Abstract
Identification of proteins that were present in a polyhydroxybutyrate (PHB) granule fraction isolated from Ralstonia eutropha but absent in the soluble, membrane, and membrane-associated fractions revealed the presence of only 12 polypeptides with PHB-specific locations plus 4 previously known PHB-associated proteins with multiple locations. None of the previously postulated PHB depolymerase isoenzymes (PhaZa2 to PhaZa5, PhaZd1, and PhaZd2) and none of the two known 3-hydroxybutyrate oligomer hydrolases (PhaZb and PhaZc) were significantly present in isolated PHB granules. Four polypeptides were found that had not yet been identified in PHB granules. Three of the novel proteins are putative α/β-hydrolases, and two of those (A0671 and B1632) have a PHB synthase/depolymerase signature. The third novel protein (A0225) is a patatin-like phospholipase, a type of enzyme that has not been described for PHB granules of any PHB-accumulating species. No function has been ascribed to the fourth protein (A2001), but its encoding gene forms an operon with phaB2 (acetoacetyl-coenzyme A [CoA] reductase) and phaC2 (PHB synthase), and this is in line with a putative function in PHB metabolism. The localization of the four new proteins at the PHB granule surface was confirmed in vivo by fluorescence microscopy of constructed fusion proteins with enhanced yellow fluorescent protein (eYFP). Deletion of A0671 and B1632 had a minor but detectable effect on the PHB mobilization ability in the stationary growth phase of nutrient broth (NB)-gluconate cells, confirming the functional involvement of both proteins in PHB metabolism.
INTRODUCTION
Polyhydroxybutyrate (PHB) and related polyhydroxyalkanoates (PHA) are storage compounds for carbon and energy and are widespread in prokaryotic species (1). PHB is deposited in the form of 200- to 500-nm particles (granules) when PHB-accumulating bacteria are cultivated in the presence of a surplus of a suitable carbon source. Ralstonia eutropha H16 (alternative designation, Cupriavidus necator) has become the model organism of PHB research, and the species is also used in industrial processes to produce PHB as a biodegradable polymer with plastic-like properties (2–4). The intensive research of the last few decades has led to a good understanding of the biochemical routes leading to PHB/PHA and of the biochemical properties of proteins with established functions in PHB/PHA metabolism. For example, the key enzymes of PHB synthesis, the PHB synthases (PhaCs), of many PHB-accumulating species have been purified; the coding genes were cloned; and the polymerization reaction has been studied (for overviews and recent results, see references 5 to 11). Meanwhile, much evidence has accumulated showing that PHB granules are not simply storage molecules but represent well-defined subcellular organelles that consist of a polymer core and a surface layer to which many proteins with specific functions are attached (12). Besides the aforementioned PHB synthase, small amphiphilic polypeptides, so-called phasin proteins (PhaPs) (13, 14) that mediate between the hydrophilic cytoplasm and the hydrophobic polymer and that can contribute to higher stress resistance of PHA-accumulating bacteria (15), and PHB depolymerase(s) (PhaZs) (16–18) have been identified in PHB granules of all investigated PHB-accumulating species. Moreover, at least two proteins were identified that can bind both to the PHB granule and to DNA. One is PhaR, a transcriptional regulator that can be titrated from its DNA binding site by binding to the PHB granule surface (19–21); the other is PhaM, which is responsible for anchoring PHB granules to the bacterial nucleoid via binding to the PHB synthase PhaC1 and to DNA (11, 22, 23). A comparable set of PHA granule-associated proteins (PGAPs) is present in the surface layer of medium-chain-length PHA in bacteria such as Pseudomonas putida (reference 24 and references therein) and in all PHA-accumulating species that have been examined, including Archaea (25, 26). At present, it is not known whether the PHB/PHA granule surface layer is a phospholipid monolayer in which the proteins mentioned above are embedded or whether the granule surface layer consist of proteins only. (For recent overviews and reports on PHB granule formation, see references 9 and 27–29.)
Analysis of the protein compositions of cell fractions by a combination of SDS-PAGE, trypsin digestion, and liquid chromatography coupled with mass spectrometry (LC-MS) is a powerful and widespread tool for the determination of expression profiles. Proteome analysis has been used to study the composition of the surface layer of isolated PHB granules and to identify PGAPs. Recently, a Brazilian group determined the proteins of PHB granules isolated from Herbaspirillum seropedicae (30). Remarkably, a histone-like protein with some features similar to those of PhaM and PhaF (11, 22–24), two outer membrane proteins (OMPs), several hypothetical proteins, and also a protein of the tricarboxylic acid (TCA) cycle (aconitase) were identified, in addition to proteins with well-established functions in PHB metabolism (PHB synthase, the phasin PhaP, and the PHB depolymerase PhaZ). Proteins with functions not obviously related to PHB metabolism had already been identified in earlier studies of PHA-accumulating bacteria. One example from R. eutropha is the chaperone GroEL, which was present in substantial amounts in the PHB granule fraction of a ΔphaP1 mutant (13). On the other hand, some phasin proteins were identified in a membrane fraction from R. eutropha by proteome analysis (31). However, PHB granules were not the subject of that study, and PHB granule-associated proteins might have contaminated the membrane fraction.
We recently determined the proteome of PHB granules isolated from R. eutropha (29). Again, several proteins with known functions in PHB metabolism were identified, but a considerable number of outer membrane proteins and many other proteins (404 in total) were also detected. It is unlikely that several hundred proteins have a specific function in PHB metabolism and are bound to PHB granules in vivo. Presumably, many of the detected proteins bind artificially to PHB during cell disruption or during the PHB granule isolation process and therefore should be considered false-positive PHB granule-associated proteins. One possible way to identify true-positive PGAPs is to construct fusions with green fluorescent protein variants and to determine the subcellular localization of the fusion by fluorescence microscopy. Only the proteins that colocalize with PHB granules are true in vivo PGAPs. This method has been used to confirm the subcellular localization of PHB synthases, phasins, and PHB depolymerases (32–39). Because of the high number (>400) of proteins identified in the PHB granule fraction by proteome analysis (29), the in vivo confirmation of their subcellular localizations would require the construction of hundreds of fusions with green fluorescent protein. This is hardly practicable, and therefore, we decided to use another approach to identify in vivo PHB granule-associated proteins; to this end, four subcellular fractions (soluble, membrane, membrane-associated, and PHB granule fractions) were prepared from R. eutropha cells, and the proteomes of all four fractions were determined and compared. Proteins specifically present only in the PHB granule fraction but absent in the other fractions should resemble “true” PHB granule-associated proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacteria and plasmids used in this study are shown in Table 1. Escherichia coli strains were grown on LB medium supplemented with the appropriate antibiotics at 37°C. R. eutropha H16 strains were routinely grown on nutrient broth (NB) (0.8% [wt/vol]) with or without addition of 0.2% (wt/vol) sodium gluconate at 30°C. Alternatively, R. eutropha was grown on gluconate (1.0% [wt/vol]) mineral salts medium (MM) (40). For proteome analysis, six parallel cultures of R. eutropha H16 were grown (each in a 3-liter Erlenmeyer flask filled with 400 ml NB medium supplemented with 0.2% sodium gluconate or 400 ml MM with 1% [wt/vol] sodium gluconate; inoculum size, 0.1 volume). The optical density at 600 nm (OD600) was followed, and after ∼8 h (NB-gluconate) or ∼30 h (MM-gluconate), the cells were at the end of exponential growth. At this time point, four parallel cultures were harvested by centrifugation, and the two remaining cultures were used to complete the growth curve. The harvested cells were suspended in 10 mM Tris-HCl, pH 8, and transferred to 50-ml Falcon tubes. The washing step with 50 ml buffer was repeated once, and the wet weight of the cell pellet was determined. The cells were stored frozen (−20°C) until use.
TABLE 1.
Strains and plasmids used in this study
| Strain/plasmid | Relevant characteristic | Source/reference |
|---|---|---|
| E. coli JM109 | Cloning strain | |
| E. coli S17-1 | Conjugation strain | 67 |
| R. eutropha H16 | R. eutropha wild-type strain | DSMZ 428 |
| H16ΔA0671 | Chromosomal deletion of A0671 | This study |
| H16ΔB1632 | Chromosomal deletion of B1632 | This study |
| H16ΔA0671ΔB1632 | Chromosomal deletions of A0671 and B1632 | This study |
| pBBR1MCS2 | Broad-host-range vector; Kmr | 68 |
| pBBR1MCS2-PphaC-eyfp-c1 | Universal vector for construction of fusions C-terminal to eYFP under the control of the PphaC promoter | 43 |
| pBBR1MCS-2-PphaC-eyfp-A0225 | N-terminal fusion of A0225 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-A0671 | N-terminal fusion of A0671 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-A2001 | N-terminal fusion of A2001 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-B1632 | N-terminal fusion of B1632 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-phaZc | N-terminal fusion of PhaZc to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-A3253 | N-terminal fusion of A3253 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-A2924 | N-terminal fusion of A2924 to eYFP | This study |
| pBBR1MCS-2-PphaC-eyfp-B0232 | N-terminal fusion of B0232 to eYFP | This study |
| pLO3 | Suicide vector; Tcr | 69 |
| pLO3-ΔA0671 | Deletion vector for A0671; fragments up- and downstream of A0671 cloned between SacI and XbaI sites of pLO3 | This study |
| pLO3-ΔB1632 | Deletion vector for B1632; fragments up- and downstream of B1632 cloned between SacI and XbaI sites of pLO3 | This study |
Cell fractioning.
Frozen cell pellets (approximately 3 g [wet weight] from 400 ml of culture) were thawed and resuspended in 20 ml of ice-cold Tris-HCl buffer (10 mM, pH 8). Shortly before cell disruption, DNase I and RNase I were added to the samples (100 μl of a 1-mg/ml solution each) to decrease the viscosity of the samples. The cells were disrupted by two passages through a cooled French press chamber (Aminco, Silver Spring, MD, USA) at 95 MPa. For separation of the PHB granules, the lysed cells were loaded onto a first glycerol gradient consisting of 5 ml 87% (vol/vol) and 10 ml 50% (vol/vol) glycerol (dilutions of glycerol in 10 mM Tris buffer, pH 8). After centrifugation for 40 min at 20,000 rpm and 4°C (Beckman Avanti-35 ultracentrifuge; SW28 swing-out rotor), the supernatant (which contained soluble proteins and membranes) was separated from the PHB granules. The PHB granules were diluted with 1 volume of Tris-HCl buffer (10 mM, pH 8); purified by application of a second glycerol gradient centrifugation step consisting of 5 ml (each) 87%, 80%, 60%, and 40% glycerol; and stored at −20°C until use. The supernatant was centrifuged at 35,000 rpm and 4°C for 90 min (TFT65.13 rotor; Kontron) to separate the membranes from the soluble proteins. To this end, each supernatant was split into two centrifuge tubes (each containing ∼7 to 8 ml of supernatant). Seven milliliters of ice-cold Tris-HCl buffer (pH 8; 10 mM) was added to the membrane pellets obtained, and the membranes were homogenized by repeated passages (≥15 times) through a Potter-Elvehjem homogenizer (a glass/Teflon homogenizer in which the mortar is made of glass and the pestle is Teflon coated). The homogenized membranes were centrifuged again (90 min; 35,000 rpm; 4°C) and subsequently suspended in 300 μl of an ice-cold Na2CO3 solution (100 mM, pH 11.5) with the aid of a glass piston. The membranes were incubated on ice overnight (carbonate extraction of peripheral membrane proteins). After centrifugation for 90 min at 35,000 rpm (4°C), the resulting supernatant was used as a carbonate extract for proteome analysis. The remaining membrane pellets were washed again as described above and then dissolved in 300 μl of 2% (wt/vol) SDS with 10 mM dithiothreitol (DTT). Part of the PHB granule fraction (0.2 to 1.0 ml, depending on the density of the PHB granule fraction) was diluted with 5 volumes of Tris-HCl buffer (10 mM, pH 8) and centrifuged for 1 to 2 min at 12,000 to 13,000 rpm in an Eppendorf centrifuge. These washing and centrifugation steps were repeated four times. The PHB pellets were then suspended in 100 μl of 2% (wt/vol) SDS and heated at 95°C for 10 min to solubilize PGAPs from the granule surface. After an additional centrifugation step to remove the insoluble PHB, the supernatant served as the PHB granule extract. The protein concentrations of the crude extract, carbonate extract, dissolved membranes, and PHB granule extract were determined with the bicinchoninic acid (BCA) assay kit (Pierce), and samples were diluted with 3× sample buffer (6% [wt/vol] SDS, 25% [vol/vol] glycerol, 3% [vol/vol] 2-mercaptoethanol, 0.15% bromphenol blue) and heated for 3 min at 95°C before being loaded onto gels for proteome analysis.
Construction of fusion proteins with eYFP.
Fusions of the gene (eyfp) for enhanced yellow fluorescent protein (eYFP) with genes of R. eutropha H16 (Table 1) were generally constructed as N-terminal fusions using a universal vector (pBBR1MCS-2-PphaC-eyfp-c1) based on the broad-host-range plasmid pBBR1MCS-2, as described previously (22, 39). All constructs were conjugatively transferred from recombinant E. coli S17-1 to R. eutropha H16, and selection was achieved by plating on mineral salts medium supplemented with 0.2% fructose and 350 μg ml−1 kanamycin.
Construction of chromosomal knockouts.
Precise chromosomal deletions of the A0671 or B1632 gene or of both genes were constructed in R. eutropha H16 using the sacB-sucrose selection method (15% sucrose was used for selection) and pLO3 as the deletion vector. The genotype of the resulting deletion mutant was verified by PCR of the respective genomic region and determination of its DNA sequence.
Proteome analysis.
Proteome analysis was performed by the proteome core facility of the Life Science Center, University of Hohenheim (Stuttgart, Germany), as described in detail previously (41), with some modifications. Three replicates, each with 50 to 200 μg of the respective protein fraction, were diluted in sample buffer (see above) and separated on a 10% SDS-polyacrylamide gel (SDS-PAGE). The gel was stained with 0.12% (wt/vol) colloidal Coomassie brilliant blue G250, 10% (wt/vol) ammonium sulfate, 10% (vol/vol) methanol, 20% (vol/vol) phosphoric acid. Each sample lane was cut into 5 slices (soluble fraction), 3 slices (membrane fraction), 3 slices (membrane-associated fraction), or 1 slice (PHB granule fraction), with only a short electrophoresis time of 20 to 25 min; digested with trypsin: and extracted with acetonitrile (10 min). The supernatants were vacuum dried and dissolved in formic acid (15 μl; 0.1% [vol/vol]). Five microliters was subjected to LC-tandem MS (MS-MS) analysis. The data were analyzed using Scaffold proteome software (version 3.4.9). Only proteins that were identified by at least two peptides and that had a protein probability of >99% were considered. Label-free quantification was performed by calculating the ion intensities using Progenesis LC-MS software 3.1 (Nonlinear Dynamics) as described elsewhere (41). Protein abundance was calculated by summing up the abundances of all peptides assigned to a given protein. Calculations of the P value by one-way analysis of variance (ANOVA) were done with the sum of the protein abundances in all runs.
Other methods.
Quantitative analysis of PHA content was done by gas chromatography (GC) after acid methanolysis of lyophilized cells according to the method of Brandl et al. (42). For determination of PHB contents in samples of growth curves, two to four biological replicates, each with two technical replicates, were performed. Only technical replicates could be compared directly, but the PHB contents of biological replicates at a given time point showed considerable variation. Therefore, the time-dependent increase during exponential growth or decrease of PHB content during the stationary growth phase was determined (slopes of graphs of PHB content versus time), and the calculated average values of all biological replicates were determined. Manipulation and construction of plasmids were done by standard molecular biological methods and according to the supplier′s instructions. Protein concentrations were routinely determined by BCA assay. Polyacrylamide gel electrophoresis was performed under denaturing (sodium dodecyl sulfate) and reducing (mercaptoethanol) conditions. Gels were stained with Coomassie brilliant blue G250. Formation of PHB granules was determined by fluorescence microscopy using Nile red (1 to 10 μg/ml dimethyl sulfoxide [DMSO], Nile red solution at 5 to 40% [vol/vol]), as described previously (22, 43).
RESULTS
Preparation of subcellular fractions.
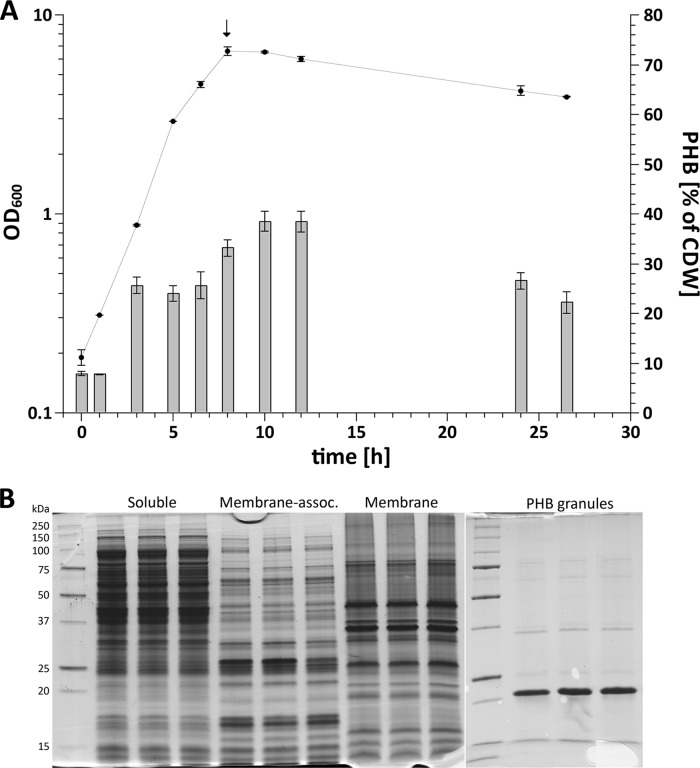
Cultures of R. eutropha H16 were grown on NB-gluconate medium. The cells were in the late exponential growth phase, as revealed by maximal OD600 values of 6 to 7 (Fig. 1A), and had a maximum PHB content of about 30% to 40% of the cellular dry weight. Cells were harvested and washed by centrifugation. Subsequently, the soluble, membrane, membrane-associated, and PHB granule fractions were prepared as described in Materials and Methods. All four subcellular fractions were separated by SDS-PAGE, each with three replicates. As shown in Fig. 1B, the protein patterns of the replicates appeared identical but clearly differed for the different subcellular fractions. This indicated that the four fractions had different protein compositions, as was expected for a successful fractioning.
FIG 1.
Growth of R. eutropha H16 and subcellular fractioning for proteome analysis. (A) Growth of R. eutropha on NB-gluconate medium. The arrow indicates the time of harvesting of part of the culture for proteome analysis. The bars show the PHB contents at different time points. The error bars indicate standard deviations. CDW, cell dry weight. (B) SDS-PAGE of subcellular fractions (three replicates for each fraction). The 1st (from the left) and the 11th lanes contain marker proteins.
Determination of the proteomes of the soluble, membrane, membrane-associated, and PHB granule fractions.
The polyacrylamide gel of each fraction and of each replicate was cut into slices as specified in Materials and Methods, and each slice was subjected to trypsin digestion and subsequent LC–MS-MS analysis. In total, 1,756, 889, 439, and 268 proteins were identified in the soluble, membrane, membrane-associated, and PHB granule fractions, respectively (see Data Set S1 in the supplemental material), if only proteins with at least two separately identified peptide fragments were considered. The total number of identified proteins was 2,264, which corresponded to 37% of the 6,116 predicted proteins (1, 2) of the R. eutropha genome. The number of identified proteins increased to 2,354 (38.5%) if the threshold was set to only one identified peptide. A considerable number of the identified proteins were strongly enriched in one cell fraction and showed no or only low abundance in the other cell fractions. For example, citrate synthase (A2627), phosphoenolpyruvate carboxylase (A2921), isocitrate dehydrogenase (A3056), glutamate synthase (A3431), and others (e.g., A3046, A3246, and B1213) are typical soluble proteins and were found exclusively in the soluble fraction. Similarly, many typical transport proteins (e.g., A0904, A1173, A1419, A1467/A1468, A2665/A2666, and B1644), other membrane proteins (A1373, B1038, and B2295), and membrane-associated proteins (PHG211, PHG271, A0808, B1229, and B1721) were found only in the membrane fraction or membrane-associated fraction (see Data Set S1 in the supplemental material). On the other hand, many proteins were present in more than one or even in all fractions. This indicated that the four cell fractions were contaminated by proteins of other fractions or that some proteins have multiple locations.
Two hundred sixty-eight proteins were identified in the PHB granule fraction. However, when we looked for proteins that were present solely in the PHB granule fraction but were absent in the other three fractions, only 12 proteins remained (Table 2). We conclude that these 12 proteins represent specifically (“true”) PHB-associated proteins. Surprisingly, many putative OMPs were identified in the PHB granule fraction, but many peptides of these OMPs were also present in great abundance in the membrane fraction (see Data Sets S1 and S2 in the supplemental material and Discussion below). We assume that the outer membrane proteins and most of the other remaining proteins of the PHB granule fraction probably represent false-positive proteins that bind artificially to PHB granules during the isolation process. However, we cannot exclude the presence of some PHB granule-specific proteins that have double or multiple in vivo localizations or that bind only very loosely to PHB granules and detach during the granule isolation process.
TABLE 2.
Abundances of PGAPs
| Granule ranka | All protein rankb | Locusc | PGAPd | Mass (kDa) | Abundance |
Fold change (NB/MM) | |
|---|---|---|---|---|---|---|---|
| NB | MM | ||||||
| 1 | 1 | A1381 | PhaP1 | 20 | 1.6 × 1010 | 9.5 × 108 | 17 |
| 2 | 2 | PHG202 | PhaP2 | 20 | 5.2 × 108 | 4.9 × 107 | 11 |
| 3 | 7 | B2021 | PhaP4 | 20 | 7.5 × 107 | 7.9 × 107 | 0.95 |
| 4 | 9 | B1632 | α/β-Hydrolase | 86 | 7.0 × 107 | 9.9 × 106 | 7.1 |
| 5 | 10 | A2172 | PhaP3 | 20 | 6.1 × 107 | 6.8 × 107 | 0.90 |
| 6 | 11 | A1437 | PhaC1 | 64 | 4.9 × 107 | 1.9 × 107 | 2.6 |
| 7 | 17 | B1934 | PhaP5 | 16 | 3.1 × 107 | 1.5 × 106 | 21 |
| 8 | 18 | A1150 | PhaZa1 | 47 | 2.4 × 107 | 3.2 × 106 | 7.5 |
| 9 | 28 | A0671 | α/β-Hydrolase | 84 | 1.3 × 107 | 3.9 × 106 | 3.3 |
| 10 | 36 | A0225 | Phospholipase | 51 | 1.0 × 107 | 1.3 × 106 | 7.7 |
| 11 | 40 | A2001 | Hypothetical protein | 18 | 9.2 × 106 | 3.9 × 106 | 2.4 |
| 12 | 136 | B2326 | PhaP7 | 16 | 1.5 × 106 | 2.2 × 105 | 6.8 |
| 13 | 323 | B1988 | PhaP6 | 23 | 2.9 × 105 | 6.4 × 104 | 4.5 |
| 14 | 332 | A2003 | PhaC2 | 65 | 2.6 × 105 | 1.3 × 105 | 2.0 |
| 15 | 21 | A1440 | PhaR | 21 | 1.8 × 107 | 3.4 × 106 | 5.3 |
| 16 | 224 | A0141 | PhaM | 27 | 5.8 × 105 | 1.1 × 105 | 5.3 |
| 22 | PHG252 | Nitrous oxide red | 70 | 1.4 × 107 | 2.4 × 107 | 0.58 | |
| 29 | A0472 | ABC transporter | 33 | 8.2 × 106 | 1.7 × 107 | 0.48 | |
| 30 | A3284 | Omp | 40 | 1.0 × 107 | 3.1 × 107 | 0.32 | |
| 37 | A3030 | ABC transporter | 41 | 7.3 × 106 | 2.9 × 107 | 0.25 | |
Rank numbers of PHB granule-associated proteins by their relative abundances in granules of NB-grown cells (averages of three replicates).
Rank numbers sorted by relative abundances of all proteins detected in PHB granules of NB-grown cells.
Italics indicates examples of proteins that had higher abundance in PHB granules of fructose mineral salts medium-grown cells. Boldface indicates novel, specifically PHB granule-associated proteins.
Note that PhaP1, PhaC1, PhaR, and PhaM are also present in other fractions.
Eight out of the 12 specifically PHB granule-bound proteins had been previously described as PGAPs or as proteins with prominent functions in PHB metabolism. These proteins are the intracellular PHB depolymerase (PhaZa1) and six phasin proteins (PhaP2 to PhaP7). The second PHB synthase-like protein, PhaC2, the function of which is unknown, was also specifically present in the PHB granule faction, although only at low abundance. The phasin PhaP1, the PHB synthase PhaC1, the transcriptional regulator PhaR, and the recently identified PhaM protein showed multiple localizations, in addition to a PHB granule localization. The presence of PhaR and PhaM in fractions other than the PHB granule fraction is explained by their multiple in vivo localizations: both proteins can bind to PHB granules and to the nucleoid (19–23). The finding of PHB synthase (PhaC1) and of the phasin PhaP1 in several cell fractions is discussed below.
A0225, A0671, A2001, and B1632 are novel PHB granule-associated proteins in R. eutropha.
The four new specifically PHB granule-associated proteins are A2001 (hypothetical protein), A0225 (patatin-like phosholipase), and A0671 and B1632 (both are putative α/β hydrolases with PHB synthase/depolymerase signatures). The A2001 gene codes for a hypothetical protein of 159 amino acids (mass, 17.8 kDa). The deduced amino acid sequence of A2001 has a UvdE (UV damage repair endonuclease) and a TIM barrel motif. The relationship of UV repair to PHB metabolism is not known. Interestingly, the A2001 gene is part of a three-gene operon (A2001 to A2003). The genes adjacent to A2001 (A2002 and A2003) code for the PhaB2 protein (acetoacetyl-coenzyme A [CoA] reductase) and the second PHB synthase, PhaC2, respectively. The function of (soluble) PhaB2 in PHB metabolism has been described previously (2-4, 44), and the colocalization of PhaC2 with PHB granules has also been shown (5-11, 38) and was confirmed in this study (see Data Set S1 in the supplemental material). A protein similar to A2001 (27% identity; 43% similarity) and in the same genomic context (Rmet5124, in an operon with an acetoacetyl-CoA reductase and a PHB synthase) is present in Ralstonia metallidurans CH34, and the protein has a phasin 2 motif.
The A0225 gene codes for a 51-kDa polypeptide, and the deduced amino acid sequence has a patatin motif and a phospholipase motif. Patatin is a storage protein in potatoes but also has phospholipase activity. Patatin-like phospholipases are frequently found in bacteria (12, 45).
The products of the A0671 and B1632 genes are predicted to be α/β-hydrolases and have a domain with unknown function (Duf 3141). This domain is also found in proteins of many proteobacteria. A0671 (84 kDa) and B1632 (86 kDa) share 46% identical and 62% similar residues (see Fig. S1 in the supplemental material). Short stretches of ∼100 residues have a relationship to type I PHB synthases and to subunit PhaC of type III PHA synthases. BLASTP analysis gives PHB synthases/depolymerases as top hits (in addition to orthologs of A0671 and B1632 in other proteobacteria).
Subcellular localization of R. eutropha proteins by construction of fusions with eYFP.
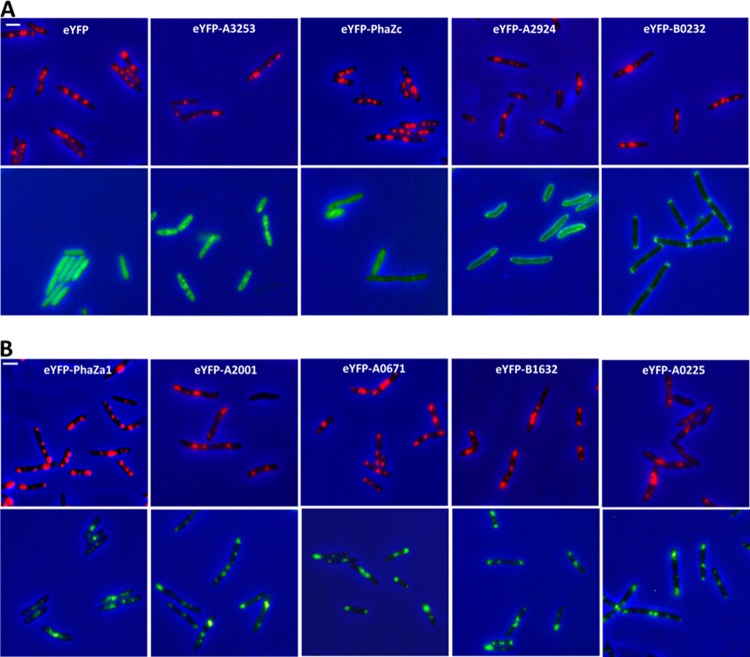
To confirm the results of comparative proteome analysis, we constructed fusion proteins of selected control proteins and of the four novel PHB-associated proteins with eYFP and determined their subcellular localizations after overexpression from the constitutive phaC1 promoter. Free eYFP is a typical soluble protein, and accordingly, expression of eYFP alone in R. eutropha resulted in uniform cellular fluorescence (Fig. 2A). The same result was obtained when a randomly selected protein (A3253; hexaprenyltrans-transferase) that localized in the soluble fraction in comparative proteome analysis (see Data Set S1 in the supplemental material) was investigated (Fig. 2A). Fusion analysis of 3-hydroxybutyryl (3HB) oligomer hydrolase (PhaZc)—a protein that was found only in the soluble fraction in this study—revealed soluble localization, and no colocalization with PHB granules could be detected (Fig. 2A). A2924 and B0232 are two predicted membrane proteins, and fusion analysis with eYFP confirmed their localization at the cell membrane. Remarkably, B0232, which is annotated as a methyl-accepting chemotaxis protein and therefore should localize in the membrane near the cell poles, indeed showed cell pole-specific membrane localization (Fig. 2A). Finally, the PHB depolymerase PhaZa1, which is a well-known PHB granule-located PHB depolymerase and was identified almost only in the PHB granule fraction, clearly colocalized with PHB granules after expression as a fusion with eYFP (Fig. 2B). Taken together, all the control proteins showed the expected subcellular localization and confirmed that fusion analysis with eYFP is a suitable tool to determine subcellular localization. Construction of fusion proteins of the four novel identified proteins that were specifically present only in the PHB granule fraction (see Data Set S1 in the supplemental material), namely, A2001, A0232, A0671, and B1632, with eYFP and constitutive expression confirmed that all of them are PHB granule bound in vivo (Fig. 2B). We conclude that A2001, A0232, A0671, and B1632 represent novel PGAPs in R. eutropha H16.
FIG 2.
Subcellular localization of R. eutropha protein fusions. (A) Subcellular localization of control proteins. Columns (from left to right): 1, cells expressing eYFP alone without a fused protein; 2, cells expressing soluble hexaprenyltranstransferase (A3253); 3, cells expressing the soluble 3HB oligomer hydrolase PhaZc; 4, cells expressing membrane-bound ECF anti-sigma factor (A2924); 5, cells expressing cell pole-localized methyl-accepting chemotaxis protein (B0232). The bar (top left) corresponds to 2 μm. (B) Subcellular localization of PHB granule-associated proteins. Columns (from left to right): 1, PhaZa1; 2, A0225; 3, A2001; 4, A0671; 5, B1632. Note the soluble, cell membrane-specific localization, or cell pole-specific localization of control proteins in panel A and colocalization of PHB granules with eYFP for all fusion proteins in panel B. The colocalization of fusion proteins with PHB granules was confirmed by identification of translucent PHB granules using phase-contrast microscopy and by observation of eYfp fluorescence of Nile red-stained cells (not shown). The bar corresponds to 2 μm. All cells were imaged after staining with Nile red (top rows) or without staining (bottom rows), showing the eYFP fluorescence without cross talk between fluorescence channels. The cells of all strains were grown in NB-gluconate medium, and after 4 h of growth, samples were taken for microscopy.
Most of the previously described intracellular PHB depolymerases and 3-hydroxybutyrate oligomer hydrolases are not part of the PHB granule proteome.
Only one of the previously described intracellular PHB depolymerases, PhaZa1 (A1150), was specifically present in high abundance in the PHB granule fraction, and 17 or 21 peptide fragments were identified there (see Data Set S1 in the supplemental material). In two of the three replicates, 2 or 4 peptides of A1150 were detected in the soluble fraction, which indicates minor contamination of the soluble fraction by the PhaZa1 protein. The other four PhaZa isoenzymes (PhaZa2 to PhaZa5) were not identified in any fraction, suggesting that none of the PhaZa2 to PhaZa5 proteins is detectably expressed under the applied conditions (late exponential phase of growth on NB-gluconate). The other two putative intracellular PHB depolymerases (PhaZd1 and PhaZd2), which have high PHB depolymerase activity when artificially expressed, were also not detected or were present only in trace amounts in some fractions, and this result is in line with previous data (13, 14, 39). In contrast, the two previously described intracellular 3-hydroxybutyrate oligomer hydrolases (PhaZb and PhaZc) were substantially expressed but were not specifically present in the PHB fraction. PhaZb was mainly in the membrane fraction (with traces in the soluble and PHB granule fractions), and PhaZc was exclusively present is the soluble fraction. The latter finding was confirmed by fusion analysis with eYFP, as described above. These data indicate that PhaZa1 is the only known PHB depolymerase that is bound to PHB granules in vivo in NB-gluconate-grown cells.
Comparison of the PHB granule proteome from NB-gluconate-grown cells with that of gluconate-mineral-medium-grown cells.
R. eutropha reproducibly accumulates PHB during growth on NB or NB-gluconate medium with a maximum between 8 h and 12 h (30 to 50% of cell dry weight) after inoculation. The accumulated PHB is reutilized (mobilized) in the stationary growth phase, and after 48 h, most of the accumulated PHB has been consumed. In contrast to this, R. eutropha accumulates much larger amounts of PHB in gluconate mineral salts medium (usually about 70 to 80% of cell dry weight), but only a portion of the accumulated PHB is mobilized in the stationary growth phase. Even incubation of PHB-rich cells in ammonium-containing fresh mineral salts solution did not lead to complete mobilization of PHB within 48 h (15, 16). One reason for this difference could be that R. eutropha synthesizes different amounts or different types (or a mixture of both) of intracellular PHB depolymerases during growth on fructose mineral salts medium in comparison to NB-gluconate medium. To determine a potential quantitative or qualitative difference in the PHB granule proteome, we grew R. eutropha in glucose mineral salts medium until the end of exponential growth and isolated PHB granules by two glycerol gradient centrifugation and subsequent washing steps. The granules isolated from mineral salts medium are referred to here as MM-PHB granules in contrast to the NB-PHB granules of NB-gluconate medium. Replicates using the same amount of proteins (∼50 μg) solubilized from the isolated MM-PHB granules compared to NB-PHB granules were subjected to SDS-PAGE and subsequent proteome analysis. To allow a direct and quantitative comparison, the determination of the NB-PHB granule proteome was repeated, and both fractions (NB-PHB and MM-PHB) were analyzed directly one after the other. Remarkably, the same 12 (plus 4) proteins that had been identified as associated with NB-PHB granules were also present in the MM-PHB granule fraction (Table 2; see Data Set S2 in the supplemental material). No other proteins with postulated functions in PHB metabolism were identified in substantial amounts in the MM-PHB fraction (e.g., PHB depolymerases other than PhaZa1 or 3HB oligomer hydrolases). However, the abundance of the MM-PHB granule-associated proteins was significantly lower than that of NB-PHB granule-associated proteins (Table 2). In particular, the most abundant proteins of NB-PHB granules, the two major phasins PhaP1 and PhaP2, were present in more than 1 order of magnitude higher abundance in NB-PHB granules than in MM-PHB granules. The differences in abundance of the other PGAPs varied from 0.9- to 7.5-fold (Table 2 shows details; see Data Set S2 in the supplemental material).
A0671 and B1632 have minor, yet detectable, functions in PHB mobilization.
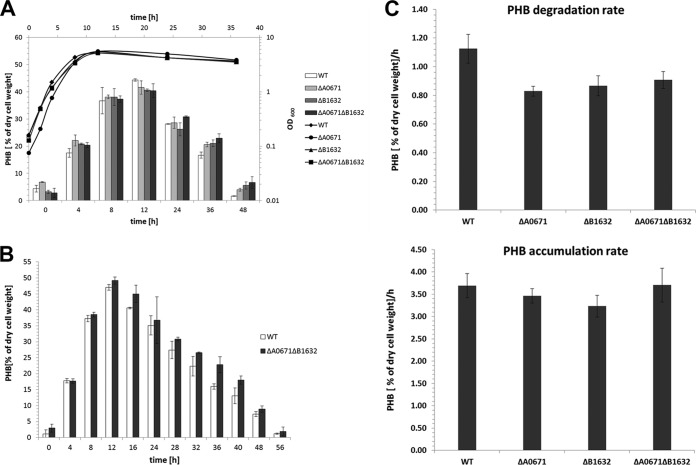
The amino acid sequences of the A0671 and B1632 gene products are similar and have a PHB synthase/depolymerase signature. Both proteins have a putative lipase box with a central cysteine (Fig. 3). It is unlikely that the A0671 and B1632 gene products have significant PHB synthase activity, as mutants in the major PHB synthase (PhaC1) are no longer able to synthesize and to accumulate PHB. We therefore speculated that A0671 and B1632 more likely represent PHB depolymerases. To find evidence for the involvement of the two proteins in PHB metabolism, we deleted A0671 and B1632 and constructed a double mutant with deletion of both genes. The wild type, the single mutants (ΔA0671 or ΔB1632), and the double mutant (ΔA0671 and ΔB1632) were grown in NB-gluconate medium, and growth and PHB contents were determined. As is evident from Fig. 4A, all the strains did not differ significantly in growth and PHB accumulation. When the PHB accumulation rates of the strains were calculated from the data during the exponential growth phase, comparable rates of 3.4 to 3.7% (cell dry weight) accumulated PHB per hour were determined for all the strains, indicating that neither A0671 nor B1632 contributed significantly to the PHB synthase activity (Fig. 4C, top). However, a small difference in PHB content during the mobilization phase (t = 12 to 48 h) was determined for the mutants: both single mutants and the double mutant were able to mobilize previously accumulated PHB, but the residual PHB contents were slightly higher than in the wild type. In particular, the PHB content of the double mutant was considerably higher than that of the wild type (Fig. 4B). When the mobilization of accumulated PHB was quantified by determination of the PHB degradation rates in the stationary growth phase, the rates for the three mutants were lower (0.82 to 0.90% [cell dry weight] mobilized PHB per hour) than for the wild type (1.12% [cell dry weight] mobilized PHB per hour) (Fig. 4C). These results show that the ability to mobilize previously accumulated PHB was reduced in the mutants and suggest that A0671 and B1632 partially contribute to PHB-mobilizing activity.
FIG 3.
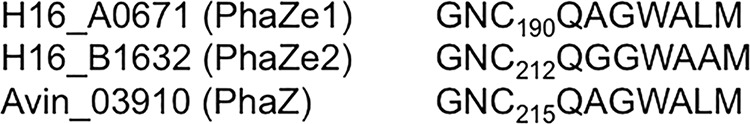
Amino acid alignment of the lipase box-like sequences of A0671 and B1632. The lipase box-like sequences and four adjacent amino acids of the A0671, B1632, and Avin03910 proteins were aligned. Note that the central amino acid of the GXSXG/A lipase box motif is a cysteine in all three sequences.
FIG 4.
Growth and PHB contents of R. eutropha wild-type (WT) and mutant cells. (A) Cells of the indicated strains were grown on NB-gluconate medium. At selected time points, samples were taken, and the OD600 and PHB content were determined. (B) Determination of PHB content was repeated for the R. eutropha H16 wild type and double mutant (ΔA0671 and ΔB1632) with more data points in the mobilization phase. Two to four biological replicates, each with two technical replicates, were performed. (C) The PHB accumulation rate (0 to 12 h) and PHB mobilization rate (12 to 48 h) were calculated from all replicates of the experiments shown in panels A and B and expressed as percent (cellular dry weight) PHB accumulated/mobilized per hour. The error bars indicate standard deviations.
DISCUSSION
A surprisingly high number (18) of PHB granule-associated proteins with proven or postulated functions in PHB metabolism were previously described or postulated for R. eutropha. These PGAPs are the PHB synthase PhaC1 (16–18, 46, 47); seven phasin proteins (PhaP1 to PhaP7) (13, 19–21, 38, 43); seven intracellular PHB depolymerases (PhaZ1 to PhaZ7 or PhaZa1 to PhaZa5, PhaZd1, and PhaZd2) (2, 11, 16, 22, 23, 37, 48–52); two 3-hydroxybutyrate oligomer hydrolases (PhaZb, PhaZc, or PhaYs) (24, 53, 54); and the second PHB synthase, PhaC2 (2, 9, 27–29, 38). The function and biological activity of PhaC2 are not known, but the protein is attached to PHB granules in vivo after overexpression as a fusion with eYFP (38). In addition, PhaR and PhaM raise the number of PGAPs to 20. The latter can also bind to DNA, in addition to PHB (19, 20, 22, 30), and therefore must have multiple in vivo localizations, as described in Results. However, for most of the PhaZa PHB depolymerase isoenzymes, except for PhaZa1 and the two 3-hydroxybutyrate oligomer hydrolases (PhaZb and PhaZc), there is no or only poor evidence that they are bound to PHB granules in vivo. We (in a recent study) and others have shown that PhaZd1 and PhaZd2 are attached to PHB granules in vivo and represent two highly active intracellular PHB depolymerases when expressed from the constitutive phaC promoter (13, 39). However, neither PhaZd1 nor PhaZd2 was significantly expressed in the wild type in NB-gluconate-grown cells. In only one of the proteome replicates in this study, three PhaZd2-specific peptide fragments were detected in the membrane fraction. In conclusion, the composition of the PHB granule proteome, i.e., the true number of PGAPs in vivo in R. eutropha, is not known, and probably, some of the postulated PHB depolymerases or 3HB oligomer hydrolases are not bound to PHB granules, at least during cultivation on NB-gluconate medium.
When we recently determined the proteome of isolated PHB granules from R. eutropha, we identified more than 400 different proteins (29). Besides previously known PHB granule-associated proteins, we found a large number of proteins that are annotated as (putative) OMPs or that are typical soluble proteins, such as malate dehydrogenase. Presumably, isolated PHB granules are contaminated by proteins of other cell fractions. On the other hand, proteins can be misannotated in databases, and, for example, a postulated membrane or postulated outer membrane protein could be part of the surface layer of PHB granules in reality. This is more likely to be the the case, since there is a scientific debate as to whether the PHB surface layer consists of a phospholipid monolayer in which proteins such as phasins, PHB synthase, and PHB depolymerase are embedded (8, 27–29, 55, 56) or whether the granule surface consists only of proteins. Moreover, recent studies on PGAPs in H. seropedicae (30), in Synechocystis sp. (W. Hauf and K. Forchhammer, personal communication), and in Azotobacter vinelandii (G. Espin, personal communication) also revealed the presence of OMPs or other proteins with no obvious function in PHB metabolism. Therefore, we could not exclude the possibility that at least some of the identified putative OMPs were misannotated and represent PGAPs. However, convincing evidence for true in vivo localization of these apparently non-PHB-related proteins on the PHB surface layer is presently not available.
In this study, we chose an extensive proteome approach for the identification of true PGAPs and performed a comparative proteome analysis of four separated subcellular fractions. In sum, of 2,264 proteins of all fractions, only 12 were specifically present in the PHB granule fraction (16 if PhaP1, PhaC1, PhaR, and PhaM are included) and were not significantly detected in any other fraction. The last four proteins all have a well-established function in PHB metabolism, and convincing data exist that they are attached to PHB granules in vivo (12, 19, 22, 36, 38, 40, 57). The PHB synthase (PhaC1) is constitutively expressed and forms an initiation complex consisting of at least PhaM, PhaC1, and the nucleoid. This explains why a portion of the PhaC1 molecules is present in other cell fractions than PHB. The detection of PhaP1-derived peptides in other fractions than the PHB granule fraction probably is a consequence of the extremely high expression level of the PhaP1 protein under PHB-permissive conditions. PhaP1 contributes approximately 5% of the total proteins and the phaP1 transcript is permanently translated under PHB-permissive conditions, and this results in a fraction of PhaP1 molecules that have not yet bound to PHB granules. Eight out of the 12 PHB granule-specific proteins represent previously identified proteins of PHB metabolism. The phasin PhaP1 was by far the most abundant protein in the PHB granule fraction, as revealed by quantification of the ion counts of all PhaP1-specific peptide fragments in the average of all replicates (Table 2; see Data Set S2 in the supplemental material). The second- and third-most-abundant proteins were also phasins (PhaP2 and PhaP4). The PHB synthase PhaC1 was only the sixth-most-abundant protein. Similar differences in the expression of PHB synthase and phasins were determined by transcriptional analysis or quantitative Western blot analysis in previous works (41, 58–60). However, the culture conditions and the time points of sampling were different from those in our study.
Four novel proteins were identified in the PHB granule fraction in this study. Two of them (B1632 and A671) are putative α/β hydrolases with PHB synthase/depolymerase signatures, and they show high amino acid similarity to each other (42% identity and 62% similarity). Interestingly, A0671 and B1632 are also similar to a putative intracellular PHB depolymerase of A. vinelandii (Avin03910; D. Segura and L. Adaya, personal communication). An insertion mutant in Avin03910 accumulates more PHB than the wild type, and this is in agreement with a putative function as an intracellular PHB depolymerase. We speculate that A0671 and B1632, as well as Avin03910, could represent a novel type of intracellular PHB depolymerase and could be responsible for the remaining PHB-degrading activity of ΔphaZa1 mutant strains (16, 42). Although B1632 and A0671 are not related in amino acid sequence to the PHB depolymerase PhaZa1, the proteins share similar lipase box-like sequences with a putative active-site cysteine (Fig. 3). Determination of the PHB content of A0671 or B1632, or both, gene deletion mutants in comparison to the wild type showed slightly decreased PHB mobilization rates (Fig. 4). In summary, our data indicate that PHB granules of NB-gluconate-grown R. eutropha cells have two other PHB depolymerases (A0671 and B1632) that could contribute to the PHB mobilization ability of R. eutropha, in addition to PhaZa1 (22, 37, 43, 61). However, B1632 and A0671 do not have amino acid similarities to PhaZa1 or to other known intra- or extracellular PHB depolymerases and probably represent—together with Avin03910—a novel type of intracellular PHB depolymerase with high molecular mass (80 to 90 kDa). We suggest the classification of B1632 and A0671 (and Avin03910) in a new subgroup of intracellular PHB depolymerases (subgroup PhaZe) according to the published literature. We tentatively designate the gene products PhaZe1 (A0671) and PhaZe2 (B1632).
The genomic context of B1632 is of special interest. The B1632 gene is located close to three other open reading frames, and B1632 to B1629 apparently form an operon (Fig. 5). The genes adjacent to B1632 are a predicted phosphate acetyltransferase gene (phosphotransacetylase; pta1; B1631), a putative acetate kinase gene (ackA; B1630), and a putative enoyl-reductase gene (fabI2; B1629). All three gene products were expressed in NB-gluconate-grown cells (see Data Set S1 in the supplemental material). A similar gene cluster with the same annotation of genes is present in many genome-sequenced proteobacterial species, including A. vinelandii. The cysteine-containing lipase box of the B1632 gene product and its similarity to an intracellular A. vinelandii PHB depolymerase suggest that B1632 is a thiolytic PHB depolymerase cleaving PHB to 3HB-CoA, similar to PhaZa1 (37, 61). Expression of the complete B1629-to-B1632 operon could constitute a pathway leading from 3HB-CoA to 3HB-phosphate (via Pta1), and subsequently, ATP could be generated via a (nonspecific) acetate kinase (AckA) reaction, assuming that Pta1 and AckA accept substrates longer than C2 compounds. 3HB would be the reaction product and could be excreted. Excretion of considerable amounts of 3HB by R. eutropha was described many years ago (62). Alternatively, PHB could be thiolytically cleaved and dehydrated to crotonyl-CoA, as was shown for PhaZa1 (61), and subsequently reduced to butyryl-CoA via FabI2 or via an acyl-CoA dehydrogenase, many of which are present in the R. eutropha genome. Butyryl-CoA could be converted to butyryl-phosphate (via nonspecific Pta1), and finally, ATP and butyrate would be formed in an acetate-kinase analogue reaction. Butyrate is also a product that can be secreted by R. eutropha in substantial amounts. Formation of ATP via the phosphate acetyltransferase and acetate-kinase reaction could also take place using the 3HB-CoA/crotonyl-CoA formed by the PhaZa1-catalyzed thiolytic reaction. In this context, “old knowledge” from the late 1970s should be recalled, namely, that growth of R. eutropha under oxygen limitation stimulates formation of PHB and expression of fermentation enzymes, such as NAD-dependent dehydrogenases (63, 64). As a consequence of oxygen limitation, fermentation products, such as ethanol, 2,3-butanediol, and lactate, are formed. It has also long been known that R. eutropha can produce and excrete acids, in particular, 3-hydroxybutyrate and butyrate (62, 65), and this finding could have its explanation in the expression of the B1632-to-B1629 operon.
FIG 5.

Genomic context of B1632 in R. eutropha H16. A part of the annotated genome of R. eutropha H16 (B1628 to B1632) is shown. B1632 to B1629 apparently form an operon.
The other two new PHB granule-specific proteins (A2001 and A0225) were present in comparably low abundance (less than the phasins PhaP1 to Pha5, the PHB synthase PhaC2, and PHB depolymerases) but had a higher abundance than PhaP6, PhaP7, and the second PHB synthase, PhaC2. The ion counts of the last three proteins were 1 to 3 orders of magnitude lower than those of the other PHB-specific proteins. The in vivo colocalization of A2001 and A0225 with PHB granules was confirmed by construction of fusion proteins with eYFP (Fig. 2), but their functions in PHB metabolism remain obscure. The A0225 protein (a putative phospholipase) could be involved in the removal/cleavage of phospholipids from PHB granules, but a function in the biosynthesis of phospholipids in the hydrophobic surroundings of the PHB surface or an involvement of A0225 in attachment of PHB granules to DNA cannot be excluded at this stage. In this context, an interesting phenotype has been described for a transposon mutant of Legionella pneumophila. In this species, a patatin-like phospholipase gene forms an operon with a 3-hydroxybutyrate dehydrogenase gene. A mutant with an insertion in the 3HB dehydrogenase gene resulted in impaired expression of both genes of the operon and in decreased phospholipase activity. Interestingly, mutant cells of L. pneumophila revealed an increased PHB content (66). It is not clear whether this finding was a consequence of reduced expression of the 3HB dehydrogenase gene, the patatin-like phospholipase gene, or both genes.
Eight of the first 20 most abundant proteins in isolated PHB granules of R. eutropha were proteins with established functions in PHB metabolism (see Data Set S2 in the supplemental material), and 8 other proteins represent putative OMPs (see above). However, none of the last eight proteins were specific to the PHB granule fraction, and they were also detected in at least one other cell fraction. These proteins presumably bind artificially to PHB granules after disruption of the cells and apparently do not belong to the PHB granule proteome sensu stricto. However, analysis of fusion proteins of outer membrane proteins with fluorescent proteins have not yet been performed to test this assumption.
Analysis of the PHB proteome of MM-gluconate cells revealed the presence of the same 14 PHB-specific proteins detected in NB-gluconate cells. However, the relative abundances of most of these proteins were significantly lower in MM-PHB granules than in NB-PHB granules (Table 2; see Data Set S2 in the supplemental material). In particular, the ion counts for peptides of the phasins PhaP1 and PhaP2 were more than 1 order of magnitude lower in MM-PHB granules. The different concentrations of proteins of the PHB granule fraction cannot be caused by differences in gel loading during electrophoresis. First, care was taken that the amounts of loaded material were the same for all samples, and second, a number of proteins identified in the PHB granule fraction from gluconate-mineral salt medium-grown cells were present in significantly higher concentrations than in NB-gluconate PHB granules. For illustration, proteins with 2- to 4-fold-higher concentrations in fructose-MM-PHB granules are listed at the bottom of Table 2 (italics). None of these proteins belonged to the proteins specifically present in the PHB granule fraction. The presence of proteins at higher and at lower concentrations in the two different PHB granule preparations shows that differences in protein concentrations are not caused by differences in the amounts of loaded proteins but reflect different protein compositions of PHB granule fractions. The generally low abundance of PHB depolymerase proteins among MM-PHB granule proteins could be one explanation of why mobilization of PHB in mineral salts medium is much slower than mobilization of PHB in NB-grown cells. Determination of the functions of the novel PHB granule-associated proteins will be the subject of future studies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft and the Landesstiftung Baden Württemberg.
We thank Libertad Adaya and Daniel Segura (Mexico), as well as Waldemar Hauf and Karl Forchhammer (Tübingen, Germany), for sharing unpublished results.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03791-14.
REFERENCES
- 1.Anderson AJ, Dawes EA. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, Eitinger T, Ewering C, Pötter M, Schwartz E, Strittmatter A, Voss I, Gottschalk G, Steinbüchel A, Friedrich B, Bowien B. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat Biotechnol 24:1257–1262. doi: 10.1038/nbt1244. [DOI] [PubMed] [Google Scholar]
- 3.Reinecke F, Steinbüchel A. 2009. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J Mol Microbiol Biotechnol 16:91–108. doi: 10.1159/000142897. [DOI] [PubMed] [Google Scholar]
- 4.Chen G-Q. 2009. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev 38:2434–2446. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- 5.Madison LL, Huisman GW. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63:21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehm BHA. 2003. Polyester synthases: natural catalysts for plastics. Biochem J 376:15–33. doi: 10.1042/BJ20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stubbe J, Tian J, He A, Sinskey AJ, Lawrence AG, Liu P. 2005. Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu Rev Biochem 74:433–480. doi: 10.1146/annurev.biochem.74.082803.133013. [DOI] [PubMed] [Google Scholar]
- 8.Pötter M, Steinbüchel A. 2006. Biogenesis and structure of polyhydroxyalkanoate granules. Microbiol Monogr 1:110–136. doi: 10.1007/7171_005. [DOI] [Google Scholar]
- 9.Cho M, Brigham CJ, Sinskey AJ, Stubbe J. 2012. Purification of a polyhydroxybutyrate synthase from its native organism, Ralstonia eutropha: implications in the initiation and elongation of polymer formation in vivo. Biochemistry 51:2276–2288. doi: 10.1021/bi2013596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Shrestha R, Buckley RM, Jewell J, Bossmann SH, Stubbe J, Li P. 2014. Mechanistic insight with HBCH2CoA as a probe to polyhydroxybutyrate (PHB) synthases. ACS Chem Biol 9:1773–1779. doi: 10.1021/cb5002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeiffer D, Jendrossek D. 2014. PhaM is the physiological activator of poly(3-hydroxybutyrate) (PHB) synthase (PhaC1) in Ralstonia eutropha. Appl Environ Microbiol 80:555–563. doi: 10.1128/AEM.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191:3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pötter M, Müller H, Reinecke F, Wieczorek R, Fricke F, Bowien B, Friedrich B, Steinbüchel A. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301–2311. doi: 10.1099/mic.0.26970-0. [DOI] [PubMed] [Google Scholar]
- 14.Steinbüchel A, Aerts K, Babel W, Follner C, Liebergesell M, Madkour MH, Mayer F, Pieper-Furst U, Pries A, Valentin HE. 1995. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can J Microbiol 41(Suppl 1):94–105. doi: 10.1139/m95-175. [DOI] [PubMed] [Google Scholar]
- 15.Mezzina MP, Wetzler DE, Catone MV, Bucci H, Di Paola M, Pettinari MJ. 2014. A phasin with many faces: structural insights on PhaP from Azotobacter sp. FA8. PLoS One 9:e103012. doi: 10.1371/journal.pone.0103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handrick R, Reinhardt S, Jendrossek D. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J Bacteriol 182:5916–5918. doi: 10.1128/JB.182.20.5916-5918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito T, Kobayashi T. 2002. Instracellular degradation of PHAs, p 23–40. In Doi Y, Steinbüchel A (ed), Biopolymers: polyesters II. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 18.Jendrossek D, Handrick R. 2002. Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol 56:403–432. doi: 10.1146/annurev.micro.56.012302.160838. [DOI] [PubMed] [Google Scholar]
- 19.Pötter M, Madkour MH, Mayer F, Steinbüchel A. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148:2413–2426. [DOI] [PubMed] [Google Scholar]
- 20.York GM, Stubbe J, Sinskey AJ. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J Bacteriol 184:59–66. doi: 10.1128/JB.184.1.59-66.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada M, Yamashita K, Wakuda A, Ichimura K, Maehara A, Maeda M, Taguchi S. 2007. Autoregulator protein PhaR for biosynthesis of polyhydroxybutyrate [P(3HB)] possibly has two separate domains that bind to the target DNA and P(3HB): functional mapping of amino acid residues responsible for DNA binding. J Bacteriol 189:1118–1127. doi: 10.1128/JB.01550-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82:936–951. doi: 10.1111/j.1365-2958.2011.07869.x. [DOI] [PubMed] [Google Scholar]
- 23.Wahl A, Schuth N, Pfeiffer D, Nussberger S, Jendrossek D. 2012. PHB granules are attached to the nucleoid via PhaM in Ralstonia eutropha. BMC Microbiol 12:262. doi: 10.1186/1471-2180-12-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galán B, Dinjaski N, Maestro B, de Eugenio LI, Escapa IF, Sanz JM, García JL, Prieto MA. 2011. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol Microbiol 79:402–418. doi: 10.1111/j.1365-2958.2010.07450.x. [DOI] [PubMed] [Google Scholar]
- 25.Cai S, Cai L, Liu H, Liu X, Han J, Zhou J, Xiang H. 2012. Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl Environ Microbiol 78:1946–1952. doi: 10.1128/AEM.07114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Luo Y, Han J, Wu J, Wu Z, Feng D, Cai S, Li M, Liu J, Zhou J, Xiang H. 2013. Proteome reference map of Haloarcula hispanica and comparative proteomic and transcriptomic analysis of polyhydroxyalkanoate biosynthesis under genetic and environmental perturbations. J Proteome Res 12:1300–1315. doi: 10.1021/pr300969m. [DOI] [PubMed] [Google Scholar]
- 27.Grage K, Jahns AC, Parlane N, Palanisamy R, Rasiah IA, Atwood JA, Rehm BHA. 2009. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules 10:660–669. doi: 10.1021/bm801394s. [DOI] [PubMed] [Google Scholar]
- 28.Beeby M, Cho M, Stubbe J, Jensen GJ. 2012. Growth and localization of polyhydroxybutyrate granules in Ralstonia eutropha. J Bacteriol 194:1092–1099. doi: 10.1128/JB.06125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jendrossek D, Pfeiffer DN. 2014. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16:2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 30.Tirapelle EF, Müller-Santos M, Tadra-Sfeir MZ, Kadowaki MAS, Steffens MBR, Monteiro RA, Souza EM, Pedrosa FO, Chubatsu LS. 2013. Identification of proteins associated with polyhydroxybutyrate granules from Herbaspirillum seropedicae SmR1—old partners, new players. PLoS One 8:e75066. doi: 10.1371/journal.pone.0075066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz E, Voigt B, Zühlke D, Pohlmann A, Lenz O, Albrecht D, Schwarze A, Kohlmann Y, Krause C, Hecker M, Friedrich B. 2009. A proteomic view of the facultatively chemolithoautotrophic lifestyle of Ralstonia eutropha H16. Proteomics 9:5132–5142. doi: 10.1002/pmic.200900333. [DOI] [PubMed] [Google Scholar]
- 32.McCool GJ, Cannon MC. 1999. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J Bacteriol 181:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters V, Rehm BHA. 2005. In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. FEMS Microbiol Lett 248:93–100. doi: 10.1016/j.femsle.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 34.Barnard GC, McCool JD, Wood DW, Gerngross TU. 2005. Integrated recombinant protein expression and purification platform based on Ralstonia eutropha. Appl Environ Microbiol 71:5735–5742. doi: 10.1128/AEM.71.10.5735-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultheiss D, Handrick R, Jendrossek D, Hanzlik M, Schüler D. 2005. The presumptive magnetosome protein Mms16 is a poly(3-hydroxybutyrate) granule-bound protein (phasin) in Magnetospirillum gryphiswaldense. J Bacteriol 187:2416–2425. doi: 10.1128/JB.187.7.2416-2425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumann L, Spinozzi F, Sinibaldi R, Rustichelli F, Pötter M, Steinbüchel A. 2008. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J Bacteriol 190:2911–2919. doi: 10.1128/JB.01486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchino K, Saito T, Gebauer B, Jendrossek D. 2007. Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J Bacteriol 189:8250–8256. doi: 10.1128/JB.00752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeiffer D, Jendrossek D. 2012. Localization of poly(3-hydroxybutyrate) (PHB) granule-associated proteins during PHB granule formation and identification of two new phasins, PhaP6 and PhaP7, in Ralstonia eutropha H16. J Bacteriol 194:5909–5921. doi: 10.1128/JB.00779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sznajder A, Jendrossek D. 2014. To be or not to be a PHB depolymerase: PhaZd1 (PhaZ6) and PhaZd2 (PhaZ7) of Ralstonia eutropha are highly active PHB depolymerases but have no detectable role in mobilization of accumulated PHB. Appl Environ Microbiol 80:4936–4946. doi: 10.1128/AEM.01056-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlegel HG, Gottschalk G, von Bartha R. 1961. Formation and utilization of poly-beta-hydroxybutyric acid by Knallgas bacteria (Hydrogenomonas). Nature 191:463–465. doi: 10.1038/191463a0. [DOI] [PubMed] [Google Scholar]
- 41.Simon O, Klaiber I, Huber A, Pfannstiel J. 2014. Comprehensive proteome analysis of the response of Pseudomonas putida KT2440 to the flavor compound vanillin. J Proteomics 109:212–227. doi: 10.1016/j.jprot.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Brandl H, Gross RA, Lenz RW, Fuller RC. 1988. Pseudomonas oleovorans as a source of poly(beta-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol 54:1977–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeiffer D, Jendrossek D. 2011. Interaction between poly(3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157:2795–2807. doi: 10.1099/mic.0.051508-0. [DOI] [PubMed] [Google Scholar]
- 44.Budde CF, Mahan AE, Lu J, Rha C, Sinskey AJ. 2010. Roles of multiple acetoacetyl coenzyme A reductases in polyhydroxybutyrate biosynthesis in Ralstonia eutropha H16. J Bacteriol 192:5319–5328. doi: 10.1128/JB.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerji S, Flieger A. 2004. Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150:522–525. doi: 10.1099/mic.0.26957-0. [DOI] [PubMed] [Google Scholar]
- 46.Peoples OP, Sinskey AJ. 1989. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J Biol Chem 264:15298–15303. [PubMed] [Google Scholar]
- 47.Schubert P, Steinbüchel A, Schlegel HG. 1988. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol 170:5837–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saegusa H, Shiraki M, Kanai C, Saito T. 2001. Cloning of an intracellular poly[D(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J Bacteriol 183:94–100. doi: 10.1128/JB.183.1.94-100.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.York GM, Lupberger J, Tian J, Lawrence AG, Stubbe J, Sinskey AJ. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[D-(−)-3-hydroxybutyrate] depolymerase genes. J Bacteriol 185:3788–3794. doi: 10.1128/JB.185.13.3788-3794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abe T, Kobayashi T, Saito T. 2005. Properties of a novel intracellular poly(3-hydroxybutyrate) depolymerase with high specific activity (PhaZd) in Wautersia eutropha H16. J Bacteriol 187:6982–6990. doi: 10.1128/JB.187.20.6982-6990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchino K, Saito T, Jendrossek D. 2008. Poly(3-hydroxybutyrate) (PHB) depolymerase PhaZa1 is involved in mobilization of accumulated PHB in Ralstonia eutropha H16. Appl Environ Microbiol 74:1058–1063. doi: 10.1128/AEM.02342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brigham CJ, Reimer EN, Rha C, Sinskey AJ. 2012. Examination of PHB depolymerases in Ralstonia eutropha: further elucidation of the roles of enzymes in PHB homeostasis. AMB Express 2:26. doi: 10.1186/2191-0855-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi T, Shiraki M, Abe T, Sugiyama A, Saito T. 2003. Purification and properties of an intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZ2) in Ralstonia eutropha H16 and its identification as a novel intracellular poly(3-hydroxybutyrate) depolymerase. J Bacteriol 185:3485–3490. doi: 10.1128/JB.185.12.3485-3490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobayashi T, Uchino K, Abe T, Yamazaki Y, Saito T. 2005. Novel intracellular 3-hydroxybutyrate-oligomer hydrolase in Wautersia eutropha H16. J Bacteriol 187:5129–5135. doi: 10.1128/JB.187.15.5129-5135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griebel R, Smith Z, Merrick JM. 1968. Metabolism of poly-beta-hydroxybutyrate. I. Purification, composition, and properties of native poly-beta-hydroxybutyrate granules from Bacillus megaterium. Biochemistry 7:3676–3681. [DOI] [PubMed] [Google Scholar]
- 56.Ruth K, de Roo G, Egli T, Ren Q. 2008. Identification of two acyl-CoA synthetases from Pseudomonas putida GPo1: one is located at the surface of polyhydroxyalkanoates granules. Biomacromolecules 9:1652–1659. doi: 10.1021/bm8001655. [DOI] [PubMed] [Google Scholar]
- 57.Wieczorek R, Pries A, Steinbüchel A, Mayer F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol 177:2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian J, Sinskey AJ, Stubbe J. 2005. Kinetic studies of polyhydroxybutyrate granule formation in Wautersia eutropha H16 by transmission electron microscopy. J Bacteriol 187:3814–3824. doi: 10.1128/JB.187.11.3814-3824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawrence AG, Schoenheit J, He A, Tian J, Liu P, Stubbe J, Sinskey AJ. 2005. Transcriptional analysis of Ralstonia eutropha genes related to poly-(R)-3-hydroxybutyrate homeostasis during batch fermentation. Appl Microbiol Biotechnol 68:663–672. doi: 10.1007/s00253-005-1969-3. [DOI] [PubMed] [Google Scholar]
- 60.Brigham CJ, Speth DR, Rha C, Sinskey AJ. 2012. Whole-genome microarray and gene deletion studies reveal regulation of the polyhydroxyalkanoate production cycle by the stringent response in Ralstonia eutropha h16. Appl Environ Microbiol 78:8033–8044. doi: 10.1128/AEM.01693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eggers J, Steinbüchel A. 2013. Poly(3-hydroxybutyrate) degradation in Ralstonia eutropha H16 is mediated stereoselectively to (S)-3-hydroxybutyryl coenzyme A (CoA) via crotonyl-CoA. J Bacteriol 195:3213–3223. doi: 10.1128/JB.00358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vollbrecht D, Schlegel HG. 1979. Excretion of metabolites by hydrogen bacteria III. D(−)-3-hydroxybutanoate. Eur J Appl Microbiol Biotechnol 7:259–266. doi: 10.1007/BF00498020. [DOI] [Google Scholar]
- 63.Schlegel HG, Vollbrecht D. 1980. Formation of the dehydrogenases for lactate, ethanol and butanediol in the strictly aerobic bacterium Alcaligenes eutrophus. J Gen Microbiol 117:475–481. [Google Scholar]
- 64.Jendrossek D, Steinbüchel A, Schlegel HG. 1987. Three different proteins exhibiting NAD-dependent acetaldehyde dehydrogenase activity from Alcaligenes eutrophus. Eur J Biochem 167:541–548. doi: 10.1111/j.1432-1033.1987.tb13371.x. [DOI] [PubMed] [Google Scholar]
- 65.Vollbrecht D. 1980. Oxygen deficiency and excretion of metabolites by strictly aerobic bacteria. Biotechnol Lett 2:49–54. [Google Scholar]
- 66.Aurass P, Pless B, Rydzewski K, Holland G, Bannert N, Flieger A. 2009. bdhA-patD operon as a virulence determinant, revealed by a novel large-scale approach for identification of Legionella pneumophila mutants defective for amoeba infection. Appl Environ Microbiol 75:4506–4515. doi: 10.1128/AEM.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 68.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 69.Lenz O, Friedrich B. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc Natl Acad Sci U S A 95:12474–12479. doi: 10.1073/pnas.95.21.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.