Abstract
Genome analysis revealed the existence of a putative transcriptional regulatory system governing CO metabolism in Thermococcus onnurineus NA1, a carboxydotrophic hydrogenogenic archaeon. The regulatory system is composed of CorQ with a 4-vinyl reductase domain and CorR with a DNA-binding domain of the LysR-type transcriptional regulator family in close proximity to the CO dehydrogenase (CODH) gene cluster. Homologous genes of the CorQR pair were also found in the genomes of Thermococcus species and “Candidatus Korarchaeum cryptofilum” OPF8. In-frame deletion of either corQ or corR caused a severe impairment in CO-dependent growth and H2 production. When corQ and corR deletion mutants were complemented by introducing the corQR genes under the control of a strong promoter, the mRNA and protein levels of the CODH gene were significantly increased in a ΔCorR strain complemented with integrated corQR (ΔCorR/corQR↑) compared with those in the wild-type strain. In addition, the ΔCorR/corQR↑ strain exhibited a much higher H2 production rate (5.8-fold) than the wild-type strain in a bioreactor culture. The H2 production rate (191.9 mmol liter−1 h−1) and the specific H2 production rate (249.6 mmol g−1 h−1) of this strain were extremely high compared with those of CO-dependent H2-producing prokaryotes reported so far. These results suggest that the corQR genes encode a positive regulatory protein pair for the expression of a CODH gene cluster. The study also illustrates that manipulation of the transcriptional regulatory system can improve biological H2 production.
INTRODUCTION
Carbon monoxide (CO) serves as a central metabolic intermediate in anaerobic metabolism (1), as an enzyme metallocenter ligand (2, 3), as a physiologically significant signal in higher organisms (4), and as a speculative component in an early mode of metabolism and the origin of life (5). CO can be utilized as carbon and energy sources for growth by numerous microorganisms containing carbon monoxide dehydrogenase (CODH), a key enzyme in CO metabolism. CODH oxidizes CO to carbon dioxide (CO2), and the electrons generated by the process are coupled to diverse reactions, such as oxygen reduction, desulfurification, hydrogenogenesis, acetogenesis, and methanogenesis (6). When CO is aerobically oxidized, as in Pseudomonas thermocarboxydovorans and Oligotropha carboxidovorans, the reducing equivalents are transferred to oxygen through a CO-insensitive respiratory chain. Under anaerobic conditions, CO oxidation is linked to acetate production through the reductive acetyl coenzyme A (acetyl-CoA) or Wood-Ljungdahl pathway in acetogenic bacteria and to methane production as a substrate of CODH/acetyl-CoA synthase in methanogenic archaea. Carboxydotrophic hydrogenogens like Rhodospirillum rubrum and Carboxydothermus hydrogenoformans oxidize CO through a water-gas shift reaction, CO + H2O → CO2 + H2 (ΔG°′ = −20 kJ/mol), which produces hydrogen gas (7–9).
There are several distinct CO regulation systems known for aerobic and anaerobic CO oxidation (1, 10). Aerobic CO oxidation systems are encoded by cox genes (coxMSL) and transcriptionally regulated by CoxC, CoxH, or RcoM (11, 12). The CoxC and CoxH proteins have a LytTR DNA-binding domain and an MHYT sensor domain of six transmembrane segments with conserved His and Met residues (13). The LytTR DNA-binding domain is unique, in that it is mainly comprised of β strands and predominantly found in pathogenic bacteria (14). RcoM has a LytTR domain and an N-terminal heme-bearing PAS sensor domain (12). The PAS domain is a well-known signal transduction module and senses various environmental signals, such as gases (O2, NO, or CO), light, redox potential, voltage, xenobiotics, and nitrogen availability (15). There is a second type of aerobic CO oxidation system which is found in the Gram-negative carboxydobacteria P. thermocarboxydovorans and Hydrogenophaga pseudoflava and a Gram-positive bacterium, Mycobacterium sp. strain JC1 DSM 3803 (16, 17). The system is encoded by cut genes (cutBCA or cutMSL) and is regulated by the CutR protein, a LysR-type transcriptional regulator (LTTR) with a plausible helix-turn-helix (HTH) motif in the N-terminal domain and a LysR substrate binding domain in the central and C-terminal domains (18). LTTRs, as some of the most common positive regulators in prokaryotes, govern very diverse genes and physiological functions (19).
Anaerobic CO oxidation systems are encoded by coo genes, which encode CODH (cooS), a ferredoxin-like protein (cooF), a multisubunit hydrogenase (cooMKLXUH), maturation proteins for CODH and hydrogenase, and a transcriptional activator, CooA (20). CooA is a homodimeric heme-binding CO sensor belonging to the cyclic AMP receptor protein (CRP) family of transcriptional regulators (21). When CO is present, CooA binds to CO and undergoes a conformational change to transcriptionally activate the required coo genes for CO oxidation (21). CooA is also a redox sensor so that CooA is not activated in the presence of oxygen (high redox potential) (10).
Two hyperthermophilic archaea, Thermococcus sp. strain AM4 and Thermococcus onnurineus NA1, can hydrogenogenically grow on CO, and CODH gene clusters are present in their genomes (22–24). The CODH gene is clustered with hydrogenase genes similar to the coo gene cluster of R. rubrum, but the primary structure and the organization of the genes are considerably different from those of the coo gene cluster (24). Additionally, Na+/H+ antiporter genes are present in the hyperthermophilic archaea but not in R. rubrum. A gene encoding a putative transcriptional regulator is also present in the upstream region of the CODH gene, but it does not encode a CooA homolog (25). We previously demonstrated that the CODH catalytic subunit and hydrogenase large subunit encoded by this CODH gene cluster are essential for the carboxydotrophic hydrogenogenic metabolism in T. onnurineus NA1 (26).
In this study, we describe a novel type of CO-responsive regulatory system (CorQR) in T. onnurineus NA1 through bioinformatic analysis, characterization of in-frame deletion mutants, and transcriptional analysis. This study also illustrates that the manipulation of the regulatory circuit can improve CO-dependent H2 production.
MATERIALS AND METHODS
Strain, media, and culture conditions.
T. onnurineus NA1 (KCTC10859) was isolated from a deep-sea hydrothermal vent area in the Papua New Guinea-Australia-Canada-Manus (PACMANUS) field (27). This strain was routinely cultured in yeast extract-peptone-sulfur (YPS) medium as previously reported (27). Modified medium 1 (MM1) (23, 28) was prepared with 1 g liter−1 yeast extract, 35 g liter−1 NaCl, 0.7 g liter−1 KCl, 3.9 g liter−1 MgSO4, 0.4 g liter−1 CaCl2·2H2O, 0.3 g liter−1 NH4Cl, 0.15 g liter−1 Na2HPO4, 0.03 g liter−1 NaSiO3, 0.5 g liter−1 NaHCO3, 0.5 g liter−1 cysteine-HCl, and 0.001 g liter−1 resazurin. One milliliter liter−1 of Holden's trace elements/Fe-EDTA solution (29) and 1 ml liter−1 of Balch's vitamin solution (30) were added as supplements to the medium. After autoclaving, the medium was kept in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) filled with an anoxic gas mixture (N2, H2, CO2, 90:5:5) for equilibration, and the final pH of the medium was adjusted to 6.5 with 2 N HCl.
For the cultures in serum bottles, the media were reduced with 0.005% Na2S·9H2O, and the headspaces were filled with 100% CO (MM1-CO) or 5 g liter−1 sodium pyruvate was provided to support the growth of the corQ and corR mutant strains (MM1-pyruvate). The serum bottles were sealed with bromobutyl rubber stoppers and aluminum crimp caps.
For the pH-stat batch culture, T. onnurineus NA1 was cultured in a 100-ml serum bottle and then 3-liter bioreactors (Fermentec, Cheongwon, Republic of Korea), and the working volumes were 50 ml and 2 liters, respectively, at 80°C. For the cultures in bioreactors, MM1 was supplemented with 10 g liter−1 yeast extract and a 10 times greater amount of Holden's trace elements/Fe-EDTA solution. Bioreactors were sparged with pure argon gas (99.999%) through a microsparger. The agitation speed was 300 rpm, and the pH was maintained at 6.1 to 6.2 using 0.2 M NaOH in 3.5% NaCl. The inlet gas of 100% CO was supplied by using a mass flow controller (MKPrecision, Seoul, Republic of Korea) at a feeding rate of 400 ml min−1. The gas outlet was open to let the H2 and CO2 gases escape and maintain the total pressure at 105 Pa.
Bioinformatic analysis.
The open reading frame (ORF) was predicted using the Glimmer (version 3.02) program (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi). An homology search was performed using a search with the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) against the nonredundant protein database from the National Center for Biotechnology Information (NCBI). Multiple-sequence alignment was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2) of the European Bioinformatics Institute (EBI).
Analytical methods.
Cell growth was monitored by measuring the optical density at 600 nm (OD600) with a BioPhotometer plus UV-visible spectrophotometer (Eppendorf, Hamburg, Germany). Biomass was determined on the basis of the fact that the unit value of the OD600 corresponded to 0.361 g (cell dry weight) liter−1. The H2 production rate was calculated on the basis of the H2 content in the gases produced from a bioreactor, and the gas flow rate was measured with a wet gas meter (Shinagawa, Tokyo, Japan). The amounts of CO, H2, and CO2 were measured by using a YL6100 gas chromatograph (GC; YL Instrument Co., Anyang, Republic of Korea) equipped with a Molsieve 5A column (Supelco, Bellefonte, PA), a Porapak N column (Supelco), a thermal conductivity detector, and a flame ionization detector. Argon was used as the carrier gas at a flow rate of 30 ml min−1.
Reverse transcription-quantitative PCR (RT-qPCR).
RNA was prepared with the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, with some modifications. Genomic DNA was eliminated by DNase I (Thermo Scientific Fermentas, St. Leon-Rot, Germany). One microgram of RNA was incubated with 1 unit of DNase I at 37°C for 30 min and purified through chloroform extraction and ethanol precipitation. RNA was quantified with a spectrophotometer, and cDNA was created using Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Thermo Scientific Fermentas, St. Leon-Rot, Germany). One microgram of RNA was incubated with 40 units of reverse transcriptase, 5 μM random hexamers, and 1 mM deoxynucleoside triphosphate (dNTP) at 37°C for 1 h in reverse transcription buffer (1×, as supplied by the enzyme manufacturer). The reaction products were serially diluted to find an adequate concentration for real-time PCR analysis, and the samples were amplified with SYBR green real-time PCR master mix (Toyobo, Osaka, Japan). Amplified signals were detected using a StepOnePlus system (Applied Biosystems, Foster City, CA), and all primers used are listed in Table S1 in the supplemental material. The relative amount of transcript for each gene was calculated from the cycle threshold (CT) values using a relative standard curve after normalization to the corresponding 16S rRNA (TON_1979) quantity.
Genetic manipulation.
Mutant strains with either complementation of corQR or an in-frame deletion of corQ or corR were constructed by modifying the gene disruption system used for a hyperthermophilic archaeon, Thermococcus kodakarensis KOD1 (31) (see Fig. S1 in the supplemental material). Both corQ and corR deletion mutants were generated by unmarked in-frame deletion through homologous recombination, and deletions were verified by PCR using the primers listed in Table S1 in the supplemental material.
For the complementation of corQ and corR deletion mutants, the pQRc vector was constructed (see Fig. S2 and Table S1 in the supplemental material). The intergenic region between TON_1126 and TON_1127 was chosen for the gene integration site because the region has a relatively low transcription level, on the basis of deep sequencing data (data not shown). PTON_0157, the promoter of TON_0157 (encoding a glutamate dehydrogenase), which shows a high transcription level in MM1-CO (data not shown), was used for high-level expression of the Pyrococcus furiosus hmg (hmgPfu) cassette. corQR genes were amplified by PCR using the primers listed in Table S1 in the supplemental material, and the amplified product was inserted in the downstream region of the hmgPfu cassette.
Western blotting.
Chemiluminescent signals created by Immun-Star horseradish peroxidase (HRP; Bio-Rad, Hercules, CA) were detected by a ChemiDocMP imaging system (Bio-Rad, Hercules, CA). Western blots were prepared and analyzed using a chemiluminescent dye with an Immun-Star HRP chemiluminescent kit (Bio-Rad, Hercules, CA). Antibodies against each protein encoded by TON_1016, TON_1018, and TON_1023, which were overexpressed in Escherichia coli Rosetta(DE3)pLysS cells (Stratagene, La Jolla, CA), were generated and purified through His-Bind resin (Novagen, Madison, WI) or through excision of the corresponding protein band after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation.
Nucleotide sequence accession number.
The DNA and deduced protein sequences have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) under accession number KM489057.
RESULTS AND DISCUSSION
Bioinformatic analysis of the putative regulatory genes.
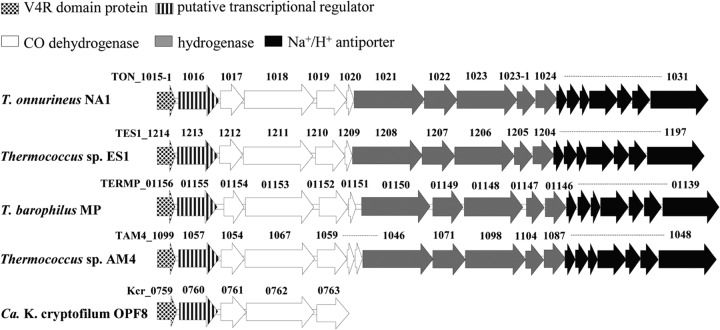
In the CODH gene cluster of T. onnurineus NA1, an open reading frame (ORF; TON_1016), namely, corR, encodes a putative transcriptional regulator belonging to the LTTR family (Fig. 1). A potential helix-turn-helix (HTH) motif, which would represent the DNA-binding domain, is present in the C-terminal part (amino acids 285 to 328) of CorR (see Fig. S3 in the supplemental material). However, a distinctive substrate-binding domain, which plays roles in coinducer recognition and/or response in the LTTR family of transcriptional regulators, was not found in the CorR protein. A BLAST analysis of the CorR protein of T. onnurineus NA1 showed that it exhibits a high degree of resemblance (50 to 55% identity and 70 to 74% similarity) to the CorR homologs of Thermococcus barophilus MP, Thermococcus sp. AM4, and Thermococcus sp. strain ES1. The CorR protein also displayed some homology (26% identity and 43% similarity) to a molybdate-binding protein of “Candidatus Korarchaeum cryptofilum” OPF8. In all cases, the corR genes were associated with CODH gene clusters (Fig. 1), implying an important role of CorR in CO oxidation. The CODH gene clusters were very similar among the strains in terms of organization and sequence, except in “Ca. Korarchaeum cryptofilum” OPF8, which has only a partial CODH gene cluster.
FIG 1.
Gene organization of CODH gene clusters in T. onnurineus NA1, Thermococcus sp. ES1, T. barophilus MP, Thermococcus sp. AM4, and “Ca. Korarchaeum cryptofilum” OPF8. TON_1015-1 was added as an ORF on the basis of the findings of this study. Locus tag information for every gene is indicated as a number above each corresponding arrow.
Intriguingly, an ORF annotated as a hypothetical protein was also conserved in the upstream region of the corR gene in the CODH gene clusters of the T. barophilus MP, Thermococcus sp. AM4, and Thermococcus sp. ES1 strains. The ORF was not annotated as a protein-coding sequence in the original genome annotation of T. onnurineus NA1 (22). Because of the conservation of the ORF in the three genomes mentioned above, a locus tag, TON_1015-1, could be assigned to a missing ORF in T. onnurineus NA1. A BLAST analysis of the deduced amino acid sequence of TON_1015-1 (corQ) revealed >70% identity (>90% similarity) to the CorQ homologs of Thermococcus strains and showed a significant sequence identity (∼40%) to the 4-vinyl reductase (V4R) proteins of various methanogens. In contrast, the ClustalW2 pairwise identity between TON_1015-1 and Kcr_0759 from “Ca. Korarchaeum cryptofilum” OPF8 was only 20%. All CorQ homologs contained a V4R domain. The V4R domain is present in many bacterial and archaeal proteins either by itself or fused with other domains, such as HTH domains or the AAA+ domains, and is primarily involved in transcription regulation and signal transduction (32). The function of the V4R domain is not well understood, but it appears to function in hydrocarbon binding or redox sensing (33–35). The V4R domains of the CorQ proteins contain three conserved cysteines (see Fig. S3 in the supplemental material). The three conserved cysteines of the CorQ proteins can coordinate a metal, as suggested for the V4R domain of MsvR of Methanothermobacter thermoautotrophicus, which plays a role in ligand recognition (32).
Importantly, the genomes of these Thermococcus strains and “Ca. Korarchaeum cryptofilum” OPF8 do not encode any previously known CO-responsive regulator, and therefore, if regulated, the expression of CODH gene clusters must be controlled by another protein(s). We hypothesize that it is the CorQR pair that regulates the CODH gene cluster in these species.
Inactivation of corR and corQ by in-frame deletion and complementation of the deletion mutants.
To investigate the importance of the corR and corQ genes in the CO metabolism of T. onnurineus NA1, each gene was inactivated by in-frame deletion. The resultant mutant strains, the ΔCorR and ΔCorQ strains, grew normally in YPS medium (heterotrophic condition) and had growth comparable to that of the wild type-strain (data not shown). However, under the lithotrophic condition where CO was supplied as the main energy source, these mutants grew poorly: the final cell densities of the ΔCorQ and ΔCorR strains were only about 30% and 20% of the final cell density of the wild-type strain, respectively (Fig. 2A). The H2 production of the mutants was severely impaired as well, with the H2 production rates being about 10% of the rate of the wild-type strain (Fig. 2B). To confirm that these CO-dependent growth defects in the mutants were caused by the loss of the corQ and corR genes and not by unexpected genetic variations that could happen during genetic manipulation, each mutant was complemented by inserting intact corQR genes at the intergenic region of TON_1126 and TON_1127 (see Fig. S2 in the supplemental material). For this complementation, the corQR genes were placed under the control of a strong promoter, PTON_0157, as described in Materials and Methods. In both complemented strains, the transcription levels of corR were increased by 5- to 6-fold compared with the level for the wild-type strain (data not shown). The ΔCorQ and ΔCorR strains complemented with integrated corQR, designated ΔCorQ/corQR↑ and ΔCorR/corQR↑, respectively, grew normally and produced H2 gas at a level comparable to that for the wild-type strain (Fig. 2).
FIG 2.
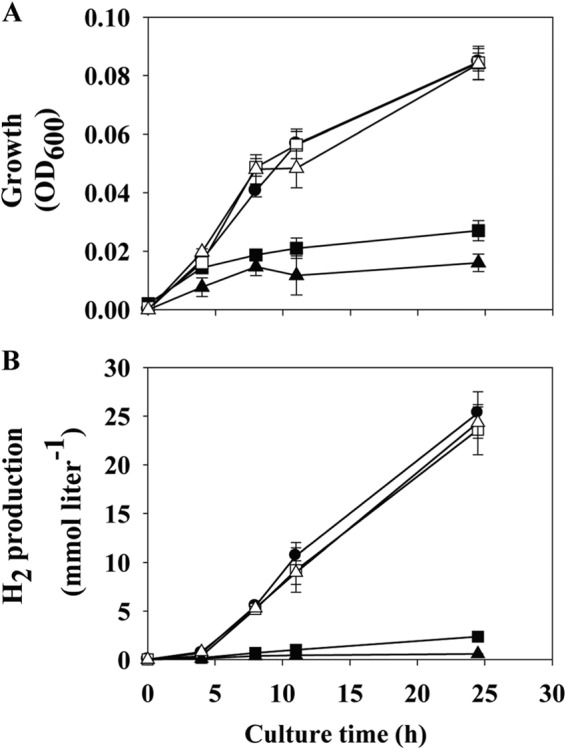
Growth (A) and H2 production (B) of the wild-type strain T. onnurineus NA1 (closed circles) and ΔCorQ (closed squares), ΔCorR (closed triangles), ΔCorQ/corQR↑ (open squares), and ΔCorR/corQR↑ (open triangles) mutants on MM1-CO. Cell growth was monitored by measuring the OD600. Average values from triplicate experiments are displayed, and error bars indicate the standard deviations from triplicate experiments.
These results indicate that the corQ and corR genes play important roles in CO-dependent growth and the resulting H2 production in T. onnurineus NA1. Also, growth defects were observed only under the CO condition, and it is likely that the functions of the corQ and corR genes are specific to CO-dependent growth.
Transcriptional analysis of the CODH gene in the wild-type and genetically manipulated strains.
To test whether corR and corQ indeed play a regulatory role in the CO metabolism of T. onnurineus NA1, the transcription levels of the catalytic subunit of CODH (TON_1018), a key CO metabolic enzyme, were monitored by RT-qPCR in the wild-type, ΔCorQ, ΔCorR, ΔCorQ/corQR↑, and ΔCorR/corQR↑ strains. While the ΔCorQ and ΔCorR mutant strains barely grew on CO, they could be grown in MM1-pyruvate until exponential phase, harvested, transferred to fresh MM1 with or without 100% CO gas, and incubated for an additional 3 h at 80°C. Total RNAs were extracted from these cells, they were subjected to RT-qPCR as described in Materials and Methods, and the fold changes in the level of CODH gene expression were calculated. The ΔCorR and ΔCorQ mutants failed to show any CO-dependent changes in the CODH mRNA levels. In contrast, for the wild-type strain and the two complemented mutant strains, there were clear and significant increases in the CODH mRNA level in response to CO (Fig. 3A). However, the two complemented mutants were dissimilar in terms of the fold difference in the level of expression. The fold difference in the level of expression by the ΔCorR/corQR↑ strain was about 20-fold higher than the differences for the ΔCorQ/corQR↑ and wild-type strains.
FIG 3.
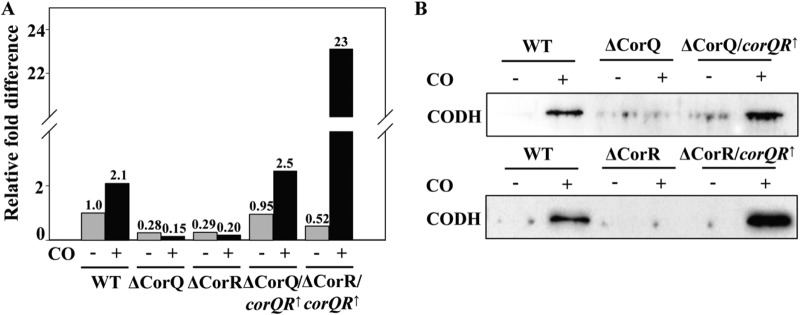
Changes of CODH gene expression at the transcriptional and translational levels after CO addition. (A) mRNA levels of the CODH gene (TON_1018) in the wild-type, ΔCorQ, ΔCorR, ΔCorQ/corQR↑, and ΔCorR/corQR↑ strains. The mRNA quantity was measured by RT-qPCR. The y axis indicates the relative fold difference when the value for the wild-type strain in the absence of CO was set equal to 1.0. (B) Protein levels of CODH in the wild-type, ΔCorQ, ΔCorR, ΔCorQ/corQR↑, and ΔCorR/corQR↑ strains. The amount of the CODH protein was monitored by Western blot analysis. WT, wild-type strain; −, no injection of CO (gray bars); +, injection of CO (black bars).
The CODH protein levels were also measured in the strains mentioned above, and the results were generally consistent with the transcript analysis data (Fig. 3B). The CODH protein was detected in the wild-type strain and the two complemented mutants, while the protein was barely visible in the ΔCorR and ΔCorQ mutant strains. Again, the amount of CODH protein was much higher in the ΔCorR/corQR↑ strain than in the wild-type and ΔCorQ/corQR↑ strains.
The observation of the difference in the CODH mRNA and protein levels between the two complemented strains is puzzling because we believe that the CorQ and CorR proteins are present at similar saturating levels in both the ΔCorQ/corQR↑ and ΔCorR/corQR↑ strains, where the corQR genes are under the control of the same strong promoter, PTON_0157. A mechanistic understanding of this phenomenon awaits further experiments. Nonetheless, our data indicate that the CorQR proteins are required for the CO-dependent transcriptional activation of the CODH gene in response to CO. This may also indicate that the proteins positively regulate the whole CODH gene cluster in T. onnurineus NA1.
Enhanced H2 production in the ΔCorR/corQR↑ strain.
We previously reported that the overexpression of the CODH gene cluster in the MC01 mutant led to 3.9-fold higher levels of H2 production (26). The increase in the amount of the CODH protein in the ΔCorR/corQR↑ strain, as shown in Fig. 3, may indicate an increase in the level of translation of the whole CODH gene cluster. If this is true, the strain was hypothesized to exhibit a higher H2 production rate as well. We therefore examined the H2 production potential of the ΔCorR/corQR↑ strain using a bioreactor with 100% CO fed stepwise at a flow rate of 20 to 400 ml min−1. The ΔCorR/corQR↑ strain indeed showed a significantly higher cell density and H2 production rate than the wild-type and ΔCorQ/corQR↑ strains (Fig. 4). The ΔCorR/corQR↑ strain displayed kinetic parameters 1.6- to 5.8-fold greater than those of the wild-type strain (see Table S2 in the supplemental material). In particular, the maximum H2 production rate and H2 productivity were much higher (over 5-fold) in the ΔCorR/corQR↑ strain than in the wild-type strain. The ΔCorR/corQR↑ strain showed a slightly higher H2 production rate (1.6-fold) than even the MC01 mutant, making the ΔCorR/corQR↑ strain the prokaryote with the best CO-dependent H2 production rate (Table 1).
FIG 4.
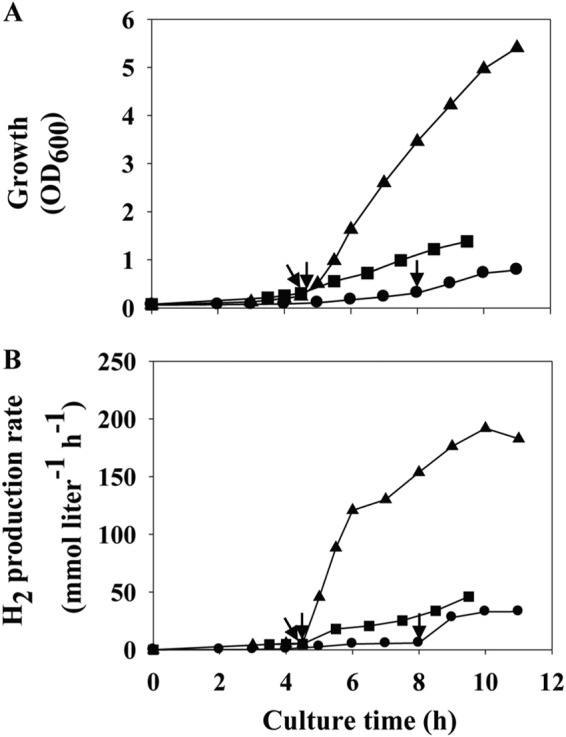
Growth (A) and H2 production (B) of the wild-type strain T. onnurineus NA1 (circles) and the ΔCorQ/corQR↑ (squares) and ΔCorR/corQR↑ (triangles) mutants in a 3-liter bioreactor with CO supplied continuously. Each experiment was performed one time. Cell growth was monitored by measuring the OD600. The initial CO flow rate of 20 ml min−1 was raised to 400 ml min−1 when the OD600 reached about 0.3, as indicated by arrows.
TABLE 1.
H2 production rates of various carboxydotrophic hydrogenogenic microbes
| Organism | Cultivation method | H2 production rate (mmol liter−1 h−1)a | Specific H2 production rate (mmol g−1 h−1)a | Reference or source |
|---|---|---|---|---|
| T. onnurineus NA1 (ΔCorR/corQR↑) | Batch | 191.9 | 249.6 | This study |
| T. onnurineus NA1 (MC01) | Batch | 123.5 | 194.7 | 26 |
| T. onnurineus NA1 (wild type) | Batch | 32.9 | 151.3 | 26 |
| Carboxydothermus hydrogenoformans | Continuous | 125 | 18.3 | 36 |
| Rhodopseudomonas palustris P4 | Batch | 41 | 41 | 37 |
| Citrobacter sp. strain Y19 | Batch | 5.7 | 27.1 | 38 |
| Rhodospirillum rubrum | Continuous | 4.7 | 11 | 39 |
| Rubrivivax gelatinosus CBS-2 | Continuous | 2.7 | 1.3–33 | 40 |
Kinetic parameters were calculated with data from the graphs in Fig. 4. The units of grams represent dry cell weight, except for the volatile suspended solid (VSS) used in C. hydrogenoformans.
In summary, we have shown that CorQR is a novel regulatory element of CO metabolism in T. onnurineus NA1. The CorQR regulatory system presented here is unique because it consists of two components, while all known CO-sensing transcriptional regulators are single-component proteins where the sensor domain is covalently linked to the response domain. Mechanistic details as to how this CorQR system senses CO and transcriptionally activates CODH genes remain to be elucidated. This study also illustrates that transcription regulation circuits can be used to improve biological H2 production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the KIOST in-house program (PE99212) and the Development of Biohydrogen Production Technology Using the Hyperthermophilic Archaea program of the Ministry of Oceans and Fisheries in the Republic of Korea. This work was also supported by NSF MCB-1020498 (to H.Y.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03019-14.
REFERENCES
- 1.Ragsdale SW. 2004. Life with carbon monoxide. Crit Rev Biochem Mol Biol 39:165–195. doi: 10.1080/10409230490496577. [DOI] [PubMed] [Google Scholar]
- 2.Drennan CL, Heo J, Sintchak MD, Schreiter E, Ludden PW. 2001. Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase. Proc Natl Acad Sci U S A 98:11973–11978. doi: 10.1073/pnas.211429998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korbas M, Vogt S, Meyer-Klaucke W, Bill E, Lyon EJ, Thauer RK, Shima S. 2006. The iron-sulfur cluster-free hydrogenase (Hmd) is a metalloenzyme with a novel iron binding motif. J Biol Chem 281:30804–30813. doi: 10.1074/jbc.M605306200. [DOI] [PubMed] [Google Scholar]
- 4.Wu L, Wang R. 2005. Carbon monoxide: endogenous production, physiological functions, and pharmacological applications. Pharmacol Rev 57:585–630. doi: 10.1124/pr.57.4.3. [DOI] [PubMed] [Google Scholar]
- 5.Miyakawa S, Yamanashi H, Kobayashi K, Cleaves HJ, Miller SL. 2002. Prebiotic synthesis from CO atmospheres: implications for the origins of life. Proc Natl Acad Sci U S A 99:14628–14631. doi: 10.1073/pnas.192568299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oelgeschläger E, Rother M. 2008. Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch Microbiol 190:257–269. doi: 10.1007/s00203-008-0382-6. [DOI] [PubMed] [Google Scholar]
- 7.Fox JD, Kerby RL, Roberts GP, Ludden PW. 1996. Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J Bacteriol 178:1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox JD, He Y, Shelver D, Roberts GP, Ludden PW. 1996. Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum. J Bacteriol 178:6200–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soboh B, Linder D, Hedderich R. 2002. Purification and catalytic properties of a CO-oxidizing:H2-evolving enzyme complex from Carboxydothermus hydrogenoformans. Eur J Biochem 269:5712–5721. doi: 10.1046/j.1432-1033.2002.03282.x. [DOI] [PubMed] [Google Scholar]
- 10.Roberts GP, Youn H, Kerby RL. 2004. CO-sensing mechanisms. Microbiol Mol Biol Rev 68:453–473. doi: 10.1128/MMBR.68.3.453-473.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santiago B, Schübel U, Egelseer C, Meyer O. 1999. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236:115–124. doi: 10.1016/S0378-1119(99)00245-0. [DOI] [PubMed] [Google Scholar]
- 12.Kerby RL, Youn H, Roberts GP. 2008. RcoM: a new single-component transcriptional regulator of CO metabolism in bacteria. J Bacteriol 190:3336–3343. doi: 10.1128/JB.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikolskaya AN, Galperin MY. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res 30:2453–2459. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicod SS, Weinzierl RO, Burchell L, Escalera-Maurer A, James EH, Wigneshweraraj S. 2014. Systematic mutational analysis of the LytTR DNA binding domain of Staphylococcus aureus virulence gene transcription factor AgrA. Nucleic Acids Res 42:12523–12536. doi: 10.1093/nar/gku1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilles-Gonzalez MA, Gonzalez G. 2004. Signal transduction by heme-containing PAS-domain proteins. J Appl Physiol 96:774–783. doi: 10.1152/japplphysiol.00941.2003. [DOI] [PubMed] [Google Scholar]
- 16.Kang BS, Kim YM. 1999. Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava. J Bacteriol 181:5581–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson DM, O'Reilly C, Colby J, Black GW. 1994. DNA sequence of the cut A, B and C genes, encoding the molybdenum containing hydroxylase carbon monoxide dehydrogenase, from Pseudomonas thermocarboxydovorans strain C2. Biochim Biophys Acta 1188:432–438. doi: 10.1016/0005-2728(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 18.Oh J-I, Park S-J, Shin S-J, Ko I-J, Han SJ, Park SW, Song T, Kim YM. 2010. Identification of trans- and cis-control elements involved in regulation of the carbon monoxide dehydrogenase genes in Mycobacterium sp. strain JC1 DSM 3803. J Bacteriol 192:3925–3933. doi: 10.1128/JB.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schell MA. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol 47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 20.Aono S. 2003. Biochemical and biophysical properties of the CO-sensing transcriptional activator CooA. Acc Chem Res 36:825–831. doi: 10.1021/ar020097p. [DOI] [PubMed] [Google Scholar]
- 21.Puranik M, Nielsen SB, Youn H, Hvitved AN, Bourassa JL, Case MA, Tengroth C, Balakrishnan G, Thorsteinsson MV, Groves JT, McLendon GL, Roberts GP, Olson JS, Spiro TG. 2004. Dynamics of carbon monoxide binding to CooA. J Biol Chem 279:21096–21108. doi: 10.1074/jbc.M400613200. [DOI] [PubMed] [Google Scholar]
- 22.Lee HS, Kang SG, Bae SS, Lim JK, Cho Y, Kim YJ, Jeon JH, Cha S-S, Kwon KK, Kim H-T, Park C-J, Lee H-W, Kim SI, Chun J, Colwell RR, Kim S-J, Lee J-H. 2008. The complete genome sequence of Thermococcus onnurineus NA1 reveals a mixed heterotrophic and carboxydotrophic metabolism. J Bacteriol 190:7491–7499. doi: 10.1128/JB.00746-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokolova TG, Jeanthon C, Kostrikina NA, Chernyh NA, Lebedinsky AV, Stackebrandt E, Bonch-Osmolovskaya EA. 2004. The first evidence of anaerobic CO oxidation coupled with H2 production by a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Extremophiles 8:317–323. doi: 10.1007/s00792-004-0389-0. [DOI] [PubMed] [Google Scholar]
- 24.Lim JK, Kang SG, Lebedinsky AV, Lee J-H, Lee HS. 2010. Identification of a novel class of membrane-bound [NiFe]-hydrogenases in Thermococcus onnurineus NA1 by in silico analysis. Appl Environ Microbiol 76:6286–6289. doi: 10.1128/AEM.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shelver D, Kerby RL, He Y, Roberts GP. 1997. CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc Natl Acad Sci U S A 94:11216–11220. doi: 10.1073/pnas.94.21.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim M-S, Bae SS, Kim YJ, Kim TW, Lim JK, Lee SH, Choi AR, Jeon JH, Lee J-H, Lee HS, Kang SG. 2013. CO-dependent H2 production by genetically engineered Thermococcus onnurineus NA1. Appl Environ Microbiol 79:2048–2053. doi: 10.1128/AEM.03298-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bae SS, Kim YJ, Yang SH, Lim JK, Jeon JH, Lee HS, Kang SG, Kim SJ, Lee JH. 2006. Thermococcus onnurineus sp. nov., a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent area at the PACMANUS field. J Microbiol Biotechnol 16:1826–1831. [Google Scholar]
- 28.Kim YJ, Lee HS, Kim ES, Bae SS, Lim JK, Matsumi R, Lebedinsky AV, Sokolova TG, Kozhevnikova DA, Cha S-S, Kim S-J, Kwon KK, Imanaka T, Atomi H, Bonch-Osmolovskaya EA, Lee J-H, Kang SG. 2010. Formate-driven growth coupled with H2 production. Nature 467:352–355. doi: 10.1038/nature09375. [DOI] [PubMed] [Google Scholar]
- 29.Holden JF, Takai K, Summit M, Bolton S, Zyskowski J, Baross JA. 2001. Diversity among three novel groups of hyperthermophilic deep-sea Thermococcus species from three sites in the northeastern Pacific Ocean. FEMS Microbiol Ecol 36:51–60. doi: 10.1111/j.1574-6941.2001.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 30.Balch WE, Wolfe RS. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol 32:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumi R, Manabe K, Fukui T, Atomi H, Imanaka T. 2007. Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J Bacteriol 189:2683–2691. doi: 10.1128/JB.01692-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anantharaman V, Koonin EV, Aravind L. 2001. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol 307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 33.O'Neill E, Ng LC, Sze CC, Shingler V. 1998. Aromatic ligand binding and intramolecular signalling of the phenol-responsive σ54-dependent regulator DmpR. Mol Microbiol 28:131–141. [DOI] [PubMed] [Google Scholar]
- 34.Karr EA. 2010. The methanogen-specific transcription factor MsvR regulates the fpaA-rlp-rub oxidative stress operon adjacent to msvR in Methanothermobacter thermautotrophicus. J Bacteriol 192:5914–5922. doi: 10.1128/JB.00816-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isom CE, Turner JL, Lessner DJ, Karr EA. 2013. Redox-sensitive DNA binding by homodimeric Methanosarcina acetivorans MsvR is modulated by cysteine residues. BMC Microbiol 13:163. doi: 10.1186/1471-2180-13-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Haddad M, Cimpoiab R, Liua Z, Guiot SR. 2013. Performance of a Carboxydothermus hydrogenoformans-immobilizing membrane reactor for syngas upgrading into hydrogen. Int J Hydrogen Energy 38:2167–2175. doi: 10.1016/j.ijhydene.2012.11.038. [DOI] [Google Scholar]
- 37.Oh Y-K, Kim Y-J, Park J-Y, Lee TH, Kim M-S, Park S. 2005. Biohydrogen production from carbon monoxide and water by Rhodopseudomonas palustris P4. Biotechnol Bioprocess Eng 10:270–274. doi: 10.1007/BF02932024. [DOI] [Google Scholar]
- 38.Jung GY, Kim JR, Park JY, Park S. 2002. Hydrogen production by a new chemoheterotrophic bacterium Citrobacter sp. Y19. Int J Hydrogen Energy 27:601–610. doi: 10.1016/S0360-3199(01)00176-8. [DOI] [Google Scholar]
- 39.Klasson KT, Lundbäck KMO, Clausen EC, Gaddy JL. 1993. Kinetics of light limited growth and biological hydrogen production from carbon monoxide and water by Rhodospirillum rubrum. J Biotechnol 29:177–188. doi: 10.1016/0168-1656(93)90049-S. [DOI] [Google Scholar]
- 40.Wolfrum EJ, Watt AS. 2002. Bioreactor design studies for a hydrogen-producing bacterium. Appl Biochem Biotechnol 98-100:611–625. doi: 10.1385/ABAB:98-100:1-9:611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.