Abstract
The gastric pathogen Helicobacter pylori must combat chronic acid and oxidative stress. It does so via many mechanisms, including macromolecule repair and gene regulation. Mitomycin C-sensitive clones from a transposon mutagenesis library were screened. One sensitive strain contained the insertion element at the locus of hp119, a hypothetical gene. No homologous gene exists in any (non-H. pylori) organism. Nevertheless, the predicted protein has some features characteristic of histone-like proteins, and we showed that purified HP119 protein is a DNA-binding protein. A Δhp119 strain was markedly more sensitive (viability loss) to acid or to air exposure, and these phenotypes were restored to wild-type (WT) attributes upon complementation of the mutant with the wild-type version of hp119 at a separate chromosomal locus. The mutant strain was approximately10-fold more sensitive to macrophage-mediated killing than the parent or the complemented strain. Of 12 mice inoculated with the wild type, all contained H. pylori, whereas 5 of 12 mice contained the mutant strain; the mean colonization numbers were 158-fold less for the mutant strain. A proteomic (two-dimensional PAGE with mass spectrometric analysis) comparison between the Δhp119 mutant and the WT strain under oxidative stress conditions revealed a number of important antioxidant protein differences; SodB, Tpx, TrxR, and NapA, as well as the peptidoglycan deacetylase PgdA, were significantly less expressed in the Δhp119 mutant than in the WT strain. This study identified HP119 as a putative histone-like DNA-binding protein and showed that it plays an important role in Helicobacter pylori stress tolerance and survival in the host.
INTRODUCTION
Helicobacter pylori infects the stomachs of approximately 50% of humans and results in a series of human gastric diseases, including gastritis, peptic ulcers, and gastric cancer (1–4). The pathogenesis of H. pylori relies on its persistence in surviving a harsh environment, including acidity, peristalsis, and attack by phagocyte cells and their released reactive oxygen species (5). H. pylori survives on the surface of the stomach lining, often for the lifetime of its host, and causes a chronic inflammatory response. Under physiological conditions, H. pylori is thought to frequently suffer oxidative and acid stress (6, 7). H. pylori is equipped with diverse oxidant detoxification enzymes (e.g., superoxide dismutase, catalase, and peroxiredoxins) (8) and potent acid avoidance mechanisms (mainly urease) (9). To survive the harsh conditions, H. pylori regulates its gene expression in response to the stress signals; however, the bacterium lacks many response regulators known to occur in model microorganisms, such as the SOS response, OxyR/SoxR, and RpoS. Our current knowledge of the stress tolerance mechanisms cannot account for the well-described dynamic survival abilities of H. pylori.
Studies in recent years have indicated that DNA recombination and repair play a significant role in H. pylori's persistent colonization of the host (10–14). In an attempt to identify additional components of the DNA recombination/repair system in H. pylori, we screened for mitomycin C (MMC)-sensitive clones from a random transposon mutagenesis library. In one of the mitomycin C-sensitive strains the transposon was shown to be inserted at the hup locus encoding a protein that is homologous to the histone-like protein (HLP) HU of Escherichia coli. Subsequently, we investigated the physiological roles of H. pylori Hup in protecting its DNA from (oxidative and acid) stress damage and its contribution to bacterial survival within the host stomach (15). In another of the mitomycin C-sensitive clones from the random transposon mutagenesis library, the transposon was shown to be inserted at the locus of hp119, a hypothetical gene.
Genomic DNA in a bacterial cell is folded into a compact structure called a nucleoid, and the proper assembly of active higher-order genome structures requires accessory proteins, termed nucleoid-associated proteins (NAPs). Several nucleoid-associated proteins, such as HU, IHF, H-NS, Fis, and Lrp, in E. coli have been studied and shown to play roles in DNA organization and protection (16, 17). These proteins are sometimes referred to as histone-like because they have roles in nucleoid compaction comparable to that described for eukaryotic histones. These proteins not only are involved in DNA supercoiling and compaction but also modulate DNA functions such as replication, recombination, repair, and transcription (18). Each bacterial species harbors a specific set of NAPs, with only HU-like proteins being ubiquitous among bacteria. In this study, we first identified HP119 as a putative histone-like protein in H. pylori. Then we investigated the physiological roles of HP119 in stress tolerance. The contribution of HP119 for bacterial survival in contact with cultured murine immune cells and within the mouse stomach was examined. In addition, a proteomic analysis was performed to identify the potential roles of HP119 in gene and protein expression.
MATERIALS AND METHODS
H. pylori culture conditions.
H. pylori was cultured on brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood or 5% fetal bovine serum (called BA plates). Chloramphenicol (50 μg/ml) or kanamycin (40 μg/ml) was added to the medium for culturing mutants. Cultures of H. pylori were grown microaerobically at 37°C in an incubator under continuously controlled levels of oxygen (4% partial-pressure O2, 5% CO2, and the balance N2).
Construction of an H. pylori hp119 mutant.
Overlapping PCR (19) and allele exchange mutagenesis was used to generate an hp119 deletion mutant. H. pylori 26695 genomic DNA was used as a template to amplify an approximately 400-bp DNA fragment both upstream and downstream of the target locus. Primers P119-1 (5′-ATGCCTGTTATAAGAGTTT-3′) and P119-2 (5′-ATCCACTTTTCAATCTATATCCCAATTCTATCCCACTCTT-3′) were used to amplify the upstream region, while primers P119-3 (5′-CCCAGTTTGTCGCACTGATAACCAAACTCTAAACAACCTC-3′) and P119-4 (5′-TTACTATAACCATAACCCG-3′) were used to amplify the downstream region. The cat cassette (encoding chloramphenicol resistance [20]) was amplified using primers Pcat-5 (5′-GATATAGATTGAAAAGTGGAT-3′) and Pcat-6 (5′-TTATCAGTGCGACAAACTGGG-3′). The cat cassette contains an upstream promoter and lacks a transcription termination sequences in order to avoid polar effects on downstream genes. Primers P119-2 and P119-3 contain 5′-end regions that anneal to either end of cat cassette sequence. Final overlapping PCRs resulted in a sandwich fusion in which the cat cassette was flanked by the upstream and downstream regions of hp119, while the hp119 gene was deleted. This PCR product was used to transform the H. pylori wild-type (WT) strain by selection on chloramphenicol (50 μg/ml)-containing BA plates that were incubated under a low-O2 (2% partial pressure) condition. Screening the transformants at low oxygen was shown to be useful for obtaining oxygen-sensitive mutants (21, 22). Successful disruption of the target allele was confirmed by PCR or gel electrophoresis and by direct sequencing of the PCR fragment (Georgia Genomics Facility).
Construction of H. pylori hp119 complementation strain.
The complemented hp119+ strain was constructed by inserting a wild-type copy of the hp119 gene in the rdxA locus of the hp119::cat chromosome. Disruption of rdxA results in metronidazole resistance that is used for selection in DNA transformation. PCR products corresponding to the upstream sequence of the rdxA gene (∼300 bp), the full-length hp119 gene sequence, and the downstream sequence of the rdxA gene (∼300 bp) were amplified in three separate PCRs and then stitched together in subsequent PCR (overlapping PCR). In the sequence of the final PCR product, the rdxA gene was exactly replaced by the intact hp119 gene; thus, hp119 gene would be expressed under the control of the rdxA promoter. The final PCR product was used to transform the hp119::cat strain, with subsequent selection for metronidazole (16 μg/ml)-resistant colonies. Through homologous DNA recombination, an intact hp119 gene was inserted at the rdxA locus of the hp119::cat strain.
Overexpression and purification of HP119 protein.
A DNA fragment containing H. pylori hp119 gene was amplified by PCR and cloned into the pET-21a vector to generate pET-Hup-6His. E. coli BL21 Origami cells harboring pET-HP119-6His were grown at 37°C to an OD at 600 nm (OD600) of 0.5 in 500 ml of LB medium with ampicillin (100 μg/ml) and kanamycin (40 μg/ml). Expression of the HP119 protein was induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the medium, followed by further incubation for 3 h; cells were then harvested by centrifugation (5,000 × g, 15 min, and 4°C). All subsequent steps were performed at 4°C. Cells were washed with 200 ml of 20 mM Na2HPO4 (pH 7.4), 500 mM NaCl, and 5 mM imidazole (buffer A) and resuspended in 5 ml of the same buffer. Cells were lysed by two passages through a cold French pressure cell at 18,000 lb/in2. Cell debris was removed by centrifugation at 20,000 × g. The supernatant was applied to a nickel-nitrilotriacetic acid (Ni-NTA) affinity column (Qiagen), and buffer A was used to wash the resin until the A280 reached the baseline. Proteins were washed with buffer B (buffer A with 30 mM imidazole) until the A280 reached the baseline and were finally eluted with buffer C (buffer A with 250 mM imidazole). Extracts of E. coli BL21 Origami containing the vector only did not result in retrievable proteins from this purification (Ni affinity) procedure. Fractions were analyzed by gel electrophoresis, and the HP119-positive fractions were pooled. Protein concentrations were determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, IL).
Assay for DNA binding activity.
Electrophoretic mobility shift assay was carried out as described by Liu et al. (23). Briefly, a supercoiled plasmid, pGEMT (6 nM), was incubated with increasing amounts (60, 600, and 6,000 nM) of purified HP119 protein or bovine serum albumin (BSA) in reaction buffer (10 mM Tris [pH 8.0], and 75 mM KCl) at 25°C for 25 min. The resulting nucleoprotein complexes were separated by 0.8% agarose gel electrophoresis in 0.5× Tris-acetate-EDTA (TAE) buffer at 10 V for 16 h and visualized by ethidium bromide staining.
Oxygen sensitivity (air survival) assay.
H. pylori strains were grown on BA plates to late log phase, and the cells were suspended in phosphate-buffered saline (PBS) at a concentration of ∼108 cells/ml. The cell suspensions were incubated at 37°C under normal atmospheric conditions (21% O2) with moderate shaking. Samples were then removed at various time points (2, 4, 6, 8, and 10 h), serially diluted, and spread onto BA plates. Colony counts were recorded after 4 days of incubation in a microaerobic atmosphere (4% partial-pressure O2) at 37°C.
Assessment for sensitivity to low-pH condition.
H. pylori strains were grown on BA plates to late log phase, and the cells were suspended in the buffer (20 mM Tris-HCl, 150 mM NaCl) with different pH levels (pH 7.0, 5.0, or 3.0) at a concentration of ∼108 cells/ml. The cell suspensions were incubated under a microaerobic condition (4% O2) at 37°C for 1 h. The samples were serially diluted and plated for CFU counts (after 4 days of incubation under a microaerobic growth condition). The percent cell survival at pH 5.0 or pH 3.0 relative to that at pH 7.0 was calculated.
Macrophage killing assay.
The survival of H. pylori cells within macrophages was investigated by following the methods published previously (14, 24, 25), with minor modifications. Briefly, RAW264.7 macrophages were seeded in 24-well plates in the culture medium (0.5 ml) and incubated at 37°C and 5% CO2 for 4 days (cell density was about 105 cells per well). The medium was replaced by fresh medium to remove the nonadherent cells. H. pylori cells were added at a ratio of 20 CFU bacteria per macrophage (the number of H. pylori cells added was separately determined by serial dilution and plate counting for CFU determinations). After addition of H. pylori cells, the coculture was maintained under a low (4%)-oxygen condition to eliminate oxygen killing effects. Phagocytosis was synchronized by centrifugation at 600 × g for 5 min and then allowed to proceed for 1 h. Extracellular bacteria were removed by washing and incubation in medium supplemented with gentamicin (100 mg/ml) for 1 h at 37°C and 5% CO2. After three washes to remove the antibiotics, the cells were further incubated in fresh medium for 2 h. After removal of the medium, the macrophages were lysed with ice-cold PBS with 0.1% saponin for 5 min. Appropriate dilutions of the supernatant were plated on BA plates and incubated at 37°C, 5% CO2, and 2% O2 for 4 days to count the surviving bacteria. The number of surviving bacteria (CFU/ml) was compared with the number of viable bacteria initially added.
Mouse colonization assay.
Mouse colonization assays were performed essentially as described previously (12, 15). Briefly, wild-type X47 or isogenic Δhp119 mutant cells were harvested after 48 h of growth on BA plates (37°C and 4% oxygen) and suspended in PBS to an OD600 of 1.7. Headspace in the tube was sparged with argon gas to minimize oxygen exposure, and the tube was tightly sealed. The bacterial suspensions were administered to C57BL/6NCr mice (3 × 108 H. pylori cells/mouse). Three weeks after the first inoculation, the mice were sacrificed and the stomachs were removed, weighed, and homogenized in argon-sparged PBS to avoid O2 exposure. Stomach homogenate dilutions (dilutions conducted in sealed tubes containing argon-sparged buffer) were plated on BA plates supplemented with bacitracin (100 μg/ml), vancomycin (10 μg/ml), and amphotericin B (10 μg/ml). The plates were rapidly transported into an incubator containing sustained 4% partial-pressure O2. After incubation for 5 to 7 days, H. pylori colonies were counted and the data expressed as CFU per gram of stomach.
Two-dimensional gel electrophoresis (2D PAGE).
Cells of H. pylori wild-type strain 26695 and its isogenic Δhp119 mutant were grown to late log phase in an atmosphere containing 4% O2, and then cells were exposed to air (20% O2) for 4 h. The cells were harvested, washed with PBS, and lysed in SDS boiling buffer (5% SDS, 10% glycerol, 60 mM Tris-HCl [pH 6.8]). The protein concentration of the cell extract was determined with a Bradford protein assay (Bio-Rad). One hundred micrograms of protein was precipitated, purified, and cleaned with a 2D-cleanup kit (GE Healthcare Life Sciences). The pellets of precipitated proteins were resuspended in 60 μl of rehydration buffer {7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 25 mM Tris-HCl (pH 8.8)}.
Twenty-five micrograms of each sample was labeled with 200 pmol of N-hydroxysuccinimidyl-ester of cyanine dye Cy3 and Cy5 (GE Healthcare Life Sciences, Piscataway, NJ), with dye swapping to eliminate labeling bias between Cy3 and Cy5. Sample buffer (7 M urea, 4 M thiourea, 4% CHAPS 2% dithiothreitol [DTT], 2% immobilized pH gradient buffer [IPG; pH 4 to 11]) and rehydration solution (7 M urea, 4 M thiourea, 4% CHAPS, 1% DTT, 1% IPG) were added to a final volume of 350 μl for each mix. In the case of preparation gel for spot picking, 150 μg of each unlabeled protein was added to labeled mix. First-dimension isoelectric focusing (IEF) was performed using 24-cm IPG strips (pH 4 to 7) in Ettan IPGphor. After IEF, the strips were equilibrated, reduced, alkylated, and stained by sequential incubation in 1.0% DTT equilibration buffer (50 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol, and 2% SDS) and 4.5% iodoacetamide equilibration buffer slightly colored with bromophenol blue for 20 min each. The second-dimension SDS-PAGE was conducted on a 10% polyacrylamide gel in the Ettan DALT II system separation unit (GE Healthcare Life Sciences) until the tracking dye reached the bottom of the gel.
After completion of 2-dimensional electrophoresis, the gel images were obtained using Typhoon Trio (GE Healthcare Life Sciences) at appropriate wavelengths for Cy3 and Cy5 dyes and analyzed with Decyder image analysis software (v. 7.0; GE Healthcare Life Sciences). The gels were then visualized by colloidal Coomassie staining (SimplyBlue; Invitrogen). Each Coomassie-stained gel was scanned again with a Typhoon Trio scanner. The Coomassie-stained gel image was matched and aligned with the previous Cy3 and Cy5 fluorescence image to generate a pick list of proteins of interest.
MS protein identification.
The generated pick list was exported to Ettan Spot Picker (GE Healthcare Life Sciences), and protein spots were excised and picked to microtiter plates by the Ettan Spot Picker. The picked gel pieces were washed first with twice-distilled H2O and subsequently with washing solution I (50% methanol, 10% acetic acid), and washing solution II (50% acetonitrile, 50 mM ammonium bicarbonate [pH 8.3]). The washed gel pieces were finally dehydrated with 100% acetonitrile and dried under SpeedVac. The dried gel pieces were either subjected to trypsin digestion or kept at −80°C until they were treated with trypsin for the mass spectrometry (MS) peptide analysis. In brief, the gel pieces were incubated with an appropriate amount of trypsin (modified Trypsin Gold) in Proteomax surfactant (Promega, Madison, WI) at 37°C for 2 to ∼3 h. After incubation, the digested peptides were extracted with 2.5% trifluoroacetic acid. The extracted peptides were further purified and concentrated by a C18 ZipTip, a micro-reverse-phase column (Millipore, Billerica, MA).
Extracted peptides were then analyzed by a 4800 MALDI TOF/TOF tandem mass spectrometer (AB Sciex, Framingham, MA) with tandem mass spectrometry (MS/MS) mode. Protein identifications were carried out by Mascot search engine (Matrix Science Inc., Boston, MA) against the Swiss-Prot or NCBI protein database.
RESULTS
Identification of HP119 as a putative HLP.
Using a random transposon mutagenesis library of H. pylori (26), we identified genes that confer resistance to mitomycin C. Mitomycin C is a DNA-damaging agent that causes predominantly DNA intrastrand cross-links, leading ultimately to DNA double-strand breaks. We identified 12 genes; the insertion of the transposon within these genes makes the strain mitomycin C sensitive. Nine out of the 12 identified genes are known to be involved in DNA recombination and repair (recA, addA, addB, recN, recR, recO, ruvC, uvrC, and mfd). One identified gene encodes a putative outer membrane protein, which awaits further investigation. Another identified gene was hup encoding a histone-like protein (HLP) (15). In another MMC-sensitive strain, the transposon was inserted within the hp119 locus that was annotated as a hypothetical gene in the published H. pylori genome sequence (27). Interestingly, there exist five paralogous genes in the H. pylori genome: hp118, hp119, and hp120 (in one locus) and hp1187 and hp1188 (in another locus). A BLAST search indicates that no homologous genes of hp119 exist in any other organism. The five hypothetical proteins contain a conserved C-terminal domain (DUF874). DUF874 (∼200 amino acid residues) is an H. pylori-specific domain of unknown function, and it is well conserved (>90% amino acids identical) across all H. pylori strains. The central regions of these proteins are not well conserved but contain repeated sequences that are highly enriched in the amino acids lysine (K), glutamic acid (E), and glutamine (Q). For example, HP119 contains three tandem repeat sequences of EQEQQKTEQEKQKTEQEKQKTEQEKQKTEQEKQKTSNIETNNQIKV.
From sequence analysis of the five HP119-related proteins, we found they have some features of histone-like proteins. We compared the percentages of KEQ residues in the known histone-like proteins (Table 1) and found that K and E are highly abundant in almost all the histone-like proteins and Q is rich in some of them. Like H. pylori Hup protein, the HP119-related proteins contain ∼16% K and ∼10% E residues. HP119-related proteins also contain 8 to 10% Q residues, similar to E. coli Fis protein.
TABLE 1.
Percentage of KEQ residues and isoelectric points of known histone-like proteins and HP119-related proteins
| Organism | Protein | % indicated amino acid |
pI | ||
|---|---|---|---|---|---|
| Lys (K) | Glu (E) | Gln (Q) | |||
| E. coli | HupA | 12.1 | 7.7 | 4.4 | 10.4 |
| IhfA | 10 | 11 | 3 | 10.1 | |
| H-NS | 8.1 | 12.6 | 5.9 | 9 | |
| Fis | 7.1 | 4 | 10.1 | 9.8 | |
| H. pylori | Hup | 18.9 | 14.7 | 2.1 | 9.7 |
| HP118 | 16 | 9.7 | 9.2 | 10.1 | |
| HP119 | 16.6 | 12.3 | 12.7 | 9.9 | |
| HP120 | 15.5 | 10 | 8.5 | 10.2 | |
| HP1187 | 15.3 | 10.1 | 9.6 | 10.2 | |
| HP1188 | 15.6 | 10 | 7.8 | 10.1 | |
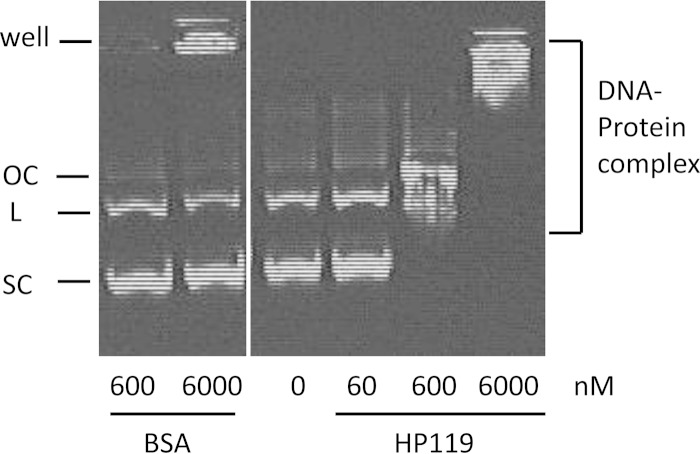
To test the hypothesis that HP119 may function as a histone-like protein, we purified His-tagged HP119 protein (∼54 kDa) and examined its DNA binding activity by use of an electrophoretic mobility shift assay. Plasmid pGEMT DNA (6 nM) was incubated with various amounts of purified HP119 protein, followed by agarose gel electrophoresis to visualize the protein-DNA complex (Fig. 1). At the lowest protein concentration (60 nM), we could not detect a DNA size shift. With higher concentrations (600 and 6,000 nM HP119 protein), we saw retarded DNA bands, and the extent of retardation depended on the protein level. As a negative control, the same amounts of BSA protein (600 and 6,000 nM) did not produce any retarded DNA bands. Note that some DNA was trapped in the well of the agarose gel with the presence of the highest concentration of the control lane (6,000 nM BSA), while more than 70% of DNA remained free. In contrast, all DNA was complexed with HP119 at the concentration of 600 or 6,000 nM protein. The HP119 protein can bind to both supercoiled and linear DNA in an apparently random manner. The DNA binding properties of HP119 are comparable to those of known bacterial HLPs characterized with similar assays (23, 28).
FIG 1.
DNA binding activity of HP119. Plasmid pGEMT DNA (6 nM, 3 kb) was incubated with various amounts (60, 600, and 6,000 nM) of purified HP0119 protein. As a negative control, pGEMT DNA was incubated with 600 or 6,000 nM BSA protein. Different forms of plasmid DNA (OC, open circular; L, linear; SC, supercoiled) are labeled on the left. The protein-DNA complex was visualized by ethidium bromide staining after agarose gel (1%) electrophoresis.
HP119 plays a significant role in stress resistance.
To study the physiological role of HP119, we constructed an hp119::cat mutant. Δhp119 mutant strains can be easily obtained if transformants are screened under a low-oxygen (2% partial pressure) condition. The Δhp119 mutants were originally constructed in genome-sequenced strain 26695 and then in the mouse-adapted strain X47. Most of the in vitro and all the in vivo assays reported herein were done with X47 and its isogenic Δhp119 mutant. The X47 Δhp119 mutant was confirmed to be mitomycin C sensitive (data not shown).
Under the normal in vitro growth condition (4% O2 and 5% CO2), the Δhp119 mutant grew similarly to the wild-type strain. During the exponential growth phase (before reaching 24 h), similar growth rates were observed for the WT and mutant strains. Upon entrance of the cells into the stationary phase, however, the survival of the Δhp119 mutant decreased much faster than that of the wild-type strain. Based on a plating assay, the Δhp119 mutant lost viability completely after 3 days, while the majority of the WT cells were still viable (data not shown). To ensure that the phenotypes (both in vitro and in vivo) observed for the Δhp119 mutant strain were completely attributed to inactivation of hp119, we introduced a functional copy of the hp119 gene back into the Δhp119 strain. The strain X47 hp119::cat-hp119+ contains a deletion of the hp119 gene at the original locus and an intact hp119 gene at the rdxA locus (see Materials and Methods). The growth characteristics of the complemented strain were similar to those of the wild type (data not shown).
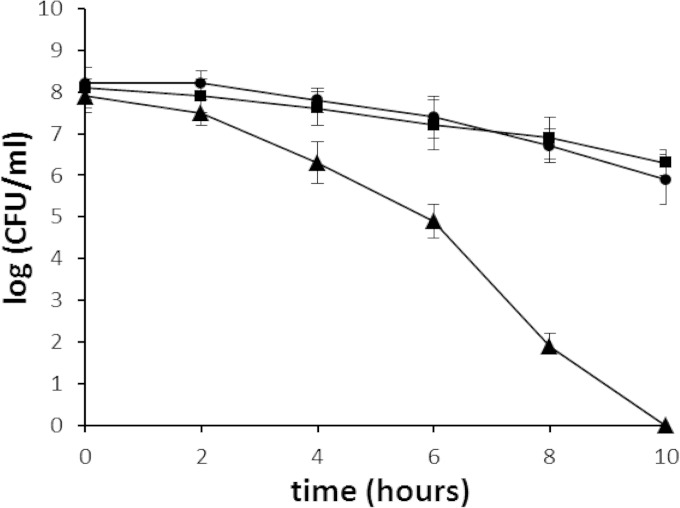
To examine the possible role of the HP119 protein in oxidative stress resistance, we examined the sensitivity of the Δhp119 mutant to oxidative stress by an air survival assay. The cell suspensions (∼5 × 108 cells/ml) were exposed to air, and the numbers of surviving cells were determined at various time points (Fig. 2). The number of wild-type cells decreased slowly; at the 10-h time point, about 5 × 106 cells (∼1% of that at the time zero) had survived. In contrast, the Δhp119 mutant showed a greater sensitivity to air exposure. Two hours after the cells were exposed to air, the number of surviving mutant cells started to decrease at a rate much higher than that of the wild-type cells. At the 10-h time point, the Δhp119 cells were eliminated (i.e., no viable cells were recovered). The sensitivity of the hp119-complemented strain to the oxygen stress condition was similar to that of the wild type.
FIG 2.
Survival of H. pylori cells upon exposure to air. H. pylori cell suspensions in PBS were incubated at 37°C under normal atmospheric conditions (21% partial-pressure O2). Samples were removed at the times indicated on the x axis and were used for plate count determinations after incubation in a 5% oxygen environment. The data are the means from three experiments, with standard deviations as indicated. Symbols: square, wild type; triangle, hp119::cat; circle, hp119 complementation strain. Based on statistical analysis (Student t test), the cell survival differences between the WT and the mutant strains are significant (P < 0.01) for all the data points except for the 2-h time point.
Next, we characterized the Δhp119 mutant for its sensitivity to low-pH conditions. The wild-type H. pylori or the Δhp119 mutant cells were treated for 1 h by suspension in the buffer at different pHs (pH 7.0, 5.0, or 3.0), and the survival rate was subsequently determined. As shown in Table 2, treatment at pH 5.0 for 1 h did not have a significant effect on survival of the wild-type cells, while the same treatment killed approximately 90% of the Δhp119 mutant cells. About 40% of the wild-type H. pylori cells survived the pH 3.0 condition for 1 h, whereas the Δhp119 mutant cells were almost completely killed (>95% lethality) by the same treatment. Thus, the Δhp119 mutant is more sensitive to acid stress than is the wild type. The complementation of hp119 function restored the acid sensitivity to the wild-type level.
TABLE 2.
Acid sensitivities of H. pylori strains
| Strain | % survival ata: |
||
|---|---|---|---|
| pH 7.0 | pH 5.0 | pH 3.0 | |
| X47 WT | 100 | 96 + 9 | 41 + 7 |
| X47 hp119::cat | 100 | 9.3 + 3.4 | 4.2 + 1.4 |
| X47 hp119 complementation strain | 100 | 93 + 7 | 39 + 5 |
The values are the percentages of cells surviving after treatment for 1 h at pH 5.0 or pH 3.0 relative to the survival at pH 7.0 (normalized to 100%). The data are the means + standard errors from three independent determinations (3 biological replicates).
The Δhp119 mutant is more sensitive to macrophage killing.
As the Δhp119 mutant is sensitive to oxidative stress, we investigated whether the HP119 protein contributes to survival of H. pylori within macrophages. A macrophage killing assay was performed using a murine macrophage line, RAW264.7, for the H. pylori WT, the Δhp119 mutant, or the complemented strain. As a control, different strains were incubated in the tissue culture medium (in the absence of macrophages) at 4% oxygen for 2 h. No significant difference was observed between the survival of the Δhp119 mutant and the WT strain (data not shown). In the macrophage killing assay, similar numbers (∼5 × 108 CFU/ml) of the H. pylori cells were inoculated to the macrophage culture. After killing of extracellular bacteria by gentamicin and further incubation for 2 h, the surviving H. pylori cells were recovered and their numbers determined. As shown in Table 3, a mean number of 2.97 × 106 CFU/ml X47 WT cells survived. In contrast, the same treatment resulted in recovery of a mean number of 2.67 × 105 CFU/ml of the Δhp119 mutant cells (approximately 10-fold less than the WT). Based on statistical analysis (Student t test), the cell survival differences between the WT and the mutant strains are significant (P < 0.01). The same assay was performed for the hp119-complemented strain; a mean number of 2.48 × 106 CFU/ml cells survived, similar to the surviving number of WT cells. These results indicated a role for HP119 in survival of H. pylori within macrophages.
TABLE 3.
Survival of H. pylori cells in RAW264.7 macrophages determined with the gentamicin killing assay
| Strain | Amt inoculated (CFU/ml)a | Amt surviving (CFU/ml)b | % surviving/inoculum |
|---|---|---|---|
| X47 WT | (5.18 + 0.45) × 108 | (2.97 + 0.34) × 106 | 0.573 |
| X47 hp119::cat | (4.10 + 0.26) × 108 | (2.67 + 0.28) × 105 | 0.065 |
| X47 hp119 compl. | (4.26 + 0.32) × 108 | (2.48 + 0.36) × 106 | 0.582 |
Similar numbers of cells of different H. pylori strains were inoculated to the macrophages. The inoculated CFU/ml of H. pylori cells was determined by serial dilution and plate counting. Data are means from three independent determinations, with standard deviations.
After extracellular killing by gentamicin and 2 h of incubation within macrophages, the numbers of surviving CFU/ml of H. pylori cells was determined by serial dilution and plate counting. Data are means from three independent determinations, with standard deviations.
The Δhp119 mutant displays a severely attenuated ability to colonize mouse stomachs.
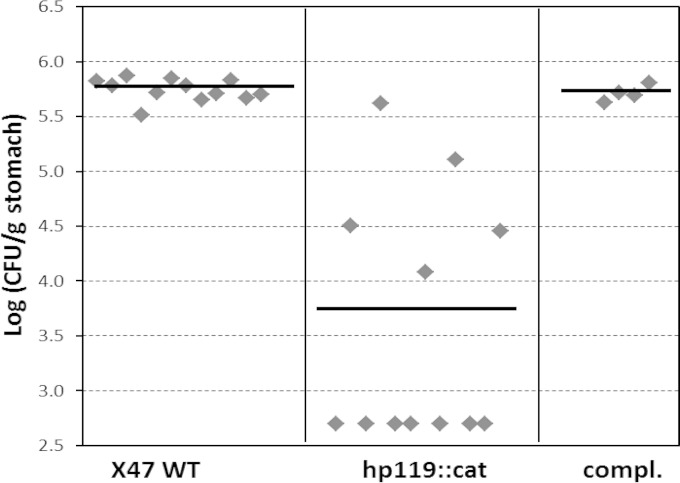
Considering the in vitro phenotypes of the Δhp119 mutant strain, we proceeded to determine the effect of the HP119 on H. pylori colonization in the host. The wild-type X47 or the isogenic Δhp119 mutant strain was inoculated into C57BL/6J mice, and the colonization of H. pylori cells in the mouse stomachs was examined 3 weeks after inoculation (Fig. 3). H. pylori colonies were recovered from all 12 mice that had been inoculated with the wild-type strain, with a mean number of 5.7 ×105 CFU/g stomach. In contrast, only 5 of 12 mice that were inoculated with the Δhp119 mutant strain were found to harbor H. pylori. The geometric mean of the colonization number for the Δhp119 strain in the 12 mice was 3.6 ×103 CFU/g of stomach. According to Wilcoxon rank test analysis, the range of colonization values of the Δhp119 strain is significantly lower than that of the wild type at the 99% confidence level (P < 0.01). These results indicate that HP119 protein plays a significant role in bacterial survival and colonization in the host.
FIG 3.
Mouse colonization results of H. pylori strains. The mice were inoculated with H. pylori at a dose of 1.5 × 108 viable cells per animal. Colonization of H. pylori in mouse stomachs was examined 3 weeks after the inoculation. Data are presented as a scatter plot (log scale) of CFU per gram of stomach as determined by plate counts. Each point represents the CFU count from one mouse stomach, and the horizontal bars represent the geometric means of the colonization numbers for each group. The detection limit of the assay is 500 CFU/g stomach, corresponding to a log10 (CFU/g) of 2.7.
The hp119-complemented strain was also examined for the mouse colonization ability. H. pylori bacteria were recovered from all 4 mice that had been inoculated with the hp119 complementation strain, with a geometric mean number of 5.1 ×105 CFU/g stomach (Fig. 3). According to Wilcoxon rank test analysis, the range of colonization values of the hp119 complementation strain is not significantly different from that of the wild type but is significantly (P < 0.01) higher than that of the Δhp119 mutant strain. This indicates that the complementation of the Δhp119 strain restored its ability to colonize mouse stomachs.
HP119 is involved in a multicomponent oxidative stress response.
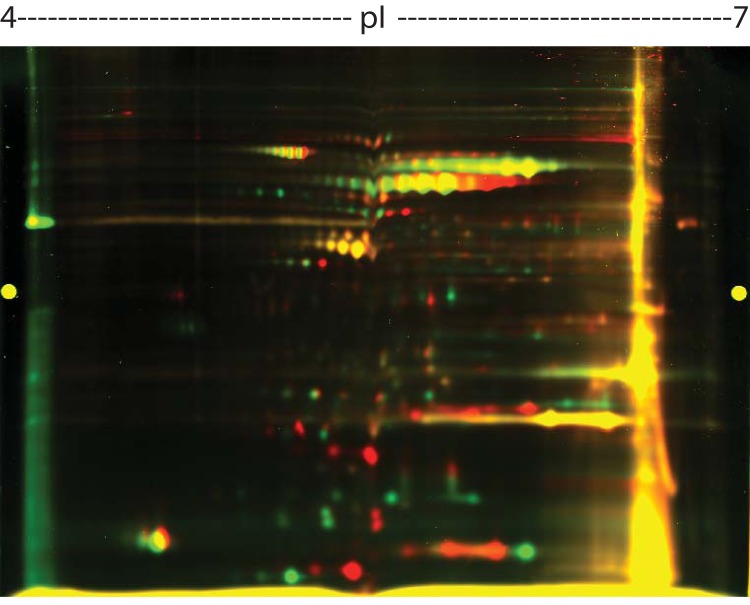
As a putative HLP, HP119 may have multiple functions that confer stress resistance and contribute to survival and persistence in the host. To investigate if it has putative regulatory functions in augmenting oxidative stress resistance, we performed a proteomic study wherein the Δhp119 mutant was compared to the WT strain under oxidative stress conditions. Cells of each strain were grown to late log phase under the 4% O2 condition and then exposed to air (20% O2) for 4 h. Under the stress condition (air exposure for 4 h), H. pylori cells stop growing, and they change protein expressions in order to survive (8, 29, 30). The protein profiles of each strain were analyzed by 2D PAGE. Most protein spots fell in the pH range of 4 to 7, and there was a small number of extremely acidic or basic proteins that were excluded from the analysis. The 2D PAGE (pH 4 to 7) was repeated once in an independent experiment, and a similar profile of proteins was obtained. A representative image is shown in Fig. 4. Approximately 200 protein spots were well resolved, and about half of them displayed yellow color (similar expression in both strains). From one gel, we picked 27 green spots (high expression in the WT) and 29 red spots (high expression in the Δhp119 mutant) for protein identification by MS analysis. Probably due to the small amount protein or mixed proteins in some spots, the MS failed to identify 23 spots. The identities of the 18 green spots and 15 red spots are listed in Table 4.
FIG 4.
Comparative 2D PAGE protein profiles of H. pylori WT and Δhp119 mutant strains grown under oxygen stress. Proteins of the WT strain were labeled with Cy3 (green), and proteins from Δhp119 mutant strain were labeled with Cy5 (red) (see Materials and Methods).
TABLE 4.
Protein expression differences between WT (26695) and the Δhp119 mutant strain under oxygen stressa
| TIGR ORF | Protein identification | Fold change (mean) | SD |
|---|---|---|---|
| HP1195 | Elongation factor, EF-G, isoform 1 | −1.75 | 0.16 |
| HP0109 | Molecular chaperone, DnaK, isoform 1 | −3.03 | 0.25 |
| HP0072 | Urease subunit B, UreB, isoform 1 | −3.32 | 0.43 |
| HP0072 | Urease subunit B, UreB, isoform 2 | −2.47 | 0.86 |
| HP0010 | Molecular chaperone, GroEL, isoform 1 | −3.15 | 0.04 |
| HP0010 | Molecular chaperone, GroEL, isoform 2 | −2.40 | 0.13 |
| HP0840 | Flagellin subunit A, FlaA | −13.82 | 4.28 |
| HP0795 | Trigger factor | −5.55 | 2.07 |
| HP1293 | RNA polymerase, RpoA, isoform 1 | −1.88 | 0.18 |
| HP1555 | Elongation factor, EF-Ts | −5.89 | 0.67 |
| HP0310 | Peptidoglycan deacetylase, PgdA, isoform 1 | −66.30 | 8.20 |
| HP0170 | Hypothetical protein | −20.38 | 2.46 |
| HP0825 | Thioredoxin reductase, TrxR isoform 1 | −3.37 | 0.29 |
| HP0068 | Urease subunit G, UreG | −3.47 | 0.38 |
| HP0691 | 3-Oxoadipate coenzyme A-transferase subunit A | −8.24 | 1.25 |
| HP0389 | Superoxide dismutase, SodB | −5.96 | 1.04 |
| HP0390 | Thiol peroxidase, TagD (Tpx) | −19.40 | 6.32 |
| HP0243 | Neutrophil-activating protein, NapA isoform 1 | −9.23 | 2.46 |
| HP0322 | Poly-E-rich protein | +7.32 | 1.07 |
| HP1195 | Elongation factor, EF-G, isoform 2 | +1.86 | 0.06 |
| HP0109 | Molecular chaperone, DnaK, isoform 2 | +5.24 | 0.38 |
| HP0010 | Molecular chaperone, GroEL, isoform 3 | +14.14 | 7.11 |
| HP0828 | ATP synthase F0F1 subunit A, AtpA | +30.8 | 4.60 |
| HP0512 | Glutamine synthase, GlnA | +2.11 | 0.02 |
| HP1205 | Elongation factor, EF-Tu | +2.17 | 0.45 |
| HP0310 | Peptidoglycan deacetylase, PgdA, isoform 2 | +2.10 | 0.07 |
| HP0154 | Enolace | +2.68 | 0.25 |
| HP1293 | RNA polymerase, RpoA, isoform 2 | +18.49 | 9.82 |
| HP1563 | Alkyl hydroperoxide reductase, AhpC | +4.91 | 0.59 |
| HP0825 | Thioredoxin reductase, TrxR, isoform 2 | +3.92 | 0.56 |
| HP1588 | Hypothetical protein | +4.26 | 0.59 |
| HP0697 | Hypothetical protein | +34.70 | 5.46 |
| HP0243 | Neutrophil-activating protein, NapA isoform 2 | +5.43 | 0.20 |
TIGR ORF (open reading frame) number refers to H. pylori 26695 genome sequence. Fold change represents the density of each protein spot derived from Δhp119 mutant (red) in comparison to that from the WT (green). Data are means from two independent experiments with standard deviations.
Table 4 shows that several proteins involved in antioxidative stress (8) were differently expressed between the two strains. Particularly, superoxide dismutase (SodB) and a thiol peroxidase (Tpx) were greatly decreased in the Δhp119 mutant, while the expression of alkyl hydroperoxide reductase (AhpC) was increased. Some antioxidative stress proteins, such as thioredoxin reductase (TrxR) and neutrophil-activating protein (NapA), are present in different isoforms. These isoforms have similar molecular masses but different isoelectric points, and they were differently expressed between the Δhp119 mutant and the WT strain. Strikingly, an oxidative stress induced peptidoglycan deacetylase (PgdA) was greatly decreased in the Δhp119 mutant compared to the WT. In addition, urease subunits UreB and UreG, flagellin subunit FlaA, elongation factor EF-Ts, 3-oxoadipate CoA-transferase subunit A, HP0795 annotated as a trigger factor, and hypothetical protein HP0170 were also expressed at a significantly lower level in the Δhp119 mutant than in the WT strain, whereas a poly-E-rich protein, molecular chaperone GroEL, ATP synthase F0F1 subunit, RNA polymerase RpoA, and hypothetical protein HP0697 were greatly increased in the Δhp119 mutant compared to the WT strain. The comparative proteomic results suggest that HP119 is involved in expression of a variety of oxidative stress-combating responses.
DISCUSSION
E. coli has many histone-like proteins (HLPs), such as HU, IHF, H-NS, and Fis; these proteins play important roles in DNA organization, in modulating DNA replication, recombination, repair, and in regulating gene expression (16, 17). From analyzing H. pylori genome sequences, only one HU-like protein, Hup, was annotated as a histone-like protein. In a bacterium harboring multiple nucleoid-associated proteins (NAPs), these NAPs usually have overlapping and complementary functions. For example, deletion of HU from the E. coli genome is not lethal unless IHF and H-NS are deleted as well (31). In contrast, the disruption of HU is lethal for organisms in which it is the only NAP available (23, 32, 33). For H. pylori, we can easily obtain hup mutant strains which show normal growth at the exponential growth phase in vitro (15). This supports the idea that other histone-like proteins may exist in H. pylori. In this study, we identified HP119 protein as a novel histone-like protein and investigated its physiological roles. We observed significant phenotypes of the Δhp119 mutant strain in stress resistance and host colonization. All other HP119-related proteins appear not to complement the function of HP119, suggesting that their functions are not redundant, although the proteins may have some overlapping functions.
Histone-like proteins have common characteristics, including small size (∼10 kDa), basic nature (pI ∼ 10), cellular abundance, and sequence-independent DNA binding capacity. It is known that lysine residues are common in histone-like proteins and are important for DNA binding. Based on the high values of pI (∼10) and the extremely high percentage of KEQ residues in HP119-related proteins (Table 1), we hypothesized that HP119-related proteins may function as HLPs. The high abundance of Q residues in HP119-related proteins resembles that in E. coli Fis, which contains 10% Q residues. Fis was originally discovered as an E. coli protein essential for the action of a bacteriophage-encoded site-specific recombinase (34). However, Fis is now known to play important phage-independent functions and affect multiple processes, including replication, recombination, and transcription (35, 36). Although HP119-related proteins have a molecular mass of 40 to 50 kDa, they contain repeated sequences that could form separate functional domains, each equivalent to an HLP.
In this study, we obtained evidence that HP119 protein is able to bind DNA. This seems to occur randomly, as non-H. pylori was the source of DNA, and more shifting was observed at the higher concentrations tested. The protein can bind to both supercoiled and linear DNAs. All known HLPs are associated with genomic DNA with low affinity due to their abundance (16, 18, 37), playing a role in nucleoid compaction. However, different HLPs may have different specific functions, like the multiple HLPs found in E. coli, playing roles in DNA recombination or repair or in regulation of transcription. For these specific roles, HLPs bind to specific DNA substrates with a high affinity. For example, E. coli Fis and H-NS can bind specifically at the dps gene promoter, downregulating Dps expression in exponentially growing cells (38). H. pylori Hup was shown to be involved in DNA recombinational repair (15), and it has a high preference for binding four-way DNA junctions (recombination intermediates) (39). The ability of HP119 to bind (with high affinity) to special DNA structures (including the recombination intermediates and lesion-containing DNA) or to specific DNA sequence motifs (e.g., promoters of specific genes) needs to be determined.
Oxidative stress is a major stress condition that H. pylori encounters in its physiological niche. H. pylori induces strong host inflammatory responses that involve recruitment of neutrophils, lymphocytes, and macrophages; these immune cells release reactive oxygen species that damage DNA. Previous studies demonstrated that oxidative stress causes damage to H. pylori genomic DNA (30, 40, 41). Further studies showed that mutant cells of ruvC (14), mutS (42), mutY (43), recN (11), addA (recB) (12, 13), or recRO (10) were more sensitive to oxidative stress, indicating important roles of DNA recombination and repair in H. pylori for the bacterial survival of oxidative damage. Our recent study on Hup, a known HLP in H. pylori, also indicated a role in oxidative stress resistance (15). The current study showed that the Δhp119 mutant displays a similar phenotype of oxygen sensitivity.
H. pylori survives in and colonizes an acidic niche on the gastric surface (7). Therefore, low pH is another stress condition that H. pylori encounters in its physiological niche. Despite the existence of sophisticated pH homeostasis systems and acid tolerance mechanisms, bacteria may still suffer DNA damage from acid stress. Previously, we showed that H. pylori DNA recombination proteins (RecN and RecRO) and the histone-like protein Hup are involved in acid stress resistance (10, 11, 15). In this study, we characterized the Δhp119 mutant for its sensitivity to low-pH conditions, and the results indicated that HP119 plays a similar role in acid stress resistance. Interestingly, HP119 transcription was shown to be regulated by the two-component system ArsRS (HP166-HP165), although functions of HP119 were completely unknown (44). The response regulator ArsR (through its phosphorylation) controls the transcription of a set of target genes, and the cognate histidine kinase ArsS senses the environmental stimulus (45). Subsequently it was shown that acidic pH is the stimulus triggering the autophosphorylation of the histidine kinase ArsS; thus, the ArsRS system controls expression of certain genes, including hp119, in response to low pH (46). In this work, we identified HP119 as a putative HLP that plays a role in resistance to acid stress as well as to oxidative stress.
Like many known HLPs, HP119 protein may have multiple functions in stress resistance. It may have an ability to physically protect DNA via nonspecific DNA binding. Notably, a Mycobacterium tuberculosis HLP (Lsr2) was shown to be able to protect DNA against reactive oxygen intermediates (47). A similar function was shown for H. pylori Hup protein (15). The possible functions of HP119 for direct protection of DNA from oxidative and acid stress damage need to be determined. As an alternative function, HP119 may regulate expression of other genes involved in stress resistance. In E. coli, HU influences the expression of genes involved in anaerobic respiration, the responses to osmotic stress and to acid stress, and the response to DNA damage (48–50). On the basis of modeling DNA topology, E. coli HU is known to play a role in modulation of transcription profiles which has important impacts on cellular physiology (28, 48). Salmonella enterica HU controls three regulons that coordinate virulence, response to stress, and general physiology (51). In this study, we performed a proteomic study showing that HP119 is involved in increasing enzyme levels that aid H. pylori survival. Particularly, several important antioxidant proteins, such as SodB, Tpx, TrxR, and NapA, were less expressed in the Δhp119 mutant than in the WT strain. Strikingly, oxidative stress induces a peptidoglycan modification enzyme (PgdA) in WT H. pylori (52), and the aconitase (AcnB) was shown to be involved in this regulation at the posttranscriptional level (53). The results in Table 4 show that the expression of PgdA was greatly decreased, although another isoform of PgdA was slightly increased, in the Δhp119 mutant compared to the WT, suggesting that HP119 may also be involved in the regulation of oxidative stress induction of PgdA. Interestingly, certain virulence factors, such as urease and flagellin proteins, were also differently expressed between the Δhp119 mutant and the WT strain. The molecular mechanisms impacting these proteins expression by HP119 await further investigation. As a putative HLP, HP119 may have effects on global gene expression, either directly by interacting with DNA at promoter regions or indirectly through its effects on expression of other transcription regulators. It is also possible that oxygen activation of proteases led to selected protein turnover, partly explaining our proteomic results.
H. pylori infection induces a strong inflammatory response by the host, with the recruitment of lymphocytes, macrophages, and polymorphonuclear cells; however, the bacterium is often able to resist this immune response and establish a persistent gastric infection. Although H. pylori is not invasive, it can be efficiently ingested by the different types of phagocytic cells, it is able to survive for prolonged periods within these cells, and it presumably is able to resist damage by free radicals derived from the phagocytic respiratory burst (25, 54). The mechanisms known to contribute to survival within macrophages include enzymatic detoxification of reactive oxygen species and DNA repair (14, 24). In this study, we showed that Δhp119 mutant H. pylori survived significantly less well than the wild-type strain; a similar phenotype was observed for the H. pylori hup mutant (15). The observed effects on macrophage survival could be ascribed either to direct protection of DNA from stress damage or to HLP-mediated gene regulation in stress response or to both mechanisms.
In agreement with its in vitro phenotypes (sensitivity to oxidative stress and acid stress and the survival defect within macrophages), the Δhp119 mutant strain showed an attenuated ability to colonize mouse stomachs. The mean colonization numbers from mouse stomachs were 158-fold less for the mutant strain than for the WT strain. This attenuation effect is more severe than that observed for the hup mutant as well as for other mutants defective in DNA recombination and repair functions. For example, the mean colonization numbers from mouse stomachs were decreased (compared to the WT) 9-, 47-, 38-, and 40-fold, respectively, for recN, recB, recR, and hup mutant strains (10–12, 15). This study highlights the role of the novel HLP in H. pylori survival and persistence in the host. As HLPs have multiple functions in DNA organization, in modulating DNA replication, recombination, and repair, and in regulating gene expression, the mouse colonization results would be the combined effects due to loss of a subset of these functions. Little is known about HLPs and their relevance to pathogen virulence at all, although an HLP from Mycobacterium leprae was implicated in adhesion to mouse epithelial cells (55) and the H. pylori Hup protein expression level was shown to be associated with gastric carcinogenesis (56). Identifying a colonization role for a novel HLP in H. pylori connects its function to stomach persistence and to pathogenesis.
ACKNOWLEDGMENTS
This work was supported by The University of Georgia Research Foundation.
We thank Sue Maier for her expertise and assistance on mouse colonization assays. We are grateful to Hyuk-Kyu Seoh at Georgia State University for support on 2D PAGE and mass spectrometric protein identification.
REFERENCES
- 1.Dunn BE, Cohen H, Blaser MJ. 1997. Helicobacter pylori. Clin Microbiol Rev 10:720–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusters JG, van Vliet AH, Kuipers EJ. 2006. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suerbaum S, Michetti P. 2002. Helicobacter pylori infection. N Engl J Med 347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 4.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. 2001. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 5.McGee DJ, Mobley HL. 1999. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr Top Microbiol Immunol 241:155–180. [DOI] [PubMed] [Google Scholar]
- 6.Ding SZ, Minohara Y, Fan XJ, Wang J, Reyes VE, Patel J, Dirden-Kramer B, Boldogh I, Ernst PB, Crowe SE. 2007. Helicobacter pylori infection induces oxidative stress and programmed cell death in human gastric epithelial cells. Infect Immun 75:4030–4039. doi: 10.1128/IAI.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. 2007. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc Natl Acad Sci U S A 104:7235–7240. doi: 10.1073/pnas.0702300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, Alamuri P, Maier RJ. 2006. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol 61:847–860. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- 9.Pflock M, Kennard S, Finsterer N, Beier D. 2006. Acid-responsive gene regulation in the human pathogen Helicobacter pylori. J Biotechnol 126:52–60. doi: 10.1016/j.jbiotec.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Lo LF, Maier RJ. 2011. The RecRO pathway of DNA recombinational repair in Helicobacter pylori and its role in bacterial survival in the host. DNA Repair (Amst) 10:373–379. doi: 10.1016/j.dnarep.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Maier RJ. 2008. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori. Infect Immun 76:153–160. doi: 10.1128/IAI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Maier RJ. 2009. A RecB-like helicase in Helicobacter pylori is important for DNA repair and host colonization. Infect Immun 77:286–291. doi: 10.1128/IAI.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. 2008. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol 69:994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loughlin MF, Barnard FM, Jenkins D, Sharples GJ, Jenks PJ. 2003. Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa. Infect Immun 71:2022–2031. doi: 10.1128/IAI.71.4.2022-2031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, Lo LF, Maier RJ. 2012. A histone-like protein of Helicobacter pylori protects DNA from stress damage and aids host colonization. DNA Repair (Amst) 11:733–740. doi: 10.1016/j.dnarep.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dame RT. 2005. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol 56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 17.Azam TA, Ishihama A. 1999. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem 274:33105–33113. [DOI] [PubMed] [Google Scholar]
- 18.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 19.Chalker AF, Minehart HW, Hughes NJ, Koretke KK, Lonetto MA, Brinkman KK, Warren PV, Lupas A, Stanhope MJ, Brown JR, Hoffman PS. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J Bacteriol 183:1259–1268. doi: 10.1128/JB.183.4.1259-1268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Taylor DE. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 21.Olczak AA, Olson JW, Maier RJ. 2002. Oxidative-stress resistance mutants of Helicobacter pylori. J Bacteriol 184:3186–3193. doi: 10.1128/JB.184.12.3186-3193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seyler RW Jr, Olson JW, Maier RJ. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun 69:4034–4040. doi: 10.1128/IAI.69.6.4034-4040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, Yumoto H, Murakami K, Hirota K, Ono T, Nagamune H, Kayama S, Matsuo T, Miyake Y. 2008. The essentiality and involvement of Streptococcus intermedius histone-like DNA-binding protein in bacterial viability and normal growth. Mol Microbiol 68:1268–1282. doi: 10.1111/j.1365-2958.2008.06232.x. [DOI] [PubMed] [Google Scholar]
- 24.Basu M, Czinn SJ, Blanchard TG. 2004. Absence of catalase reduces long-term survival of Helicobacter pylori in macrophage phagosomes. Helicobacter 9:211–216. doi: 10.1111/j.1083-4389.2004.00226.x. [DOI] [PubMed] [Google Scholar]
- 25.Odenbreit S, Gebert B, Puls J, Fischer W, Haas R. 2001. Interaction of Helicobacter pylori with professional phagocytes: role of the cag pathogenicity island and translocation, phosphorylation and processing of CagA. Cell Microbiol 3:21–31. doi: 10.1046/j.1462-5822.2001.00088.x. [DOI] [PubMed] [Google Scholar]
- 26.Salama NR, Shepherd B, Falkow S. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol 186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomb J-F, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 28.Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S. 2006. Right-handed DNA supercoiling by an octameric form of histone-like protein HU: modulation of cellular transcription. J Biol Chem 281:40144–40153. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- 29.Olczak AA, Wang G, Maier RJ. 2005. Up-expression of NapA and other oxidative stress proteins is a compensatory response to loss of major Helicobacter pylori stress resistance factors. Free Radic Res 39:1173–1182. doi: 10.1080/10715760500306729. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Conover RC, Olczak AA, Alamuri P, Johnson MK, Maier RJ. 2005. Oxidative stress defense mechanisms to counter iron-promoted DNA damage in Helicobacter pylori. Free Radic Res 39:1183–1191. doi: 10.1080/10715760500194018. [DOI] [PubMed] [Google Scholar]
- 31.Yasuzawa K, Hayashi N, Goshima N, Kohno K, Imamoto F, Kano Y. 1992. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene 122:9–15. doi: 10.1016/0378-1119(92)90026-L. [DOI] [PubMed] [Google Scholar]
- 32.Micka B, Marahiel MA. 1992. The DNA-binding protein HBsu is essential for normal growth and development in Bacillus subtilis. Biochimie 74:641–650. doi: 10.1016/0300-9084(92)90136-3. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen HH, de la Tour CB, Toueille M, Vannier F, Sommer S, Servant P. 2009. The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Mol Microbiol 73:240–252. doi: 10.1111/j.1365-2958.2009.06766.x. [DOI] [PubMed] [Google Scholar]
- 34.Koch C, Kahmann R. 1986. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem 261:15673–15678. [PubMed] [Google Scholar]
- 35.Travers A, Schneider R, Muskhelishvili G. 2001. DNA supercoiling and transcription in Escherichia coli: the FIS connection. Biochimie 83:213–217. doi: 10.1016/S0300-9084(00)01217-7. [DOI] [PubMed] [Google Scholar]
- 36.Finkel SE, Johnson RC. 1992. The Fis protein: it's not just for DNA inversion anymore. Mol Microbiol 6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 37.Grove A. 2011. Functional evolution of bacterial histone-like HU proteins. Curr Issues Mol Biol 13:1–12. [PubMed] [Google Scholar]
- 38.Grainger DC, Goldberg MD, Lee DJ, Busby SJ. 2008. Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol Microbiol 68:1366–1377. doi: 10.1111/j.1365-2958.2008.06253.x. [DOI] [PubMed] [Google Scholar]
- 39.Chen C, Ghosh S, Grove A. 2004. Substrate specificity of Helicobacter pylori histone-like HU protein is determined by insufficient stabilization of DNA flexure points. Biochem J 383:343–351. doi: 10.1042/BJ20040938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park AM, Li Q, Nagata K, Tamura T, Shimono K, Sato EF, Inoue M. 2004. Oxygen tension regulates reactive oxygen generation and mutation of Helicobacter pylori. Free Radic Biol Med 36:1126–1133. doi: 10.1016/j.freeradbiomed.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 41.O'Rourke EJ, Chevalier C, Pinto AV, Thiberge JM, Ielpi L, Labigne A, Radicella JP. 2003. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc Natl Acad Sci U S A 100:2789–2794. doi: 10.1073/pnas.0337641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, Alamuri P, Humayun MZ, Taylor DE, Maier RJ. 2005. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol Microbiol 58:166–176. doi: 10.1111/j.1365-2958.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- 43.Eutsey R, Wang G, Maier RJ. 2007. Role of a MutY DNA glycosylase in combating oxidative DNA damage in Helicobacter pylori. DNA Repair (Amst) 6:19–26. doi: 10.1016/j.dnarep.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dietz P, Gerlach G, Beier D. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J Bacteriol 184:350–362. doi: 10.1128/JB.184.2.350-362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beier D, Frank R. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J Bacteriol 182:2068–2076. doi: 10.1128/JB.182.8.2068-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pflock M, Dietz P, Schar J, Beier D. 2004. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol Lett 234:51–61. doi: 10.1111/j.1574-6968.2004.tb09512.x. [DOI] [PubMed] [Google Scholar]
- 47.Colangeli R, Haq A, Arcus VL, Summers E, Magliozzo RS, McBride A, Mitra AK, Radjainia M, Khajo A, Jacobs WR Jr, Salgamea P, Alland D. 2009. The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc Natl Acad Sci U S A 106:4414–4418. doi: 10.1073/pnas.0810126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kar S, Edgar R, Adhya S. 2005. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc Natl Acad Sci U S A 102:16397–16402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oberto J, Nabti S, Jooste V, Mignot H, Rouviere-Yaniv J. 2009. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One 4:e4367. doi: 10.1371/journal.pone.0004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bi H, Sun L, Fukamachi T, Saito H, Kobayashi H. 2009. HU participates in expression of a specific set of genes required for growth and survival at acidic pH in Escherichia coli. Curr Microbiol 58:443–448. doi: 10.1007/s00284-008-9340-4. [DOI] [PubMed] [Google Scholar]
- 51.Mangan MW, Lucchini S, Fitzgerald TOCS, Hinton JC, Dorman CJ. 2011. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiology 157:1075–1087. doi: 10.1099/mic.0.046359-0. [DOI] [PubMed] [Google Scholar]
- 52.Wang G, Olczak A, Forsberg LS, Maier RJ. 2009. Oxidative stress-induced peptidoglycan deacetylase in Helicobacter pylori. J Biol Chem 284:6790–6800. doi: 10.1074/jbc.M808071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Austin CM, Maier RJ. 2013. Aconitase-mediated posttranscriptional regulation of Helicobacter pylori peptidoglycan deacetylase. J Bacteriol 195:5316–5322. doi: 10.1128/JB.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramarao N, Gray-Owen SD, Meyer TF. 2000. Helicobacter pylori induces but survives the extracellular release of oxygen radicals from professional phagocytes using its catalase activity. Mol Microbiol 38:103–113. doi: 10.1046/j.1365-2958.2000.02114.x. [DOI] [PubMed] [Google Scholar]
- 55.Silva CA, Danelishvili L, McNamara M, Berredo-Pinho M, Bildfell R, Biet F, Rodrigues LS, Oliveira AV, Bermudez LE, Pessolani MC. 2013. Interaction of Mycobacterium leprae with human airway epithelial cells: adherence, entry, survival, and identification of potential adhesins by surface proteome analysis. Infect Immun 81:2645–2659. doi: 10.1128/IAI.00147-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khoder G, Yamaoka Y, Fauchere JL, Burucoa C, Atanassov C. 2009. Proteomic Helicobacter pylori biomarkers discriminating between duodenal ulcer and gastric cancer. J Chromatogr B Analyt Technol Biomed Life Sci 877:1193–1199. doi: 10.1016/j.jchromb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]