Abstract
Listeria monocytogenes is involved in food-borne illness with a high mortality rate. The persistence of the pathogen along the food chain can be associated with its ability to form biofilms on inert surfaces. While most of the phenotypes associated with biofilms are related to their spatial organization, most published data comparing biofilm formation by L. monocytogenes isolates are based on the quantitative crystal violet assay, which does not give access to structural information. Using a high-throughput confocal-imaging approach, the aim of this work was to decipher the structural diversity of biofilms formed by 96 L. monocytogenes strains isolated from various environments. Prior to large-scale analysis, an experimental design was created to improve L. monocytogenes biofilm formation in microscopic-grade microplates, with special emphasis on the growth medium composition. Microscopic analysis of biofilms formed under the selected conditions by the 96 isolates revealed only weak correlation between the genetic lineages of the isolates and the structural properties of the biofilms. However, a gradient in their geometric descriptors (biovolume, mean thickness, and roughness), ranging from flat multilayers to complex honeycomb-like structures, was shown. The dominant honeycomb-like morphotype was characterized by hollow voids hosting free-swimming cells and localized pockets containing mixtures of dead cells and extracellular DNA (eDNA).
INTRODUCTION
Listeria monocytogenes still represents an important risk for public health; 1,740 listeriosis cases were reported in the European Union (EU) in 2011 with a mortality rate of 12.7% (1). Listeriosis is particularly dangerous for pregnant women and elderly or immunocompromised people. Persistence of L. monocytogenes strains on food plant surfaces can occur due to maladapted design of equipment and biofilm formation (2, 3). L. monocytogenes is able to attach to and colonize various surfaces, such as stainless steel, glass, and polystyrene, and to contaminate food products during processing (4–6). Biofilms of L. monocytogenes are associated with important ecological advantages, such as protection against biocide action (7). Several molecular determinants, such as flagella, biofilm-associated proteins (Bap), SecA2, and cell-cell communication systems, have been shown to be involved in biofilm construction within the species (8, 9). While no exopolysaccharidic components have been evidenced in the L. monocytogenes biofilm matrix (8), extracellular DNA (eDNA) has been shown to participate in initial cellular adhesion and biofilm organization under specific growth conditions (10). Biofilm formation by the species is highly dependent on environmental conditions, such as variations in temperature, pH, and nutrients (11, 12). L. monocytogenes is structured into four major phylogenetic lineages, each of which is genetically heterogeneous and substructured into highly recognizable clonal complexes as defined by multilocus sequence typing (MLST) (13, 14). Attempts to relate biofilm formation to strain origin, lineage, or persistence status led to contradictory results. Currently, the association of biotype structure with lineages or clonal complexes of L. monocytogenes is unknown.
Limited data are available on the intraspecific diversity of the architecture of L. monocytogenes biofilms. Indeed, most published reports focusing on the biofilm formation of several strains are based on global quantitative measurements (15–19).
The few studies focusing on the structure of the L. monocytogenes biofilm showed a variety of architectures, including a monolayer of adherent cells, flat unstructured multilayers, and a knitted-chain network, depending on the strains and experimental setup used (5, 9, 19–22). Early characterization by scanning electron microscopy (SEM) evidenced multilayers and honeycomb-like organizational structures of L. monocytogenes biofilms (21). However, this ultrastructural technique is time-consuming and involves drastic artifactual preparation steps, like chemical fixation and dehydration, that can alter the native spatial organization. So far, reports on the investigation of the three-dimensional (3D) structures of L. monocytogenes biofilms by confocal laser scanning microscopy (CLSM) are scarce. The coupling of CLSM with flow cell devices has highlighted the formation of a complex structure by the strain EGD-e, composed of ball-shaped microcolonies surrounded by a network of knitted chains (22). Recently, a high-throughput method based on CLSM combined with the use of 96-well microtiter plates was successfully applied in our laboratory to explore the biofilm architecture of 60 pathogens (23). In this study, we selected culture conditions adapted to the growth of static L. monocytogenes biofilms and deciphered the diversity of the architecture of the biofilms formed by a selection of 96 L. monocytogenes strains collected from diverse origins (food, animals, humans, and soil).
MATERIALS AND METHODS
Bacterial strains.
The 96 L. monocytogenes isolates used in this study were selected according to their diverse origins and are listed in Table S1 in the supplemental material. The collection, named ListRA (Listeria monocytogenes reference collection A) is constituted of 37 human isolates (13 from healthy human carriage and 24 from patients), 8 strains isolated from animals, 40 from the food industry, and 11 from soil samples. L. monocytogenes 10403S wild type (WT) and its isogenic ΔflaA (HEL-304) mutant (24) were used to evaluate the role of flagella in biofilm architecture. For real-time confocal observation, autofluorescent variants (25) harboring the pNF8 plasmid encoding GFPmut1 (26) or pJEBAN6 encoding DsRedExpress (27) were used. All strains were stored at −80°C in tryptone soya broth (TSB) (Oxoid, France) containing 20% (vol/vol) glycerol.
L. monocytogenes biofilm formation in microscopic-grade microplates.
Different factors, including the medium dilution, glucose supplementation, and buffer solution addition, were analyzed to select growth conditions allowing L. monocytogenes static-biofilm formation in microscopic-grade microplates. As the nutrient concentration is a critical parameter for L. monocytogenes biofilm formation (28), nutrient-rich and nutrient-poor media were tested using, respectively, TSB and 10×-diluted TSB. Glucose supplementation was also tested, as it has been shown previously to increase biofilm biomass (29). In order to avoid acidification of the medium, buffering with MOPS [3-(N-morpholino)propanesulfonic acid], pH 7.4, was also tested. For all conditions, frozen stocks of the strain EGD-e were subcultured twice in TSB at 25°C under vigorous orbital shaking (180 rpm). The subcultures were diluted to approximately 5 × 106 CFU/ml in the medium used for the growth analysis, prepared by combinations of three factors with two levels: dilution of TSB (1 or 1:10), addition of a buffered solution of MOPS (0 or 0.1 M), and glucose supplementation (0 or 1% [wt/vol]). These suspensions (250 μl) were used to inoculate the wells of a 96-well polystyrene microtiter plate with a μclear base (Greiner Bio-one, France). After 1 h of adhesion at 25°C, the supernatants containing nonadherent cells were removed and the wells were refilled with 250 μl of the medium. Biofilms were analyzed after 48 h of static incubation at 25°C. A complete 24 factorial design was constructed and analyzed with the dedicated module of Statgraphics (Manugistics, Rockville, MD, USA) to identify the combination of factors enabling the best biofilm formation under the tested conditions.
CLSM.
Fluorescent labeling of the biofilm was performed at 25°C for 20 min with a combination of two dyes: Syto 9 (3 μM), a green cell-permeant nucleic acid marker, and propidium iodide (20 μM), a red impermeant nucleic acid marker (LIVE/DEAD viability kit from Molecular Probes). After biofilm staining, image acquisition was performed using a Leica SP2 AOBS Confocal Laser Scanning Microscope (Leica, Leica-Microsystems, France) on the MIMA2 microscopy platform (http://www6.jouy.inra.fr/mima2). All biofilms were scanned at 800 Hz using a 63× oil immersion objective lens with a 488-nm argon laser set at 25% intensity. The emitted fluorescence was recorded within the range of 500 to 600 nm to collect Syto 9 emission fluorescence and 610 to 710 nm to collect propidium iodide-emitted fluorescence. Two stacks of horizontal-plane images with a z-step of 1 μm were acquired per well. The assays were all repeated on a different day for independent cultures (4 image series for each strain). Three-dimensional projections of the biofilms were constructed from the CLSM acquisitions using the easy 3D function of the IMARIS 7.1 software (Bitplane, Switzerland). Trajectories of motile bacteria from time series acquisition were analyzed with the IMARIS tracking function. Quantitative structural parameters (biovolume, thickness, and roughness) were extracted from confocal image series with PHLIP (30), a freely available Matlab-based image analysis toolbox (http://sourceforge.net/projects/phlip/).
SEM.
L. monocytogenes biofilms were prepared for scanning electron microscopy by immersing glass coupons in the wells of a 24-well polystyrene plate with 107 CFU. After a 1-h adhesion, supernatant containing nonadherent cells was removed and the wells were refilled with 1 ml of TSB supplemented with 1% (wt/vol) glucose and 0.1 M MOPS, pH 7.4. After 48 h of static incubation at 25°C, the biofilms were fixed for 24 h at 4°C in a solution containing 2.5% glutaraldehyde, 0.1 M sodium cacodylate (pH 7.4). Then, the coupons were positioned into a new plate and washed three times for 10 min with a solution containing 0.1 M sodium cacodylate. After transfer into 50% ethanol, samples were progressively dehydrated by passage through a graded series of ethanol solutions from 50% to 100%. The samples were then critical-point dehydrated (Emitech K850; United Kingdom) using carbon dioxide as the transition fluid and finally coated with gold-palladium in an automatic sputter coater (Polaron SC7640; United Kingdom). The samples were observed with a scanning electron microscope (FE-SEM Hitachi S4500; Hitachi, Tokyo, Japan).
Genotyping of the ListRA isolates.
In order to detect potential correlation between the genotype and the biofilm architecture, genotyping was achieved for the 96 isolates by sequencing two housekeeping genes, cat and dapE (13). These two genes were selected because their allele variation is strongly associated with major lineages and clonal complexes (14). The combination of cat and dapE alleles was used to deduce the clonal complex of the isolates, based on knowledge of the allelic variation of the two genes (http://www.pasteur.fr/mlst).
Statistical analysis.
All statistical analyses (experimental design, discriminant analysis, and analysis of variance [ANOVA]) were performed using Statgraphics v16.1 software (Manugistics, Rockville, MD, USA).
RESULTS
All 96 L. monocytogenes strains from the ListRA collection were characterized by sequencing of the MLST genes cat and dapE (see Table S1 in the supplemental material). Thirty-six isolates were grouped in lineage I, 59 isolates in lineage II, and one strain in lineage III. Eighteen different clusters were distinguished, and their correspondence with previously described clonal complexes (14) was established (see Table S1 in the supplemental material).
Using TSB as a basis for the growth medium, a complete 24 factorial design was applied to determine the influence of three parameters on the EGD-e strain biofilm formation as evaluated by the biofilm biovolume extracted from CLSM images, namely (i) medium dilution, (ii) glucose supplementation, and (iii) buffer concentration. Maximum biofilm biovolumes were obtained when glucose and MOPS were simultaneously added to undiluted TSB (Fig. 1).
FIG 1.
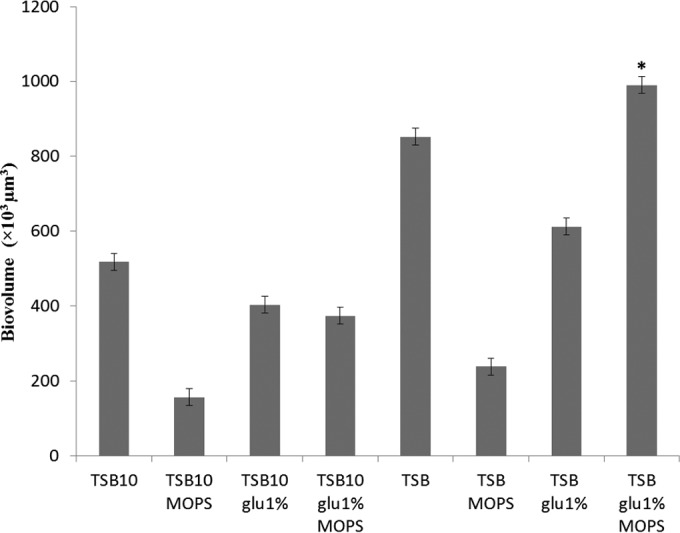
Biovolumes of L. monocytogenes EGD-e biofilms depending on their growth conditions. Biofilms were grown for 48 h at 25°C. The media used are shown on the x axis (TSB10, 10× dilution of TSB; glu 1%, addition of 10 g/liter of glucose; MOPS, addition of 0.1 M 3-(N-morpholino)propanesulfonic acid, pH 7.4). The error bars indicate the standard errors from four experiments. *, optimal combination as determined by the experimental design.
Structural diversity of the L. monocytogenes biofilms.
The 96 L. monocytogenes isolates were investigated for static biofilm formation with the selected growth protocol (TSB supplemented with 1% glucose and 0.1 M MOPS). CLSM image acquisition showed that all the strains were able to form three-dimensional structures after 48 h of incubation (Fig. 2). The biofilm architecture ranged from a flat homogeneous layer of cells to a honeycomb-like structure. Within the full data set, diversity was particularly evident when considering biofilm roughness and biovolume (Fig. 3). Statistical analysis showed a positive correlation between the thickness of biofilms and their biovolumes (P < 0.05), which are both anticorrelated with biofilm roughness (P < 0.05). Of note, two of the collection strains (CIP103574 and CIP104794) exhibited the highest biofilm thickness and biovolume, whereas the two nonmotile strains (CIP82110 and H6) formed flat multilayer structures. A discriminant analysis was used to classify strains in the MLST clonal complexes using the biofilm structural parameters (thickness, roughness, and biovolume) (see Fig. S1 in the supplemental material). The developed discriminant function significantly improved correct classification of the strains in the right clonal complex from 5.6% (at random) to 25.0% (P < 0.05). However, no discriminant function helped predict the origin of the strains using the biofilm structural parameters (P > 0.05).
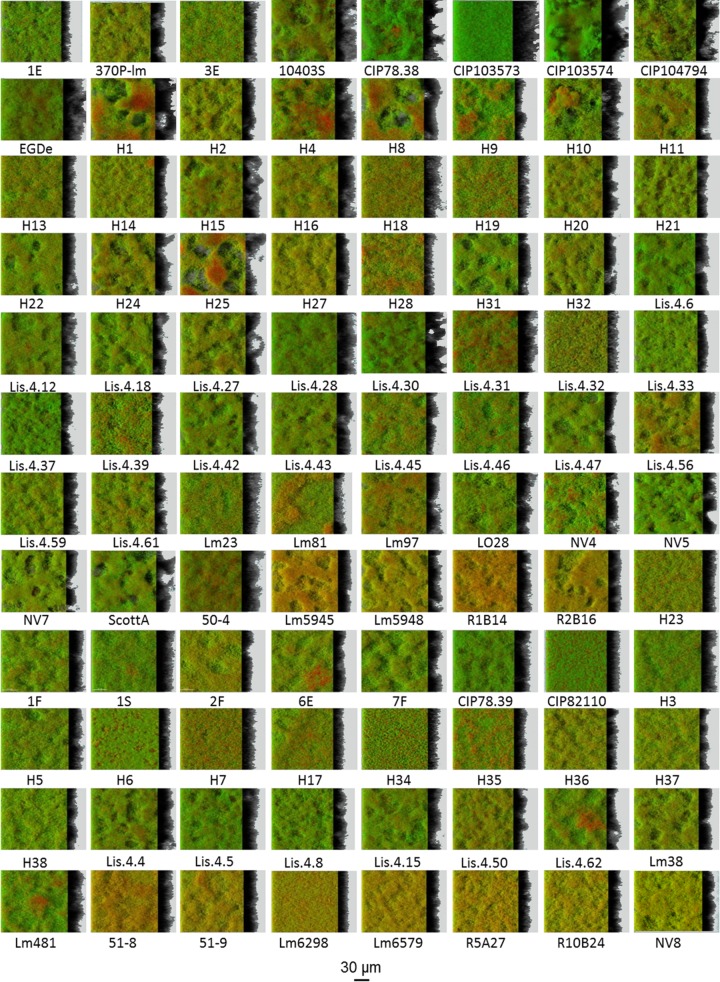
FIG 2.
IMARIS easy 3D projections from CLSM images of the biofilms formed by the 96 isolates of the ListRA collection showing the predominance of the honeycomb-like morphotype. The biofilms were labeled in green with Syto 9 and in red with propidium iodide. All the biofilms were grown at 25°C using the selected medium: TSB supplemented with 1% glucose and 0.1 M MOPS.
FIG 3.
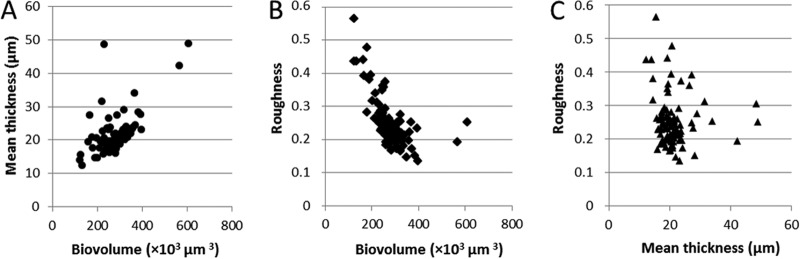
Correlation between the biofilm structural parameters of the ListRA collection. Shown are the distributions of the mean thickness (A) and roughness (B) as a function of the biovolume and the distribution of roughness as a function of the mean thickness (C) for the 96 isolates. All the values were extracted from CLSM images with the PHLIP Matlab routine and were averaged from 4 values for each case from 2 independent sets of experiments.
Flat multilayer biofilms.
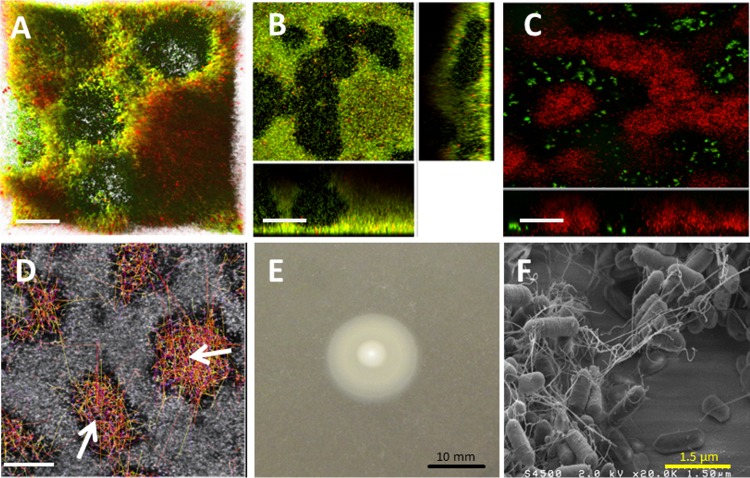
Within the collection, only the two nonmotile strains (CIP82110 and H6) formed flat biofilms characterized by low roughness and relatively high biovolume. The isosurface representation and the section view of a representative strain for this type of architecture (CIP82110) showed dense and homogeneous biofilm with scattered damaged or dead cells as stained by propidium iodide (Fig. 4A and B). In accordance with the nonmotile phenotype determined using a soft-agar assay (Fig. 4C), no visible flagella were observed with SEM on the bacteria forming these biofilms (Fig. 4D).
FIG 4.
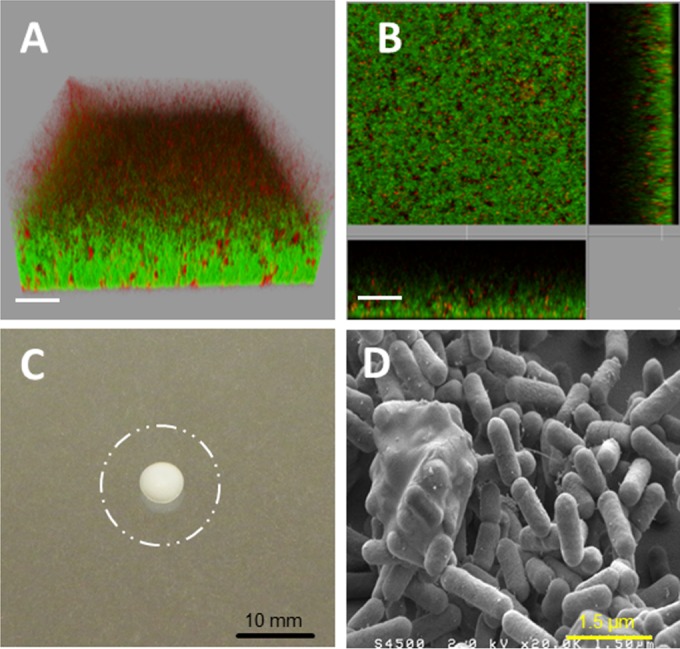
Strain CIP82110 as a representative of nonmotile strains forming flat biofilms. (A and B) IMARIS isosurface representation (A) and section view (B) of the CLSM images of the biofilms stained with Syto 9 and propidium iodide. The scale bars represent 20 μm. (C) Twenty-four-hour swimming plate (TSB plus 0.25% agar) of the nonmotile strain CIP82110 compared to the swimming phenotype of strain H25, represented by the dashed circle. (D) SEM observation of the biofilm formed, at 2 × 104 magnification.
Honeycomb-like biofilms.
The vast majority of the tested strains formed complex honeycomb-like architectures decorated with hollow voids. The structures with the largest hollow voids generally presented the greatest roughness and the smallest bacterial biovolume. L. monocytogenes H25 is a telling example of strains forming a honeycomb-like biofilm (Fig. 5; see Videos S1 and S2 in the supplemental material). Large holes or channels were scattered in the biofilm, as seen in the 3D reconstruction (Fig. 5A) and the section view (Fig. 5B). To investigate whether those “hollow voids” were filled with unseen materials, 2-μm green-fluorescent latex beads were deposited at the tops of the biofilms stained in red with Syto 61, and their sedimentation was followed in time. After less than 10 min, the beads were observed at the bottoms of the voids, showing the absence of a compact matrix in the voids (Fig. 5C). Staining the biofilm with propidium iodide showed the presence of red pockets of materials likely formed by a mixture of dead cells and eDNA (Fig. 5A; see Video S1 in the supplemental material). Direct time series observations showed the presence of swimming bacteria in the hollow voids, with an average speed of 2.3 μm/s (see Video S2 in the supplemental material). The trajectories of the swimming bacteria (Fig. 5D) showed that motile cells were almost exclusively located in the holes. A low-agar swimming test confirmed the high motility of the strain (Fig. 5E). SEM observations of this honeycomb-like biofilm showed many filamentous materials between the cells (Fig. 5F). The filaments were presumed to be flagella, as they were also observed in the biofilms formed by the strain 10403S WT, but not in the biofilms formed by its isogenic flagellum-deficient mutant 10403S ΔflaA (Fig. 6). Biofilms formed by the nonmotile mutant exhibited a flat, unstructured architecture compared to the honeycomb-like biofilms formed by the motile WT strain (Fig. 6), suggesting a role of flagella in the honeycomb-like architecture. Taking advantage of an L. monocytogenes 10403S mutant expressing green fluorescent protein (GFP), the dynamics of honeycomb-like formation were continuously visualized for 48 h by real-time CLSM (see Video S3 in the supplemental material). Over time, surface-associated bacteria expanded as clusters, while the number of planktonic motile cells decreased until 48 h, when motile cells became undetectable in the bulk.
FIG 5.
Strain H25 as a representative of motile strains forming biofilms with a honeycomb morphotype. (A and B) IMARIS isosurface representation (A) and section view (B) of CLSM images from biofilms forming honeycomb-like structures stained in green with Syto 9 and in red with propidium iodide. (C) IMARIS 3D projection of the same biofilm stained with the red Syto 61 in the presence of green-fluorescent 2-μm latex microbeads. (D) Tracking of motile bacteria in the hollow voids. The arrows indicate the cell trajectories. (E) Twenty-four-hour swimming plate (TSB plus 0.25% agar) of strain H25. (F) SEM image at 2 × 104 magnification. The scale bars represent 30 μm.
FIG 6.
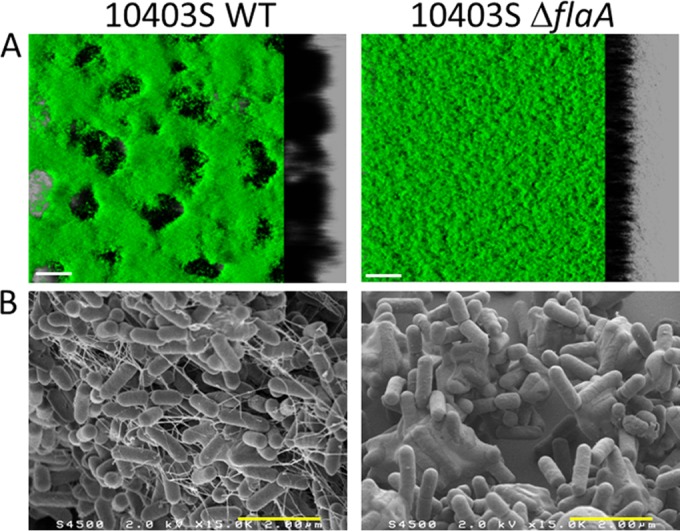
Microscopic observations of the biofilms formed by the motile L. monocytogenes 10403S WT strain and its isogenic nonmotile 10403S ΔflaA mutant. (A) Isosurface representation obtained from the confocal image series using the IMARIS software (green Syto 9 staining). (B) SEM image at 1.5 × 104 magnification. The white scale bars correspond to 30 μm and the yellow bars to 2 μm.
DISCUSSION
The objective of this study was to evaluate the biofilm structural diversity within the species L. monocytogenes. To this end, a medium for static biofilm growth in microscopic-grade microplates was selected from a 24 experimental design. Using this medium (TSB supplemented with glucose and MOPS), the biofilm architecture of the 96 isolates of the ListRA collection was analyzed by CLSM.
Various 3D structures of static L. monocytogenes biofilm have been reported: unorganized architectures with multicellular layers or aggregates (17, 22), clustered biofilms (31), and a 3D network of cells (19, 21). However, differences in the protocols and data analyses used render comparisons between reports impossible. Here, with the same protocol, all strains were able to form 3D structures. The architectures formed by the 96 isolates were quite diverse, as highlighted by the quantitative biofilm roughness and biovolume parameters. This intraspecies diversity in the ability to form biofilms is in accordance with previous works, where quantitative methods were used to evaluate biofilm formation (19, 32, 33). The 3D reconstruction of CLSM images showed that the majority of strains formed honeycomb-like structures consisting of layers of cohesive cells decorated with hollow voids with diameters ranging from 5 to 50 μm. This type of spatial organization was described only once for biofilms of L. monocytogenes (21). Honeycomb-like biofilm architectures were previously described for other species, including Staphylococcus epidermidis (34) and Staphylococcus aureus (23). Surprisingly, hollow voids were maintained up to 72 h and were not colonized by sessile bacteria. Time course observations of 48-h honeycomb-like biofilms showed progressive invasion of the surface by the bacteria resulting from cell multiplication. These hollow voids likely originate from both privileged colonization locations and localized cell death, as previously described for Pseudomonas aeruginosa (35). Bacteria in movement were visible in the hollow voids. The high average speed of the motile cells (2.3 μm/s) excluded Brownian motion as the driving force for this subpopulation. Coexistence of sessile and motile cells on the surface has been described previously in different species and was often associated with active biofilm dispersal phenomena (36–38).
The observation of the presence of eDNA in the matrix is in accordance with previously published reports about the key role of DNA for L. monocytogenes cell adhesion and biofilm structure (10, 33). The origin of the eDNA composing the L. monocytogenes matrix remains unclear but could involve cellular lysis or the release of small vesicles, as for other species (39). DNA pockets observed in the case of honeycomb-like structures are similar to previously described localized cell death resulting from quorum-sensing-driven processes (40). This phenomenon can provide nutrients for the starved surviving subpopulation. The release of eDNA can also influence the spatial organization of the biofilm and contributes to the stability of its structure. The observation of bacteriophages associated with sessile cells using SEM and in biofilm supernatants using transmission electron microscopy (TEM) (see Fig. S2 in the supplemental material) suggests that localized cell death could result from prophage activity, as has been observed with some strains of P. aeruginosa (41).
No polysaccharides could be detected in the honeycomb-like biofilms when tested with two different fluorescent lectins (wheat germ agglutinin [WGA] and concanavalin A). SEM observations showed the presence of extracellular fibrils in the honeycomb-like structures. These filaments are likely flagella, as they were not observed in a biofilm formed by the nonmotile strain CIP82110 or with the 10403S mutant defective in flagellum production. Previous studies reported that L. monocytogenes strains whose motility was affected displayed a reduced capacity to form static biofilms (42, 43). Under our specific experimental conditions, lack of flagella did not impede biofilm formation but resulted in a flat, unstructured architecture. These results suggested that flagella play a structural role in the complex architecture of L. monocytogenes honeycomb-like biofilms.
Genotyping of the 96 isolates from the ListRA collection revealed that most isolates were grouped into lineages I and II (with only 1 strain from lineage III). Indeed, lineage III strains are scarce and poorly represented in collections (19, 28, 44). Previous work showed variable capacities of the strains to form biofilms depending on their lineage (28). Some works showed that lineage I strains produced more biofilms than lineage II strains (15, 16), and others reached the opposite conclusion (17, 19). In our study, a weak correlation was detected between the strain lineage and the structure of the biofilms. In accordance with a recent report (33), we observed no correlation between strain origin and the structure of the biofilms formed (see Table S1 in the supplemental material).
Altogether, we have shown in this report that most of the L. monocytogenes isolates form a spatially structured honeycomb-like biofilm morphotype under static conditions. This morphotype involves both eDNA and flagella as structural components. The link between this biofilm architecture and the persistence of L. monocytogenes on surfaces remains to be elucidated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Thierry Meylheuc and Christine Longin for electron microscopy observations (INRA MIMA2 imaging center). We are grateful to Jens Bo Andersen and Tine Rask Licht (Technical University of Denmark, Soeborg, Denmark) for kindly providing a multiple-fluorescence labeling system for L. monocytogenes, as well as Hélène Marquis (Cornell University, Ithaca, NY, USA) for the nonflagellated mutant strain and Abdelkader Boubetra (Institut Scientifique d'Hygiène et d'Analyse) for L. monocytogenes food strains. The “Essonne department” contributed to the acquisition of the confocal microscope.
This work was supported by Institut National de la Recherche Agronomique funding.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03173-14.
REFERENCES
- 1.European Food Safety Authority. 2011. European Union summary report on trends and sources of zoonoses. Zoonotic agents and food-borne outbreak. European Food Safety Authority, Parma, Italy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bridier A, Sanchez-Vizuete P, Guilbaud M, Piard JC, Naïtali M, Briandet R. 2015. Biofilm-associated persistence of food-borne pathogens. Food Microbiol 45:167–178. doi: 10.1016/j.fm.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Carpentier B, Cerf O. 2011. Review—persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol 145:1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Blackman IC, Frank JF. 1996. Growth of Listeria monocytogenes as a biofilm on various food-processing surfaces. J Food Prot 59:827–831. [DOI] [PubMed] [Google Scholar]
- 5.Chae MS, Schraft H. 2000. Comparative evaluation of adhesion and biofilm formation of different Listeria monocytogenes strains. Int J Food Microbiol 62:103–111. doi: 10.1016/S0168-1605(00)00406-2. [DOI] [PubMed] [Google Scholar]
- 6.Briandet R, Meylheuc T, Maher C, Bellon-Fontaine M-N. 1999. Listeria monocytogenes Scott A: cell surface charge, hydrophobicity, and electron donor and acceptor characteristics under different environmental growth conditions. Appl Environ Microbiol 65:5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank JF, Koffi RA. 1990. Surface-adherent growth of Listeria monocytogenes is associated with increased resistance to surfactant sanitizers and heat. J Food Prot 53:550–554. [DOI] [PubMed] [Google Scholar]
- 8.Renier S, Hebraud M, Desvaux M. 2011. Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic Gram-positive foodborne pathogen. Environ Microbiol 13:835–850. doi: 10.1111/j.1462-2920.2010.02378.x. [DOI] [PubMed] [Google Scholar]
- 9.Renier S, Chagnot C, Deschamps J, Caccia N, Szlavik J, Joyce SA, Popowska M, Hill C, Knøchel S, Briandet R, Hébraud M, Desvaux M. 2014. Inactivation of the SecA2 protein export pathway in Listeria monocytogenes promotes cell aggregation, impacts biofilm architecture and induces biofilm formation in environmental condition. Environ Microbiol 16:1176–1192. doi: 10.1111/1462-2920.12257. [DOI] [PubMed] [Google Scholar]
- 10.Harmsen M, Lappann M, Knochel S, Molin S. 2010. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl Environ Microbiol 76:2271–2279. doi: 10.1128/AEM.02361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moltz AG. 2005. Formation of biofilms by Listeria monocytogenes under various growth conditions. J Food Prot 68:92–97. [DOI] [PubMed] [Google Scholar]
- 12.Nilsson RE, Ross T, Bowman JP. 2011. Variability in biofilm production by Listeria monocytogenes correlated to strain origin and growth conditions. Int J Food Microbiol 150:14–24. doi: 10.1016/j.ijfoodmicro.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantinelli T, Chenal-Francisque V, Diancourt L, Frezal L, Leclercq A, Wirth T, Lecuit M, Brisse S. 2013. “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J Clin Microbiol 51:3770–3779. doi: 10.1128/JCM.01874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djordjevic D, Wiedmann M, McLandsborough LA. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol 68:2950–2958. doi: 10.1128/AEM.68.6.2950-2958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi H, Miya S, Igarashi K, Suda T, Kuramoto S, Kimura B. 2009. Biofilm formation ability of Listeria monocytogenes isolates from raw ready-to-eat seafood. J Food Prot 72:1476–1480. [DOI] [PubMed] [Google Scholar]
- 17.Combrouse T, Sadovskaya I, Faille C, Kol O, Guerardel Y, Midelet-Bourdin G. 2013. Quantification of the extracellular matrix of the Listeria monocytogenes biofilms of different phylogenic lineages with optimization of culture conditions. J Appl Microbiol 114:1120–1131. doi: 10.1111/jam.12127. [DOI] [PubMed] [Google Scholar]
- 18.Kalmokoff M, Austin J, Wan X-D, Sanders G, Banerjee S, Farber J. 2001. Adsorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J Appl Microbiol 91:725–734. doi: 10.1046/j.1365-2672.2001.01419.x. [DOI] [PubMed] [Google Scholar]
- 19.Borucki MK, Peppin JD, White D, Loge F, Call DR. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol 69:7336–7342. doi: 10.1128/AEM.69.12.7336-7342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavant P, Martinie B, Meylheuc T, Bellon-Fontaine MN, Hebraud M. 2002. Listeria monocytogenes LO28: surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl Environ Microbiol 68:728–737. doi: 10.1128/AEM.68.2.728-737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh EJ, Luo H, Wang H. 2003. A three-tiered approach to differentiate Listeria monocytogenes biofilm-forming abilities. FEMS Microbiol Lett 228:203–210. doi: 10.1016/S0378-1097(03)00752-3. [DOI] [PubMed] [Google Scholar]
- 22.Rieu A, Briandet R, Habimana O, Garmyn D, Guzzo J, Piveteau P. 2008. Listeria monocytogenes EGD-e biofilms: no mushrooms but a network of knitted chains. Appl Environ Microbiol 74:4491–4497. doi: 10.1128/AEM.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bridier A, Dubois-Brissonnet F, Boubetra A, Thomas V, Briandet R. 2010. The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J Microbiol Methods 82:64–70. doi: 10.1016/j.mimet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 24.O'Neil HS, Marquis H. 2006. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect Immun 74:6675–6681. doi: 10.1128/IAI.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortinea N, Trieu-Cuot P, Gaillot O, Pellegrini E, Berche P, Gaillard JL. 2000. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res Microbiol 151:353–360. doi: 10.1016/S0923-2508(00)00158-3. [DOI] [PubMed] [Google Scholar]
- 27.Andersen JB, Roldgaard BB, Lindner AB, Christensen BB, Licht TR. 2006. Construction of a multiple fluorescence labelling system for use in co-invasion studies of Listeria monocytogenes. BMC Microbiol 6:86. doi: 10.1186/1471-2180-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valderrama WB, Ostiguy N, Cutter CN. 2014. Multivariate analysis reveals differences in biofilm formation capacity among Listeria monocytogenes lineages. Biofouling 30:1199–1209. doi: 10.1080/08927014.2014.980818. [DOI] [PubMed] [Google Scholar]
- 29.Zameer F, Gopal S, Krohne G, Kreft J. 2009. Development of a biofilm model for Listeria monocytogenes EGD-e. World J Microbiol Biotechnol 26:1143–1147. doi: 10.1007/s11274-009-0271-4. [DOI] [Google Scholar]
- 30.Xavier J, White D, Almeida J. 2003. Automated biofilm morphology quantification from confocal laser scanning microscopy imaging. Water Sci Technol 47:31–37. [PubMed] [Google Scholar]
- 31.Mosquera-Fernández M, Rodríguez-López P, Cabo ML, Balsa-Canto E. 2014. Numerical spatio-temporal characterization of Listeria monocytogenes biofilms. Int J Food Microbiol 182-183:26–36. doi: 10.1016/j.ijfoodmicro.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Di Bonaventura G, Piccolomini R, Paludi D, D'Orio V, Vergara A, Conter M, Ianieri A. 2008. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: relationship with motility and cell surface hydrophobicity. J Appl Microbiol 104:1552–1561. doi: 10.1111/j.1365-2672.2007.03688.x. [DOI] [PubMed] [Google Scholar]
- 33.Kadam SR, den Besten HM, van der Veen S, Zwietering MH, Moezelaar R, Abee T. 2013. Diversity assessment of Listeria monocytogenes biofilm formation: impact of growth condition, serotype and strain origin. Int J Food Microbiol 165:259–264. doi: 10.1016/j.ijfoodmicro.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 34.Schaudinn C, Stoodley P, Kainovic A, Keeffe OT, Costerton B, Robinson D, Baum M, Ehrlich G, Webster P. 2007. Bacterial biofilms, other structures seen as mainstream concepts. Microbe 2:231–237. [Google Scholar]
- 35.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luijten E, Parsek MR, Wong GC. 2013. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 497:388–391. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchal M, Briandet R, Halter D, Koechler S, DuBow MS, Lett M-C, Bertin PN. 2011. Subinhibitory arsenite concentrations lead to population dispersal in Thiomonas sp. PLoS One 6:e23181. doi: 10.1371/journal.pone.0023181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houry A, Gohar M, Deschamps J, Tischenko E, Aymerich S, Gruss A, Briandet R. 2012. Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc Natl Acad Sci U S A 109:13088–13093. doi: 10.1073/pnas.1200791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klausen M, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 39.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 40.Bayles KW. 2007. The biological role of death and lysis in biofilm development. Nat Rev Microbiol 5:721. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 41.Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol 185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemon KP, Higgins DE, Kolter R. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol 189:4418–4424. doi: 10.1128/JB.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todhanakasem T, Young GM. 2008. Loss of flagellum-based motility by Listeria monocytogenes results in formation of hyperbiofilms. J Bacteriol 190:6030–6034. doi: 10.1128/JB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts A, Nightingale K, Jeffers G, Fortes E, Kongo JM, Wiedmann M. 2006. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152:685–693. doi: 10.1099/mic.0.28503-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.