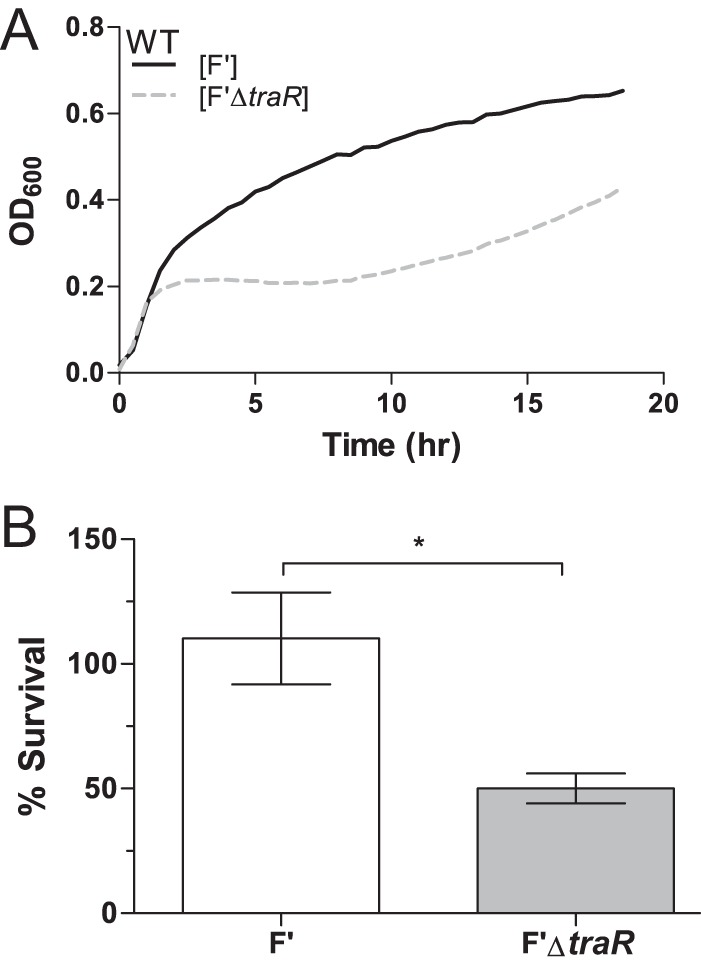
Presence of traR on the F′ plasmid increases resistance to stress. (A) Growth curves of wild-type culture carrying F′ and F′ΔtraR plasmid show that there is a fitness cost associated with the loss of TraR at 42°C in LB. Data shown are a representation of 3 independent experiments (R2 > 0.90). (B) F′ΔtraR cells are more sensitive to 10% ethanol than wild-type F′ cells. Exponential-phase cultures were exposed to 10% ethanol for 1 h, and plating efficiency was measured by comparing CFU/ml on LB plates with or without ethanol (P < 0.05). The data were normalized to preexposure CFU, with 100% set as the CFU at time zero. Because wild-type strains continued to grow, the final CFU counts were above 100%.
We also tested the effects of TraR in an additional assay for activation of transcription by EσE. Wild-type E. coli cells containing an F′ plasmid or an F′ plasmid without traR were incubated in the presence of 10% ethanol. As shown in Fig. 5B, in the absence of TraR viability is reduced by 50% after a 1-h exposure to 10% ethanol compared to the level for the wild-type F′ plasmid. Together, these data suggest that TraR provides a growth advantage for cells carrying an F′ plasmid under protein stress.
DISCUSSION
Conjugative plasmids play a major role in bacterial evolution by promoting the active exchange of genetic material between bacteria, but they also come with a fitness cost. Synthesis and maintenance of the conjugative apparatus causes envelope and cellular stress (6, 7). Here, we show that TraR, a transcription factor proposed to interact with RNAP and bind within the secondary channel, increases σE-dependent transcription independently of the signal transduction cascade and in the absence of periplasmic stress. In addition, removing traR from the conjugative F′ plasmid decreases tolerance to both heat and ethanol stress. These results suggest that TraR helps the bacterium to anticipate the periplasmic stress created by the biosynthesis of the sex pilus.
TraR is active during conjugation.
Our previous observations suggested that TraR was active continuously from the F′ plasmid. To look at the activity of traR from another conjugative plasmid, we examined if the pR1 conjugative plasmid also is able to reverse the ΔdksA auxotrophic phenotype, like strains that carry an inducible TraR. Under laboratory conditions, the conjugation frequency of wild-type pR1 is about 10−4 and is 10−2 for the F′ plasmid, because expression of the F′ transfer operon (tra) is constitutive due to the inactivation of a repressor element (finO) by a transposon (25). When the ΔdksA pR1 strain was plated on minimal medium, no rescue was observed after 2 days, whereas the pR1drd19 mutant (which constitutively expresses the tra operon) was able to compensate for the growth of the ΔdksA mutant on minimal medium (Fig. 1A and B). This result suggests that TraR normally is functional when the tra operon is actively expressed, such as when conjugation is occurring.
Role of TraR in σE regulation is direct and independent of degS and rseA.
To date, TraR has been demonstrated to act similarly to DksA/ppGpp by inhibiting promoters involved in rRNA synthesis and activating promoters involved in amino acid synthesis (20). Since DksA has been shown to regulate the activity of the σE extracytoplasmic stress in the presence of ppGpp, we looked at the role of TraR in σE activation. We observed upregulation of the P3rpoH-lacZ fusion (Fig. 2A) upon TraR induction in vivo, and we showed that TraR activated a σE-dependent promoter in a purified system in vitro in the absence of ppGpp, and that ppGpp did not further stimulate the effect of TraR (Fig. 3C and D). We conclude that direct activation of EσE transcription initiation by TraR accounts, at least in part, for its effects on expression from EσE promoters in vivo. In addition, we showed that the effects of TraR are not mediated through the anti-sigma factor-dependent regulatory pathway (RseA and DegS) (Fig. 2C and D).
Role of TraR in stress survival in the presence of the F′ plasmid.
TraR's effects on σE induction are further supported by the in vivo acquired thermotolerance assay, in which the E. coli strain containing the F′ plasmid grew better at 42°C than the F′ΔtraR strain (Fig. 5A). Additionally, the F′ΔtraR strain has decreased viability with ethanol stress (Fig. 5B). Both types of stress affect envelope homeostasis and induce the extracytoplasmic stress response. The reduced viability of cells carrying a conjugative plasmid without TraR during stress suggests that a conjugative plasmid could generate stress transiently in the periplasm, and this ultimately could be harmful for the bacterium. Furthermore, we suggest that this response, like that of the cpx system (6, 7), would provide a better folding environment for phenomena at the cell surface, such as pilus synthesis. Although we have not tested TraR's effects on pilus formation directly, we show that TraR increases transcription from a σE-dependent promoter, an indicator of periplasmic stress. In addition, activation of the cpxAR envelope stress response also would reduce expression of the transfer operon to potentially alleviate the problem (8).
We have demonstrated that TraR functions independently of ppGpp in vitro. We suggest that TraR evolved to function without ppGpp, a necessity if conjugative stress does not mirror ppGpp-inducing starvation stress. The starvation-induced stress conditions that result in elevated ppGpp levels likely differ from the stress conditions that might ensue following conjugation. We suggest that TraR-RNAP interactions cause an allosteric change in RNAP that is similar to that produced by DksA and ppGpp acting together on RNAP. This mechanism could account for the observation that TraR by itself upregulates EσE-dependent transcription at P3rpoH (Fig. 3), whereas both DksA and ppGpp are required for upregulation of transcription at the Eσ70-dependent amino acid biosynthesis promoters (30). Thus, the study of TraR may be useful for determining the details of the conformational change in RNAP structure associated with positive control of transcription by ppGpp and DksA together.
We propose that the presence of TraR could prepare the bacterium to overcome the stressful act of conjugation and possibly prevent an overload of unfolded protein coming from the synthesis of pili during conjugation, as well as possibly increasing cellular resources for the plasmid (Fig. 6). Because many bacterial pathogens spread antibiotic resistance genes and virulence factor genes via conjugative plasmids (31), these observations may provide a basis for new studies designed to combat antibiotic resistance and virulence in emerging pathogens.
FIG 6.
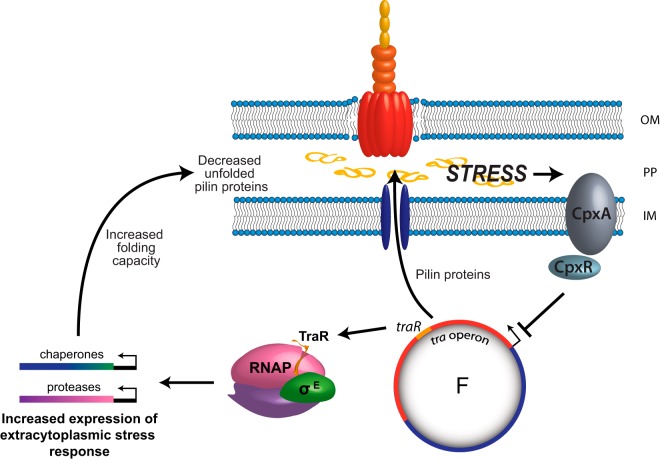
Model for the role of extracytoplasmic stress response during conjugation. During conjugation and subsequent induction of the transfer operon, pilus biosynthesis proteins are expressed and transported through the inner membrane (IM) to the periplasm (PP), where they ultimately are inserted into the outer membrane (OM). This leads to a massive amount of unfolded pilin proteins in the periplasmic space. To anticipate this stress in the periplasm, TraR interacts with RNAP and stimulates σE-dependent transcription to induce the extracytoplasmic stress response. Ultimately, the downstream chaperones and proteases produced allow the cell to deal with the stress of unfolded proteins during conjugation. If the extracytoplasmic folding capacity is overloaded, the CpxAR stress response is activated, leading to the repression of the tra operon and decreased pilin synthesis. TraR and CpxAR work together as a surveillance mechanism to help the bacterium cope with stress during conjugation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Halliday, Priya Sivaramakrishnan, and Janet Gibson for critically reviewing the manuscript and Virgil Rhodius, Jean-Marc Ghigo, and Carol Gross for sharing strains and plasmids. This project was supported by the expert assistance of Joel M. Sederstrom.
This work was funded by NIH 1R01GM088653 to C.H., NIH R37 GM37048 to R.L.G., and an NSF-K12 fellowship (NSF 0440525) to E.D.G. This project was supported by the Cytometry and Cell Sorting Core at BCM with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02279-14.
REFERENCES
- 1.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 2.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 4.Anthony KG, Sherburne C, Sherburne R, Frost LS. 1994. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol 13:939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 5.Frost LS, Ippen-Ihler K, Skurray RA. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58:162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahrl D, Wagner M, Bischof K, Koraimann G. 2006. Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli. J Bacteriol 188:6611–6621. doi: 10.1128/JB.00632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidlack JE, Silverman PM. 2004. An active type IV secretion system encoded by the F plasmid sensitizes Escherichia coli to bile salts. J Bacteriol 186:5202–5209. doi: 10.1128/JB.186.16.5202-5209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubbins MJ, Lau I, Will WR, Manchak JM, Raivio TL, Frost LS. 2002. The positive regulator, TraJ, of the Escherichia coli F plasmid is unstable in a cpxA* background. J Bacteriol 184:5781–5788. doi: 10.1128/JB.184.20.5781-5788.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones CH, Danese PN, Pinkner JS, Silhavy TJ, Hultgren SJ. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J 16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alba BM, Gross CA. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol 52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 11.Rouvière PE, De Las Peñas A, Mecsas J, Lu CZ, Rudd KE, Gross CA. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J 14:1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Las Peñas A, Connolly L, Gross CA. 1997. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol Microbiol 24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 13.Kanehara K, Ito K, Akiyama Y. 2002. YaeL (EcfE) cytometry the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev 16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alba BM, Zhong HJ, Pelayo JC, Gross CA. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma (E) activity. Mol Microbiol 40:1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- 15.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev 16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costanzo A, Ades SE. 2006. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J Bacteriol 188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown L, Gentry D, Elliott T, Cashel M. 2002. DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol 184:4455–4465. doi: 10.1128/JB.184.16.4455-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spira B, Hu X, Ferenci T. 2008. Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology 154:2887–2895. doi: 10.1099/mic.0.2008/018457-0. [DOI] [PubMed] [Google Scholar]
- 19.Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. 2008. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol 67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 20.Blankschien MD, Potrykus K, Grace E, Choudhary A, Vinella D, Cashel M, Herman C. 2009. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet 5:e1000345. doi: 10.1371/journal.pgen.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maneewannakul K, Ippen-Ihler K. 1993. Construction and analysis of F plasmid traR, trbJ, and trbH mutants. J Bacteriol 175:1528–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY. [Google Scholar]
- 23.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strozen TG, Langen GR, Howard SP. 2005. Adenylate cyclase mutations rescue the degP temperature-sensitive phenotype and induce the sigma E and Cpx extracytoplasmic stress regulons in Escherichia coli. J Bacteriol 187:6309–6316. doi: 10.1128/JB.187.18.6309-6316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshioka Y, Ohtsubo H, Ohtsubo E. 1987. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J Bacteriol 169:619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 27.Meynell E, Datta N. 1967. Mutant drug resistant factors of high transmissibility. Nature 214:885–887. doi: 10.1038/214885a0. [DOI] [PubMed] [Google Scholar]
- 28.Dionisio F, Matic I, Radman M, Rodrigues OR, Taddei F. 2002. Plasmids spread very fast in heterogeneous bacterial communities. Genetics 162:1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mutalik VK, Nonaka G, Ades SE, Rhodius VA, Gross CA. 2009. Promoter strength properties of the complete sigma E regulon of Escherichia coli and Salmonella enterica. J Bacteriol 191:7279–7287. doi: 10.1128/JB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci U S A 102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nogueira T, Rankin DJ, Touchon M, Taddei F, Brown SP, Rocha EPC. 2009. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr Biol 19:1683–1691. doi: 10.1016/j.cub.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.