Abstract
The diversity of enterococcal populations from fecal samples from hospitalized (n = 133) and nonhospitalized individuals (n = 173) of different age groups (group I, ages 0 to 19 years; group II, ages 20 to 59 years; group III, ages ≥60 years) was analyzed. Enterococci were recovered at similar rates from hospitalized and nonhospitalized persons (77.44% to 79.77%) of all age groups (75.0% to 82.61%). Enterococcus faecalis and Enterococcus faecium were predominant, although seven other Enterococcus species were identified. E. faecalis and E. faecium (including ampicillin-resistant E. faecium) colonization rates in nonhospitalized persons were age independent. For inpatients, E. faecalis colonization rates were age independent, but E. faecium colonization rates (particularly the rates of ampicillin-resistant E. faecium colonization) significantly increased with age. The population structure of E. faecium and E. faecalis was determined by superimposing goeBURST and Bayesian analysis of the population structure (BAPS). Most E. faecium sequence types (STs; 150 isolates belonging to 75 STs) were linked to BAPS groups 1 (22.0%), 2 (31.3%), and 3 (36.7%). A positive association between hospital isolates and BAPS subgroups 2.1a and 3.3a (which included major ampicillin-resistant E. faecium human lineages) and between community-based ampicillin-resistant E. faecium isolates and BAPS subgroups 1.2 and 3.3b was found. Most E. faecalis isolates (130 isolates belonging to 58 STs) were grouped into 3 BAPS groups, BAPS groups 1 (36.9%), 2 (40.0%), and 3 (23.1%), with each one comprising widespread lineages. No positive associations with age or hospitalization were established. The diversity and dynamics of enterococcal populations in the fecal microbiota of healthy humans are largely unexplored, with the available knowledge being fragmented and contradictory. The study offers a novel and comprehensive analysis of enterococcal population landscapes and suggests that E. faecium populations from hospitalized patients and from community-based individuals differ, with a predominance of certain clonal lineages, often in association with elderly individuals, occurring in the hospital setting.
INTRODUCTION
Enterococci are relatively minor constituents of the human gastrointestinal microbiota (less than 1%) but are able to cause a wide diversity of infections, mostly in patients with underlying diseases (1, 2). High-density colonization by antibiotic-resistant enterococci increases the risk of bacteremia and transmission; however, the population structure and ecological and evolutionary forces influencing the population dynamics of gut colonizers largely remain unknown (3–5). Next-generation sequencing has provided a wealth of data about the influence of characteristics of the host (age, diet, health status, and antibiotic treatment) on the diversity and population frequency of different bacterial groups, including enterococci (6–10). However, the information provided by current metagenomic analysis, based on 16S rRNA (11, 12), or by the traditional culture-based studies (1, 13, 14) precludes any possible analysis of enterococci at the subspecies level. Furthermore, the available information about the frequency and diversity of enterococcal species in the fecal microbiota by host age is fragmented and contradictory (1, 15).
Different methods based on multilocus sequence typing (MLST), comparative genomic hybridization, and whole-genome sequencing revealed intraspecies diversity for Enterococcus faecalis and Enterococcus faecium, which are the predominant enterococcal species colonizing the human gastrointestinal tract (16–25). E. faecium has a population structure that has split into two major phylogenomic clusters, designated clade B, which includes community-based human isolates, and clade A, which comprises isolates from humans and animals, with a clade A1 being enriched with isolates from hospitalized patients. Specifically, strains belonging to the sequence type (ST) 17 (ST17), ST18, and ST78 lineages, which are found within clade A1, are often resistant to antibiotics and are the most frequently associated with the hospital environment (22, 26, 27). E. faecalis, on the other hand, seems to lack such a clear clade structure, probably because this species occupies a larger variety of ecological microniches, thus having access to a more heterogeneous spectrum of alleles than E. faecium (28). As a result, no clear genotypic differences are observed between hospital and community isolates (25, 28, 29), even though some clonal complexes (CCs) are more prevalent either among hospitalized patients, e.g., CC6-ST6, CC9-ST9, CC28-ST87, and CC40-ST40, or among community healthy volunteers, e.g., ST16 and CC58 (30–33). Recombination, which was previously detected in enterococci (17, 34, 35), may have a considerable impact on patterns of evolutionary descent, as displayed by sequence-based gene trees or even by popular allele-based population snapshots provided by eBURST analysis. This may obscure the genetic relatedness of strains and clones and, as such, interfere with epidemiological and clinical investigations, in particular, when strains are assigned to specific CCs. In addition, knowledge about the population structure of enterococcal species is biased by an overrepresentation of contemporary multidrug-resistant (MDR) clinical isolates belonging to a few high-risk clonal complexes often associated with nosocomial outbreaks and frequently associated with elderly individuals (36–38). Studies analyzing early isolates have documented a more diverse enterococcal population able to cause disease acquired either nosocomially or in the community and often associated with nonelderly adults and children. Isolates causing infections or colonizing these populations have less frequently been analyzed at the molecular level (33, 39–41).
The objective of this study was to assess for the first time the population structure of enterococci in the feces of both hospitalized and nonhospitalized individuals within different age groups. In addition, Bayesian analysis of the population structure (BAPS), a nonphylogenetic method able to find the best partition of a set of isolates into subpopulations, was applied, broadening former results obtained for E. faecium and providing the first analysis to probabilistically assign E. faecalis strains to evolutionary groups.
MATERIALS AND METHODS
Bacterial samples.
Three hundred six fecal samples were collected between April 2009 and April 2011 at the Ramón y Cajal University Hospital (HRyC) and its community care area of influence. HRyC is a tertiary care public hospital with 1,155 beds that provides specialized attention to a population of about 600,000 habitants in the northern area of Madrid, Spain, who primarily attend 20 primary health centers (PHCs) of the Madrid Health Service (SERMAS). This study was conducted according to applicable government regulations and approved studies by institutional research policies (e.g., reference CEIC-106/09 [A. M. Sánchez-Díaz, C. Cuartero, J. D. Rodríguez, S. Lozano, J. M. Alonso, M. J. Rodríguez-Domínguez, A. P. Tedim, R. del Campo, J. López, R. Cantón, and P. Ruiz-Garbajosa, unpublished data]).
The samples analyzed were recovered from 173 patients with nonsevere diseases that attended a PHC or had a consultation at HRyC (and for whom no hospitalization was registered in the 6 months prior to sample collection) and from 133 hospitalized patients admitted to HRyC. The fecal samples were submitted to HRyC for stool culture with or without a specific request for Clostridium difficile or parasite detection and were anonymously processed so that the patients' demographic information was kept confidential. Hospitalized patients were mostly located in medical wards (78.2%), surgical wards (8.3%), and intensive care units (ICUs; 9.8%). All but 20 samples from hospitalized patients were collected after more than 48 h of hospital admission. However, these 20 patients had a history of several recent previous hospitalizations (see Tables S1 and S2 in the supplemental material).
Samples were also classified into three age groups according to the host's age. These three groups are designated with roman numerals as group I (young people 0 to 19 years old, n = 92 [30%], 57 nonhospitalized persons and 35 hospitalized patients), group II (adults 20 to 59 years old, n = 108 [35%], 62 nonhospitalized persons and 46 hospitalized patients), and group III (elderly individuals ≥60 years old, n = 106 [35%], 54 nonhospitalized persons and 52 hospitalized patients). Only one sample per patient was analyzed (see Tables S1 and S2 in the supplemental material).
Sample processing.
About 0.5 g of each fecal sample was suspended in 1 ml of saline solution, plated on plain m-Enterococcus agar (Difco, Detroit, MI, USA) or m-Enterococcus agar supplemented with either ampicillin (10 μg/ml) or vancomycin (6 μg/ml), and incubated for 48 h at 37°C. For each sample, one colony per morphology type and plate was selected (28) for further studies. In order to enhance the recovery of minority populations of ampicillin-resistant enterococci and vancomycin-resistant enterococci (VRE), 0.1 ml of the original suspension of each sample was preenriched in brain heart infusion (BHI) broth (Difco, Detroit, MI, USA) supplemented with 2 μg/ml of ampicillin or 2 μg/ml of vancomycin, incubated for 24 h at 37°C, and subsequently plated on m-Enterococcus agar (Difco, Detroit, MI, USA) containing ampicillin (10 μg/ml) or vancomycin (6 μg/ml), respectively.
Identification, antibiotic susceptibility, and virulence traits.
Bacterial identification was performed by the amplification of species-specific genes, d-alanine–d-alanine ligase (ddl) for E. faecalis and aac(6′)-Ii for E. faecium, as previously described (42, 43), and by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker, Daltonics, Bremen, Germany). Susceptibility to ampicillin, vancomycin, teicoplanin, streptomycin, gentamicin, ciprofloxacin, levofloxacin, erythromycin, tetracycline, and chloramphenicol (Oxoid, Basingstoke, United Kingdom) was determined by the disc diffusion method according to CLSI guidelines (44).
The presence of putative virulence genes encoding the E. faecium enterococcal surface protein (esp), glycosyl hydrolase (hylE. faecium), and collagen-binding adhesin (acm) and the E. faecalis enterococcal surface protein (esp), hyaluronidase (hylE. faecalis), cytolysin/hemolysin (cylA), gelatinase (gelE), and aggregation substance (asa1) was investigated by PCR and sequencing, as described before (45, 46).
Clonal relatedness.
The clonal relationship among isolates of each enterococcal species was established by pulsed-field gel electrophoresis (PFGE) and MLST as previously described (16, 47), and the relationships are detailed in Tables S1 and S2 in the supplemental material. Clusters of related STs for E. faecalis (differing in no more than two of the seven MLST loci) were considered to belong to the same CC using the goeBURST algorithm (48, 49). CCs were defined on the basis of goeBURST analysis of the 524 STs present in the E. faecalis MLST database (http://efaecalis.mlst.net/).
Analysis of population structure.
A BAPS software was used to probabilistically assign E. faecalis and E. faecium STs to nonoverlapping evolutionary groups (27, 50). BAPS clustering was performed with the second-order Markov model and the standard MLST data input option in a hierarchical manner. For E. faecium, the major clusters identified at the first stage were reanalyzed after excluding the remaining data. The rationale for this approach is to increase the statistical power to detect the more fine-scale genetic structure of a population when analyzing particular lineages separately from the remaining population. In all BAPS analyses, 10 runs of the estimation algorithm were performed using a priori upper bounds (10 to 30 for the major group analysis and 2 to 10 for the subgroup analysis) for the number of clusters over the interval, and in each case the runs converged to a nearly identical partition of the data in question, indicating a high level of peakedness of the posterior distribution (estimated P = 1.000).
The accuracy of BAPS for establishing the E. faecium population structure was determined using different sample sizes and discarding the inclusion of E. faecalis as the outgroup (see Fig. S1 to S3 in the supplemental material) (27). Correlation analysis was performed for each of the comparisons mentioned above using Microsoft Excel 2010 software. This study constitutes the first application of BAPS to investigate E. faecalis population and evolutionary genetics by the same approach that was previously used for E. faecium (27).
Statistical analysis.
The statistical significance of the results was calculated by the chi-square test; P values of <0.05 were considered statically significant.
The STATA generalized estimating equations (GEE) model (which takes into account clone-related data) (51) was used for calculating odd ratios (ORs) and 95% confidence intervals (CIs) related to the colonization isolates. The analyses were done by comparison with major BAPS subgroup 3.3a (BAPS 3.3a) for E. faecium and BAPS group 1 (BAPS 1) for E. faecalis.
For the analysis of all isolates available in MLST databases, ORs between BAPS groups and different sources (hospitalized patients, nonhospitalized persons, and animals) were calculated. Environmental, food, and other sources were also considered, but due to the low number of isolates in these categories, OR analysis was not performed.
RESULTS
Prevalence and diversity of enterococcal species in human fecal samples.
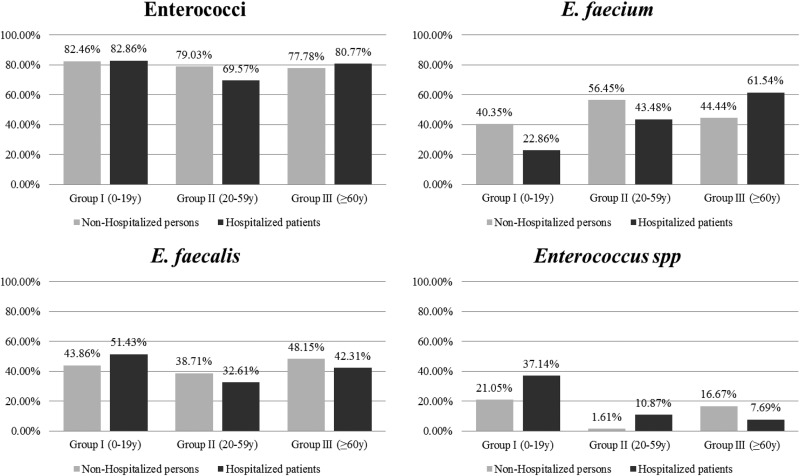
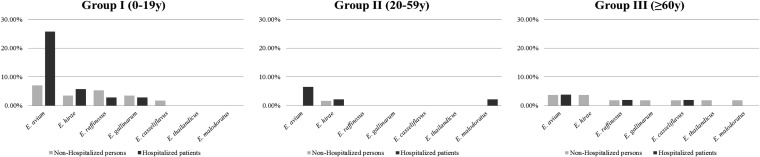
Enterococci were recovered by culture from 78.8% of the individuals analyzed (n = 241/306) and at similar rates between hospitalized and nonhospitalized individuals (77.4% versus 79.8%) and among all age groups (75.0 to 82.6%). The enterococci recovered corresponded to three of the five groups of enterococci previously described by Facklam et al. (52) on the basis of phenotypic and genotypic characteristics, which used to be designated by roman numerals (1). The rate at which individuals were colonized by different species varied in each age group, with E. faecalis and E. faecium being the predominant species identified (Fig. 1 and 2). Among nonhospitalized persons, E. faecalis and E. faecium colonization rates were age independent (E. faecalis/E. faecium ratios, 1.14, 0.71, 1.12 for age groups I, II, and III, respectively). The E. faecalis colonization rate among hospitalized patients was also age independent, but the E. faecium colonization rate and particularly the ampicillin-resistant E. faecium colonization rate significantly (P < 0.01) increased with age (E. faecalis/E. faecium ratios, 1.90, 0.71, and 0.65 for age groups I, II, and III, respectively) (Fig. 1 and 3). Besides E. faecium and E. faecalis, both classified within enterococcal group II described by Facklam et al. (52), other species within enterococcal groups I (20 E. avium, 7 E. raffinosus, 2 E. malodoratus isolates), II (4 E. casseliflavus isolates, 3 E. gallinarum isolates, 1 E. thailandicus isolate), and group III (8 E. hirae isolates) were identified (Fig. 2).
FIG 1.
Proportion of nonhospitalized and hospitalized individuals colonized with enterococci by age group. The E. faecalis/E. faecium colonization ratios in nonhospitalized persons by age group were as follows: group I (0 to 19 years old), 1.08; group II (20 to 59 years old), 0.68; and group III (≥60 years old), 1.08. The E. faecalis/E. faecium colonization ratios in hospitalized patients were as follows: group I (0 to 19 years old), 2.25; group II (20 to 59 years old), 0.75; and group III (≥60 years old), 0.68.
FIG 2.
Proportion of nonhospitalized and hospitalized individuals colonized by different Enterococcus spp. by age group.
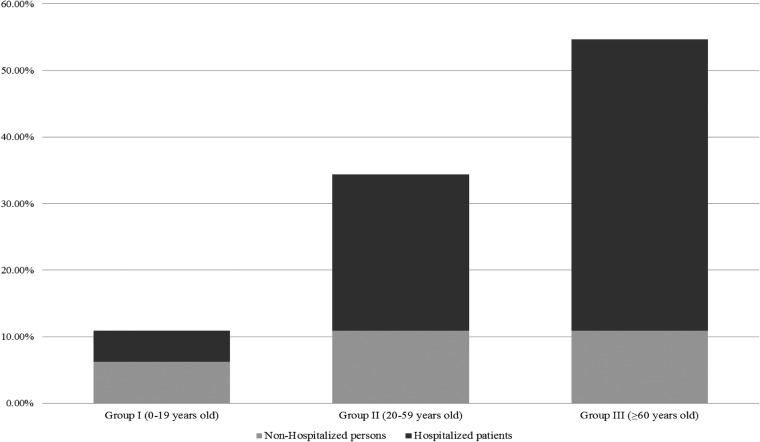
FIG 3.
Proportion of nonhospitalized and hospitalized individuals colonized by ampicillin-resistant E. faecium by age group. The proportions of hospitalized/nonhospitalized individuals colonized by ampicillin-resistant E. faecium were as follows for the different age groups: group I (0 to 19 years old), 0.75; group II (20 to 59 years old), 2.14; and group III (≥60 years old), 4.
Colonization by more than one enterococcal species was a frequent event. The simultaneous recovery of both E. faecalis and E. faecium (13.7%, n = 42/306) was increasingly observed as age increased (P < 0.01) (for age groups I, II, and III, 6.5% [n = 6/92], 13.9% [n = 15/108], and 19.8% [n = 21/106], respectively), suggesting that the increased rate of E. faecium colonization in hospitalized patients in age groups II and III did not interfere with the E. faecalis colonization rate, although it might have influenced the E. faecalis clonal composition. Low rates of cocolonization by E. faecalis and Enterococcus spp. (5.23%, 16/306), E. faecium and Enterococcus spp. (4.25%, 13/306), or E. faecalis, E. faecium, and other enterococcal species (0.65%, 2/306) were also detected.
Ampicillin resistance (22.2%, n = 68/306) was detected among E. faecium (94.1%, n = 64/68) and E. raffinosus (5.9%, n = 4/68) isolates. Ampicillin-resistant E. faecium isolates were more significantly associated with hospitalized patients (44.7%, n = 46/103) than with nonhospitalized individuals (13.0%, n = 18/138). A low number of individuals colonized with VRE, all of which were identified to be vancomycin-resistant E. faecalis isolates, was also detected (1.6%, n = 5/306, consisting of 2 nonhospitalized and 3 hospitalized individuals of different ages) (see Table S2 in the supplemental material).
The population structure of the E. faecium and E. faecalis isolates is detailed below. For other enterococci, isolates of the same species exhibited different PFGE types, with the exception of some E. avium isolates (data not shown). All these species were resistant to quinupristin-dalfopristin, were often resistant to erythromycin (E. avium, E. hirae, E. raffinosus, E. gallinarum, E. casseliflavus) and tetracycline (E. raffinosus), and eventually became resistant to levofloxacin (E. raffinosus, E. gallinarum) and high concentrations of streptomycin (E. avium, E. raffinosus, E. gallinarum) and gentamicin (E. avium, E. raffinosus).
BAPS analysis of E. faecium population structure.
A BAPS analysis was used to infer the population structure of the E. faecium isolates according to previous findings that demonstrated that eBURST analysis is not sufficient to reliably delineate the patterns of recent evolutionary descent of E. faecium (27, 53). The analysis was repeated by taking into account the significant enlargement of the MLST database (http://efaecium.mlst.net/), in which the number of STs increased from 492 to 837 in the 2 years since the time of publication of the original 2012 study (27).
A hierarchical BAPS clustering analysis of the currently available 837 E. faecium STs representing 2,402 isolates of different origins yielded 8 BAPS groups. The majority of STs grouped in BAPS 1, BAPS 2, BAPS 3, and BAPS 7 (15.1%, 39.7%, 31.5%, and 8.5%, respectively), while BAPS 4, BAPS 5, BAPS 6, and BAPS 8 were much more infrequently detected (1.3%, 1.9%, 0.8% and 1.2%, respectively). BAPS nested analysis subdivided BAPS 1 into six subgroups (BAPSs 1.1 to 1.6) and BAPS 2 (BAPSs 2.1a, 2.1b, 2.3a, 2.3b), BAPS 3 (BAPSs 3.1, 3.2, 3.3a, 3.3b), and BAPS 7 (BAPSs 7.1 to 7.4) into four subgroups each (Table 1). The original BAPS subgroups 2.1, 2.3, and 3.3 described by Willems et al. (27) have now been split into two subgroups each, and here these are arbitrarily designated BAPSs 2.1a and 2.1b, BAPSs 2.3a and 2.3b, and BAPSs 3.3a and 3.3b, for backwards compatibility (see Fig. S3 in the supplemental material).
TABLE 1.
E. faecium BAPS analysis data
| BAPS group | BAPS subgroup | No. of STs | % STs | No. of isolates |
|---|---|---|---|---|
| BAPS 1 | 1.1 | 9 | 1.08 | 11 |
| 1.2 | 61 | 7.29 | 100 | |
| 1.3 | 12 | 1.43 | 16 | |
| 1.4 | 2 | 0.24 | 2 | |
| 1.5 | 36 | 4.30 | 41 | |
| 1.6 | 6 | 0.72 | 6 | |
| Subtotal | 126 | 15.05 | 176 | |
| BAPS 2 | 2.1a | 88 | 10.51 | 577 |
| 2.1b | 133 | 15.89 | 321 | |
| 2.3a | 78 | 9.32 | 135 | |
| 2.3b | 33 | 3.94 | 49 | |
| Subtotal | 332 | 39.67 | 1,082 | |
| BAPS 3 | 3.1 | 72 | 8.60 | 122 |
| 3.2 | 28 | 3.35 | 59 | |
| 3.3a | 107 | 12.78 | 679 | |
| 3.3b | 57 | 6.81 | 92 | |
| Subtotal | 264 | 31.54 | 952 | |
| BAPS 4 | 11 | 1.31 | 11 | |
| BAPS 5 | 16 | 1.91 | 19 | |
| BAPS 6 | 7 | 0.84 | 9 | |
| BAPS 7 | 7.1 | 54 | 6.45 | 120 |
| 7.2 | 6 | 0.72 | 6 | |
| 7.3 | 10 | 1.19 | 14 | |
| 7.4 | 1 | 0.12 | 2 | |
| Subtotal | 71 | 8.48 | 142 | |
| BAPS 8 | 10 | 1.19 | 11 | |
| Total | 837 | 2,402 |
Next, we analyzed the congruence between the BAPS grouping of the 492 STs using the BAPS assignment previously described by Willems et al. (27) and the BAPS grouping from this study. A correlation coefficient of 0.5958 indicates some discrepancies between the partitioning of the 492 STs. These discrepancies, probably related to the presence of an E. faecalis outgroup in the BAPS analysis of Willems et al. (27), are mostly due to STs that moved from BAPS 2 (15 STs), BAPS 3 (25 STs), and BAPS 5 (1 ST) in the 2012 study (27) to BAPS 7 in our study (see Fig. S1 in the supplemental material). Subsequently, the 492 E. faecium STs included in the work of Willems et al. (27) were compared to the BAPS grouping of the same 492 STs obtained using the extended E. faecium MLST database of 837 STs in order to infer the influence of the sample size on BAPS assignment. The correlation coefficient analysis revealed almost perfect correlations for the classification of BAPS groups (0.9996) and BAPS subgroups (0.9988) on the basis of 492 and 837 STs (see Fig. S2 and S3 in the supplemental material) and that only a small number of changes in BAPS assignment (44/837 STs, 5.2%) occurred at either the group or the subgroup level when the number of STs analyzed was significantly increased. This further indicates that, for E. faecium, BAPS analysis is both reproducible and robust and may accurately describe the E. faecium population structure.
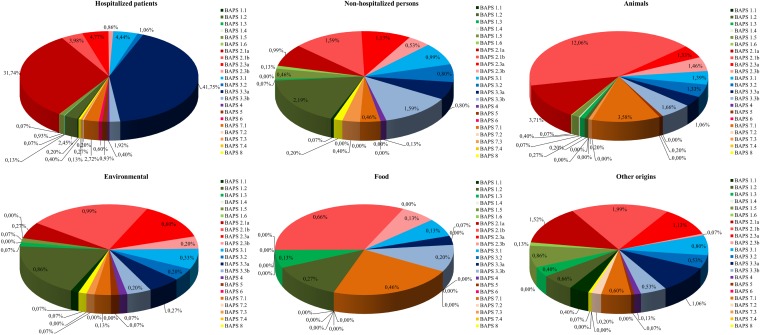
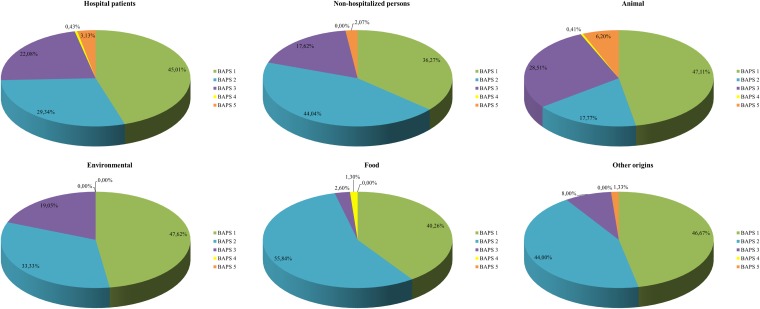
Since the extended data set of 837 STs slightly changed the BAPS grouping of STs, we decided to recalculate the ORs to assess the significance of the association between the BAPS groups and the origins of the isolates (see Table S3 in the supplemental material). This revealed that isolates from hospitalized individuals were positively associated with BAPSs 2.1a and 3.3a and negatively associated with all other BAPS groups. Conversely, isolates of all BAPS groups from nonhospitalized individuals were negatively associated with BAPSs 2.1a and 3.3a but positively associated with BAPS 1.2 and BAPS 3.3b (Fig. 4).
FIG 4.
E. faecium BAPS group distribution by isolate origin. This distribution by isolate origin was calculated by inclusion of all isolates present in the E. faecium MLST database (http://efaecium.mlst.net/) in August 2013.
Isolates of animal origin were negatively associated with BAPS 3.3a and BAPS 1.2 but showed a positive association with BAPSs 1.5, 2.1a, 2.1b, 2.3a, 2.3b, 3.1, 3.2, and 7.1 (Fig. 4; see also Table S3 in the supplemental material).
Genotypic relatedness of E. faecium isolates colonizing different age groups.
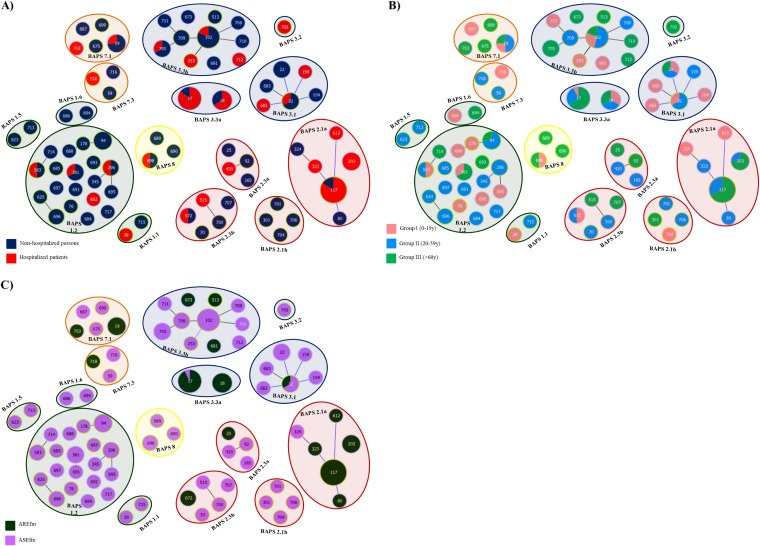
The 150 E. faecium isolates obtained from 142 samples in this study corresponded to 75 distinct STs. Forty-seven STs, representing 62.7% of the studied isolates, were STs first reported here (see Table S1 in the supplemental material). The remaining ones corresponded to globally spread STs, like ST78 (n = 34, 7 STs), ST17 (n = 14, 1 ST), and ST18 (n = 6, 1 ST), and also ST102 (n = 20, 7 STs), ST22 (n = 13, 9 STs), ST94 (n = 12, 7 STs), ST9 (2, 2 STs), and ST5 (1 ST), which were previously detected among community-based isolates (see Table S1 in the supplemental material). The 75 STs were partitioned into BAPS 1 (24 STs, 22.0% of isolates), BAPS 2 (19 STs, 31.3% of isolates), BAPS 3 (20 STs, 36.7% of isolates), BAPS 7 (8 STs, 7.3% of isolates), and BAPS 8 (3 STs, 2.7% of isolates) (Fig. 5).
FIG 5.
Population structure of E. faecium colonization by origin (A), by age group (B), and by susceptibility to ampicillin (C). AREfm, ampicillin-resistant E. faecium; ASEfm, ampicillin-susceptible E. faecium.
STs classified as BAPS 1 mainly corresponded to subgroup 1.2 (n = 27 [81.2%], 19 STs). The proportion of isolates with STs that grouped in BAPS 1 steadily decreased with age (Fig. 5 and 6), but isolates of this group were still prevalent among the adults of group II (15/27). All strains within BAPS 1 were ampicillin susceptible and were mainly recovered from nonhospitalized persons (23/27, P < 0.01).
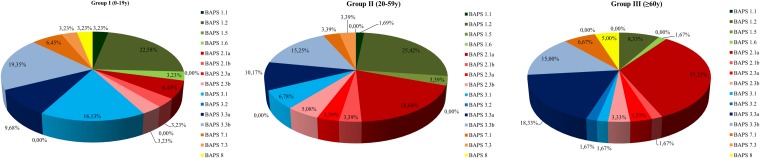
FIG 6.
E. faecium BAPS group distribution in human colonization by age group.
Within BAPS 2, subgroup 2.1a was predominant (70.2%, 33/47) and increasingly detected with age, constituting the predominant group in elderly patients (Fig. 5 and 6). Most isolates were recovered in hospitals, exhibited ampicillin resistance, and harbored genes encoding adhesive surface protein (Esp) and collagen-adhesin (Acm) (27/33 and 31/33, respectively), which are associated with colonization and pathogenicity (see Table S1 in the supplemental material). STs contained within BAPS 2.1a were ST117 (n = 25), ST203 (n = 4), ST80 (n = 1), ST323 (n = 1), ST324 (n = 1), and ST612 (n = 1). E. faecium ST117 (strain showing PFGE type CEfm1), apart from predominantly being a colonizing clone, was also frequently associated with severe infections in patients in HRyC (54). The other BAPS 2 subgroups, namely, BAPS 2.3b (n = 6 [12.8%], 5 STs), BAPS 2.3a (n = 4 [8.5%], 4 STs), and BAPS 2.1b (n = 4 [8.5%], 4 STs) were detected in both hospitalized and nonhospitalized individuals (Fig. 5 and 6).
BAPS 3 was represented by subgroups 3.1, 3.2, 3.3a, and 3.3b. Most isolates in BAPS 3.1 (6 STs, 18.2% of isolates) were ampicillin-susceptible E. faecium isolates (9/10) from nonhospitalized individuals (7/10) (P < 0.01) in age groups I and II (Fig. 5 and 6; see also Table S1 in the supplemental material). Isolates in BAPS 3.3a and BAPS 3.3b, previously described to be BAPS 3.3, differed in their susceptibility to ampicillin. BAPS 3.3a (n = 20 [36.4%], 14 ST17 and 6 ST18 isolates) comprised ampicillin-resistant E. faecium isolates (19/20, P < 0.01) that contained hyl from E. faecium (16/20) and that were predominantly recovered from hospitalized patients (16/20, P < 0.01). Conversely, BAPS 3.3b (11 STs, 43.6% of isolates) was significantly associated with ampicillin-susceptible E. faecium isolates (21/24, P < 0.01), mostly from nonhospitalized persons (18/24, P < 0.01).
BAPS 7.1 was the most predominant subgroup within BAPS 7 and comprised 5 STs representing 8 isolates. Finally, 3 STs comprising BAPS 8 and representing 4 isolates (2 ST698 isolates, 1 ST689 isolate, and 1 ST690 isolate), all of which were ampicillin-susceptible E. faecium isolates (of which 3 were esp+), were recovered from nonhospitalized persons (see Table S1 in the supplemental material).
Differences in the rates of recovery of ampicillin-resistant enterococci (56.3%, 36/64) but not in the rates of recovery of vancomycin-resistant enterococci (100%, 5/5) were noticed when samples were cultured without or with selective enrichment. Ampicillin-resistant E. faecium isolates that were cultured only after enrichment mostly belonged to BAPS 2.1a (n = 14; 9 ST117 isolates, 3 ST203 isolates, 1 ST80 isolate, and 1 ST323 isolate) and BAPS 3.3a (n = 8; 6 ST17 and 2 ST18 isolates), and the majority of these isolates were recovered from hospitalized patients. Isolates of other BAPS groups were also found and are described in Table S1 in the supplemental material.
BAPS analysis of E. faecalis population structure.
Previous studies based on MLST have suggested that recombination may play an important role in the diversification of E. faecalis (17, 20, 21). As methods to infer evolutionary descent are highly influenced by recombination, we analyzed the E. faecalis population structure using Bayesian-based population genetic modeling implemented in BAPS software, in addition to goeBURST analysis. The sample included 1,310 isolates corresponding to 523 STs available in a public database (http://efaecalis.mlst.net/).
A maximum likelihood-based phylogenetic reconstruction of STs using concatenated MLST gene sequences placed ST80 far apart from all other STs. When ST80 (amounting to only 1 isolate from the MLST database) was excluded from the analysis to better observe differences among tree features, practically all clades showed low bootstrap support, which supports previous analyses indicating that recombination may obscure the phylogenetic signal in nucleotide-based phylogenetic reconstructions in E. faecalis. A hierarchical BAPS clustering analysis subdivided the E. faecalis population into 5 BAPS groups (Fig. 7). Most of the STs and isolates were distributed among BAPSs 1, 2, and 3 (44.7%, 27.5%, and 20.6%, respectively), while BAPSs 4 (1.0%) and 5 (6.1%) represented only a small fraction of the STs analyzed (see Table S4 in the supplemental material).
FIG 7.
E. faecalis BAPS group distribution by origin. This analysis of the distribution by origin included all isolates present in the E. faecalis MLST database (http://efaecalis.mlst.net/) in August 2013.
OR calculations revealed that isolates from hospitalized patients were not significantly associated with any of the BAPS groups, while BAPS 2 was positively associated with isolates from nonhospitalized persons (OR = 1.8507, P < 0.01) and negatively associated with animal isolates (OR = 0.4659, P < 0.01) (Fig. 7; see also Table S5 in the supplemental material). Although signals of microevolutionary hospital specialization within the different BAPS groups were not found, some STs were enriched in isolates from hospitalized patients: ST6 (107/123), ST64 (12/18), ST9 (22/25), ST28 (16/17), ST87 (15/16), ST49 (4/4), ST88 (4/4), and ST159 (4/4). Furthermore, isolates from animals were frequently found to be ST58 (8/8), ST82 (25/27), and ST174 (11/11).
We also analyzed the E. faecalis population for traces of significant admixtures, as recombination is the driving force of admixture dynamics and it might influence the evolvability of specific amplified lineages. Admixtures were significantly present in some STs from animal and community-based hosts. However, additional analyses revealed that admixtures were not significantly found in STs that were unique or shared between hosts, STs from hospital or nonhospital origin, STs from human and nonhuman origin, or STs that represented antibiotic-resistant isolates (data not shown). The combination of these results suggests that the majority of E. faecalis isolates seem to belong to a single recombining population that exchanges alleles regardless of the genetic background (BAPS groups), ecological origin (isolation source, hospital or nonhospital, human or nonhuman), or antibiotic resistance phenotype.
The influence of the sample size (and, therefore, the underlying diversity) in the accuracy of BAPS for establishing the E. faecalis population structure was assessed using two data sets (see Fig. S4 in the supplemental material). The first data set consisted of 433 STs available in the MLST database (http://efaecalis.mlst.net/) before, including the new E. faecalis STs found in this study. The second data set included 523 STs available in the MLST database at the end of 2013. In both analyses, ST80 was excluded. The negative correlation coefficient of −0.6439 obtained when comparing ST assignments to BAPS groups of the set with 433 STs and those to BAPS groups of the set with 523 STs was due to the split of BAPS 1 and BAPS 2 and the existence of three more BAPS groups when using the second, larger data set (see Fig. S4 in the supplemental material). These results indicate that in E. faecalis, BAPS analysis is highly influenced by the sample size, as larger samples contain a higher diversity of strains of different spatial-temporal origins.
Genotypic relatedness of E. faecalis isolates colonizing different age groups.
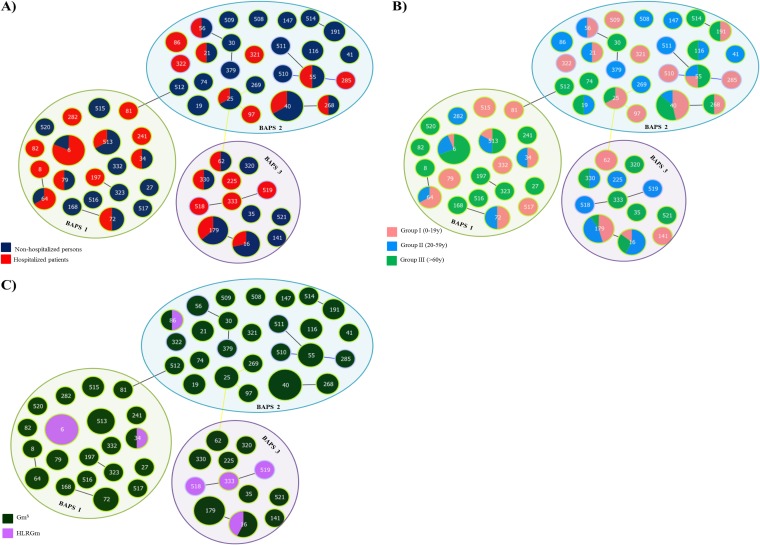
The 130 E. faecalis isolates identified in this study represented 58 STs (see Table S2 in the supplemental material) that were partitioned into E. faecalis BAPS 1 (36.9%), BAPS 2 (40.0%), and BAPS 3 (23.1%). OR calculations revealed that none of the three BAPS groups were significantly associated with a particular source or age group, as all the BAPS groups contained isolates from both hospitalized and nonhospitalized individuals of all ages in more or less equal numbers (Fig. 8).
FIG 8.
Population structure of E. faecalis colonization by origin (A), by age group (B), and by susceptibility to gentamicin (C). GmS, gentamicin susceptible; HLRGm, high-level resistance to gentamicin.
Within BAPS 1 (n = 48/130 [36.9%], 20 STs), ST6 (n = 16) was predominant and mainly comprised isolates from hospitalized patients (13/16) and elderly individuals (11/13) (Fig. 8; see also Table S2 in the supplemental material). All were multidrug resistant (MDR), showing high levels of resistance to gentamicin or streptomycin and also to erythromycin (100% of isolates), tetracycline (93.8%, 15/16), and levofloxacin (87.5%, 14/16) and exhibiting a highly similar PFGE profile (the ST6-H10 profile) identical to that of the widespread international mid-Atlantic clone, which also causes bacteremia in HRyC (55). The 5 vancomycin-resistant E. faecalis isolates (vanA; data not shown) found in this study were also ST6-H10. Putative virulence factors asa1 (100%) and gelE (81.3%) were identified in most ST6 isolates, while cylA (56.3%) and esp (37.5%) were less frequently identified. Other STs were represented by a very small number of isolates, which were usually susceptible to antibiotics and which had a highly variable presence of virulence factors.
Within BAPS 2 (n = 52/130 [40.0%], 26 STs), ST40 isolates (n = 15) were predominant. These isolates were recovered from both nonhospitalized and hospitalized individuals of different ages, often harbored gelE (88.2%) and, less frequently, asa1 (41.2%) and esp (47.1%), and were resistant to tetracycline (70.6%) and erythromycin (47.1%). Similar to the findings for BAPS 1, other STs were represented by a single isolate or very few isolates that often contained esp (see Table S2 in the supplemental material). Among them were STs that were identified over several decades to be ST55, ST30, or ST19 (31, 33).
Finally, BAPS 3 (n = 30/130, 12 STs) was predominantly comprised of ST16 and ST179, previously classified to be CC16 by goeBURST analysis (7 ST16 and 11 ST179 isolates). These STs also included isolates from both nonhospitalized and hospitalized individuals of different ages that often harbored asa1 (99.4%), esp (77.8%), or gelE (61.1%) and that were often resistant to different antibiotics (see Table S2 in the supplemental material). Other STs classified before as CC28 by goeBURST analysis (1 isolate each of ST333, ST518, and ST519) and recovered from adults or elderly hospitalized patients were also enriched in isolates that harbored putative virulence factors (all harbored asa1, gelE, esp, and cylA) and that were also MDR (with all isolates showing high levels of resistance to gentamicin and streptomycin, tetracycline, erythromycin, and levofloxacin) (see Table S2 in the supplemental material).
DISCUSSION
This study describes a consistently high rate of recovery of enterococci from feces from both hospitalized and nonhospitalized individuals and individuals in different age groups, similar to the findings reported in other studies, in which the rates of recovery ranged from 71% to 80% (1, 56, 57). These equilibrated constant rates of colonization indicate a major resiliency of the genus Enterococcus under heterogeneous conditions imposed by age, changing environments, and highly variable host niches. Previous studies (6, 58–60) have described changes in the rates of recovery of the genus Enterococcus in fecal microbiota with aging, which was not confirmed in our work, and a consistent predominance of the species E. faecalis in the fecal flora of young and elderly individuals, which is essentially consistent with our findings, with the important exception of the growing predominance of E. faecium in elderly individuals, particularly hospitalized elderly patients. Other studies yielded contradictory information about the frequency and diversity of E. faecium and other enterococcal species in the fecal microbiota (1, 15). Shifts in the prevalence of Enterococcus populations might result from fluctuating changes in the environmental conditions over time as a result of changes in diet (10) or changes in health status or antibiotic treatment (1, 5, 61–64), all of which delineate particular selective landscapes in hospitals (58, 61). Aging interacts with these conditions, and age-dependent enterococcal colonization dynamics have also been demonstrated for chickens and calves (1, 65), probably as an interaction with antibiotic consumption (1, 66, 67).
Considering the currently available diversity of known genotypes, the superimposing of goeBURST analysis of the clonal relationship among multiple isolates with BAPS allowed the detection of a low number of presumptive evolutionarily and functionally heterogeneous clades for the E. faecium species (22, 25–27). BAPS 1 E. faecium isolates, associated with the clade B phylogenetic lineage (a clade with isolates with pathways of complex carbohydrate utilization linked to host diet and with a majority of ampicillin-susceptible E. faecium strains), were highly represented in the different age groups, although their incidence was slightly reduced in the elderly (26, 27). Conversely, BAPS 2.1a and BAPS 3.3a (containing most of the ampicillin-resistant E. faecium strains), associated with clade A1 (26, 27) and mostly found in elderly hospitalized patients, represented E. faecium strains that are spreading in hospitals and causing clinical infections. The rates of these populations in the nosocomial and the community settings might be underestimated, as we have demonstrated here that if you do not preenrich the sample, some of the clones that are even more widespread might escape screening, probably due to low colonization densities. The observed population structure of E. faecium indicates a certain specialization of subpopulations for the colonization of particular age groups, which is usually associated with several other host-associated factors and also differences in the selectable characteristics harbored by the isolates, such as antibiotic resistance genes. Interestingly, some groups evolve independently from the acquisition of ampicillin resistance, suggesting a certain genetic isolation, which seems to be the case for different lineages within BAPS 3.3b, BAPS 1, and BAPS 2. These results further confirm a population structure comprised of ecotypes representing specialization in different hosts (16, 68).
E. faecalis populations showed a considerable level of genetic diversity. Because of that and in contrast to the findings for E. faecium, no BAPS groups were significantly associated with aging, hospital exposure, or host species, and with the exception of BAPS 2, none of the BAPS groups showed a positive association with nonhospitalized individuals. The wide recovery of certain STs (e.g., ST6, ST16, ST40, or ST55) able to colonize hospitalized and nonhospitalized humans (this study) and also animals (30, 31, 69) may be related to the more generalist lifestyle of this enterococcal species, which weakens the possibility of the recognition of ecotypes associated with a particular environment, at least by using the same approach that was so useful with E. faecium. However, despite possible limitations in the methods available for analysis of the E. faecalis population structure, it is now clear that certain multihost E. faecalis subpopulations, such as ST6 or ST16, have developed different strategies of adaptation to harsh and fluctuating habitats (31, 33). Among these are the lack of loci for cluster regulatory interspaced short palindromic repeats (CRISPR; a bacterial defense system against foreign DNA that facilitates the acquisition of foreign DNA, such as antibiotic resistance and virulence genes) (70) and the frequent acquisition of phages (71).
Other enterococcal species have largely been recognized to be part of the human fecal microbiota (1), and this was confirmed in our study. The inverse parallel trends in the frequencies of populations of these species and the frequency of E. faecium are of particular interest. The dynamics of colonization by these species might reflect differences in the functional requirements of the host with age and deserve further analysis.
This study provides a novel, integrated, and comprehensive image of the landscape of Enterococcus populations in a balanced amount of nonhospitalized and hospitalized individuals of different ages and suggests that a number of enterococcal lineages might be predominant in certain age groups and/or hospital environments. However, a number of clones are spread in different types of individuals and their prevalence is reduced in others in a kind of source-sink dynamics (72–74), with frequent cases of coexistence and the preservation of rare clonal populations being found. This suggests a frequency-dependent evolution of enterococcal populations which prevents the extinction of different genotypes that do not play equivalent ecological roles (75–78).
The work also illustrates the high degree of plasticity of E. faecium and E. faecalis genomes, as reflected by admixture analysis (27; this study), which showed variable intraclonal PFGE patterns (31, 63) (see Tables S1 and S2 in the supplemental material) and recombination of large fragments of the chromosome (79–83; our unpublished results). The consequences of such a high degree of variability have scarcely been explored from a population-based perspective. However, it can be expected that genome plasticity would contribute to the variation and selection of genes from a common intraspecies genetic pool needed for adaptation to environments imposing different stress conditions. Future progress in understanding enterococcal population biology will require a global analysis combining many ecological features, population dynamics, and population genetics (78, 84, 85).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Spanish Ministry of Economy and Competitiveness (PI12-01581 to T.M.C.), by the Seventh Framework Program (EvoTAR-282004 to T.M.C., F.B., and R.J.W.; R-GNOSIS-282512 to R.C. and P.R.-G.), and by the Regional Government of Madrid, Spain (PROMPT-S2010/BMD2414 to F.B.). A.P.T. is funded by the European Union (EvoTAR-282004). We are grateful to the Spanish Network for the Study of Plasmids and Extrachromosomal Elements (REDEEX) for funding cooperation among Spanish microbiologists working on the biology of mobile genetic elements (Spanish Ministry of Science and Innovation BFU2011-14145-E). J.C. is funded by ERC grant no. 239784 and AoF grant no. 251170.
We thank Victor Abraira for the contribution to the biostatistics analysis and Val F. Lanza for technical assistance in the comparison of MLST databases.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03661-14.
REFERENCES
- 1.Lebreton F, Willems RJL, Gilmore MS, Lebreton F, Willems RJL. 2014. Enterococcus diversity, origins in nature, and gut colonization. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA: http://www.ncbi.nlm.nih.gov/books/NBK190427/. [PubMed] [Google Scholar]
- 2.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales M-A, Jenq RR, van den Brink MRM, Pamer EG. 2012. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonten MJ, Willems R, Weinstein RA. 2001. Vancomycin-resistant enterococci: why are they here, and where do they come from? Lancet Infect Dis 1:314–325. doi: 10.1016/S1473-3099(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 5.Donskey CJ. 2004. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis 39:219–226. doi: 10.1086/422002. [DOI] [PubMed] [Google Scholar]
- 6.Woodmansey EJ, McMurdo MET, Macfarlane GT, Macfarlane S. 2004. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl Environ Microbiol 70:6113–6122. doi: 10.1128/AEM.70.10.6113-6122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enck P, Zimmermann K, Rusch K, Schwiertz A, Klosterhalfen S, Frick J-S. 2009. The effects of ageing on the colonic bacterial microflora in adults. Z Gastroenterol 47:653–658. doi: 10.1055/s-0028-1109055. [DOI] [PubMed] [Google Scholar]
- 8.Tiihonen K, Ouwehand AC, Rautonen N. 2010. Human intestinal microbiota and healthy ageing. Ageing Res Rev 9:107–116. doi: 10.1016/j.arr.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Petersen C, Round JL. 2014. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol 16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firmesse O, Rabot S, Bermúdez-Humarán LG, Corthier G, Furet J-P. 2007. Consumption of Camembert cheese stimulates commensal enterococci in healthy human intestinal microbiota. FEMS Microbiol Lett 276:189–192. doi: 10.1111/j.1574-6968.2007.00933.x. [DOI] [PubMed] [Google Scholar]
- 11.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. 2005. The microbial pan-genome. Curr Opin Genet Dev 15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Dobrindt U, Hacker J. 2001. Whole genome plasticity in pathogenic bacteria. Curr Opin Microbiol 4:550–557. doi: 10.1016/S1369-5274(00)00250-2. [DOI] [PubMed] [Google Scholar]
- 13.Finegold SM, Sutter VL, Methisen GE. 1983. Normal indigenous intestinal flora, p 3–32. In Hentges D. (ed), Human intestinal microflora in health and disease. Academic Press, New York, NY. [Google Scholar]
- 14.Tannock GW, Cook G. 2002. Enterococci as members of the intestinal microflora of humans, p 101–132. In Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB (ed), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC. [Google Scholar]
- 15.Silva N, Igrejas G, Gonçalves A, Poeta P. 2011. Commensal gut bacteria: distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann Microbiol 62:449–459. doi: 10.1007/s13213-011-0308-4. [DOI] [Google Scholar]
- 16.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, Van Embden JDA, Willems RJL. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol 40:1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz-Garbajosa P, Bonten MJM, Robinson DA, Top J, Nallapareddy SR, Torres C, Coque TM, Cantón R, Baquero F, Murray BE, del Campo R, Willems RJL. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol 44:2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leavis HL, Willems RJL, van Wamel WJB, Schuren FH, Caspers MPM, Bonten MJM. 2007. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog 3:e7. doi: 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Schaik W, Willems RJL. 2010. Genome-based insights into the evolution of enterococci. Clin Microbiol Infect 16:527–532. doi: 10.1111/j.1469-0691.2010.03201.x. [DOI] [PubMed] [Google Scholar]
- 20.Nallapareddy SR, Duh R-W, Singh KV, Murray BE. 2002. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J Clin Microbiol 40:868–876. doi: 10.1128/JCM.40.3.868-876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nallapareddy SR, Wenxiang H, Weinstock GM, Murray BE. 2005. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J Bacteriol 187:5709–5718. doi: 10.1128/JB.187.16.5709-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galloway-Peña J, Roh JH, Latorre M, Qin X, Murray BE. 2012. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS One 7:e30187. doi: 10.1371/journal.pone.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin X, Galloway-Peña JR, Sillanpaa J, Roh JH, Nallapareddy SR, Chowdhury S, Bourgogne A, Choudhury T, Muzny DM, Buhay CJ, Ding Y, Dugan-Rocha S, Liu W, Kovar C, Sodergren E, Highlander S, Petrosino JF, Worley KC, Gibbs RA, Weinstock GM, Murray BE. 2012. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol 12:135. doi: 10.1186/1471-2180-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3(1):e00318-11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howden BP, Holt KE, Lam MMC, Seemann T, Ballard S, Coombs GW, Tong SYC, Grayson ML, Johnson PDR, Stinear TP. 2013. Genomic insights to control the emergence of vancomycin-resistant enterococci. mBio 4(4):e00412-13. doi: 10.1128/mBio.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4(4):e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willems RJL, Top J, van Schaik W, Leavis H, Bonten M, Sirén J, Hanage WP, Corander J. 2012. Restricted gene flow among hospital subpopulations of Enterococcus faecium. mBio 3(4):e00151-12. doi: 10.1128/mBio.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz-Garbajosa P, Cantón R, Pintado V, Coque TM, Willems R, Baquero F, del Campo R. 2006. Genetic and phenotypic differences among Enterococcus faecalis clones from intestinal colonisation and invasive disease. Clin Microbiol Infect 12:1193–1198. doi: 10.1111/j.1469-0691.2006.01533.x. [DOI] [PubMed] [Google Scholar]
- 29.Waar K, Muscholl-Silberhorn AB, Willems RJL, Slooff MJH, Harmsen HJM, Degener JE. 2002. Genogrouping and incidence of virulence factors of Enterococcus faecalis in liver transplant patients differ from blood culture and fecal isolates. J Infect Dis 185:1121–1127. doi: 10.1086/339682. [DOI] [PubMed] [Google Scholar]
- 30.Kuch A, Willems RJL, Werner G, Coque TM, Hammerum AM, Sundsfjord A, Klare I, Ruiz-Garbajosa P, Simonsen GS, van Luit-Asbroek M, Hryniewicz W, Sadowy E. 2012. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. J Antimicrob Chemother 67:551–558. doi: 10.1093/jac/dkr544. [DOI] [PubMed] [Google Scholar]
- 31.Freitas AR, Novais C, Ruiz-Garbajosa P, Coque TM, Peixe L. 2009. Clonal expansion within clonal complex 2 and spread of vancomycin-resistant plasmids among different genetic lineages of Enterococcus faecalis from Portugal. J Antimicrob Chemother 63:1104–1111. doi: 10.1093/jac/dkp103. [DOI] [PubMed] [Google Scholar]
- 32.Kawalec M, Pietras Z, Daniłowicz E, Jakubczak A, Gniadkowski M, Hryniewicz W, Willems RJL. 2007. Clonal structure of Enterococcus faecalis isolated from Polish hospitals: characterization of epidemic clones. J Clin Microbiol 45:147–153. doi: 10.1128/JCM.01704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride SM, Fischetti VA, Leblanc DJ, Moellering RC, Gilmore MS. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. doi: 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willems RJL, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJM. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 11:821–828. doi: 10.3201/1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Been M, van Schaik W, Cheng L, Corander J, Willems RJ. 2013. Recent recombination events in the core genome are associated with adaptive evolution in Enterococcus faecium. Genome Biol Evol 5:1524–1535. doi: 10.1093/gbe/evt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Safdar N, Maki DG. 2002. The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, Enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Ann Intern Med 136:834–844. doi: 10.7326/0003-4819-136-11-200206040-00013. [DOI] [PubMed] [Google Scholar]
- 37.Willems RJL, Hanage WP, Bessen DE, Feil EJ. 2011. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev 35:872–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leavis HL, Bonten MJM, Willems RJL. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol 9:454–460. doi: 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Chowdhury SA, Nallapareddy SR, Arias CA, Murray BE. 2014. The majority of a collection of U.S. endocarditis Enterococcus faecalis isolates obtained from 1974 to 2004 lack capsular genes and belong to diverse, non-hospital-associated lineages. J Clin Microbiol 52:549–556. doi: 10.1128/JCM.02763-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galloway-Peña JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. 2009. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J Infect Dis 200:1566–1573. doi: 10.1086/644790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler KM. 2006. Enterococcal infection in children. Semin Pediatr Infect Dis 17:128–139. doi: 10.1053/j.spid.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Coque TM, Murray BE. 1995. Identification of Enterococcus faecalis strains by DNA hybridization and pulsed-field gel electrophoresis. J Clin Microbiol 33:3368–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Depardieu F, Perichon B, Courvalin P. 2004. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J Clin Microbiol 42:5857–5860. doi: 10.1128/JCM.42.12.5857-5860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.CLSI. 2013. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI, Wayne, PA. [Google Scholar]
- 45.Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. 2008. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect Immun 76:4110–4119. doi: 10.1128/IAI.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. 2004. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol 42:4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coque TM, Willems RJL, Fortún J, Top J, Diz S, Loza E, Cantón R, Baquero F. 2005. Population structure of Enterococcus faecium causing bacteremia in a Spanish university hospital: setting the scene for a future increase in vancomycin resistance? Antimicrob Agents Chemother 49:2693–2700. doi: 10.1128/AAC.49.7.2693-2700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francisco AP, Bugalho M, Ramirez M, Carriço JA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carriço JA. 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corander J, Marttinen P, Sirén J, Tang J. 2008. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hardin JW, Hilbe JH. 2002. Generalized estimating equations. Chapman and Hall/CRC, London, United Kingdom. [Google Scholar]
- 52.Facklam RR, Carvalho MG, Teixeira LM. 2002. Enterococcus, p 1–54. In Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB (ed), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC. [Google Scholar]
- 53.Turner KME, Hanage WP, Fraser C, Connor TR, Spratt BG. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol 7:30. doi: 10.1186/1471-2180-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tedim AP, Ruiz-Garbajosa P, Rodriguez CM, Derdoy L, Cardenas G, Loza E, Cantón R, Baquero F, Coque TM. 2014. Evolution of healthcare bacteraemic episodes of Enterococcus faecium (1995-2012) reflects changes in phylogenomic groups with internal clonal epidemic waves, poster P1239, p 203 Abstr 24th Eur Congr Clin Microbiol Infect Dis., Barcelona, Spain. European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland. [Google Scholar]
- 55.Tedim AP, Miranda AC, Ruiz-Garbajosa P, Freitas AR, Francia MV, Baquero F, Coque TM. 2010. Plasmid diversity among Enterococcus faecalis clinical isolates from Spain (2001-2009). Abstr 3rd Int Am Soc Microbiol Conf Enterococci, Portland, OR American Society for Microbiology, Washington, DC. [Google Scholar]
- 56.Mundt JO. 1963. Occurrence of enterococci in animals in a wild environment. Appl Microbiol 11:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coque TM, Arduino RC, Murray BE. 1995. High-level resistance to aminoglycosides: comparison of community and nosocomial fecal isolates of enterococci. Clin Infect Dis 20:1048–1051. doi: 10.1093/clinids/20.4.1048. [DOI] [PubMed] [Google Scholar]
- 58.Woodmansey EJ. 2007. Intestinal bacteria and ageing. J Appl Microbiol 102:1178–1186. doi: 10.1111/j.1365-2672.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- 59.Stewart CJ, Marrs ECL, Magorrian S, Nelson A, Lanyon C, Perry JD, Embleton ND, Cummings SP, Berrington JE. 2012. The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta Paediatr 101:1121–1127. doi: 10.1111/j.1651-2227.2012.02801.x. [DOI] [PubMed] [Google Scholar]
- 60.Kirtzalidou EI, Mitsou EK, Pramateftaki P, Kyriacou A. 2012. Screening fecal enterococci from Greek healthy infants for susceptibility to antimicrobial agents. Microb Drug Resist 18:578–585. doi: 10.1089/mdr.2012.0028. [DOI] [PubMed] [Google Scholar]
- 61.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berg RD. 1996. The indigenous gastrointestinal microflora. Trends Microbiol 4:430–435. doi: 10.1016/0966-842X(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 64.O'Sullivan O, Coakley M, Lakshminarayanan B, Conde S, Claesson MJ, Cusack S, Fitzgerald AP, O'Toole PW, Stanton C, Ross RP. 2013. Alterations in intestinal microbiota of elderly Irish subjects post-antibiotic therapy. J Antimicrob Chemother 68:214–221. doi: 10.1093/jac/dks348. [DOI] [PubMed] [Google Scholar]
- 65.Devriese LA, Laurier L, De Herdt P, Haesebrouck F. 1992. Enterococcal and streptococcal species isolated from faeces of calves, young cattle and dairy cows. J Appl Bacteriol 72:29–31. doi: 10.1111/j.1365-2672.1992.tb04877.x. [DOI] [PubMed] [Google Scholar]
- 66.Aarestrup FM, Butaye P, Witte W. 2002. Non-human reservoirs of enterococci, p 55–99. In Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB (ed), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC. [Google Scholar]
- 67.Devriese LA, Pot B, Collins MD. 1993. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol 75:399–408. doi: 10.1111/j.1365-2672.1993.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 68.Willems RJ, Top J, van Den Braak N, van Belkum A, Endtz H, Mevius D, Stobberingh E, van Den Bogaard A, van Embden JD. 2000. Host specificity of vancomycin-resistant Enterococcus faecium. J Infect Dis 182:816–823. doi: 10.1086/315752. [DOI] [PubMed] [Google Scholar]
- 69.Freitas AR, Coque TM, Novais C, Hammerum AM, Lester CH, Zervos MJ, Donabedian S, Jensen LB, Francia MV, Baquero F, Peixe L. 2011. Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. J Clin Microbiol 49:925–931. doi: 10.1128/JCM.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1(4):e00227-10. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duerkop BA, Clements CV, Rollins D, Rodrigues JLM, Hooper LV. 2012. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci U S A 109:17621–17626. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pulliam H. 1988. Sources, sinks, and population regulation. Am Nat 132:652–661. doi: 10.1086/284880. [DOI] [Google Scholar]
- 73.Perron GG, Gonzalez A, Buckling A. 2007. Source-sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc Biol Sci 274:2351–2356. doi: 10.1098/rspb.2007.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hermsen R, Hwa T. 2010. Sources and sinks: a stochastic model of evolution in heterogeneous environments. Phys Rev Lett 105:248104. doi: 10.1103/PhysRevLett.105.248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levin BR. 1988. Frequency-dependent selection in bacterial populations. Philos Trans R Soc Lond B Biol Sci 319:459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- 76.Dugatkin LA, Perlin M, Lucas JS, Atlas R. 2005. Group-beneficial traits, frequency-dependent selection and genotypic diversity: an antibiotic resistance paradigm. Proc Biol Sci 272:79–83. doi: 10.1098/rspb.2004.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonzalez A, Ronce O, Ferriere R, Hochberg ME. 2013. Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Philos Trans R Soc Lond B Biol Sci 368:20120404. doi: 10.1098/rstb.2012.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cordero OX, Polz MF. 2014. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12:263–273. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- 79.Quintiliani R, Courvalin P. 1994. Conjugal transfer of the vancomycin resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol Lett 119:359–363. doi: 10.1111/j.1574-6968.1994.tb06913.x. [DOI] [PubMed] [Google Scholar]
- 80.Laverde Gomez JA, Hendrickx AP, Willems RJ, Top J, Sava I, Huebner J, Witte W, Werner G. 2011. Intra- and interspecies genomic transfer of the Enterococcus faecalis pathogenicity island. PLoS One 6:e16720. doi: 10.1371/journal.pone.0016720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manson JM, Hancock LE, Gilmore MS. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci U S A 107:12269–12274. doi: 10.1073/pnas.1000139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Regt MJA, van Schaik W, van Luit-Asbroek M, Dekker HAT, van Duijkeren E, Koning CJM, Bonten MJM, Willems RJL. 2012. Hospital and community ampicillin-resistant Enterococcus faecium are evolutionarily closely linked but have diversified through niche adaptation. PLoS One 7:e30319. doi: 10.1371/journal.pone.0030319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Novais C, Escada R, Freitas AR, Silveira E, Coque TM, Peixe L. 2011. High incidence of horizontal transfer of ampicillin resistance (pbp5) among CC17 Enterococcus faecium, p S129 Abstr 21st Eur Congr Clin Microbiol Infect Dis, Milan, Italy. European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland. [Google Scholar]
- 84.Luck GW, Daily GC, Ehrlich PR. 2003. Population diversity and ecosystem services. Trends Ecol Evol 18:331–336. doi: 10.1016/S0169-5347(03)00100-9. [DOI] [Google Scholar]
- 85.Urban MC, Skelly DK. 2006. Evolving metacommunities: toward an evolutionary perspective on metacommunities. Ecology 87:1616–1626. doi: 10.1890/0012-9658(2006)87[1616:EMTAEP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.