Abstract
In ecological studies, microbial diversity is nowadays mostly assessed via the detection of phylogenetic marker genes, such as 16S rRNA. However, PCR amplification of these marker genes produces a significant amount of artificial sequences, often referred to as chimeras. Different algorithms have been developed to remove these chimeras, but efforts to combine different methodologies are limited. Therefore, two machine learning classifiers (reference-based and de novo CATCh) were developed by integrating the output of existing chimera detection tools into a new, more powerful method. When comparing our classifiers with existing tools in either the reference-based or de novo mode, a higher performance of our ensemble method was observed on a wide range of sequencing data, including simulated, 454 pyrosequencing, and Illumina MiSeq data sets. Since our algorithm combines the advantages of different individual chimera detection tools, our approach produces more robust results when challenged with chimeric sequences having a low parent divergence, short length of the chimeric range, and various numbers of parents. Additionally, it could be shown that integrating CATCh in the preprocessing pipeline has a beneficial effect on the quality of the clustering in operational taxonomic units.
INTRODUCTION
A powerful method to assess microbial diversity of different environments is the identification of specific phylogenetic marker genes like the 16S rRNA genes (rRNA) for Bacteria and Archaea or the internal transcribed spacer (ITS) for Fungi. However, because of the limitations of most current sequencing technologies, PCR amplification of the targeted DNA is an unavoidable step in this approach. During this PCR amplification, chimeras might be created due to incomplete extension. If an incomplete PCR fragment acts as a primer in the next round of the PCR (by binding to the template DNA of a different species), a chimeric sequence originates which consists of two or more fragments amplified from the DNA of distinct species (1–6). Chimeras introduced via this mechanism are propagated through the PCR as any other DNA sequence (4, 7–9). Consequently, these chimeras result in an artificial inflation of the microbial diversity in a biodiversity analysis using next-generation sequencing approaches on marker genes like the 16S or 18S rRNA gene or fungal ITS (10). Indeed, since chimeras are PCR errors and not sequencing errors, they cannot be recovered using regular data denoising approaches (11, 12) and can be falsely interpreted as unique sequences representing novel species. A significant number of chimeras has been identified in curated databases in a proportion that can reach up to 46% (4, 6, 13). Even after treatment with traditional chimera detection tools, chimeras are continuously detected in highly valuable and frequently consulted databases like RDP (14), SILVA (15), and Greengenes (16). Likewise, the percentage of chimeric sequences in the unique amplicon pool of PCR-amplified samples might reach values higher than 70% (5, 11).
In general, two classes of chimera detection tools can be distinguished: reference-based and the more recent de novo methods. Reference-based methods basically screen the sequences potentially containing chimeras against a curated reference database with chimera-free sequences. This approach has been implemented in the first generation of chimera detection tools such as Pintail (4) and Bellerophon (13). A major improvement of the reference-based approaches was achieved via the introduction of ChimeraSlayer (17), which uses 30% of each end as a seed for searching a reference data set, finding the closest parent (if any), performing alignments, and scoring to the candidate parents. Although ChimeraSlayer was found to outperform all of the previous tools, it was not able to detect chimeras with a small chimeric range (i.e., the shortest region produced by one of the parents) (18). The reference-based mode of a new chimera detection tool called UCHIME was built upon the implementation of ChimeraSlayer and outperformed it on almost all investigated data sets (10). In reference-based UCHIME, query sequences are divided into four nonoverlapping segments and searched against a reference database. Both of the three-way alignment tools (ChimeraSlayer and reference-based UCHIME) were reported to have a lower accuracy than that of DECIPHER (18) in cases where the algorithms were challenged with a data set containing chimeric sequences with a short chimeric range and long sequence lengths. The DECIPHER algorithm is a search-based algorithm that splits the query sequence into different fragments and analyzes whether those fragments are uncommon in the reference phylogenetic group where the query sequence is classified. If a significant amount of fragments is assigned to a phylogenetic group different from the complete query sequence, the sequence is classified as chimeric.
However, in general, an objective comparison between different chimera detection tools is difficult, as each of the algorithms is biased toward its own test data and has been proven to outperform other tools when applied to their own dedicated data.
De novo methodologies are generally based on the fact that parents of any chimeric sequence have gone through at least one more PCR cycle than chimeric sequences (11). Three such tools, Perseus (11), de novo UCHIME (10), and the de novo ChimeraSlayer implementation, are integrated in mothur (19). Perseus was trained using real 454 pyrosequencing data, while de novo UCHIME used simulated data. Recently, the UPARSE pipeline was released, combining in one step chimera detection with clustering of sequencing reads into operational taxonomic units (OTUs) (20).
Both reference-based and de novo methodologies have their advantages and disadvantages. In situations dealing with well-studied environments, the reference-based approaches were found to be very effective in distinguishing between chimeras and chimera-free (parent) sequences. This efficiency is assumed to be lower when dealing with less well-known environments, which is where the need for de novo approaches originated. However, most of the de novo approaches depend on redundancy differences between chimeras and parents, assuming that the number of parent sequences has to be at least one time more redundant than their corresponding chimeric sequences. This requires data abundances to have been reported with high accuracy. Practically this might be problematic, as some of the sequences are removed due to noise removal, score filtration, or any other preprocessing step. As each approach has its pros and cons, reference-based as well as de novo approaches were taken into account in the analyses described below. A schematic overview comparing the various chimera detection tools is available in the supplemental material (see Table S1).
In this work, we present a chimera detection tool consisting of a machine learning classifier called CATCh (Combining Algorithms to Track Chimeras), which is able to discriminate between chimeric and nonchimeric sequences based on a specific set of input data (called features in the context of machine learning). With this tool, we use as input data not the sequence read characteristics but rather the results (e.g., scores) of different individual chimera detection tools mentioned above and integrate them into one prediction. All different tools are run separately, and their output values are combined and processed by the classifier in order to give a prediction of whether a read is a chimera or not. This machine learning method consists of three stages. First, the necessary input features (i.e., output values of the different chimera detection tools) are identified. In the second step, the classifier is trained via a supervised learning approach. In this step, the classifier learns to make a correct prediction based on example input data; in our case, training data consist of the output features of a set of sequences reads obtained from different chimera detection tools, together with their correct classification (i.e., whether this read is a chimeric sequence or not). In the third step, the trained classifier can be used to predict chimeric sequences in new, previously unseen data (i.e., data that did not belong to the training data). By feeding the outputs of the different individual chimera detection tools into the classifier, CATCh is able to classify them into chimeric and chimera-free subsets. As two different types of chimera detection tools exist, either reference based or de novo, we also developed two different versions of CATCh. In order to illustrate its performance, CATCh (reference based as well as de novo) was benchmarked against other chimera detection tools using various publicly available benchmark data.
MATERIALS AND METHODS
Included chimera detection algorithms.
As mentioned in the introduction, the strategy of CATCh consists of consolidating the output of different chimera detection tools into one highly reliable prediction. Therefore, a wide range of different reference-based and de novo tools has been integrated into CATCh; each of the tools used is discussed below.
When running the reference-based algorithms, the implementations of the tools as available in mothur (version 1.28.0) (19) were used for UCHIME (reference mode) (10), ChimeraSlayer (reference mode) (17), and Pintail (4), each of them using the default parameters. For DECIPHER (version 1.10.0), which does not support parallelization, we increased the speed by implementing a forking approach—as already suggested in the original paper—making it possible to split up the run of DECIPHER over an arbitrary number of cores (18). All these reference-based algorithms use a database of chimera-free reads as a reference: UCHIME, ChimeraSlayer, and Pintail use the Gold reference database (available at the Broad Microbiome Utilities website, version microbiomeutil-r20110519), and DECIPHER uses a “good quality” filtered and chimera-curated version of RDP (release 10, update 22) (18). DECIPHER has two modes, depending on the length of the sequences, either full length (fs_decipher) or short length (ss_decipher). In our experiments, we adjusted these parameters according to the length of the input sequences of each database.
Similarly, for the de novo-based algorithms, the implementation of the algorithms as available in mothur (19) was used for UCHIME (10) (de novo), ChimeraSlayer (de novo) (17), and Perseus (11). For integrating the different de novo-based algorithms into the CATCh de novo classifier, the default parameter settings for each of the three tools were used.
Building CATCh.
Two different CATCh classifiers were developed, one reference and one de novo model, each of them integrating the results (called features below) of different individual chimera detection tools into a chimera prediction tool with higher performance.
For the reference-based CATCh classifier, we included the following input features: (i) the calculated score and the final decision (i.e., chimeric or nonchimeric) for UCHIME, (ii) the calculated score and final decision for ChimeraSlayer, (iii) the score, standard deviation, and final decision for Pintail, and (iv) the final decision for DECIPHER. The final decision (i.e., whether a read is predicted as chimeric or not) for the individual chimera detection tools depends on the chosen cutoff value, for which the developer's default value was used. For the de novo CATCh classifier, both the score and the final decision for (i) de novo UCHIME, (ii) de novo ChimeraSlayer, and (iii) Perseus were selected as input parameters. Based on the chimera prediction results for each of these individual tools, two classifiers were built (reference and de novo) integrating the different output values produced by these tools into one final score.
Different mathematical functions, called kernels, have already been developed for solving classification problems. To develop the CATCh classifier, we had to select the most optimal kernel based on their performance on the training data. For this purpose, the training data—consisting of the output of different chimera detection tools, together with the chimeric or nonchimeric label—were split into two subsets: a subset to train and a subset to test the classifier. The first one is used during the learning step applied for each of the kernels to build a classifier, and the second data set is used for evaluating the performance of the different kernels. An overview of the tested kernels available in WEKA (21) is given in Table S2 in the supplemental material for the reference-based and the de novo implementations.
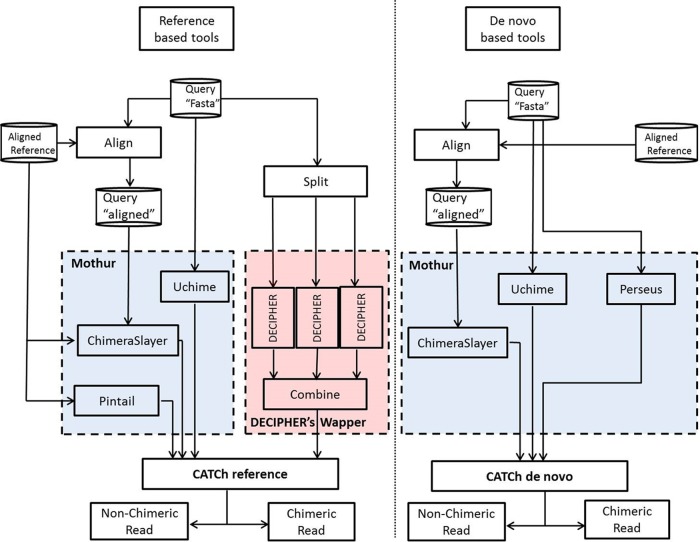
For all model parameters, the WEKA standard values, version 3.7.1, were used (21). A schematic overview of the developed methodology (reference based and de novo) is given in Fig. 1. The CATCh software and accompanying documentation are available via http://science.sckcen.be/en/Institutes/EHS/MCB/MIC/Bioinformatics/CATCh. The implementation has been tested on Mac and Linux (RHEL-derived distributions). On the CATCh website a manual together with the training data can be downloaded; the manual explains how CATCh was trained and tested using the WEKA software. This software is distributed under the terms of the GNU General Public License as published by the Free Software Foundation.
FIG 1.
Work flow diagram illustrating different steps and tools included in reference-based and de novo CATCh.
Training and testing data sets.
As a training data set to build the CATCh classifiers, the denoised output of the Titanium data set as published by Quince et al. (11) was used. These data are grouped into 91 parent sequences (chimera free) and 176 chimeric sequences (obtained directly from C. Quince). For the reference-based model, we used the Titanium data set (see below) to train as well as test different classifiers (splitting to a 3:1 ratio). For the de novo classifier, the whole Titanium data set was used for training and a separate data set (F01QS4Z01_rep2_v13, a subset of Mock2-b; see below) was used for testing, as complete information on the redundancy of the reads is essential for all de novo algorithms.
Validation data sets.
For benchmarking our tool against other available chimera detection tools, two types of data were used, (i) simulated data containing artificial chimeric sequences generated using tools like CHSIM (10) and (ii) publicly available real sequencing data (454 pyrosequencing and Illumina MiSeq and PacBio SMRT sequencing).
For the first data set, we randomly selected sequences from the GOLD database (described in reference 17) and used them as parent sequences to artificially produce chimeras using the CHSIM tool (10). By trimming these parent and chimeric sequences to a randomly selected length within 50 to 99% of the actual length, we obtained a data set consisting of 1,532 chimera-free (parent) and 1,308 chimeric sequences, referred to as Simu1.
Additionally, three publicly available data sets were used for the evaluation of different previously published reference-based chimera detection tools—SIMM (10), DECIPHER (18), and SIM2 (17) (for details, see Table 1) called Simu2, Simu3, and Simu4, respectively, in the rest of the paper. To challenge the chimera detection tools to predict chimeras in more experimentally relevant situations obtained via high-throughput sequencing technologies (sequencing errors, which can rise to 1%) as well as to simulate biological variation of reads corresponding to novel species, we used the previously published SIMM data sets with 1% and 5% insertions and deletions and SIM2 data sets with 1% and 5% insertions, deletions, indels, and substitutions (referred to as Simu2_Mutation and Simu4_Mutation, respectively).
TABLE 1.
Detailed description of each of the data sets used in this work either for training or testing, illustrating the number of chimeric and nonchimeric (parent) sequences and the processing steps applied by us or the original authors to these data
| Name | No. of sequences |
Sequencing platform | No. of samples | No. of genomes | Gene | Region | Data treatment | Reference(s) | |
|---|---|---|---|---|---|---|---|---|---|
| Nonchimeric | Chimeric | ||||||||
| Training | 91 | 176 | 454 Titanium | 1 | 91 | 16S rRNA | V45 | AmpliconNoise | 11 |
| Simu1 | 1,532 | 1,308 | Simulated | 1 | 16S rRNA | This work | |||
| Simu2 | 86 | 900 | Simulated | 1 | 16S rRNA | 10 | |||
| Simu3 | 11,000 | 4,000 | Simulated | 1 | 16S rRNA | 18 | |||
| Simu4 | 4,769 | 2,500 | Simulated | 1 | 16S rRNA | 17 | |||
| Mock1 | 246 | 2,565 | 454 GS-FLX | 3 | 67 | 16S rRNA | V2 | AmpliconNoise | 11, 22 |
| Mock2-a | 5,447 | 13,205 | 454 Titanium | 90 | 21 | 16S rRNA | V13/V35/V69 | mothur 454 SOP | 23 |
| Mock2-b | 8,302 | 22,131 | 454 Titanium | 90 | 21 | 16S rRNA | V13/V35/V69 | AmpliconNoise | 23 |
| Mock3 | 11,958 | 13,612 | MiSeq | 8 | 21 | 16S rRNA | V34/V4 | mothur MiSeq SOP | 24 |
| Mock4 | 2,050 | 5,326 | 454 GS-FLX | 30 | 12/24/48 | 18S rRNA | V13 | AmpliconNoise | 26 |
| Mock5 | 82,426 | 16,317 | 454 | 30 | 16 | 18S rRNA | V13/V46 | mothur 454 SOP | 27 |
For benchmarking our de novo classifier, the uneven mock data set as reported in the work of Quince et al. (11) and initially introduced in the work of Turnbaugh et al. (22) was used. This data set, covering the V2 region of the 16S rRNA gene, is sequenced in triplicate and consists of 3,036 sequences, from which those sequences labeled as anomalies (n = 225) were removed, leaving 2,565 chimeric sequences and 246 nonchimeric sequences in this data set, called Mock1.
The HMP-MOCK data set, part of Project SRP002397 in the NCBI Sequence Read Archive (SRA) (23), contains 454 pyrosequencing data of 16S rRNA amplicons of 21 bacteria, consisting of 10 runs covering 3 regions (V13, V35, and V69), with 3 replicates per region per run, resulting in a total of 90 samples. This data set was treated using mothur (19) (version 1.28.0) with two different denoising strategies, quality trimming (23) and flow denoising (AmpliconNoise [11]), producing two separate data sets (P. Schloss, personal communication). These data sets were named Mock2-a and Mock2-b for quality-trimmed and AmpliconNoise-treated data sets, respectively.
The MiSeq mock data set (called Mock3) consists of the 16S rRNA amplicons of a mock community containing 21 bacterial isolates and covering the V34 and V4 regions (each region sequenced in quadruplicate) (24), which were extracted from the Sequence Read Archive (SRA082708). The sequencing data were preprocessed following the mothur MiSeq SOP procedure (version online on 3 August 2014) (24).
For assessing the computational resources required for different sequencing platforms, also a (nonmock) PacBio SMRT sequencing data set was analyzed. This data set covers the V13 and V23 regions of the 16S rRNA and was extracted from the Sequence Read Archive (SRA accession number SRA056302) (25). All 16 samples were preprocessed using mothur (19) (version 1.28.0) as described in the corresponding paper (25), i.e., removing sequences with an average quality score of lower than 25, an anomalous length (<300 or >615 bp), an ambiguous base, homopolymers with a length higher than 8, or more than one mismatch in the barcode or primer.
In addition to testing the performance of chimera detections tools on 16S rRNA amplicon sequencing data, the performance on two data sets covering regions in the eukaryotic 18S rRNA gene was included in this work. The first data set, called Mock4, consists of a mock community of either 12, 24, or 48 species from closely or distantly related nematodes, as described by Fonseca et al. (26). Each sample was sequenced in five replicates, resulting in a total of 30 samples (SRA accession number SRA043810). The data were preprocessed as described in the initial paper (26) using AmpliconNoise.
A second 18S rRNA mock community (Mock5), covering the V13 and V46 regions, was obtained as described in the publication of Morgan et al. (27). The data downloaded from http://research.csiro.au/software/amplicon-pyrosequencing-denoising-program/ consist of 30 sequencing data sets and were preprocessed by removing reads with one or more ambiguous nucleotides and homopolymers longer than 8 nucleotides (nt).
An overview of all simulated and mock data sets used in this study is given in Table 1.
Evaluation parameters.
For evaluation of the performance of the different chimera detection algorithms, we adopted four parameters: sensitivity [TP/(TP + FN)], specificity [TN/(TN + FP)], accuracy [(TP + TN)/(TP + FN + TN + FP)], and Matthews correlation coefficient (MCC), determined as
where false positives (FP) and false negatives (FN) represent the number of sequences falsely predicted as chimeric and nonchimeric, respectively, while true positives (TP) and true negatives (TN) stand for the number of sequences correctly predicted as chimeric and nonchimeric, respectively. MCC is a correlation coefficient between the observed and predicted binary classifications, giving an idea on the balance between the specificity and sensitivity. It returns a value between −1 (negative correlation, i.e., performing worse than a random prediction), 0 (no correlation, i.e., performing as random predictor), and +1 (positive correlation, i.e., perfect prediction).
RESULTS
In this work, two classifiers were developed, one for reference-based approaches and a second one integrating all de novo approaches. For both CATCh classifiers, we used the output of the individual chimera detection tools as input features for the calculation of a combined chimera prediction score. Constructing a machine learning classifier generally consists of three stages: (i) identifying the necessary input data, called feature selection, (ii) training and testing the classifier, and (iii) evaluating the performance of the classifier. In the first two sections of Results, we describe the feature selection and training and testing of the classifier. The remaining sections are dedicated to the evaluation of the CATCh classifier.
Feature selection.
The feature selection step is required to identify the optimal set of features needed for the development of a robust classifier. The question answered here is whether all input features as described in Materials and Methods (and, by extension, all individual chimera prediction tools) are needed to obtain a good classifier. This step is important, as a reduction of the number of input features might have a beneficial effect on the accuracy of the classifier as well as the computational calculation time. However, based on the principal component analysis (PCA) of the training data (see Fig. S1 and Table S3 in supplemental material), all input features were needed to explain 95% of the variation in the data. This also implies that the results of all individual chimera detection tools need to be retained as input data for the classifier. This observation was further confirmed by a dedicated experiment in which we left out one of the chimera prediction models from CATCh (see Table S4 in the supplemental material). Even removing a tool with a low individual performance had a major impact on the performance of the classifier (e.g., a drop in sensitivity of reference-based CATCh when tested on Mock2-a from 0.82 when including Pintail to 0.69 when excluding Pintail in the classifier). Similarly, when calculating the overlap of correctly predicted chimeric sequences between different individual algorithms, the added effect between the different individual tools was highlighted; e.g., 1,060 out of the 13,205 chimeras in the Mock2-a data set could exclusively be predicted by Pintail (see Fig. S2 in the supplemental material).
Training and testing CATCh.
As mentioned in Materials and Methods, a wide range of mathematical functions (i.e., kernels) has been developed to address classification problems. Different kernel functions were compared by training and testing them on the training data (i.e., data containing the features extracted from the results (e.g., scores) of the different individual chimera detection tools together with the information on whether the read is chimeric or nonchimeric. As mentioned in Materials and Methods, the training data were split into two parts: the first part was used for training classifiers using different kernel functions, and the second set was used to measure the performance of different kernels. This performance of the different trained classifiers on the test data was assessed by applying a cutoff score on the output of the classifier, which guarantees a predefined level of specificity (0.90 for the reference-based method and 0.93 for the de novo method), and subsequently comparing the sensitivities for each of them. For both the reference-based and the de novo implementations of CATCh, the Support Vector Machine with Pearson VII Universal Kernel (SVM-PUK) obtained the highest accuracy, with sensitivities of 0.85 and 0.92 and specificities of 0.90 and 0.93, respectively, on the test data (see Table S2 in the supplemental material). These results were noticeably better than the performance of each individual tool as well as the union of chimeric predictions of all individual tools (see Table S5 in the supplemental material).
To test the dependency of the CATCh classifier on the accuracy of the training data (i.e., whether a read is correctly identified as being chimeric or not in the training data), we artificially introduced a mislabeling of the chimeric reads in the training data. Randomly switching the label of a read from chimeric to nonchimeric or vice versa was tolerated up to a level of 5% for the reference-based classifier and up to 10% for the de novo implementation. Exceeding these thresholds leads to a dramatic decrease in sensitivity. However, since we restricted the training data to high-quality predictions—all reads where the chimeric prediction was not clear were removed from the data—the fraction of mislabeled reads would be very low.
To test whether it was possible to integrate both approaches in one powerful classifier, a classifier combining both de novo and reference-based algorithms was built and tested on Mock2-a and Mock2-b. However, compared with the separate de novo or reference based classifier, no improvement (Mock2-a) or only a marginal improvement (Mock2-b) in the sensitivity with the same cost in the specificity was observed (see Table S6 in the supplemental material), as well as a major increase in the running time.
Performance of reference-based algorithms.
For benchmarking the different reference-based chimera detection algorithms, all available tools were run on different data sets, each of them already used to optimize one of the existing chimera detection tools. When comparing our ensemble algorithm with the best-performing alternative, the CATCh classifier was found to obtain a higher sensitivity on Simu1, Simu2, and Simu3 data (respectively, 14%, 12%, and 11% higher) and an equally high sensitivity on Simu4 data (95% for both UCHIME and CATCh), while showing a small drop in specificity (respectively, 4%, 2%, 1%, and 3%) (Table 2).
TABLE 2.
Sensitivity and specificity values of all tested chimera prediction toolsa
| Parameter | Data set | Value for indicated reference-based tool |
Data set | Value for indicated de novo tool |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UCHIME | ChimeraSlayer | Pintail | DECIPHER | CATCh | UCHIME | ChimeraSlayer | Perseus | CATCh | |||
| Sensitivity | Mock2-a | 0.79 | 0.66 | 0.58 | 0.61 | 0.82 | Mock2-a | 0.74 | 0.66 | 0.76 | 0.85 |
| Specificity | 0.94 | 0.95 | 0.42 | 0.93 | 0.92 | 0.94 | 0.94 | 0.92 | 0.91 | ||
| Sensitivity | Mock2-b | 0.90 | 0.77 | 0.58 | 0.70 | 0.93 | Mock2-b | 0.87 | 0.80 | 0.87 | 0.95 |
| Specificity | 0.94 | 0.96 | 0.46 | 0.93 | 0.92 | 0.94 | 0.94 | 0.93 | 0.91 | ||
| Sensitivity | Mock3 | 0.75 | 0.63 | 0.06 | 0.61 | 0.81 | Mock3 | 0.64 | 0.65 | 0.73 | 0.81 |
| Specificity | 0.95 | 0.99 | 1.00 | 0.98 | 0.94 | 0.98 | 0.97 | 0.98 | 0.96 | ||
| Sensitivity | Simu1 | 0.56 | 0.51 | 0.15 | 0.45 | 0.70 | Mock1 | 0.92 | 0.71 | 0.94 | 0.97 |
| Specificity | 1.00 | 1.00 | 0.88 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | ||
| Sensitivity | Simu2 | 0.69 | 0.53 | 0.21 | 0.48 | 0.81 | Mock4 | 0.37 | 0.27 | 0.27 | 0.47 |
| Specificity | 1.00 | 0.99 | 0.80 | 0.99 | 0.98 | 0.96 | 0.98 | 0.94 | 0.91 | ||
| Sensitivity | Simu3 | 0.83 | 0.66 | 0.27 | 0.87 | 0.94 | Mock5 | 0.08 | 0.11 | 0.14 | 0.16 |
| Specificity | 0.99 | 1.00 | 0.86 | 0.98 | 0.97 | 1.00 | 0.95 | 0.99 | 0.98 | ||
| Sensitivity | Simu4 | 0.95 | 0.90 | 0.17 | 0.27 | 0.95 | |||||
| Specificity | 1.00 | 0.99 | 0.86 | 1.00 | 0.97 | ||||||
| Sensitivity | Avg | 0.78 | 0.67 | 0.29 | 0.57 | 0.85 | Avg | 0.60 | 0.53 | 0.62 | 0.70 |
| Specificity | 0.97 | 0.98 | 0.75 | 0.97 | 0.96 | 0.97 | 0.96 | 0.96 | 0.95 | ||
| Difference | 7/−2 | 18/−2 | 56/21 | 28/−1 | 10/−2 | 17/−1 | 8/−1 | ||||
The first three data sets were tested with both reference-based and de novo tools. The second part of the table contains the data sets analyzed using either reference or de novo tools. The last three rows give the average sensitivity and specificity and the average increase or decrease in sensitivity/specificity compared with that of CATCh.
In order to simulate real-life situations, where sequence reads are divergent from sequences in the reference database due to factors like natural variation (i.e., predict chimera that are produced by 16S rRNA sequences which are not present in the currently used reference databases) or sequencing errors (sequences containing insertions, deletions, or mismatches can rise to 1%), we challenged the chimera detection algorithms with adapted versions of the Simu2 and Simu4 data sets in which 1% and 5% indels and mutations were added (see Table S7 in the supplemental material). While some of the individual tools, like UCHIME, could tolerate 1% mutations, the decrease in sensitivity was much more pronounced at the 5% level than that of CATCh. Indeed, the drop in sensitivity at 5% mutations varied between 10% and 20% for UCHIME (the best-performing individual tool in this test) for the different types of mutations, while CATCh showed a drop in sensitivity between 4% and 9%. Overall, CATCh shows the highest sensitivity over Simu2 as well as Simu4 data sets, with the clearest difference when the average percentage of indels and mutations rises to 5%.
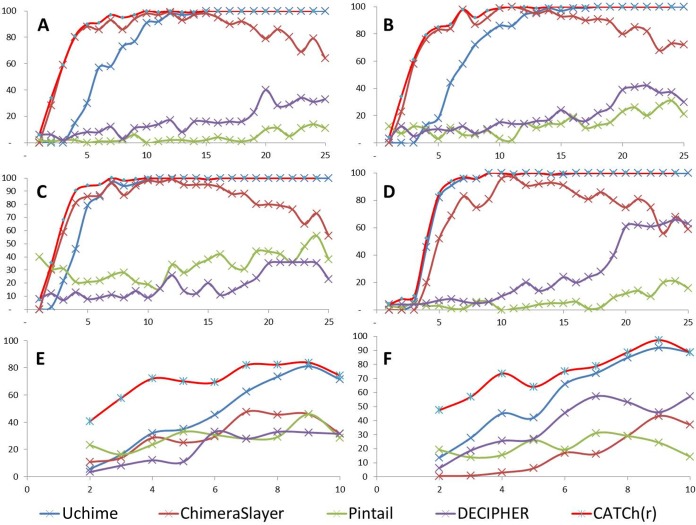
As an additional test, the chimeras in Simu2 and Simu4 were separated according to their level of divergence as reported in the original files (Simu2 data from 1 to 10% and Simu4 data from 1 to 25%). As shown in Fig. 2, CATCh appeared to endure those challenges, with the best sensitivity reported among the tools. Indeed, it combines the advantages of ChimeraSlayer—having a high sensitivity at low divergence—and maintains a good performance at higher divergence, as observed with UCHIME. This also confirms the finding that DECIPHER performs at its best with high divergence; however, the sensitivity is still lower than those of UCHIME and ChimeraSlayer.
FIG 2.
Effect of the divergence of the chimeric sequences (x axis) on the sensitivity of different reference-based tools (y axis) applied to data sets with deletions (A), indels (B and E), insertions (C), and mismatches (D and F). Both the Simu4 (A to D) and Simu2 (E and F) data sets were used, having chimeras with a divergence ranging from 1 to 25% and from 1 to 10%, respectively. Five tools were involved in this comparative study: UCHIME, ChimeraSlayer, Pintail, DECIPHER, and CATCh.
In order to test the robustness of the different tools against the number of segments in chimeric sequences (di-, tri-, and tetrameras) in sequencing data, a comparison of the performances on Simu2 and Simu3 data (respectively, 300 and 1,000 chimeras for each chimera type) was performed. In general, CATCh appeared to have the highest sensitivity for all of the classes of chimeras in both data sets (see Fig. S3 in the supplemental material).
Similarly, the evolution of the performance of the reference-based algorithms was tested in relation to the chimeric range (i.e., representing the shortest region originating from one of the parents) in a way similar to that described by Wright et al. (18). Plotting the sensitivity versus the chimeric range (see Fig. S4 in the supplemental material) emphasizes that the most challenging cases to detect are those in which one of the parents participated with less than 200 nt, a condition under which DECIPHER obtained very good results. This result is, in turn, reflected in a high sensitivity of the CATCh classifier. CATCh achieved the highest average sensitivity over all ranges, 0.95, while DECIPHER, UCHIME, ChimeraSlayer, and Pintail achieved sensitivities of 0.89, 0.88, 0.80, and 0.26, respectively.
Performance of de novo-based algorithms.
For the de novo algorithms, a similar comparative study was performed combining de novo UCHIME, de novo ChimeraSlayer, and Perseus, testing them against de novo CATCh using the Mock1 data set as described in Materials and Methods. CATCh outperformed Perseus and UCHIME in the de novo mode by achieving a sensitivity of 0.97, compared to 0.94 and 0.92 for the other tools, respectively, without any reduction in the specificity (Table 2). A second analysis took into account the different levels of divergence in the Mock1 data set (ranging from 1 to 15%). Just like the three other tools, CATCh in the de novo mode showed sustainable performance, with the clearest increase in sensitivity at lower divergence levels (see Fig. S5 in the supplemental material).
Similar to the case for reference-based algorithms, the robustness of the different tools against the number of chimeras (di-, tri-, and tetrameras) was compared for de novo chimera detection tools. The number of parents in the Mock1 data was identified as described in detail in the original paper by Quince et al. (11) (2,328 bimeras, 234 trimeras, and 3 tetrameras). As seen from these outputs, a high sensitivity is obtained for all algorithms for detection of bimeras, and our classifier showed an increase in bimera detection (0.97, compared to 0.91, 0.70, and 0.94 for UCHIME, ChimeraSlayer, and Perseus, respectively).
Benchmarking on 454 pyrosequencing data.
To illustrate in a single experiment the differences in performance between algorithms (reference and de novo based), recently published mock sequencing data (Mock2) were used (23). The data were preprocessed using two different pipelines as described in Materials and Methods, resulting in two different data sets (Mock2-a and Mock2-b), each of those preprocessing methods differing in both their computational cost and accuracy (23). The CATCh reference was found to have the highest sensitivity and maintained a similar specificity as the best-performing individual reference-based chimera detection tool on both data sets. Moreover, when comparing de novo CATCh to the best-performing individual method, a major increase in the sensitivity (11% and 8% for the Mock2-a and Mock2-b data sets, respectively) was found, with a reduction in specificity of 3% for both data sets (Table 2).
As extra evidence for the increased performance of CATCh, the cutoff score of each individual chimera detection algorithm was tuned in such way that the same specificity as obtained with CATCh was reached. Subsequently, the corresponding sensitivities for the different algorithms were compared at this fixed specificity level. Note that DECIPHER was excluded from this analysis, as it does not produce a score which can be used as a cutoff. Using Mock2-a and Mock2-b, we found that the CATCh reference had the highest sensitivity, with an improvement of 4% (on average) over the best-performing individual tool. The same behavior was observed with de novo CATCh, with an average of 3.5% improvement over the best individual de novo tool (Table 3).
TABLE 3.
Illustration of the performance difference between CATCh (default score) and each individual tool after applying a cutoff which results in the same specificity as the corresponding CATCh outputa
| Parameter | Value for indicated reference-based method |
Value for indicated de novo method |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| UCHIME | Chimera Slayer | Pintail | DECIPHER | CATCh | UCHIME | Chimera Slayer | Perseus | CATCh | |
| Mock2-a | |||||||||
| Sensitivity | 0.77 | 0.77 | 0.03 | 0.61 | 0.82 | 0.78 | 0.78 | 0.80 | 0.85 |
| Specificity | 0.92 | 0.92 | 0.92 | 0.93 | 0.92 | 0.91 | 0.91 | 0.91 | 0.91 |
| MCC | 0.64 | 0.65 | −0.12 | 0.49 | 0.70 | 0.64 | 0.64 | 0.66 | 0.72 |
| Accuracy | 0.82 | 0.83 | 0.30 | 0.72 | 0.86 | 0.83 | 0.82 | 0.83 | 0.87 |
| Cutoff | 0.44 | 72.5 | 31.5 | 0.50 | 0.62 | 0.21 | 74.0 | 0.40 | 0.70 |
| Difference | 5 | 5 | 79 | 19 | 7 | 7 | 5 | ||
| Mock2-b | |||||||||
| Sensitivity | 0.90 | 0.89 | 0.02 | 0.70 | 0.93 | 0.90 | 0.90 | 0.93 | 0.95 |
| Specificity | 0.92 | 0.92 | 0.91b | 0.93 | 0.92 | 0.91 | 0.91 | 0.91 | 0.91 |
| MCC | 0.79 | 0.77 | −0.18 | 0.56 | 0.82 | 0.78 | 0.76 | 0.82 | 0.84 |
| Accuracy | 0.91 | 0.90 | 0.26 | 0.76 | 0.92 | 0.90 | 0.90 | 0.93 | 0.94 |
| Cutoff | 0.3 | 44 | 30 | 0.5 | 0.62 | 0.17 | 73.0 | 0.23 | 0.70 |
| Difference | 3 | 4 | 91 | 13 | 5 | 5 | 2 | ||
The only exception is DECIPHER, as it does not allows the usage of a cutoff score. The difference is the increase in sensitivity obtained when using the CATCh classifier compared to that of each individual tool.
Pintail could not reach a specificity of 0.92 with any of the cutoff scores.
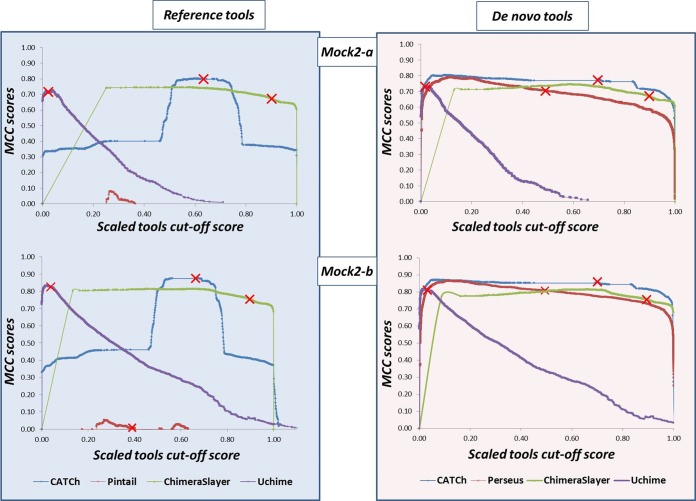
To further illustrate the improvement brought by both CATCh classifiers, and to show that this improvement is independent of the threshold score applied to any of the other individual tools, a parameter sweep using different cutoff scores was applied for the different chimera detection tools (except for DECIPHER, which does not provide a cutoff score). For each parameter setting, the MCC was calculated for all algorithms and both CATCh classifiers. This approach was applied to both Mock2 data sets (Mock2-a and Mock2-b), where in each data set reads were randomly sampled in such a way that the number of chimeric sequences equaled the nonchimeric sequences. This was not to bias the plot toward the sensitivity or the specificity (as the chimeric sequences were more abundant than nonchimeric ones in Mock2). This led to 10,894 and 16,604 reads for Mock2-a and Mock2-b, respectively, each consisting of an equal ratio of chimeric and nonchimeric reads.
From the graphs it can be determined that CATCh had the highest MCC score over a wide range of different cutoff scores (Fig. 3). For the de novo implementations, the graphs for de novo CATCh and Perseus in some regions showed similar trends. However, when tracing back the sensitivity and specificity values at these points, we noticed that these peaks are situated in the left region of the graph (i.e., corresponding to a low cutoff score), resulting in a rather exceptional low specificity. For example, for Mock2-a these high MCC values are produced by gaining a high sensitivity (e.g., 0.93 for CATCh and 0.91 for Perseus on Mock2-a), at the cost, however, of a considerable reduction of the specificity (0.87 for both tools on Mock2-a), denoting a dramatic increase in the number of false-positive chimeras. Moreover, using the default cutoff score of each tool (indicated with a red cross in Fig. 3), CATCh automatically returns those predictions that reflect the most optimal performance, while other tools might perform suboptimally with their default cutoff scores (e.g., Perseus). This behavior of CATCh is a major advantage when dealing with real-life data, as it is far from trivial to change the cutoff score to obtain an optimal performance when dealing with nonmock data.
FIG 3.
Plots illustrating the performance of each tool (reference and de novo) using the Matthews correlation coefficient (MCC) (y axis) when changing their (scaled) cutoff parameters in the x axis, using Mock2-a and Mock2-b. The scores are scaled to fit the same range between 0.00 and 1.00. The plot shows that the performance of CATCh (reference and de novo) was the highest based on results obtained for different cutoff scores for each individual tool.
Benchmarking on Illumina Miseq data.
Next to 454 pyrosequencing data, the performance of chimera detection algorithms was also tested on Illumina MiSeq data using the Mock3 community. In general, the same trend was observed regarding the performance at the level of sensitivity and specificity. For the reference-based implementations, an increase in sensitivity of 6% was observed compared with the best-performing individual tool (UCHIME), with a drop in the specificity of 1%. Similarly, for de novo implementations, the 2% drop in specificity of CATCh compared to that of Perseus was largely compensated by an increase in sensitivity of 8% (Table 2).
Benchmarking on 18S rRNA amplicon sequencing data.
The main results described in this article were obtained for 16S rRNA amplicon sequencing data. However, the de novo implementations have been suggested to work also for other marker genes (10). Comparing all de novo implementations on Mock4 and Mock5—both containing 18S rRNA amplicon sequencing data—resulted in the same conclusion as for Mock2 and Mock3, i.e., an increase in sensitivity combined with a drop in specificity (Table 2; see also Table S8 in the supplemental material). More importantly, the sensitivity values were in general much lower than the performance obtained for 16S rRNA sequencing data. The best-performing individual algorithm on these data sets (UCHIME) obtained sensitivity values of only 37% and 8% for Mock4 and Mock5, respectively, while CATCh obtained sensitivity values of 47% and 16%, respectively.
Impact of CATCh on OTU clustering.
Apart from showing the improved accuracy of CATCh on the sensitivity and specificity statistics, the effect of our algorithm on the OTU clustering was assessed. Indeed, even when working with mock communities, the number of OTUs in most cases exceeds the theoretical number of species in the mock communities. This inflation of the number of OTUs can partially be explained by the presence of undetected chimeric sequences. Consequently, chimera detection tools can reduce the number of OTUs by removing those chimeras before OTU clustering is performed. When assessing this feature on the Mock1 community, CATCh clearly had the most beneficial effect, as it reduced the number of OTUs 23% and 35% compared to the best available tools for reference (UCHIME) and de novo (Perseus) applications, respectively. This effect can also been visualized by plotting the rarefaction curves after applying all tested chimera detection tools (see Fig. S6 in the supplemental material). The same trend was observed for Mock2-a and Mock2-b (samples rarefied to 10,000 reads; see Fig. S7 in the supplemental material), leading to drops of 8% and 11%, respectively, in the numbers of OTUs using the reference-based CATCh implementation. Running de novo CATCh on Mock2-a and Mock2-b had an even more dramatic effect by reducing the number of OTUs 22% and 18%, respectively, compared with the second-best tool. It should be noted within this context that the diversity of the Mock2 community is much lower than that of Mock1 (20 species versus 91 species), which might explain the smaller improvement observed for Mock2. However, in general, by using CATCh the number of OTUs returned is closer to the expected number of species in the mock communities. This reduction in number of OTUs is also confirmed when running CATCh on real biological data, like the 11 samples used in the SOP as proposed by Schloss et al. (23) (downloaded from http://www.mothur.org/w/images/a/a1/SOPData.zip). Indeed, integrating CATCh (reference based or de novo) into the processing pipeline led to reductions of 23% and 30% of the OTUs compared with UCHIME reference or Perseus, respectively.
Computational resources.
The computational cost of the different chimera detection algorithms was assessed by examining the running times for all chimera detection algorithms on three different data sets, each of them produced by a separate sequencing technology, i.e., the Mock2-a data set obtained via 454 pyrosequencing (23), Mock3 via Illumina MiSeq sequencing technology (24), and an additional data set via PacBio SMRT sequencing (25).
The execution time—consisting of CPU time and input/output operations—differs significantly between the different tools and is also largely dependent on the number of unique reads and average read length produced per sample (Table 4). In general, de novo tools outperform reference-based tools at the level of execution time. However, as the execution time for de novo tools increases exponentially with the number of input sequences, while for reference-based algorithms this is only linear, the difference between both approaches diminishes with an increasing number of unique reads. For example, de novo UCHIME performs 130 times faster on Mock2-a than the UCHIME reference, while its performance is reduced to 6 times faster with the PacBio data set. For ChimeraSlayer, execution times are almost the same for the reference and de novo implementations on the PacBio data set.
TABLE 4.
Execution times of different reference-based and de novo chimera detection tools, tested on 16S rRNA amplicon sequencing data obtained from three different sequencing platforms
| Detection tool | Parameter | Value for indicated data set |
||
|---|---|---|---|---|
| Mock2-a | Mock3 | PacBio | ||
| Sequencing platform | 454 Titanium | MiSeq | PacBio | |
| Total data set statistics | No. of reads | 1,433,524 | 2,676,388 | 136,327 |
| No. of unique reads | 18,652 | 25,570 | 100,154 | |
| No. of samples or replicates | 90 | 8 | 16 | |
| Avg/sample/replicate | Length | 197 | 259 | 423 |
| No. of reads | 15,928 | 334,549 | 8,520 | |
| No. of unique reads | 207 | 3,196 | 6,260 | |
| Execution time (h:min:s) for reference-based methoda | UCHIME | 0:02:10 | 0:34:39 | 2:07:05 |
| ChimeraSlayer | 0:00:41 | 0:11:35 | 0:32:15 | |
| Pintail | 0:05:50 | 1:15:20 | 3:40:56 | |
| DECIPHER | 0:20:39 | 0:35:30 | 1:04:10 | |
| CATCh (classifier) | 0:00:02 | 0:00:23 | 0:00:49 | |
| CATCh (total) | 0:29:22 | 2:37:27 | 7:25:15 | |
| Execution time for de novo methoda | UCHIME | 0:00:01 | 0:01:25 | 0:21:11 |
| ChimeraSlayer | 0:00:40 | 0:11:39 | 0:28:19 | |
| Perseus | 0:00:10 | 0:34:19 | 3:12:48 | |
| CATCh (classifier) | 0:00:01 | 0:00:20 | 0:00:38 | |
| CATCh (total) | 0:00:52 | 0:47:43 | 4:02:56 | |
For CATCh the execution time of the classifier is mentioned (CATCh classifier), as well as the total runtime required for the CATCh pipeline, being the sum of the execution times of different individual tools and the classifier.
Regarding the reference-based tools, reference-based UCHIME performs very fast on small data sets (i.e., with a low number of unique reads) in comparison with DECPIHER. The longer execution time for DECIPHER for small data sets is largely due to reading in the large reference database supporting the algorithm. However, once this database is read in, DECIPHER can quite easily withstand an increase in sequencing data, leading only to a relatively small increase in execution time when running on large data sets, clearly outperforming UCHIME.
For de novo algorithms, UCHIME outperforms Perseus and, to a lesser extent, also ChimeraSlayer. However, important to note within this context is the exceptional performance of Perseus on larger data sets like Mock3: it is the best-performing individual chimera detection tool (10% higher sensitivity than UCHIME), largely making up for the longer execution time.
The added computation burden of the CATCh classifier—integrating and processing the outputs of the individual tools—is rather limited, not exceeding 1 min even for the largest data set. However, as all individual tools need to be run separately before CATCh can be applied, the execution time of our proposed pipeline is dependent to the slowest-performing algorithm (Pintail in reference mode and Perseus in de novo mode). For data sets containing a low number of unique reads (like Mock2-a), the total execution time of reference-based and de novo CATCh is significantly longer than for the best-performing individual tools (reference-based UCHIME and Perseus, respectively), i.e., 29 min versus 2 min for the reference-based implementation and 52 s versus 10 s for the de novo implementation. However, on large data sets this effect is less pronounced: the reference mode of CATCh takes around 3.5 times more time than UCHIME. For the de novo mode, this effect is even more modest, extending the execution time only 25% compared with that of the best individual tool, Perseus.
DISCUSSION
The first goal of this work was to perform a comparative study between different chimera detection algorithms. For each of the implementations, an obvious bias was observed toward the data sets that were used to fine-tune their respective algorithms, as almost each algorithm outperformed the other tools on its own subset of data. Nevertheless, important trends in chimera detection tools were observed: UCHIME showed robustness against the presence of mutations, ChimeraSlayer clearly outperformed other tools when dealing with sequences containing indels at a low divergence level, and DECIPHER performed very efficiently on chimeras with a short chimeric range.
In order to combine the advantages of all of these tools, we used a machine learning approach to develop a classifier that combines the output of all of these chimera prediction tools in an intelligent way. Our reference-based classifier obtained in almost all cases a better sensitivity. The same trend has been observed with de novo chimera detection algorithms. Where the improvement in sensitivity of our classifier was less pronounced for the Mock1 data set, a significant increase in the sensitivity was observed for the Mock2-a and Mock2-b 454 pyrosequencing read data sets as well as the Mock3 Illumina MiSeq data set. On the other hand, CATCh frequently reported a higher number of false positives, resulting in a lower specificity. However, when forcing all algorithms to produce the same specificity by tuning the cutoff scores, both CATCh implementations (reference and de novo) showed a significant increase in sensitivity compared with the best-scoring individual algorithm. In summary, when comparing the results of CATCh with the second-best-performing tool (UCHIME for reference and Perseus for de novo tools) for all possible mock data sets tested in this study, it can be seen that CATCh detects, on average, 7% and 8% more chimeras for the reference and de novo implementations, respectively. This is at the cost of only 1% at the specificity level in both cases. This beneficial effect on the detection chimera is translated into more accurate results after OTU clustering (up to 35% fewer OTUs predicted), since data processed by CATCh are for Mock1 and Mock2 consistently closer to the theoretical number of OTUs expected in the community (i.e., the number of species in the mock community). Caution should be used when extrapolating those conclusions to real biological samples, since they will have a greater diversity than most mock communities. However, mock communities are the only way to reliably assess the difference in performance when benchmarking different algorithms.
While in the initial study the database-dependent algorithms slightly outperformed the de novo tools (sensitivity of 79% versus 74%) (18), the opposite was observed when comparing our reference-based classifier with the de novo classifier (84% versus 94%). Regardless of the fact that in their study the reference-based algorithms outperformed the de novo ones, it has already been suggested that the de novo (database-independent) approach be used over the reference-based ones, first due to the independence on incomplete reference databases and second because of a faster execution time (23). Furthermore, we show here that the de novo approach developed in this study is also at the performance level the preferred approach over reference-based methodologies. Indeed, even in an area where it would be expected that reference-based algorithms would be favored—since we are dealing with mock communities containing well-studied bacterial isolates known to be present in the reference databases—the de novo classifier has a better performance than the reference-based approached. This conclusion corresponds with the work by Edgar et al. (10) calculating a higher performance of de novo UCHIME on a mock data set than for the reference-based UCHIME.
Where the de novo approach also results in faster execution times when dealing with small data sets, this effect is less clear with data sets containing a large number of unique reads (>3,000 sequences, i.e., samples with a high sequencing coverage or highly diverse samples). The best-performing individual algorithms, UCHIME and Perseus for reference-based and de novo methods, respectively, require comparable execution times in such a large-scale setup. When extending this to CATCh, the additional computational burden is larger in the reference-based mode, resulting in a significant increase in execution times for small and large sequencing data sets. However, for the de novo mode this effect is much more modest, extending the runtime only 25% for large sequencing data sets.
Important to note is that this significant increase in performance of our de novo classifier was obtained by combining only three de novo chimera detection tools (de novo UCHIME, de novo ChimeraSlayer, and Perseus). Our classifier would benefit from integrating more de novo algorithms once they are available. This also highlights the power of such ensemble algorithms, allowing the integration of as many useful tools as available. When new algorithms become available in the future, they can easily be integrated in the classifier, most probably leading to even better chimera prediction results than the ones presented here.
Additionally, when combining the output of different algorithms, a machine learning approach is preferred over a rudimentary approach such as taking the union or intersection of the predictions of different algorithms. Indeed, taking the union of ChimeraSlayer, UCHIME, and DECIPHER would result in sensitivity values comparable to results obtained with CATCh, with, however, a dramatic decrease of the specificity (88%, 78%, 83%, and 84% for the Simu1, Simu2, Simu3, and Simu4 data sets, respectively, versus 96%, 98%, 97%, and 97% for CATCh). These data agree with the observation that the number of false-positive chimera predictions is minimal between DECIPHER and UCHIME, leading to a decrease in specificity when simply combining both outputs in a straightforward way (18). For the de novo implementation, the opposite trend is observed, as the sensitivity of our classifier is higher than the union while maintaining a similar specificity, suggesting an overlap of Perseus and de novo UCHIME in the false-positive predictions while having an added effect in the detection of true chimeras (data not shown).
The majority of the conclusions discussed in this article are derived based on 16S rRNA sequencing data. Certainly for the de novo implementations, it is tempting to assume that the area of application could easily be extended to other biodiversity marker genes like 18S rRNA, 23S rRNA, or ITS regions. However, a small-scale test case using two different 18S rRNA mock communities suggests that additional analyses might lead to unexpected results and a rethinking of the chimera detection methods currently used in eukaryotic biodiversity assessments. While the basic concept of de novo tools should also be applicable for assessing chimeric sequencing in eukaryotic biodiversity studies, precaution is needed when shifting away from the 16S rRNA marker gene to eukaryotic marker genes, as a straightforward application of de novo chimera detection tools on 18S rRNA results in significantly lower sensitivity values.
In conclusion, a comparison between different chimera prediction tools was performed, pointing out each tool's strengths and weaknesses. Based on this information, an ensemble classifier was developed for reference-based as well as de novo chimera detection tools, which is able to produce stable results over various mock data sets. Moreover, since the classifier combines the strengths of various individual chimera detection tools, it shows an increased robustness against different confounding factors, like a low parent divergence, short length of the chimeric range, and a varying number of parents. The beneficial effect of CATCh is highlighted by improved OTU clustering results in mock data sets, returning OTU numbers closer to the true number of species in the mock community.
Supplementary Material
ACKNOWLEDGMENTS
Mohamed Mysara was supported by an SCK•CEN Ph.D. grant.
We thank R. C. Edgar, E. S. Wright, P. D. Schloss, and M. J. Morgan for their help.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02896-14.
REFERENCES
- 1.Wang GC, Wang Y. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142:1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- 2.Wang GC, Wang Y. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl Environ Microbiol 63:4645–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson JR, Marcelino LA, Polz MF. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR.’ Nucleic Acids Res 30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lahr DJ, Katz LA. 2009. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques 47:857–866. doi: 10.2144/000113219. [DOI] [PubMed] [Google Scholar]
- 6.Porazinska DL, Giblin-Davis RM, Sung W, Thomas WK. 2012. The nature and frequency of chimeras in eukaryotic metagenetic samples. J Nematol 44:18–25. [PMC free article] [PubMed] [Google Scholar]
- 7.Odelberg SJ, Weiss RB, Hata A, White R. 1995. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res 23:2049–2057. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Judo MS, Wedel AB, Wilson C. 1998. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res 26:1819–1825. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smyth RP, Schlub TE, Grimm A, Venturi V, Chopra A, Mallal S, Davenport MP, Mak J. 2010. Reducing chimera formation during PCR amplification to ensure accurate genotyping. Gene 469:45–51. doi: 10.1016/j.gene.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logares R, Haverkamp TH, Kumar S, Lanzen A, Nederbragt AJ, Quince C, Kauserud H. 2012. Environmental microbiology through the lens of high-throughput DNA sequencing: synopsis of current platforms and bioinformatics approaches. J Microbiol Methods 91:106–113. doi: 10.1016/j.mimet.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 14.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright ES, Yilmaz LS, Noguera DR. 2012. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78:717–725. doi: 10.1128/AEM.06516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski Ra, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 21.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. 2009. The WEKA data mining software: an update. SIGKDD Explor Newsl 11:10–18. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- 22.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, Gordon JI. 2010. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A 107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall CW, Ross DE, Fichot EB, Norman RS, May HD. 2012. Electrosynthesis of commodity chemicals by an autotrophic microbial community. Appl Environ Microbiol 78:8412–8420. doi: 10.1128/AEM.02401-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fonseca VG, Nichols B, Lallias D, Quince C, Carvalho GR, Power DM, Creer S. 2012. Sample richness and genetic diversity as drivers of chimera formation in nSSU metagenetic analyses. Nucleic Acids Res 40:e66. doi: 10.1093/nar/gks002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan MJ, Chariton AA, Hartley DM, Court LN, Hardy CM. 2013. Improved inference of taxonomic richness from environmental DNA. PLoS One 8:e71974. doi: 10.1371/journal.pone.0071974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.