Abstract
The hybrid sensor kinase BarA and its cognate response regulator UvrY, members of the two-component signal transduction family, activate transcription of CsrB and CsrC noncoding RNAs. These two small RNAs act by sequestering the RNA binding protein CsrA, which posttranscriptionally regulates translation and/or stability of its target mRNAs. Here, we provide evidence that CsrA positively affects, although indirectly, uvrY expression, at both the transcriptional and translational levels. We also demonstrate that CsrA is required for properly switching BarA from its phosphatase to its kinase activity. Thus, the existence of a feedback loop mechanism that involves the Csr and BarA/UvrY global regulatory systems is exposed.
INTRODUCTION
The BarA/UvrY two-component signal transduction system (TCS) of Escherichia coli consists of the membrane-bound sensor kinase BarA and its cognate response regulator UvrY (1). BarA, which belongs to the subfamily of tripartite sensor kinases (2, 3), senses and responds to the presence of formate and acetate but also to that of other short-chain fatty acids (4). Phosphorylated BarA catalyzes the transphosphorylation of UvrY (1), a typical response regulator of the FixJ family (1, 5), which in turn activates expression of the noncoding RNAs of the carbon storage regulation (Csr) system, CsrB and CsrC. These small regulatory RNAs possess repeated sequence elements that allow them to interact with multiple copies of the RNA binding protein CsrA and thereby prevent its regulatory interaction with its mRNA targets (6).
CsrA is a small, dimeric RNA binding protein that coordinates gene expression by positively or negatively regulating the translation, stability, and/or elongation of its target transcripts (7, 8). CsrA directly interacts with the 5′ untranslated leaders of target mRNAs at sites characterized by a GGA sequence that is often located within the loop of a short stem-loop structure (9–11). In this way, CsrA activates exponential-phase processes while it represses several stationary-phase functions (12). CsrA is widely distributed among eubacteria (13) and regulates expression of genes for virulence factors (14, 15), quorum sensing (16, 17), motility (18, 19), carbon metabolism (20, 21), biofilm formation (22, 23), cyclic di-GMP synthesis (24), and peptide uptake (10).
Curiously, activation of csrB transcription, which depends directly on UvrY-P, does not take place in a csrA mutant strain (25). Therefore, it has been suggested that CsrA has a positive effect on the BarA/UvrY TCS. In this study, we confirmed and extended these results by examining the effects of CsrA on either the expression or the activity of BarA and UvrY. Our results demonstrate that CsrA, apparently indirectly, is required for proper uvrY expression and also for activation of the BarA kinase activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this work are listed in Table 1. Strains IFC5010 (csrA::Kanr csrB-lacZ) and IFC5016 (uvrY::Camr csrA::Kanr csrB-lacZ) were constructed by P1vir transduction of the csrA::Kanr allele from strain TR1-5 (25) into strains KSB837 (csrB-lacZ) (26) and UYKSB837 (uvrY::Camr csrB-lacZ) (25), respectively. Strain IFC5015 (ackA::tetR::pta csrA::Kanr csrB-lacZ) was constructed by P1vir transduction of the ackA::tetR::pta allele from strain ECL5336 (27) into strain IFC5010. For strain IFC5017 (barA::Camr csrA::Kanr csrB-lacZ) construction, the barA gene was deleted by homologous recombination using the lambda Red recombinase system (28). Briefly, a fragment amplified by PCR, using primers barAdel-Fw (5′-ATTTAACAGTGTGACCTTAATTGTCCCATAACGGAACTCCGTGTAGGCTGGAGCTGCTTC-3′) and barAdel-Rv (5′-CATAAACACAGGCACTTTGTCACCAATCTGAAACCAGCGTATGAATATCCTCCTTAGTTCC-3′) and plasmid pKD3 (28) as the template, was used to replace the barA allele with a chloramphenicol cassette in strain IFC5010.
TABLE 1.
E. coli strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| CF7789 | MG1655 ΔlacIZ (MluI) | Michael Cashel |
| TR1-5CF7789 | CF7789 csrA::Kanr | 25 |
| KSB837 | CF7789 ϕ(csrB-lacZ) | 26 |
| IFC5010 | KSB837 csrA::Kanr | This work |
| BAKSB837 | KSB837 barA::Kanr | 25 |
| UYKSB837 | KSB837 uvrY::Camr | 25 |
| ECL5336 | MC4100 ackA::Tetr::pta | 27 |
| IFC5011 | CF7789 ϕ(uvrY-lacZ) operon fusion | This work |
| IFC5012 | TR1-5CF7789 ϕ(uvrY-lacZ) operon fusion | This work |
| IFC5013 | CF7789 ϕ(PlacUV5-uvrY22′-′lacZ) leader fusion | This work |
| IFC5014 | TR1-5CF7789 ϕ(PlacUV5-uvrY22′-′lacZ) leader fusion | This work |
| IFC5015 | KSB837 ackA::tetR::pta csrA::Kanr | This work |
| IFC5016 | KSB837 uvrY::Camr csrA::Kanr | This work |
| IFC5017 | KSB837 barA::Camr csrA::Kanr | This work |
| Plasmids | ||
| pACT3 | Low-copy-no. vector, Camr | 31 |
| pEXT21 | Low-copy-no. vector, Spr | 31 |
| pBA29 | barA in blunt-ended VspI site of pBR322, Tetr | 25 |
| pAH125 | CRIM vector for transcriptional lacZ fusions, Kanr | 29 |
| pINT-ts | CRIM integration vector, Ampr | 29 |
| pUV5 | CRIM vector for translational fusions, Ampr | 30 |
| pAH125-bla | CRIM vector for transcriptional lacZ fusions, Ampr | This work |
| pINT-cat | CRIM integration vector, Camr | This work |
| pMX539 | uvrY in SmaI site of pACT3, Camr | This work |
| pMX540 | uvrY D54Q in SmaI site of pACT3, Camr | This work |
| pMX541 | uvrY in blunt-ended NdeI site of pBA29, Tetr | This work |
| pMX542 | barA promoter in pEXT21, Spr | This work |
| pMX543 | uvrY under the control of barA promoter in pEXT21, Spr | This work |
| pMX544 | csrA in HindIII and PstI sites of pEXT21, Spr | This work |
| pMX545 | csrB in pGEMT-Easy, Ampr | This work |
| pAH-uvrY | uvrY-lacZ operon fusion, Ampr | This work |
| pUV-uvrY22 | uvrY-lacZ leader (plus 22 codons) fusion under the control of the lacUV5 promoter Ampr | This work |
To construct Ampr-linked lacZ operon fusions, plasmid pAH125-bla was first generated by replacing the kanamycin resistance cassette of pAH125 (29) with an ampicillin resistance cassette. To this end, a bla PCR product was generated, using primers Amp-Prom-Fw (5′-GCGGCGCCTTCAAATATGTATCCGCTCATG-3′) and Amp-Rv (5′-GCGCGGCCGCGGTCTGACAGTTACCAATGC-3′) and plasmid pUC18 as the template, and cloned into the NarI-NotI sites of pAH125. Helper plasmid pINT-cat was constructed by replacing the ampicillin resistance cassette of plasmid pINT-ts (29) with the chloramphenicol resistance cassette. A 1.1-kb DNA fragment containing the cat gene, obtained from plasmid pKD3 (28) by HindIII digestion, was blunt ended and cloned into blunt-ended XmnI-BsaI sites of pINT-ts. Plasmid pAH-uvrY, containing an uvrY-lacZ operon fusion, was constructed by cloning a PCR-amplified fragment containing the upstream noncoding region through the first 4 codons (nucleotides [nt] −409 to +12 relative to the start of translation) (using primers uvrYP-fw-Pst [5′-AACTGCAGGGCGGCGGAGTATACCATAAG-3′] and uvrYP-Rv-BamHI [5′-CGGGATCCAGAACGTTGATCAAAGGAATATC-3′]) into the PstI-BamHI sites of pAH125-bla. Plasmid pUV-uvrY22, containing a translational uvrY-lacZ fusion under the control of the lacUV5 promoter, was constructed by cloning the region from nt −47 to +66 relative to the start of uvrY translation, amplified by PCR using primers uvrY-lead-Fw (5′-GGAATTCAATGACTAACTATCAGTAGC-3′) and uvrY-lead22-Rv (5′-CGGGATCCTCTTCCAGAATGCGTCG-3′), into the EcoRI-BamHI sites of plasmid pUV5 (30). All fusions were integrated into the CF7789 and TR1-5CF7789 chromosomes, as previously described (29), to generate strains IFC5011 to IFC5014.
To construct plasmids pMX539 and pMX541, the uvrY open reading frame and its promoter region were PCR amplified using primers uvrY-Prom549-Fw (5′-GGAATTCGCAGCATCAGCGTCAGC-3′) and UvrY-Rv-HindIII (5′-CCCAAGCTTCCGTACCACCAGCATCG-3′) and chromosomal DNA from strain MG1655 as the template and cloned into the SmaI site of plasmid pACT3 (31) and into the blunt-ended NdeI site of pBA29 (25), respectively. Plasmid pMX540, carrying the uvrYD54Q mutant allele, was generated by site-directed mutagenesis of plasmid pMX539, using the QuikChange kit (Stratagene, CA) and the mutagenic primers uvrY-D54Q-Fw (5′-GTTGACGTGGTGCTAATGCAGATGAGTATGCCGGGC-3′) and uvrY-D54Q-Rv (5′-GCCCGGCATACTCATCTGCATTAGCACCACGTCAAC-3′). The correct amino acid replacement was confirmed by DNA sequencing. To construct plasmid pMX544, a fragment containing the csrA open reading frame and its promoter region was PCR amplified, using primers csrA1-Fw (5′-CCCAAGCTTGCCAGTGTGAAAGGCTGG-3′) and csrA1-Rv (5′-AACTGCAGGAATGAACGGGAGTAAAGCG-3′) and chromosomal DNA from strain MG1655 as the template, and cloned into the HindIII and PstI sites of plasmid pEXT21 (31). To construct plasmid pMX543, which carries the uvrY gene under the control of the barA promoter and 5′ untranslated regions (5′-UTRs), a 1-kb fragment containing the barA promoter and 5′-UTRs was first PCR amplified, using the primers BarA-Forwd (5′-GGAATTCCCGACCACACTGGCAGC-3′) and 5′-TGCTCTAGAAGATCTGATCATATGGAGTTCCGTTATGGG-3′ and chromosomal DNA from strain MG1655 as the template, and cloned into EcoRI and XbaI sites of plasmid pEXT21 (31) to generate plasmid pMX542. Then, a 762-bp PCR-amplified fragment (using primers uvrY [5′-CCCGGATCCCATATGATCAACGTTCTACTTGTTGATGACCACG-3′] and UvrY-Rv-HindIII [5′-CCCAAGCTTCCGTACCACCAGCATCG-3′] and chromosomal DNA as the template) containing the uvrY open reading frame was cloned into the NdeI-HindIII sites of pMX542 to generate pMX543.
To construct plasmid pMX545, a 376-bp DNA fragment containing the csrB gene was PCR amplified from chromosomal DNA of strain MG1655 and introduced by T/A cloning into pGEMt-Easy vector (Promega).
Bacteria were routinely cultured at 37°C in lysogeny broth (LB) medium. Media were supplemented with antibiotics at the following concentrations: chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; tetracycline, 10 μg/ml; streptomycin, 100 μg/ml; and spectinomycin, 50 μg/ml. P1vir transduction was performed as previously described (32).
RNA extraction and Northern blotting.
Total RNA was purified from samples taken at the indicated times by the hot-phenol extraction method, as described previously (33). Northern blot analysis was performed by fractionation of the purified RNA samples (5 μg) on 1.2% agarose-formaldehyde gels and transfer onto nitrocellulose membranes (Amersham XL) by capillary transfer by using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Membranes were cross-linked using a cross-linking device (Stratalinker; Stratagene) and prehybridized for 3 h at 42°C in a buffer containing 5× Denhardt solution (34), 5× SSC, 0.2% SDS, 50% formamide, and 250 mg of sheared salmon sperm DNA per ml. Subsequently, a radiolabeled csrB-specific DNA probe, denatured at 90°C for 5 min, was added to the prehybridization buffer, and the membranes were incubated at 42°C overnight. The csrB-specific probe was obtained by digestion of plasmid pMX545 with EcoRI, separation of the fragments on agarose gels, and purification of the csrB-specific band by using the Qiagen agarose purification kit. Probe labeling was performed by using [α-32P]dCTP and the Radprime kit (Invitrogen), according to the manufacturer's instructions. Membranes were washed twice with 50 ml of 2× SSC and 0.1% SDS at 37°C and twice with 0.2× SSC and 0.1% SDS at 42°C. Images were obtained using phosphorimager screens and analyzed using the Typhoon image scanner (Amersham Biosciences).
β-Galactosidase activity.
β-Galactosidase activity was assayed and expressed in Miller units as described previously (32). Cells were grown in LB broth, or LB with the pH adjusted and buffered to pH 5.0 with 0.1 M homopiperazine-N,N′-bis(2-ethanesulfonic acid) (HOMOPIPES). When indicated, acetate or formate was used at a concentration of 7 mM.
Immunoblotting and generation of polyclonal anti-UvrY and anti-BarA.
Cultures for Western blot analyses were grown aerobically at 37°C and harvested by centrifugation during mid-exponential growth. The cell pellet was resuspended in 100 μl lysis buffer (50 mM Tris-HCl, 4% SDS, pH 6.8) and boiled for 5 min. Aliquots of 10 μl were separated by SDS-PAGE (15% polyacrylamide gels for UvrY and 8% polyacrylamide gels for BarA), and the proteins were transferred to a Hybond-ECL filter (Amersham Biosciences). The filter was equilibrated in TTBS buffer (25 mM Tris, 150 mM NaCl, and 0.05% Tween 20) for 10 min and incubated in blocking buffer (1% milk in TTBS) for 1 h at room temperature. Polyclonal antibodies against UvrY and BarA, raised by subcutaneous immunization of rabbits with His6-UvrY and His6-BarA, were added at dilutions of 1:2,000 and 1:10,000, respectively, to the filter and incubated for 1 h at room temperature. The bound antibody was detected by using anti-rabbit IgG antibody conjugated to horseradish peroxidase and the ECL detection system (Amersham Biosciences).
RESULTS AND DISCUSSION
CsrA is required for activation of csrB transcription.
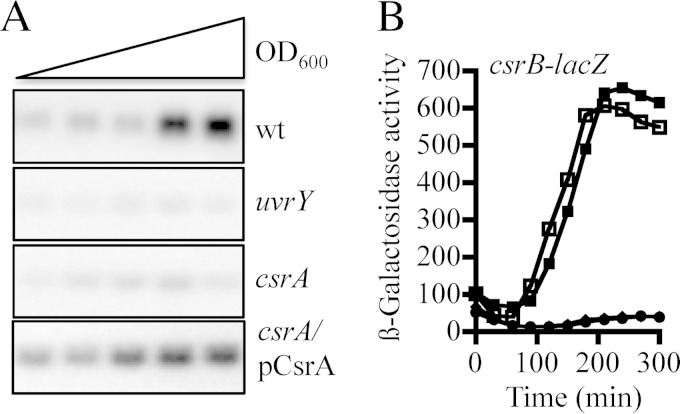
It has been previously reported that acetate and formate, as well as short-chain fatty acids, act as stimuli for the sensor kinase BarA (4), leading to its autophosphorylation and transphosphorylation of the cognate response regulator UvrY (1). Phosphorylated UvrY (UvrY-P), in turn, activates transcription of the CsrB and CsrC small untranslated RNAs, which act by sequestering the CsrA global regulatory protein and antagonizing its regulatory activity (25, 35). Curiously, UvrY-dependent activation of csrB transcription, which takes place at the transition from exponential to stationary growth phase (25), was found not to occur in a csrA mutant as judged by Northern blotting and by csrB-lacZ (located at attλ) reporter expression (Fig. 1A and B), in agreement with a previous report (25). Moreover, it was found that csrB expression in the csrA mutant was fully restored by ectopic expression of CsrA using plasmid pMX544 (Fig. 1A and B). Because CsrA does not affect csrB stability (26) and because csrB transcription is activated directly by UvrY-P, it can be inferred that CsrA affects the activity and/or the expression of the components of the BarA/UvrY signaling system.
FIG 1.
Effects of csrA and uvrY on csrB transcription. (A) Northern blot analysis of CsrB levels in the isogenic strains KSB837 (wild type [wt]), UYKSB837 (uvrY), IFC5010 (csrA), and IFC5010 carrying the csrA-expressing plasmid pMX544 (indicated as pCsrA). Cultures of these strains were grown in LB medium, and total RNA isolated from samples that were harvested throughout the growth curve (optical density at 600 nm [OD600] of 0.3 to 2.0) was probed for the CsrB transcript. Experiments were repeated three times in their entirety with essentially identical results. (B) Overnight cultures of the wild-type strain (squares) and its isogenic uvrY (circles) and csrA (diamonds) mutant strains and the csrA mutant strain carrying the csrA expressing plasmid pMX544 (closed squares), all carrying the csrB-lacZ transcriptional fusion, were diluted to an OD600 of ∼0.05 in LB medium, and the β-galactosidase activity was followed for 300 min. Note that the circles and diamonds extensively overlap. The average from four independent experiments is presented (standard deviations were less than 5% from the mean).
Acetate and formate are unable to activate BarA in a csrA mutant.
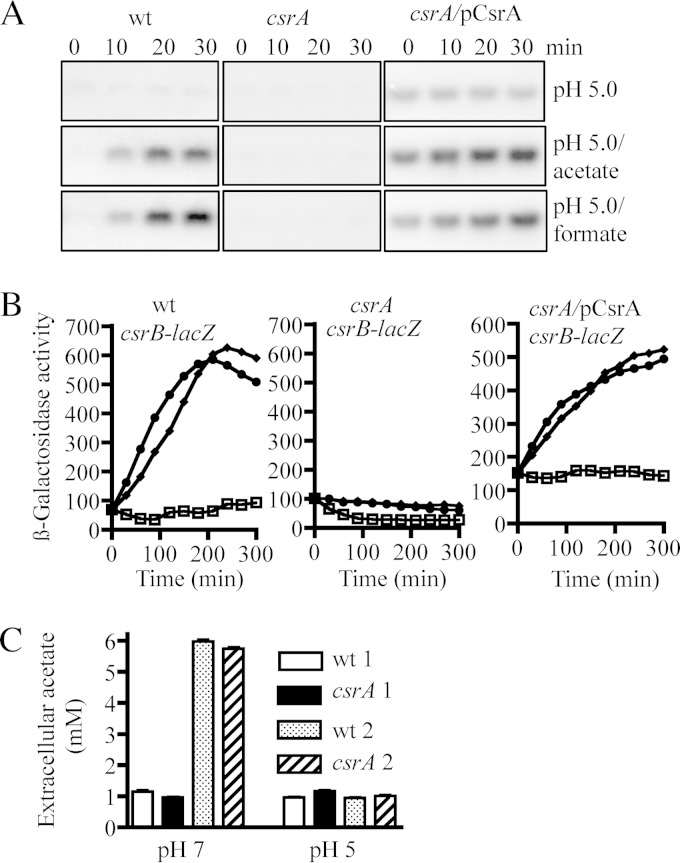
Considering that a role of CsrA, an RNA binding protein, in the control of the BarA/UvrY phosphorelay cascade as not very probable, we hypothesized that CsrA might be required either for the production of the BarA-specific stimulus and thereby activation of the BarA/UvrY signaling cascade or for the expression of the barA and/or uvrY genes. To test the first possibility, we took advantage of the fact that although the BarA/UvrY TCS remains inactive when cells are grown at pH 5.0 (36), addition of acetate or formate to the growth medium results in the immediate activation of BarA/UvrY and thereby activation of csrB transcription (4). Therefore, strains KSB837 (wild type), IFC5010 (the isogenic csrA mutant), and IFC5010 carrying the csrA-expressing plasmid pMX544 were grown in LB buffered at pH 5.0 in the absence or presence of acetate and formate, and the expression of csrB was monitored by Northern blotting and by using a csrB-lacZ transcriptional fusion as a reporter. As expected, no activation of csrB transcription was observed in either of the tested strains at pH 5.0 (Fig. 2A and B). However, addition of acetate or formate to the growth medium resulted in the immediate activation of csrB transcription in the wild-type strain but not in the csrA mutant strain (Fig. 2A and B). Finally, ectopic expression of CsrA, using plasmid pMX544, in the csrA mutant strain restored csrB transcription almost to wild-type levels (Fig. 2A and B). It can therefore be concluded that a possible insufficiency of the BarA specific stimulus is not the cause of the above-described phenotype. This was further supported by the finding that the same increase in the concentration of extracellular acetate was attained by both the wild-type and the csrA mutant strains when grown at pH 7.0 and that no increase of extracellular acetate was observed in any of the two strains at pH 5.0 (Fig. 2C), in agreement with previously reported results (4, 30). Therefore, we concluded that the requirement for CsrA in the BarA/UvrY signaling cascade does not involve the synthesis of acetate, which acts as a physiological stimulus for BarA.
FIG 2.
The synthesis of the BarA stimulus does not appear to be affected in the csrA mutant strain. (A) The isogenic strains KSB837 (wild type), IFC5010 (csrA), and IFC5010 carrying the csrA-expressing plasmid pMX544 (indicated as pCsrA) were grown in LB medium, the pH of which had been adjusted and buffered to 5.0 using 0.1 M homopiperazine-N,N′-bis-2-(ethanesulfonic acid) (HOMOPIPES). At an OD600 of 0.2, a sample was withdrawn, 7 mM acetate or formate was added to the medium, and samples were withdrawn every 10 min. Total RNA isolated from these samples was analyzed by Northern blotting using a CsrB-specific probe. The experiment was repeated three times in its entirety with essentially identical results. (B) Cultures of the isogenic strains KSB837 (wild type), IFC5010 (csrA), and IFC5010 harboring plasmid pMX544 (indicated as pCsrA), all carrying the csrB-lacZ transcriptional fusion, were diluted to an OD600 of ∼0.05 in LB medium at pH 5.0 as described for Fig. 1B alone (squares) or in the presence of acetate (circles) or formate (diamonds), and the β-galactosidase activity was followed. The average from four independent experiments is presented (standard deviations were less than 5% from the mean). (C) Concentration of extracellular acetate. The KSB837 wild-type strain and IFC5010, its isogenic csrA mutant strain, were grown in LB at pH 7.0 or 5.0. Samples were withdrawn either at early exponential growth phase (designated 1) or at late exponential phase (designated 2), and the concentration of acetate was determined with the R-Biopharm acetic acid determination kit (Boehringer Mannheim). The averages from three independent experiments are presented and the standard deviations are indicated.
CsrA is required for proper uvrY expression.
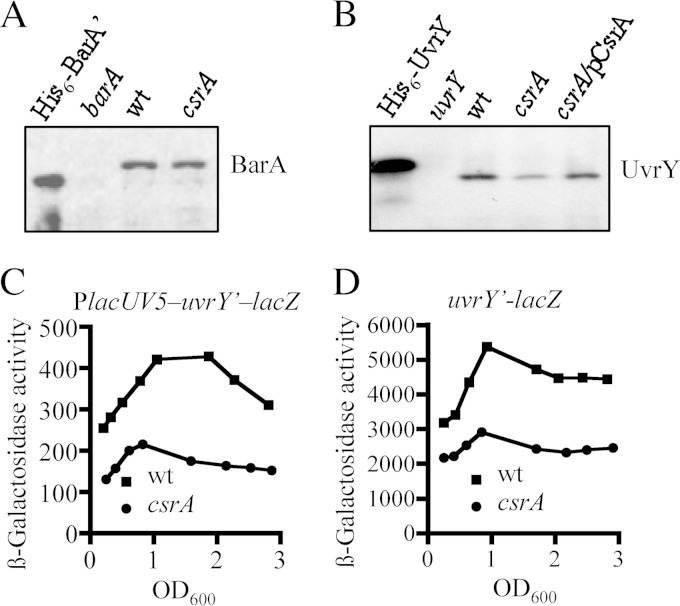
Next, we tested the possibility that CsrA affects the expression of barA and/or uvrY. To this end, the amounts of BarA and UvrY proteins in the wild-type and csrA mutant strains were compared by Western blotting, using specific BarA and UvrY polyclonal antibodies. The two strains expressed similar amounts of BarA (Fig. 3A). On the other hand, the amount of UvrY protein was significantly lower in the csrA mutant than in the wild-type strain (Fig. 3B). Moreover, wild-type levels of UvrY were expressed in the csrA mutant complemented with the csrA-expressing plasmid pMX544 (Fig. 3B), indicating that CsrA affects, directly or indirectly, uvrY expression.
FIG 3.
CsrA is required for proper uvrY expression. (A) Levels of BarA protein (102.550 Da) in the KSB837 (wild-type) and IFC5010 (csrA) strains as determined by Western blot analyses using BarA polyclonal antibodies. (B) Levels of UvrY protein (23.890 Da) in the KSB837 (wild-type) and IFC5010 (csrA) strains and in IFC5010 complemented with the csrA-expressing plasmid pMX544 (indicated as pCsrA), as determined by Western blot analyses using UvrY polyclonal antibodies. Purified His6-tagged Bar′ (81.550 Da), a BarA version lacking the 198 first amino acid residues, and His6-tagged UvrY (25.380 Da) (25), and extracts from barA (BAKSB837) and uvrY (UYKSB837) mutant cells were used in the first lanes of each Western blot. The molecular mass differences between purified and wild-type BarA and UvrY proteins are due to the His6 tag and the absence of the first 198 amino acids in BarA′. (C and D) Effects of csrA on uvrY transcription and translation. Wild-type (wt) and csrA mutant cells carrying either a chromosomal PlacUV5-uvrY′-′lacZ translational fusion (strains IFC5013 and IFC5014, respectively) (C) or a chromosomal uvrY′-′lacZ operon fusion (strains IFC5011 and IFC5012, respectively) (D) were harvested at various times throughout growth and assayed for β-galactosidase activity. The β-galactosidase activity is presented as a function of growth density (OD600). The averages from four independent experiments are presented (standard deviations were less than 5% from the mean).
Subsequently, we asked whether CsrA affects uvrY expression at the translational level and/or at the transcriptional level, e.g., by modulating the expression of a transcriptional regulator. To this end, the β-galactosidase activity from a chromosomal lacUV5-uvrY′–lacZ translational reporter fusion, in which the constitutive lacUV5 promoter replaced the native promoter, was monitored in the wild-type and csrA mutant strains. In this case, β-galactosidase activity in the csrA mutant was found to be approximately 50% of that in the wild-type strain (Fig. 3C). As mentioned above, CsrA regulates translation of its target mRNAs by interacting with their 5′ untranslated regions (9–11). However, analyses in silico did not reveal any apparent CsrA binding sites in the 5′-UTR of the uvrY transcript, which is suggestive of an indirect effect of CsrA on uvrY expression. The effect of CsrA on uvrY transcription was tested by using a chromosomal uvrY′-lacZ transcriptional fusion. It was observed that the β-galactosidase activity in the wild-type strain was almost 2-fold higher than the in the csrA mutant strain (Fig. 3D), suggesting that CsrA indirectly affects the transcription of uvrY. It should be noted that the csrA mutation, an insertion of a kanamycin cassette 10 amino acid residues before the stop codon of CsrA (21, 37), decreases but does not entirely eliminate CsrA activity (38). Therefore, the actual effect of CsrA on uvrY transcription and translation may be significantly greater than observed. Nonetheless, the above results suggest that CsrA is required for proper uvrY expression, affecting both transcription and translation of uvrY.
Overexpression of UvrY alone but not concurrently with BarA restores csrB transcription in a csrA mutant.
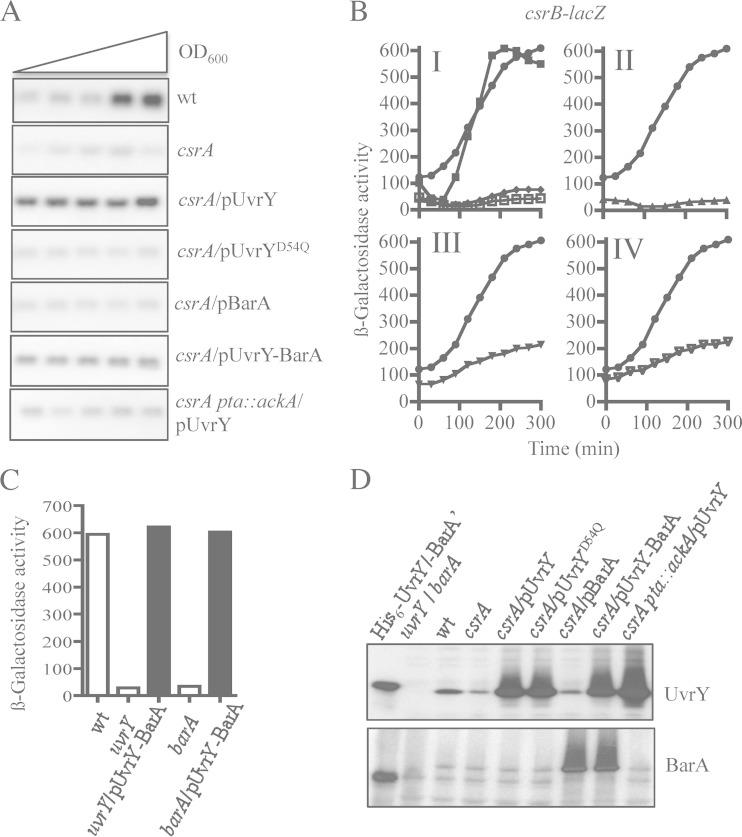
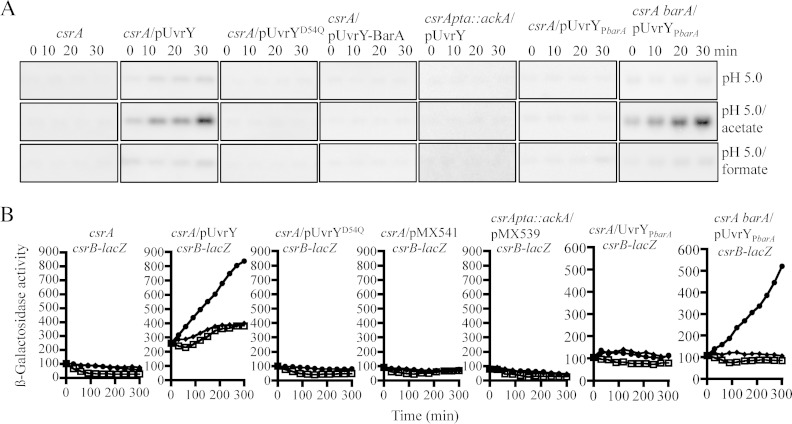
We then asked whether ectopic expression of uvrY and/or barA from plasmids pMX539 and pBA29 (25) restores csrB expression in the csrA mutant strain. It was found that expression of uvrY but not barA did restore csrB expression (Fig. 4A and B), in agreement with a previous report (25). Notably, the presence of the uvrY-expressing plasmid in the csrA mutant strain resulted in elevated amounts of CsrB RNA even at the early exponential phase of growth (Fig. 4A), an effect that could be attributed to the vast overexpression of UvrY (Fig. 4D). Moreover, overexpression of an UvrYD54Q mutant protein, in which the phosphorylatable aspartate residue was replaced with a glutamine residue, using plasmid pMX540 was not able to restore csrB transcription, indicating that phosphorylation of UvrY is required for its activation as a transcriptional regulator (Fig. 4A and B). In this respect, it is relevant to mention that phosphorylation of UvrY has been shown to occur either by BarA-P or directly by acetyl-P. Unexpectedly, it was found that, in contrast to the UvrY-expressing plasmid pMX539, plasmid pMX541, which expresses both BarA and UvrY, failed to activate csrB transcription in the csrA mutant strain (Fig. 4A and B). To ensure that UvrY and BarA were readily expressed by plasmid pMX541, we examined whether this plasmid was able to complement an uvrY mutant and a barA mutant for csrB expression. It was found that csrB expression was restored in both mutant strains (Fig. 4C), indicating that pMX541 expresses functional BarA and UvrY proteins. Moreover, Western blot analyses, using specific UvrY and BarA polyclonal antibodies, revealed that similar amounts of UvrY protein were expressed by pMX539, pMX540, and pMX541 and that similar amounts of BarA were expressed by pBA29 and pMX541 (Fig. 4D). Thus, expression and functionality of the pMX541 expressed BarA and UvrY does not provide an explanation for the above result. The above results, in combination with the fact that BarA, like other tripartite two-component sensors, has been shown to be capable of having both a kinase and a phosphatase activity on it is cognate regulator (39–41), prompted us to speculate that BarA may remain locked in its phosphatase state and fail to be activated as a kinase in the csrA mutant. In such a scenario, phosphorylation of the pMX539-expressed UvrY in the csrA mutant strain should rely on acetyl-P. Moreover, the vast overexpression of UvrY (Fig. 4D) should overwhelm the phosphatase activity of the chromosomally expressed BarA, permitting the accumulation of significant amounts of UvrY-P and culminating in activation of csrB transcription. On the other hand, when comparable amounts of BarA and UvrY proteins are expressed, i.e., when pMX541 was used, the phosphatase activity of BarA should dephosphorylate the acetyl-P-dependent UvrY-P and thereby cancel its transcriptional regulation.
FIG 4.
Acetyl-P, but not BarA, is responsible for UvrY phosphorylation in a csrA mutant. Effects of ectopic expression of uvrY, uvrYD54Q, barA, and uvrY barA on csrB expression in isogenic csrA and csrA pta::ackA mutant strains are shown. (A) Cultures of the KSB837 wild-type strain and IFC5010, its isogenic csrA mutant strain, harboring or not plasmid pMX539 (expressing uvrY and indicated as pUvrY), pMX540 (expressing uvrYD54Q, having Q substituted for the conserved phosphorylatable D, and indicated as pUvrYD54Q), pBA29 (expressing barA and indicated as pBarA), or pMX541 (expressing both uvrY and barA and indicated as pUvrY-BarA), and IFC5015, the isogenic csrA pta::ackA triple mutant strain, harboring pMX539 (indicated as pUvrY) were grown in LB medium. Total RNA isolated from samples that were harvested throughout the growth curve (OD600 of 0.3 to 2.0) was probed for the CsrB transcript. (B) Overnight cultures of the csrB-lacZ transcriptional fusion-carrying wild type (panel I, closed squares), the csrA mutant strain (panel I, open squares), and the same mutant strain harboring plasmid pMX539 (expressing uvrY) (panels I to IV, circles), pBA29 (expressing barA) (panel I, diamonds), pMX540 (expressing uvrYD54Q) (panel II, triangles), pMX541 (expressing both uvrY and barA) (panel III, triangles), and the isogenic csrA pta::ackA triple mutant strain harboring pMX539 (expressing uvrY) (panel IV, triangles) were diluted to an OD600 of ∼0.05 in LB medium, and the β-galactosidase activity was followed for 300 min. The average from four independent experiments is presented (standard deviations were less than 5% from the mean). (C) Ectopic expression of uvrY and barA by pMX541 (indicated as pUvrY-BarA) restores csrB expression in uvrY and barA mutant strains. Cultures of the csrB-lacZ transcriptional fusion-carrying wild type (KSB837) and the isogenic uvrY (UYKSB837) and barA (BAKSB837) mutant strains both carrying or not the uvrY- and barA-expressing plasmid pMX541 were grown to an OD600 of ∼2.0 in LB medium, and the β-galactosidase activity was assayed. The average from two independent experiments is presented. (D) Levels of UvrY (upper panel) and BarA (lower panel) proteins in KSB837 (wild type), IFC5010 (csrA), and IFC5010 harboring either of the following plasmids: pMX539 (indicated as pUvrY), pMX540 (indicated as pUvrYD54Q), pBA29 (indicated as pBarA), pMX541 (indicated as pUvrY-BarA), and csrA pta::ackA (IFC5015) harboring pMX539 (indicated as pUvrY) as determined by Western blot analyses. Purified His6-tagged BarA′ and UvrY proteins and extracts from barA (BAKSB837) and uvrY (UYKSB837) mutant cells were used in the first lanes of each Western blot.
CsrA is required for activation of the BarA kinase activity.
The above hypothesis, that UvrY is autophosphorylated at the expense of acetyl-P and is not transphosphorylated by BarA in the csrA mutant, was then tested. To this end, a pta::ackA mutation was inserted into the csrA mutant strain in order to block the synthesis of acetyl-P (42), and the effect of the UvrY-overexpressing plasmid pMX539 on csrB expression was probed. It was found that although similar amounts of UvrY were expressed in the csrA pta::ackA triple mutant strain and in the csrA mutant (Fig. 4D), no activation of csrB transcription occurred, as judged by Northern blotting and by the csrB-lacZ reporter fusion (Fig. 4A and B). This suggests that in the csrA mutant strain, UvrY is phosphorylated exclusively at the expense of acetyl-P rather than being transphosphorylated by BarA.
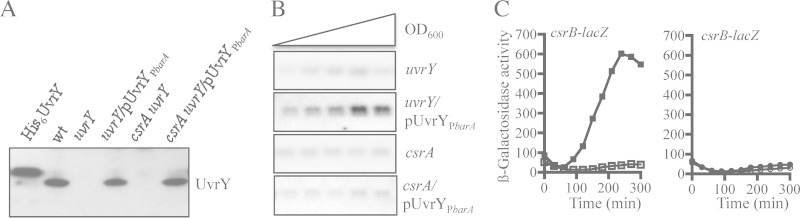
Subsequently, we explored the intriguing possibility that BarA remains inactive as a kinase in the csrA mutant. We argued that if the activity of BarA is not affected by CsrA, then reestablishing the levels of UvrY protein in the csrA mutant should restore csrB transcription. Therefore, we constructed a uvrY gene carrying low-copy-number plasmid (pMX543), where the promoter and 5′-UTR sections of uvrY were replaced with the ones of barA (Fig. 5A), the expression of which was not affected by CsrA (Fig. 3A). This plasmid was transformed into UVKSB837, a uvrY mutant, or IFC5016, a csrA uvrY double mutant strain, and the amount of UvrY was examined by Western blotting (Fig. 5A). It was found that similar amounts of UvrY protein were expressed from the plasmid in both of these strains. Subsequently, we tested whether pMX543 was able to complement csrB expression in these mutant strains. Interestingly, pMX543 restored csrB expression in the uvrY mutant but not in the csrA uvrY mutant (Fig. 5B and C). A possible explanation for this finding is that CsrA is also required for the kinase activity of BarA.
FIG 5.
Release of uvrY expression from csrA control is not sufficient for activation of csrB transcription in a csrA mutant. (A) Levels of UvrY protein in KSB837 (wild type), UYKSB837 (uvrY), UYKSB837 carrying plasmid pMX543 (having uvrY under control of the barA promoter and 5′-UTR, and indicated as pUvrYPbarA), IFC5016 (uvrY csrA), and IFC5016 carrying plasmid pMX543 (indicated as pUvrYPbarA) as determined by Western blot analyses. Purified His6-tagged UvrY protein was used in the first lane of the Western blot. (B) Northern blot analysis of CsrB levels in UYKSB837 (uvrY), UYKSB837 harboring pMX543 (indicated as pUvrYPbarA), IFC5010 (csrA), and IFC5016 harboring pMX543 (indicated as pUvrYPbarA). Cultures of these above strains were grown in LB medium, and RNA isolated from samples that were harvested throughout the growth curve (OD600 of 0.3 to 2.0) was probed for the CsrB transcript. (C) Overnight cultures of UYKSB837 (uvrY) (open squares), UYKSB837 harboring pMX543 (closed squares), IFC5010 (csrA) (open circles), and IFC5010 harboring pMX543 (closed circles), all carrying the csrB-lacZ transcriptional fusion, were diluted to an OD600 of ∼0.05 in LB medium, and the β-galactosidase activity was followed for 300 min. The average from four independent experiments is presented (standard deviations were less than 5% from the mean).
To provide further support to the above conclusions, we argued that ectopic expression of UvrY, but not that of the mutant UvrYD54Q, which is unable to be phosphorylated, in a csrA mutant should restore csrB expression when the cells are grown at pH 5.0 in the presence of acetate, which results in the production of elevated amounts of acetyl-P (43). On the other hand, the addition of formate, which acts exclusively via BarA, should be without effect. Accordingly, overexpression of UvrY in the csrA pta::ackA triple mutant, which is not able to convert acetate to acetyl-P, should not restore csrB expression when cells are grown at pH 5.0 in the presence of either acetate or formate. Indeed, csrB transcription in the csrA mutant strain grown at pH 5.0 was restored by the ectopic expression of UvrY in the presence of only acetate but not formate, whereas overexpression of UvrYD54Q was without effect (Fig. 6A and B). Also, no activation of csrB transcription was observed in the csrA pta::ackA mutant strain transformed with pMX539 when the cells were grown at pH 5.0 in the presence of either acetate or formate (Fig. 6A and B). We therefore concluded that when UvrY is overexpressed in the csrA mutant, acetyl-P-dependent phosphorylation of UvrY is responsible for the activation of csrB transcription.
FIG 6.
Effect of csrA on the activity of BarA. (A) Cultures of IFC5010 (csrA) harboring or not plasmid pMX539 (expressing uvrY and indicated as pUvrY), pMX540 (expressing uvrYD54Q and indicated as pUvrYD54Q), pMX541 (expressing both uvrY and barA and indicated as pUvrY-BarA), or pMX543 (having uvrY under control of the barA promoter and 5′-UTR and indicated as pUvrYPbarA), IFC5017 (csrA barA) harboring pMX543 (indicated as pUvrYPbarA), and IFC5015 (csrA pta::ackA) harboring pMX539 (indicated as pUvrY) were grown in LB medium the pH of which had been adjusted and buffered to 5.0, using 0.1 M homopiperazine-N,N′-bis-2-(ethanesulfonic acid) (HOMOPIPES). At an OD600 of 0.2, a sample was withdrawn, 7 mM acetate or formate was added to the medium, and samples were withdrawn every 10 min. Total RNA isolated from these samples was analyzed by Northern blotting using a CsrB-specific probe. The experiment was repeated three times in its entirety with essentially identical results. (B) Overnight cultures of the csrB-lacZ transcriptional fusion carrying IFC5010 (csrA), IFC5010 harboring plasmid pMX539 (indicated as pUvrY), pMX540 (indicated as pUvrYD54Q), pMX541 (indicated as pUvrY-BarA), or pMX543 (indicated as pUvrYPbarA), IFC5017 (csrA barA) harboring pMX543 (indicated as pUvrYPbarA), and IFC5015 (csrA pta::ackA) harboring pMX539 (indicated as pUvrY) were diluted to an OD600 of ∼0.05 in LB medium at pH 5.0 as described for Fig. 1B alone (squares) or in the presence of acetate (circles) and formate (diamonds). The average from four independent experiments is presented (standard deviations were less than 5% from the mean).
Finally, we reasoned that if CsrA was required for switching BarA from its phosphatase to its kinase activity, the simultaneous overexpression of UvrY and BarA in the csrA mutant, using plasmid pMX541, should not restore csrB expression when cells are grown at pH 5.0 in the presence of acetate or formate. The same result should be expected when wild-type levels of UvrY are reestablished in the csrA mutant by plasmid pMX543. This is because BarA should remain locked on as a phosphatase, dephosphorylating the acetyl-P-dependent UvrY-P and thereby cancelling its regulatory effect. In fact, no activation of csrB transcription was detected in the pMX541 or pMX543 carrying csrA mutant strain (Fig. 6A and B). On the other hand, reestablishing the wild-type levels of UvrY by plasmid pMX543 in a csrA barA double mutant, where no UvrY-P-dephosphorylating activity is present, should restore csrB expression in the presence of acetate but not formate. Indeed, csrB transcription in the pMX43-carrying csrA barA double mutant grown at pH 5.0 was restored to wild-type levels in the presence of acetate but not formate (Fig. 6A and B). Taken together, these results indicate that in the csrA mutant, BarA fails to be activated as a kinase but functions as a phosphatase even in the presence of its stimulus. It thus appears that one or more genes, whose expression is regulated by CsrA, may be needed for proper activation of BarA. Thus far, we have been unsuccessful in screening a plasmid-based genomic library for genes that would complement csrB expression in the csrA mutant.
Conclusions.
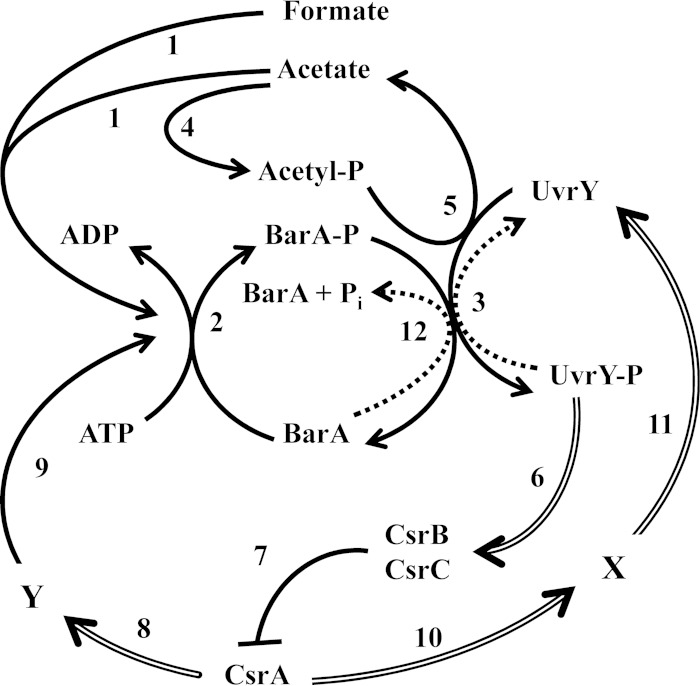
In this study, we investigated the effect of the CsrA global regulator on the expression and activity of the components of the BarA/UvrY signaling system. This was motivated by the earlier observation that csrB expression, which relies on the BarA-to-UvrY phosphorelay, did not take place in a csrA mutant (25). Our results demonstrate that the CsrA protein is required for the proper expression of the UvrY response regulator and also for the adequate switch from the phosphatase to the kinase activity of the BarA sensor kinase (Fig. 7), providing an explanation for the above observation.
FIG 7.
Model for the regulatory circuitry of the BarA/UvrY TCS and Csr system. Under stimulatory conditions, acetate and formate act as physiological signals that activate BarA (1), leading to its autophosphorylation at the expense of ATP (2) and transphosphorylation of UvrY (3). UvrY can also autophosphorylate at the expense of acetyl-P (5), which is produced from acetate (4). Phosphorylated UvrY (UvrY-P) activates expression of the noncoding CsrB and CsrC RNAs (6), which bind and sequester the CsrA protein (7) and thereby prevent its regulatory interaction with the mRNA targets. On the other hand, free CsrA regulates the expression of factor Y (8), which is required for properly switching BarA from its phosphatase to its kinase activity (9). At the same time, CsrA positively affects uvrY expression (11) by controlling the expression of the regulator(s) (X) (10). Finally, under nonstimulatory growth conditions or in a csrA mutant strain, BarA acts as a UvrY-P phosphatase (12), enabling the silencing of the system. Reactions under stimulatory and nonstimulatory conditions are indicated with solid and dotted lines, respectively. Double lines indicate effects on gene expression.
We provided evidence that CsrA positively affects uvrY expression at both the transcriptional and posttranscriptional levels. The effect of CsrA, an RNA binding protein, on uvrY transcription plausibly may be mediated via the regulation of expression of a transcriptional factor. Previously, SdiA and Crp have been reported to activate, respectively, uvrY transcription in E. coli and csrB transcription in Yersinia pseudotuberculosis (44, 45). However, CsrA modestly represses sdiA translation in E. coli (46), and Crp does not activate csrB expression (A. Pannuri and T. Romeo, unpublished data). Thus, these two regulators cannot account for the positive effects of CsrA on uvrY transcription. Another candidate is LexA, which coordinates the SOS response (47, 48), because a LexA binding site is predicted to be located between nt −120 and −139 upstream of the uvrY start site (49). CsrA-dependent modulation of uvrY translation might also be indirect, because no apparent CsrA binding sequences are present in the 5′-UTR of the uvrY transcript. In fact, the RNA DEAD box helicase DeaD was recently shown to be required for uvrY translation (50), although no link between CsrA and deaD expression is known at this time. Therefore, it is of great importance to clarify how these proteins are integrated into the Csr/UvrY circuitry, which allows for a global response through the CsrA protein.
Finally, we provide evidence that CsrA plays a significant role in the mechanism that enables BarA to switch from its phosphatase activity to its kinase activity. It is therefore tempting to speculate that, in addition to the BarA stimulus, a protein whose expression is regulated by CsrA is needed for proper activation of BarA. In this respect, it is relevant to mention that proper regulation of the kinase activity of GacS, the BarA homolog in Pseudomonas aeruginosa, requires the presence of the hybrid sensor kinases RetS and LadS (51–53). However, no homologs of these proteins exist in E. coli. Hence, identification of the protein(s) or other factors involved in the regulation of BarA signaling would greatly enhance our understanding of the Csr/BarA-UvrY regulatory network.
ACKNOWLEDGMENTS
We thank Claudia Rodriguez, Maria Luisa Tabche, and Gerardo Guzman Cordova for technical assistance and the Unidad de Biología Molecular of the Instituto de Fisiología Celular, Universidad Nacional Autónoma de México, for oligonucleotide synthesis and sequencing.
This work was partially supported by grants 178033 from the Consejo Nacional de Ciencia y Tecnología (CONACyT), IN206412 from DGAPA, UNAM, and GM059969 and R01AI097116 from the National Institutes of Health.
REFERENCES
- 1.Pernestig AK, Melefors O, Georgellis D. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J Biol Chem 276:225–231. doi: 10.1074/jbc.M001550200. [DOI] [PubMed] [Google Scholar]
- 2.Ishige K, Nagasawa S, Tokishita S, Mizuno T. 1994. A novel device of bacterial signal transducers. EMBO J 13:5195–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagasawa S, Tokishita S, Aiba H, Mizuno T. 1992. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol Microbiol 6:799–807. doi: 10.1111/j.1365-2958.1992.tb01530.x. [DOI] [PubMed] [Google Scholar]
- 4.Chavez RG, Alvarez AF, Romeo T, Georgellis D. 2010. The physiological stimulus for the BarA sensor kinase. J Bacteriol 192:2009–2012. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahn D, Ditta G. 1991. Modular structure of FixJ: homology of the transcriptional activator domain with the −35 binding domain of sigma factors. Mol Microbiol 5:987–997. doi: 10.1111/j.1365-2958.1991.tb00774.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu MY, Gui G, Wei B, Preston JF, Oakford L, Yüksel U, Giedroc DP, Romeo T. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem 272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 7.Romeo T, Vakulskas CA, Babitzke P. 2013. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol 15:313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueroa-Bossi N, Schwartz A, Guillemardet B, D'Heygère F, Bossi L, Boudvillain M. 2014. RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev 28:1239–1251. doi: 10.1101/gad.240192.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu MY, Romeo T. 1997. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J Bacteriol 179:4639–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, Babitzke P. 2003. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol 185:4450–4460. doi: 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH-T. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 12.Babitzke P, Romeo T. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 13.White D, Hart ME, Romeo T. 1996. Phylogenetic distribution of the global regulatory gene csrA among eubacteria. Gene 182:221–223. doi: 10.1016/S0378-1119(96)00547-1. [DOI] [PubMed] [Google Scholar]
- 14.Fortune DR, Suyemoto M, Altier C. 2006. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun 74:331–339. doi: 10.1128/IAI.74.1.331-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhatt S, Edwards AN, Nguyen HT, Merlin D, Romeo T, Kalman D. 2009. The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect Immun 77:3552–3568. doi: 10.1128/IAI.00418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, Chatterjee A, Liu Y, Dumenyo CK, Chatterjee AK. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol 177:5108–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol 58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 18.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 19.Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. 2007. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol Microbiol 64:1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- 20.Sabnis NA, Yang H, Romeo T. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J Biol Chem 270:29096–29104. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- 21.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol 56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 23.Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol 184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonas K, Edwards AN, Simm R, Romeo T, Romling U, Melefors O. 2008. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, Babitzke P, Romeo T. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol 184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. 2001. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol 183:6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Pena Sandoval GR, Wanner BL, Jung WS, Georgellis D, Kwon O. 2009. Evidence against the physiological role of acetyl phosphate in the phosphorylation of the ArcA response regulator in Escherichia coli. J Microbiol 47:657–662. doi: 10.1007/s12275-009-0087-9. [DOI] [PubMed] [Google Scholar]
- 28.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. 2011. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 80:1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dykxhoorn DM, St Pierre R, Linn T. 1996. A set of compatible tac promoter expression vectors. Gene 177:133–136. doi: 10.1016/0378-1119(96)00289-2. [DOI] [PubMed] [Google Scholar]
- 32.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 33.Georgellis D, Arvidson S, von Gabain A. 1992. Decay of ompA mRNA and processing of 9S RNA are immediately affected by shifts in growth rate, but in opposite manners. J Bacteriol 174:5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denhardt DT. 1966. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun 23:641–646. doi: 10.1016/0006-291X(66)90447-5. [DOI] [PubMed] [Google Scholar]
- 35.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol 48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 36.Mondragon V, Franco B, Jonas K, Suzuki K, Romeo T, Melefors O, Georgellis D. 2006. pH-dependent activation of the BarA-UvrY two-component system in Escherichia coli. J Bacteriol 188:8303–8306. doi: 10.1128/JB.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmermans J, Van Melderen L. 2009. Conditional essentiality of the csrA gene in Escherichia coli. J Bacteriol 191:1722–1724. doi: 10.1128/JB.01573-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P. 2013. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol 87:851–866. doi: 10.1111/mmi.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomenius H, Pernestig AK, Mendez-Catala CF, Georgellis D, Normark S, Melefors O. 2005. Genetic and functional characterization of the Escherichia coli BarA-UvrY two-component system: point mutations in the HAMP linker of the BarA sensor give a dominant-negative phenotype. J Bacteriol 187:7317–7324. doi: 10.1128/JB.187.21.7317-7324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georgellis D, Kwon O, De Wulf P, Lin EC. 1998. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem 273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 41.Peña-Sandoval GR, Kwon O, Georgellis D. 2005. Requirement of the receiver and phosphotransfer domains of ArcB for efficient dephosphorylation of phosphorylated ArcA in vivo. J Bacteriol 187:3267–3272. doi: 10.1128/JB.187.9.3267-3272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanner BL, Wilmes-Riesenberg MR. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J Bacteriol 174:2124–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei Y, Lee JM, Smulski DR, LaRossa RA. 2001. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol 183:2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heroven AK, Sest M, Pisano F, Scheb-Wetzel M, Steinmann R, Böhme K, Klein J, Münch R, Schomburg D, Dersch P. 2012. Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front Cell Infect Microbiol 2:158. doi: 10.3389/fcimb.2012.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yakhnin H, Baker CS, Berezin I, Evangelista MA, Rassin A, Romeo T, Babitzke P. 2011. CsrA represses translation of sdiA, which encodes the N-acylhomoserine-l-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. J Bacteriol 193:6162–6170. doi: 10.1128/JB.05975-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.d'Ari R. 1985. The SOS system. Biochimie 67:343–347. doi: 10.1016/S0300-9084(85)80077-8. [DOI] [PubMed] [Google Scholar]
- 48.Butala M, Klose D, Hodnik V, Rems A, Podlesek Z, Klare JP, Anderluh G, Busby SJW, Steinhoff H-J, Zgur-Bertok D. 2011. Interconversion between bound and free conformations of LexA orchestrates the bacterial SOS response. Nucleic Acids Res 39:6546–6557. doi: 10.1093/nar/gkr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark T, Moses RE. 1989. Interaction of the LexA repressor and the uvrC regulatory region. FEBS Lett 258:39–41. doi: 10.1016/0014-5793(89)81610-2. [DOI] [PubMed] [Google Scholar]
- 50.Vakulskas CA, Pannuri A, Cortés-Selva D, Zere TR, Ahmer BM, Babitzke P, Romeo T. 2014. Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol Microbiol 92:945–958. doi: 10.1111/mmi.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A 103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]