Abstract
The aim of this study was to determine if rumen protozoa could form large amounts of reserve carbohydrate compared to the amounts formed by bacteria when competing for glucose in batch cultures. We separated large protozoa and small bacteria from rumen fluid by filtration and centrifugation, recombined equal protein masses of each group into one mixture, and subsequently harvested (reseparated) these groups at intervals after glucose dosing. This method allowed us to monitor reserve carbohydrate accumulation of protozoa and bacteria individually. When mixtures were dosed with a moderate concentration of glucose (4.62 or 5 mM) (n = 2 each), protozoa accumulated large amounts of reserve carbohydrate; 58.7% (standard error of the mean [SEM], 2.2%) glucose carbon was recovered from protozoal reserve carbohydrate at time of peak reserve carbohydrate concentrations. Only 1.7% (SEM, 2.2%) was recovered in bacterial reserve carbohydrate, which was less than that for protozoa (P < 0.001). When provided a high concentration of glucose (20 mM) (n = 4 each), 24.1% (SEM, 2.2%) of glucose carbon was recovered from protozoal reserve carbohydrate, which was still higher (P = 0.001) than the 5.0% (SEM, 2.2%) glucose carbon recovered from bacterial reserve carbohydrate. Our novel competition experiments directly demonstrate that mixed protozoa can sequester sugar away from bacteria by accumulating reserve carbohydrate, giving protozoa a competitive advantage and stabilizing fermentation in the rumen. Similar experiments could be used to investigate the importance of starch sequestration.
INTRODUCTION
A diverse assemblage of protozoa, bacteria, methanogens, and fungi inhabit the rumen of ruminant livestock (1). Although they may account for as little as 5% of the microbial biomass (2), protozoa have an important role in stabilizing fermentation (3). Animals with protozoa absent have higher concentrations of short-chain fatty acids (SCFAs) and lower mean pHs (3–5). Furthermore, SCFAs and pH may fluctuate more when protozoa are absent (4, 6).
Protozoa have been proposed to stabilize rumen fermentation in part by consuming sugar and starch, preventing the rapid fermentation of these substrates by bacteria (3). According to this proposed mechanism, protozoa synthesize reserve carbohydrate after consuming sugar, and protozoa ferment this reserve carbohydrate and intracellular starch more slowly than do bacteria. This prevents the buildup of SCFAs, depression of pH, and onset of lactic acid acidosis that are detrimental to animal performance (7, 8). Sugar consumption has been attributed to protozoa of the family Isotrichidae, and starch consumption has been attributed to protozoa of the family Orphryoscolecidae (3, 9).
Besides its importance for animal performance, this sequestration of carbohydrate in reserve carbohydrate and intracellular starch would give protozoa a competitive advantage over bacteria. It would deprive bacteria of a substrate for growth, and it might explain why protozoa can persist alongside bacteria in the rumen, even though bacteria grow much faster than protozoa in culture (10–13).
Although protozoa are often claimed to accumulate more reserve carbohydrate than bacteria (3), quantitative support for this claim remains sparse because of the inability to culture protozoa axenically. Under a microscope, isotrichid protozoa seem to accumulate prodigious amounts of reserve carbohydrate (enough to turn opaque) (3), but this method is qualitative and difficult to apply to bacteria. Previous studies that sampled protozoa from the rumen have shown that protozoa usually contain more carbohydrate than do bacteria (14, 15), but samples were taken at only two time points (15) or samples were compiled across time points (14). Accumulation of carbohydrate may be inferred, but the dynamics of this accumulation are poorly resolved. A recent study compared glycogen accumulation in protozoa and bacteria in batch culture, but that method was indirect because it relied on selectively lysing bacteria before measurement of glycogen concentrations (16). Until now, a more direct method to measure accumulation by protozoa in batch culture, in which conditions can be controlled and samples can be taken easily and repeatedly, has not been developed.
Our objective was to quantify how much reserve carbohydrate is accumulated by protozoa compared to that accumulated by bacteria when competing for glucose in batch cultures. We developed a method that efficiently separates large protozoa from small bacteria at intervals after glucose dosing, enabling us to directly compare the dynamics of reserve carbohydrate accumulation by these 2 groups. These novel competition experiments directly show that protozoa sequester glucose in reserve carbohydrate and away from bacteria, giving protozoa a competitive advantage and stabilizing fermentation in the rumen.
MATERIALS AND METHODS
Preparation of protozoal and bacterial mixtures.
In glucose competition experiments, rumen fluid was collected from 4 Jersey cows (1 cow per experiment). The Ohio State University Institutional Animal Care and Use Committee approved all animal procedures. Cows were fed a lactation diet ad libitum in two equal meals. The diet composition was 45.3% corn silage, 13.8% legume silage, 12.5% ground corn, 8.6% soybean meal, 6.4% whole cottonseed, 3.8% distiller grains, 2.8% wheat middlings, 2.1% Amino Plus (Ag Processing Inc., Hiawatha, KS), 1.0% Megalac (Church & Dwight, Princeton, NJ), 0.3% direct-fed microbial product (XP DFM; Diamond V, Cedar Rapids, IA), and 3.3% vitamins and minerals. In experiments investigating recoveries of protozoa and bacteria during their separation, rumen fluid was collected from as many as 8 cows (1 cow per experiment). The diet composition was either that described above or, for earlier experiments, when the farm fed a different diet, 50% corn silage, 4.5% alfalfa hay, 21% corn wet milling product (Cargill Corn Milling, Dayton, OH), 9.05% ground corn, 4.64% soybean meal, 1.30% Amino Plus, 1.30% soyhulls, 0.38% fat, and 2.01% vitamins and minerals.
At 2.5 h after feeding, rumen contents were strained through 4 layers of cheesecloth. The strained fluid was diluted 1:1 with N-free buffer (Simplex type, pH 6.8 [3]) and added to a separatory funnel. All glassware was prewarmed to 39°C and pregassed with O2-free CO2. Plant particles, which rose to the top, were removed by aspiration after 1 h of incubation at 39°C.
After removal of plant particles, mixtures of protozoa and bacteria were prepared from the clarified fluid (Fig. 1). Large protozoa were isolated on a nylon cloth with a 20-μm pore size (14% open area) (catalog number 7050-1220-000-10; Sefar, Buffalo, NY) and washed with Simplex buffer. Buffer was prewarmed to 39°C to preserve viability, and the cloth was kept under a stream of O2-free CO2 by using an assembly described previously (3, 17). At the same time that protozoa were isolated, small bacteria were isolated by centrifugation on a prewarmed rotor (JA-17 rotor and J2-21 centrifuge; Beckman, Brea, CA).
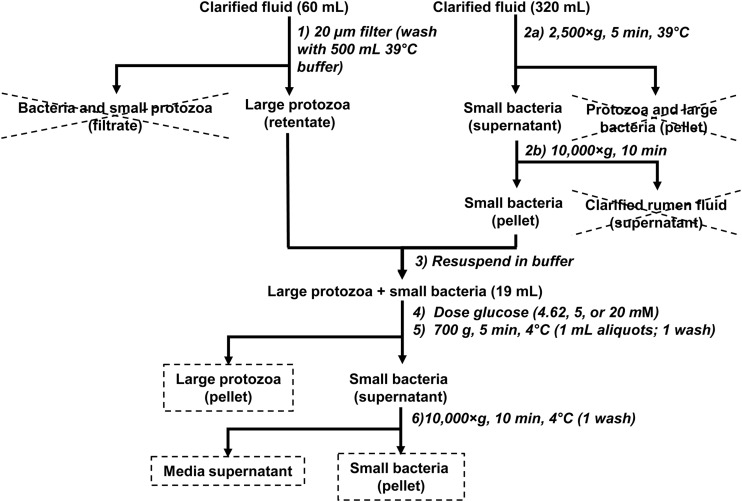
FIG 1.
Flowchart for preparation of a mixture of protozoa and bacteria, as well as harvesting of the protozoal and bacterial pellets from that mixture. Fractions saved for chemical analysis are marked by stippled boxes. Fractions discarded are crossed out. Steps are numbered as described in the text. Methodological details are reported in Materials and Methods, and recoveries are reported in Results.
After separation of large protozoa and small bacteria, these groups were recombined by resuspending them in Simplex buffer and transferring them to a culture bottle. The optical density (in conjunction with a calibration curve) was used to give a preliminary estimate of protein concentrations and to ensure that approximately equal protein masses were being combined. The culture bottle was capped with a butyl rubber stopper and incubated at 39°C. Cells were dosed with moderate (4.62 and 5 mM) (n = 2 each) or high (20 mM) (n = 4) concentrations of glucose. At intervals after dosing with glucose, the cell suspension (one 1-ml aliquot per time point) was harvested to obtain cell pellets (F45-24-11 rotor and 54515 D centrifuge; Eppendorf, Hauppauge, NY) (Fig. 1), washed once in 0.9% NaCl, and stored at −20°C. During harvesting, the suspension was placed on ice and centrifuged as quickly as possible to limit further uptake and metabolism of glucose. Pellets were harvested at intervals to give 3 time points prior to glucose dosing, at least 3 time points during glucose excess, and at least 2 time points after glucose was exhausted. The cell-free supernatant was prepared by combining the supernatants from the cell harvesting and washing steps.
Chemical and other analyses.
Chemical analyses, direct counts of protozoa and bacteria, and calculation of carbon recovery were done as previously described (18, 19). Briefly, pellets were analyzed for reserve carbohydrate by using the anthrone method and for protein by using the Pierce BCA assay kit (product number 23227; Thermo Scientific, Rockford, IL). The cell-free supernatant was analyzed for d-/l-lactic acid with a kit from R-Biopharm (product code 11112821035; R-Biopharm, Marshall, MI), for other short-chain fatty acids by gas chromatography, and for free glucose by the glucose oxidase-peroxidase method. Carbon recovery was calculated from concentrations of glucose, reserve carbohydrate, short-chain fatty acids, CO2, and CH4 both before and after glucose dosing. CO2 and CH4 concentrations were determined by reaction stoichiometry (18, 19).
To estimate the mass of bacteria contaminating large protozoa, bacterial counts in the large-protozoon fraction were determined and then multiplied by bacterial protein mass. Bacterial protein mass was assumed to be 1.790 × 10−13 g/cell (standard error of the mean [SEM], 0.098 × 10−13 g/cell); this was determined for separate samples (n = 16 across 8 cows) prepared according to the flowchart shown in Fig. S1A in the supplemental material. A similar approach was used to estimate the mass of protozoa contaminating small bacteria. Protozoal protein mass was assumed to be 1.29 × 10−8 g/cell (SEM, 0.080 × 10−8 g/cell); this was determined for separate samples (n = 12 across 5 cows) prepared according to the flowchart shown in Fig. S1B in the supplemental material.
Statistics.
Data were analyzed by using PROC MIXED of SAS (SAS Institute Inc., Cary, NC), using the model Yijk = μ + ci + Fj + Dk + Fj × Dk + εijk, where Yijk is the observation, μ is the overall mean, ci is the random effect of cow (where i indicates cow 472, 478, 491, or 492), Fj is the fixed effect of microbial type (where j indicates bacteria or protozoa), Dk is the fixed effect of glucose concentration (where k indicates a moderate or high concentration), Fj × Dk is the interaction of Fj and Dk, and εijk is the residual error. When the F test for a main effect or interaction term was significant (P < 0.05), means were separated by using Tukey's test. For determining if cell recoveries differed from 100%, an unpaired t test was used instead of the model and procedures described above.
Local regression (LOCFIT package of R [20]) was used to fit time series data to smooth curves, as described previously (18, 19). Original data are presented alongside the smooth curves in the figures, and smooth curves were used for calculations (e.g., carbon recovery and changes in reserve carbohydrate) and statistical analysis. First-order rates of exponential increase of cellular protein levels were calculated by using PROC NLIN of SAS.
RESULTS
Protozoal and bacterial mixtures.
Mixtures of protozoa and bacteria were prepared to (i) have approximately equal protein masses of protozoa and bacteria and (ii) enable quick separation during harvesting for chemical analysis. To accomplish these goals, protozoa and bacteria were preseparated from rumen fluid and then recombined to form the mixture used in subsequent experiments (Fig. 1).
Large protozoa were preseparated by filtering rumen fluid through a 20-μm nylon cloth (step 1) (Fig. 1). Based on direct counts, the extent of recovery of total protozoa was 70.8% (SEM, 4.1%) in clarified rumen fluid (n = 4 across 2 cows) and was <100% (P = 0.039). Incomplete recovery resulted from the loss of small Entodinium spp. through the cloth. The extent of recovery of Entodinium on the cloth was only 69.4% (SEM, 3.4%) (n = 4), but preliminary experiments showed that the extent of recovery on the cloth and filtrate combined was >95% (data not shown). The rate of recovery of larger, non-Entodinium spp. was 94.9% (SEM, 7.8%) and did not differ (P = 0.560) from 100% (n = 4). The final composition was 90.0% (SEM, 2.6%) Entodinium species, 4.4% (SEM, 1.1%) Isotricha species, 2.27% (SEM, 0.95%) Dasytricha species, 0.72% (SEM, 0.26%) Epidinium species, 1.18% (SEM, 0.20%) Diplodininae, and 1.44% (SEM, 0.60%) Ophryoscolex species (n = 14 across 6 cows). Based on direct counts of bacteria, the level of contamination of protozoa with bacteria was 0.77% (0.13%) of total protein (n = 8 across 3 cows). In earlier experiments, a cloth with smaller pore size (10 μm) was used to minimize losses of small protozoa, but the filtration time was long (>5 min), and the level of bacterial contamination was high (>5% of total protein).
Small bacteria were preseparated by 2 centrifugation steps (steps 2a and b) (Fig. 1) and then combined with preseparated large protozoa. Once preseparated bacteria and protozoa were combined, cell pellets were harvested for chemical analysis by 2 centrifugation steps (steps 5 and 6) (Fig. 1). The rate of recovery of protozoal cells in the protozoal pellet after the first, low-speed centrifugation step (step 5) was 92.4% (SEM, 4.9%), which did not differ (P = 0.163) from 100% (n = 8 across 2 cows). The recovery of cells of Entodinium (102.4% [SEM, 0.97%]) exceeded (P = 0.041) 100%, whereas recovery of non-Entodinium cells (70.8% [SEM, 10.8%]) was far more variable and was <100% (P = 0.030) (n = 8). For non-Entodinium species, the rates of recovery of Isotricha (101.8% [SEM, 15.3%]) and Dasytricha (84.0% [SEM, 24.2%]) were generally high and did not differ (P = 0.531) from 100% (n = 8). However, rates of recovery of Diplodininae (26.2% [SEM, 12.3%]), Ophryoscolex (17.7% [SEM, 6.3%]), and Epidinium (0%) were low for reasons that could not be immediately explained. The final composition was 95.65% (SEM, 0.50%) Entodinium, 3.16% (SEM, 0.39%) Isotricha, 0.59% (SEM, 0.18%) Dasytricha, 0% Epidinium, 0.44% (SEM, 0.20%) Diplodininae, and 0.16% (SEM, 0.11%) Ophryoscolex (n = 8). The level of contamination with bacteria was 7.10% (SEM, 0.52%) of total protein (n = 8).
The extent of recovery of bacterial cells in the bacterial pellet after the second, high-speed centrifugation step (step 6) was 93.0% (SEM, 8.9%), which did not differ (P = 0.449) from 100% (n = 8 across 2 cows). Based on direct counts of protozoa, the level of contamination with protozoa was 0.190% (SEM, 0.051%) of total protein (n = 8).
Glucose competition experiments.
Mixtures of large protozoa and small bacteria were prepared as described above for glucose competition experiments using rumen fluid from 4 cows. Recovery and contamination of cells in protozoal and bacterial pellets were not formally quantified but appeared similar to those in the experiments reported above.
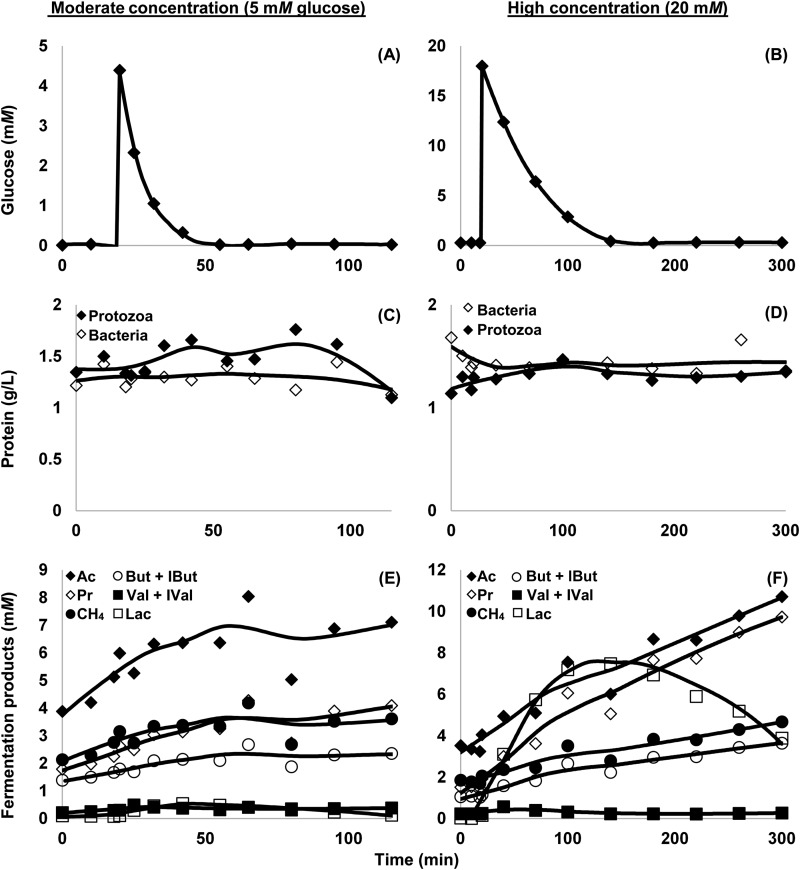
When pulse-dosed with glucose, protozoa and bacteria responded by rapidly consuming glucose, accumulating reserve carbohydrate, and producing short-chain fatty acids (Fig. 2 and 3). After glucose was exhausted, the amount of reserve carbohydrate began to decline, the level of lactate was already declining, and the production of other short-chain fatty acids slowed (Fig. 3). In comparison, protein levels remained relatively stable (Fig. 2C and D).
FIG 2.
Response of a mixture of rumen protozoa and bacteria to a pulse dose of glucose at 20 min. Data are from cow 491 and represent 1 experiment per concentration of glucose; 3 other cows (representing 3 additional experiments per concentration of glucose) had similar responses (data not shown). (A and B) Glucose in media. (C and D) Cell protein of protozoa and bacteria. (E and F) Fermentation products, including acetate (Ac), methane (CH4), propionate (Pr), butyrate and isobutyrate (But + IBut), valerate and isovalerate (Val + IVal), and lactate (Lac). Mixtures of protozoa and bacteria were prepared and harvested according to the flowchart shown in Fig. 1. Reserve carbohydrate accumulation is shown in Fig. 3. Each datum point represents one sample replicated in triplicate.
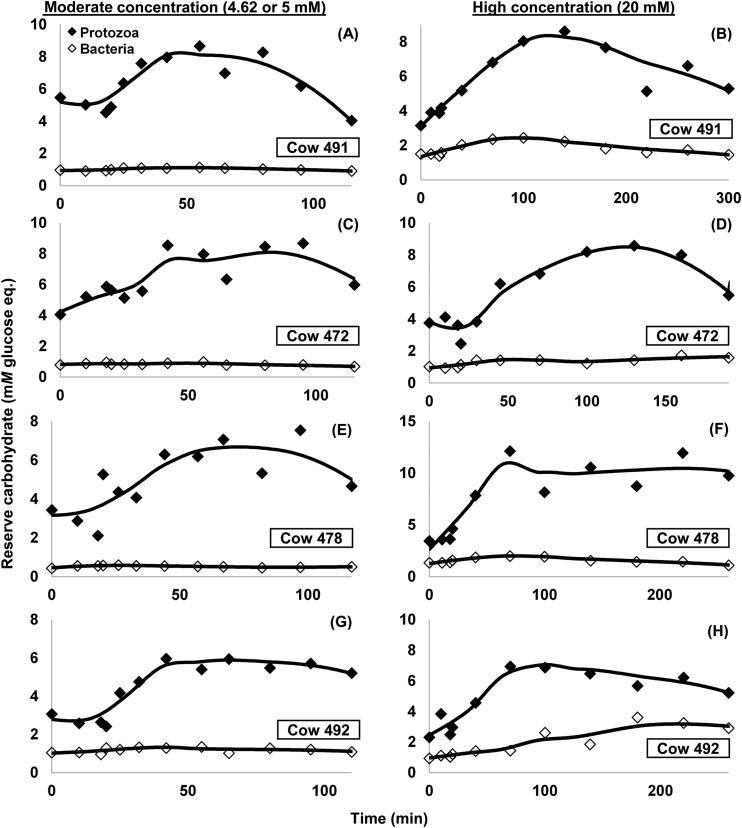
FIG 3.
Reserve carbohydrate of a mixture of rumen protozoa and bacteria after a pulse dose of glucose at 20 min. (A and C) Five millimolar glucose; (E and G) 4.62 mM glucose; (B, D, F, and H) 20 mM glucose. Shown are separate values for bacteria and protozoa for cows 491 (A and B), 472 (C and D), 478 (E and F), and 492 (G and H). Mixtures of protozoa and bacteria were prepared and harvested according to the flowchart shown in Fig. 1. Reserve carbohydrate is expressed as mM monomeric glucose equivalents to be in same units as glucose in the medium. Each panel represents data from one experiment, and each datum point represents one sample replicated in triplicate.
Protozoa accumulated more reserve carbohydrate than did bacteria (Fig. 3 and Table 1). This was found regardless of whether the concentration of glucose dosed was moderate (4.62 or 5 mM; n = 2 each; P < 0.001) or high (20 mM; n = 4; P < 0.001). At the time of peak accumulation, protozoa had incorporated nearly 60% of the total glucose carbon into reserve carbohydrate when dosed with a moderate glucose concentration (Table 1). This percentage was 34.5-fold higher than that for bacteria. For the high glucose concentration, protozoa also incorporated a high percentage of the total glucose carbon into reserve carbohydrate (Fig. 3), although this percentage was lower (P < 0.001) than when a moderate concentration was given (Table 1). When given the high dose, bacteria accumulated numerically more reserve carbohydrate than with the moderate dose (Table 1), although this difference was not significant (P = 0.694), and the accumulation of reserve carbohydrate was much lower than that for protozoa at either dose.
TABLE 1.
Net incorporation of glucose carbon into reserve carbohydrate at the time of peak carbohydrate accumulation
| Fraction | Glucose dose (concn [mM]) | Recovery of carbon in reserve carbohydrate (% carbon in glucose dose)a |
|---|---|---|
| Protozoa | Moderate (4.62 or 5) | 58.7a |
| High (20) | 24.2b | |
| Bacteria | Moderate (4.62 or 5) | 1.7c |
| High (20) | 5.0c |
Rows with different superscripts indicate significant differences (P < 0.001). The SEM for recovery of carbon in reserve carbohydrate was 2.1%.
Over the duration of the individual experiments (n = 8), protozoal and bacterial protein levels averaged 1.534 and 1.377 g/liter (SEM, 0.084 g/liter), respectively, and did not differ (P = 0.105) from each other. Cells were washed with N-free buffer to limit growth, and bacterial protein levels did not increase in experiments with moderate concentrations of glucose (−4.4% h−1 [SEM, 3.4% h−1]; P = 0.222) or high concentrations of glucose (3.8% h−1 [SEM, 3.4% h−1]; P = 0.287). Similarly, protozoal protein levels did not increase for experiments with a high concentration of glucose (1.7% h−1 [SEM, 3.4% h−1]; P = 0.627; n = 4). Unexpectedly, protozoal protein levels increased exponentially for experiments with a moderate concentration of glucose (14.1% h−1 [SEM, 3.4% h−1]; P = 0.003; n = 4).
Lactate initially accumulated, and concentrations peaked at 1.5 mM (SEM, 1.6 mM) and 5.9 mM (SEM, 1.8 mM) for moderate and high glucose concentrations, respectively. Peak concentrations were highly variable, and they did not differ (P = 0.209) across concentrations of glucose. By the end of the incubation period, concentrations had fallen to 0.93 mM (SEM, 0.82 mM) (n = 7) and did not vary (P = 0.518) by glucose concentration. Due to an insufficient amount of sample, lactate was not measured in one incubation in which 4.62 mM glucose was given.
Across all experiments (n = 7), the rate of carbon recovery was 94.0% (SEM, 3.8%) and did not differ (P = 0.230) from 100%. The single experiment in which the lactate concentration was not measured could not be included in the calculation of carbon recovery.
DISCUSSION
Protozoa have been proposed to stabilize rumen fermentation by synthesizing reserve carbohydrate from sugar and by consuming starch, sequestering rapidly fermentable substrates from bacteria. This proposal attempts to explain the observation that protozoon-free ruminants show lower pH and higher concentrations of SCFAs than do conventional animals (3–5). Such sequestration not only would be important for stabilizing rumen fermentation but also may explain how protozoa are able to persist in the rumen alongside bacteria that grow faster in culture (10–13).
We previously found that mixed rumen microbes as a whole can accumulate large amounts of reserve carbohydrate. When we washed mixed rumen microbes with N-free buffer and dosed them with 5 or 20 mM glucose, we recovered >50% of the glucose in reserve carbohydrate at the time of peak accumulation (19). Characterization of the reserve carbohydrate identified it as glycogen. The anthrone method detected changes in reserve carbohydrate accumulation quantitatively, whereas more specific methods based on amyloglucosidase hydrolysis did not (18). The anthrone method is reliable as long as cross-reacting material (microbial cell wall, microbial exopolysaccharide, and plant particles) remains constant (18). Whether protozoa or bacteria were responsible for reserve carbohydrate accumulation remains unclear in these studies, requiring a method to separate these two groups.
In this study, we developed a novel method to monitor reserve carbohydrate accumulation by bacteria and protozoa individually when in competition for glucose (Fig. 1). Using this method, we found that protozoa accumulated prodigious amounts of reserve carbohydrate. Protozoa incorporated nearly 60 and 25% of the glucose carbon into reserve carbohydrate when dosed with moderate (4.62 or 5 mM) and high (20 mM) concentrations of glucose, respectively. These levels of incorporation were ∼35- and 5-fold higher than those in bacteria, respectively. This lends weight to the idea that protozoa can sequester sugar from bacterial fermentation.
In this study, the amount of glucose incorporated into reserve carbohydrate was broadly similar to that which we previously observed for mixed rumen microbes (19), but it was more similar for moderate glucose concentrations than for high glucose concentrations. For the moderate glucose concentration (4.62 or 5 mM), ∼60% of glucose carbon was recovered in the reserve carbohydrate of bacteria and protozoa combined (Table 1). This value compares with the 60% of carbon recovered for mixed rumen microbes given 5 mM glucose (19). For the high glucose concentration (20 mM), ∼30% was recovered in the reserve carbohydrate of bacteria and protozoa combined (Table 1), which is lower than the 53% observed for mixed rumen microbes given 20 mM glucose (19). The reason for the lower rate of recovery for the high glucose concentration in this study is unclear but may be due to differences in the compositions of the microbial samples between studies. The microbial mixtures in this study contained approximately equal protein masses of protozoa and bacteria, whereas the mixed rumen microbes in the previous study would be expected to contain more bacteria than protozoa (see below). Also, the mixture in this study lacked small protozoa and large bacteria (Fig. 1). Finally, preparation of the mixtures in this study required more time than for preparation of mixed rumen microbes, and we cannot exclude that the long preparation time had some impact.
Beyond glucose incorporated into reserve carbohydrate, protozoa must have taken up additional glucose to fuel reserve carbohydrate synthesis. For the moderate concentration of glucose, protozoa would have fermented at least 23% of the glucose dose to generate ATP for synthesis, meaning that protozoa took up at least 81% of the total glucose. This calculation assumes that the synthesis of reserve carbohydrate (glycogen) requires 2 ATP molecules/mol glucose (21), and fermentation pathways for rumen protozoa yield a maximum of 5 ATP molecules/mol glucose (see references 1, 22, and 23). The rate of uptake would have been higher still if glycogen were degraded during synthesis (cycled), as found for isotrichid protozoa (24, 25). Protozoa can thus take up large amounts of glucose for direct incorporation into reserve carbohydrate or for fueling the synthesis of those reserves.
This use of the substrate for reserve carbohydrate may explain how protozoa can persist in the rumen. Generation times of protozoa are at least 6 h (growth rate, ∼12% h−1) in batch culture, and they are at least 11.3 h (growth rate, ∼6% h−1) for sequentially transferred cultures (12, 13). Growth rates of rumen bacteria, in comparison, often exceed 40% h−1 when grown on simple carbohydrates (10, 11). Predation is one mechanism by which protozoa can keep bacterial growth in check (3), but they likely also remain competitive by accumulating reserve carbohydrate and starch, depriving bacteria of a substrate for growth.
Our results agree with less direct or extensive methods for comparing reserve carbohydrate accumulation of protozoa and bacteria. Data reported previously by Czerkawski (15) show that protozoa isolated from the rumen accumulated between 2.2- and 6.5-fold more carbohydrate than did bacteria between 0 and 2 h after feeding (assuming, as did that author, that the ratio of small to large bacteria was 5:1). Volden et al. (14) found that protozoa isolated from the rumen and pooled across various sample times contained (i) 3.8- to 7.3-fold more “starch” than did liquid-associated bacteria and (ii) 12.3- to 16.2-fold more of this component than did solid-associated bacteria. In a previously reported batch culture study, rumen fluid was dosed with 16.5 mM glucose, and glycogen accumulation was measured at 3 h (16). Before glycogen measurement, cell pellets were hydrolyzed with NaOH and boiled in water to reportedly lyse both bacteria and protozoa, or pellets were boiled in water alone to reportedly lyse protozoa alone. Comparison of hydrolyzed and nonhydrolyzed treatments suggests that protozoa were responsible for 48.5% of the total glycogen accumulation. In additional treatments, isotrichids were destroyed (by blending) prior to the addition of glucose, and these treatments suggested that isotrichids were the protozoal group primarily responsible for glycogen accumulation. That study was indirect, and it is reliable insofar as treatments selectively and completely lysed target cells. Our study bolsters those previous findings by measuring reserve carbohydrate directly, under controlled batch culture conditions, and over the entire time course of glucose utilization.
Our method removed small protozoa (which could not be separated from bacteria without a high level of bacterial contamination), but the large protozoa remaining were still diverse. These protozoa included isotrichids (Isotricha and Dasytricha) and ophryoscolecids (Entodinium, Diplodininae, and Ophryoscolex). Cells of the ophryoscolecid genus Epidinium were also found in low concentrations immediately after preseparation, but they were not recovered later during harvesting of the cell pellet for chemical analysis. Entodinium species was numerically the most abundant group, but it might not have been the most important group for the observed responses to glucose: isotrichids are thought to consume sugar more actively than Entodinium and other ophryoscolecids (3, 9).
We chose to wash cells with N-free buffer before administering glucose to (i) prevent confounding effects of growth and (ii) stimulate the accumulation of reserve carbohydrate. Nonetheless, this approach has limitations. First, we found that growth can be difficult to limit even with washing, as protozoal protein concentrations still increased for experiments with moderate glucose concentrations. A tempting explanation for this increase is that protozoa preyed upon bacteria, which served as a source of N for protozoal growth. However, bacterial protein concentrations did not decrease, nor did protozoal protein concentrations increase in experiments with high glucose concentrations. A second limitation is that washing with N-free buffer reduces the rate of glucose uptake, as shown previously for pure cultures of Streptococcus bovis (26). Indeed, N limitation reduces rates of uptake in both prokaryotes and eukaryotes (27). Even so, the rate of uptake was still considerable for both Streptococcus bovis (26) and mixed rumen microbes (19) after washing with N-free buffer. A third limitation is that washing with N-free buffer reduces confounding effects of growth, but growth itself may be an important factor in competition between bacteria and protozoa. Complementary experiments could include a source of N.
Conditions in our experiments were chosen largely for a proof of concept, but they are representative of the rumen in certain cases. Our mixtures contained approximately equal protein masses of protozoa and bacteria. Under typical dairy feeding conditions, protozoa make up 5 to 13% of the total microbial N (2), although values exceeding 50% have been reported when diets were fed in restricted quantities (3). In our experiments, we used ∼5 and 20 mM glucose, although concentrations in the rumen rarely exceed ∼2.5 mM (28, 29). Still, concentrations of ∼5 mM can be reached after feeding unadapted animals large amounts of grain (6, 30), and soluble sugar concentrations of 69 mM have been recorded after feeding beet pulp (31). Furthermore, concentrations of soluble sugars may be high around feed particles due to the accumulation of hydrolysis products from primary colonizers (28, 32), and protozoa (both isotrichids and ophryoscolecids) display chemotaxis toward sugars (33). In our experiments, we intensified carbohydrate excess by removing N (by washing cells with N-free buffer). For animals fed grain, N in the rumen is present but at low concentrations, creating a carbohydrate excess (34). For such animals, N is primarily in the form of ammonia-N, and microbes grow more slowly with ammonia-N than with amino-N (35, 36). Thus, our conditions fall within or close to the measured extremes of the rumen.
Our results suggest that mixed protozoa, even those which are predominantly Entodinium species, can accumulate large amounts of reserve carbohydrate and sequester sugar away from bacteria. Our work therefore suggests one competitive advantage of protozoa over bacteria. Additional work is needed to answer whether such sequestration would also effectively stabilize rumen fermentation, as has been proposed. For such sequestration to be effective, protozoa would need to ferment reserve carbohydrate more slowly than bacteria themselves would ferment free sugar. Sequestration of starch, in addition to sugar, is also likely important, and our novel competition experiments provide a framework to investigate this idea.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhongtang Yu (The Ohio State Univerity) and Shujin Hackmann (University of Florida) for reviewing the manuscript, Josie Plank (The Ohio State University) for technical assistance with short-chain fatty acid analysis, and Reagen Bluel and the farm staff at Waterman Dairy Farm (The Ohio State University) for animal care.
Research was supported by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. The paper was assigned manuscript number 23/14AS. Additional funds were from the OARDC Director's Associateship Program Award, University Distinguished Fellowship, OARDC Graduate Research Competition Competitive Grants Program 2010-114, and USDA Cooperative State Research, Education, and Extension Service USDA/NRICGP grant 2008-35206-18847.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03736-14.
REFERENCES
- 1.Russell J. 2002. Rumen microbiology and its role in ruminant nutrition. James B. Russell, Ithaca, NY. [Google Scholar]
- 2.Sylvester JT, Karnati SK, Yu Z, Newbold CJ, Firkins JL. 2005. Evaluation of a real-time PCR assay quantifying the ruminal pool size and duodenal flow of protozoal nitrogen. J Dairy Sci 88:2083–2095. doi: 10.3168/jds.S0022-0302(05)72885-X. [DOI] [PubMed] [Google Scholar]
- 3.Williams A, Coleman G. 1992. The rumen protozoa. Springer-Verlag, New York, NY. [Google Scholar]
- 4.Whitelaw FG, Eadie JM, Mann SO, Reid RS. 1972. Some effects of rumen ciliate protozoa in cattle given restricted amounts of a barley diet. Br J Nutr 27:425–437. doi: 10.1079/BJN19720108. [DOI] [PubMed] [Google Scholar]
- 5.Nagaraja TG, Towne G, Beharka AA. 1992. Moderation of ruminal fermentation by ciliated protozoa in cattle fed a high-grain diet. Appl Environ Microbiol 58:2410–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackie R, Gilchrist F, Robberts A, Hannah P, Schwartz H. 1978. Microbiological and chemical changes in the rumen during the stepwise adaptation of sheep to high concentrate diets. J Agric Sci 90:241–254. doi: 10.1017/S0021859600055313. [DOI] [Google Scholar]
- 7.Nocek JE. 1997. Bovine acidosis: implications on laminitis. J Dairy Sci 80:1005–1028. doi: 10.3168/jds.S0022-0302(97)76026-0. [DOI] [PubMed] [Google Scholar]
- 8.Williams AG, Coleman GS. 1997. The rumen protozoa, p 73–139. In Hobson PN, Stewart CS (ed), The rumen microbial ecosystem, 2nd ed Blackie Academic & Professional, New York, NY. [Google Scholar]
- 9.Dehority BA. 2003. Rumen microbiology. Nottingham University Press, Nottingham, United Kingdom. [Google Scholar]
- 10.Russell JB, Baldwin R. 1979. Comparison of maintenance energy expenditures and growth yields among several rumen bacteria grown on continuous culture. Appl Environ Microbiol 37:537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell JB, Delfino FJ, Baldwin RL. 1979. Effects of combinations of substrates on maximum growth rates of several rumen bacteria. Appl Environ Microbiol 37:544–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehority BA. 2010. Physiological characteristics of several rumen protozoa grown in vitro with observations on within and among species variation. Eur J Protistol 46:271–279. doi: 10.1016/j.ejop.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Sylvester JT, Karnati SK, Dehority BA, Morrison M, Smith GL, St-Pierre NR, Firkins JL. 2009. Rumen ciliated protozoa decrease generation time and adjust 18S ribosomal DNA copies to adapt to decreased transfer interval, starvation, and monensin. J Dairy Sci 92:256–269. doi: 10.3168/jds.2008-1417. [DOI] [PubMed] [Google Scholar]
- 14.Volden H, Mydland LT, Harstad OM. 1999. Chemical composition of protozoal and bacterial fractions isolated from ruminal contents of dairy cows fed diets differing in nitrogen supplementation. Acta Agr Scand A Anim Sci 49:235–244. [Google Scholar]
- 15.Czerkawski JW. 1976. Chemical composition of microbial matter in the rumen. J Sci Food Agric 27:621–632. doi: 10.1002/jsfa.2740270707. [DOI] [PubMed] [Google Scholar]
- 16.Hall MB. 2011. Isotrichid protozoa influence conversion of glucose to glycogen and other microbial products. J Dairy Sci 94:4589–4602. doi: 10.3168/jds.2010-3878. [DOI] [PubMed] [Google Scholar]
- 17.Williams A, Yarlett N. 1982. An improved technique for the isolation of holotrich protozoa from rumen contents by differential filtration with defined aperture textiles. J Appl Microbiol 52:267–270. [Google Scholar]
- 18.Hackmann TJ, Keyser BL, Firkins JL. 2013. Evaluation of methods to detect changes in reserve carbohydrate for mixed rumen microbes. J Microbiol Methods 93:284–291. doi: 10.1016/j.mimet.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Hackmann TJ, Diese LE, Firkins JL. 2013. Quantifying the responses of mixed rumen microbes to excess carbohydrate. Appl Environ Microbiol 79:3786–3795. doi: 10.1128/AEM.00482-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loader C. 1999. Local regression and likelihood. Springer-Verlag, New York, NY. [Google Scholar]
- 21.Stouthamer AH. 1973. A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek 39:545–565. doi: 10.1007/BF02578899. [DOI] [PubMed] [Google Scholar]
- 22.Yarlett N, Lloyd D, Williams AG. 1985. Butyrate formation from glucose by the rumen protozoon Dasytricha ruminantium. Biochem J 228:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mertens E, Van Schaftingen E, Müller M. 1989. Presence of a fructose-2,6-bisphosphate-insensitive pyrophosphate:fructose-6-phosphate phosphotransferase in the anaerobic protozoa Tritrichomonas foetus, Trichomonas vaginalis and Isotricha prostoma. Mol Biochem Parasitol 37:183–190. doi: 10.1016/0166-6851(89)90150-3. [DOI] [PubMed] [Google Scholar]
- 24.Van Hoven W, Prins RA. 1977. Carbohydrate fermentation by the rumen ciliate Dasytricha ruminantium. Protistologica 13:599–606. [Google Scholar]
- 25.Prins RA, Van Hoven W. 1977. Carbohydrate fermentation by the rumen ciliate Isotricha prostoma. Protistologica 13:549–556. [Google Scholar]
- 26.Russell JB, Strobel HJ. 1990. ATPase-dependent energy spilling by the ruminal bacterium, Streptococcus bovis. Arch Microbiol 153:378–383. doi: 10.1007/BF00249009. [DOI] [PubMed] [Google Scholar]
- 27.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. 2011. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat Chem Biol 7:894–901. doi: 10.1038/nchembio.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kajikawa H, Amari M, Masaki S. 1997. Glucose transport by mixed ruminal bacteria from a cow. Appl Environ Microbiol 63:1847–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saleem F, Ametaj BN, Bouatra S, Mandal R, Zebeli Q, Dunn SM, Wishart DS. 2012. A metabolomics approach to uncover the effects of grain diets on rumen health in dairy cows. J Dairy Sci 95:6606–6623. doi: 10.3168/jds.2012-5403. [DOI] [PubMed] [Google Scholar]
- 30.Ryan R. 1964. Concentrations of glucose and low-molecular-weight acids in the rumen of sheep changed gradually from a hay to a hay-plus-grain diet. Am J Vet Res 25:653–659. [PubMed] [Google Scholar]
- 31.Clapperton JL, Czerkawski JW. 1969. Methane production and soluble carbohydrates in the rumen of sheep in relation to the time of feeding and the effects of short-term intraruminal infusions of unsaturated fatty acids. Br J Nutr 23:813–826. doi: 10.1079/BJN19690092. [DOI] [PubMed] [Google Scholar]
- 32.Cotta MA. 1992. Interaction of ruminal bacteria in the production and utilization of maltooligosaccharides from starch. Appl Environ Microbiol 58:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz HL, Karnati SK, Lyons MA, Dehority BA, Firkins JL. 2014. Chemotaxis toward carbohydrates and peptides by mixed ruminal protozoa when fed, fasted, or incubated with polyunsaturated fatty acids. J Dairy Sci 97:2231–2243. doi: 10.3168/jds.2013-7428. [DOI] [PubMed] [Google Scholar]
- 34.Russell JB. 1998. Strategies that ruminal bacteria use to handle excess carbohydrate. J Anim Sci 76:1955–1963. [DOI] [PubMed] [Google Scholar]
- 35.Van Kessel JS, Russell JB. 1996. The effect of amino nitrogen on the energetics of ruminal bacteria and its impact on energy spilling. J Dairy Sci 79:1237–1243. doi: 10.3168/jds.S0022-0302(96)76476-7. [DOI] [PubMed] [Google Scholar]
- 36.Argyle JL, Baldwin RL. 1989. Effects of amino acids and peptides on rumen microbial growth yields. J Dairy Sci 72:2017–2027. doi: 10.3168/jds.S0022-0302(89)79325-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.