Abstract
The increased awareness of the role of environmental matrices in enteric disease transmission has resulted in the need for rapid, field-based methods for fecal indicator bacteria and pathogen detection. Evidence of the specificity of β-glucuronidase-based assays for detection of Escherichia coli from environmental matrices relevant to enteric pathogen transmission in developing countries, such as hands, soils, and surfaces, is limited. In this study, we quantify the false-positive rate of a β-glucuronidase-based E. coli detection assay (Colilert) for two environmental reservoirs in Bangladeshi households (hands and soils) and three fecal composite sources (cattle, chicken, and humans). We investigate whether or not the isolation source of E. coli influences phenotypic and genotypic characteristics. Phenotypic characteristics include results of biochemical assays provided by the API-20E test; genotypic characteristics include the Clermont phylogroup and the presence of enteric and/or environmental indicator genes sfmH, rfaI, and fucK. Our findings demonstrate no statistically significant difference in the false-positive rate of Colilert for environmental compared to enteric samples. E. coli isolates from all source types are genetically diverse, representing six of the seven phylogroups, and there is no difference in relative frequency of phylogroups between enteric and environmental samples. We conclude that Colilert, and likely other β-glucuronidase-based assays, is appropriate for detection of E. coli on hands and in soils with low false-positive rates. Furthermore, E. coli isolated from hands and soils in Bangladeshi households are diverse and indistinguishable from cattle, chicken, and human fecal isolates, using traditional biochemical assays and phylogrouping.
INTRODUCTION
Environmental matrices (e.g., soils, surfaces, and people's hands) in households in low-income countries are heavily contaminated with fecal indicator bacteria, including Escherichia coli (1–3). For example, E. coli concentrations on surfaces (i.e., brooms, plates, toys, and wash basins) are 10- to 100-fold higher in developing countries like Cambodia, Peru, and Tanzania than on similar surfaces in developed regions like the United States and European Union (1, 3, 4). Similarly, in periurban Tanzania, dirt floors in households average 2.1 log10 CFU E. coli per gram of dry soil (1). Studies of E. coli concentrations on hands in Tanzania and Bangladesh report concentrations exceeding 103 CFU/2 hands compared to typical concentrations of less than 10 CFU/2 hands in the United States (4–6).
The presence of E. coli in households may present a health risk. In previous studies, multiple pathogenic strains of E. coli were detected on surfaces and in soils within households, as well as on hands and in stored drinking water in low-income countries (1, 5, 7). Specifically, both enteropathogenic and enterotoxigenic E. coli virotypes were detected. This is notable, as the recent findings by Kotloff et al. highlight both virotypes as etiological agents responsible for deaths related to diarrheal disease in children under five globally (8). Also, the presence of pathogenic E. coli in the household environment indicates a new infection risk or a recent infection among household members.
The increased awareness of the role of environmental matrices in enteric disease transmission has resulted in the need for rapid, field-based methods for fecal indicator bacteria and pathogen detection and quantification. Microbial water quality monitoring, for example, leverages many different field-based assays for the detection of E. coli. Examples include the Coliscan Easygel (Microbiology Laboratories, Indiana, USA), compartment bag test (Aquagenx, North Carolina, USA), IDEXX Quanti-Tray/2000 system with Colilert reagents (IDEXX Laboratories, Maine, USA), and Petrifilm E. coli plates (3M, Minnesota, USA) (9, 10). Among the most widely used tests are those that rely on the metabolism of 4-methyl-umbelliferyl-β-d-glucuronide (MUG) to generate fluorescence. E. coli metabolizes MUG via the β-glucuronidase enzyme, which then forms both an available carbon source and 4-methyl-umbelliferone. The latter fluoresces when exposed to UV light. Nontarget microorganisms do not have access to another available carbon source and, in theory, cannot grow. The specificity of β-glucuronidase-based assays for the detection of E. coli for fecal samples has been demonstrated previously (11). As a result, β-glucuronidase-based assays are used for detection of E. coli in diverse environmental matrices despite limited data on the assay specificity (12, 13).
The most-probable-number (MPN) IDEXX Quanti-Tray/2000 system with Colilert reagents (Colilert) is one of the more common β-glucuronidase-based assays used for the detection and quantification of E. coli. Colilert is capable of quantifying the number of viable E. coli in 100-ml water samples over a range of <1 to >2419.6 MPN E. coli/100 ml. Colilert has been shown to be at least as sensitive as membrane filtration methods (14–17) and other field-based E. coli detection methods (9). Colilert is widely used for field-based detection of E. coli because it is easy to use, rapid, and accurate (9). Drawbacks to Colilert include the requirement of large incubators to accommodate bulky trays, difficulty transporting tray sealers, trays, and reagents to field sites, and high costs of capital equipment (sealing machine) and consumables (tray and reagents) (9).
Multiple studies have demonstrated the specificity of Colilert and other β-glucuronidase-based assays for the detection of E. coli in drinking and recreational waters (18–21 and http://www.idexx.fr/pdf/fr_fr/water/7537-01-colilert-18-report-eng2.pdf). The false-positive rate of Colilert for drinking waters reportedly ranges between 1 and 8% (21 and http://www.idexx.fr/pdf/fr_fr/water/7537-01-colilert-18-report-eng2.pdf). Causes of false positives are related to the presence of the β-glucuronidase enzyme in nontarget microorganisms, such as Klebsiella oxytoca, Aeromonas spp., Providencia spp., and microalgae (18–20 and http://www.idexx.fr/pdf/fr_fr/water/7537-01-colilert-18-report-eng2.pdf). Notably, the β-glucuronidase-based assay typically is selective against non-E. coli species within the Escherichia genus, although at least one exception has been documented (22, 23). Additionally, β-glucuronidase-based assays do not necessarily detect all E. coli isolates in a sample. As one example, E. coli O157:H7 is β-glucuronidase negative; therefore, it would not be detected (24).
Despite the efficacy of β-glucuronidase-based assays for the detection of E. coli in water, the assays do not necessarily function well in other matrices. For example, the acidity in fruit juice impairs the fluorescence of 4-methyl-umbelliferone (25). Similarly, milk contains native β-glucuronidase that cleaves MUG to provide β-glucuronidase-negative bacteria access to glucuronic acid as a carbon source (26). Therefore, the specificity of β-glucuronidase-based detection assays can be impacted by the characteristics of both the microbiota within the matrix and the matrix itself. Evidence of the specificity of β-glucuronidase-based assays for detection of E. coli from other matrices, particularly environmental matrices relevant to enteric pathogen transmission in developing countries (e.g., surfaces, soils, and hands), is limited.
Historically, E. coli was thought to be native only to intestinal tracts of warm-blooded animals. E. coli was perceived to be a useful indicator of fecal contamination in environmental matrices (drinking water, recreational water, and soil), because they were specific to feces, abundant, easily detected and/or quantified, and safe to handle (27). However, E. coli has been shown to persist and grow in the environment, suggesting that E. coli is endemic to and/or naturalized in water and/or soil (28–30). Naturalized E. coli has been shown to be phenotypically distinguishable from enteric E. coli (31). Luo et al. examined E. coli genomes from enteric and environmental sources and identified 204 genes that were differentially present in isolated genomes from the two sources (32). Among the genes Luo et al. identified, sfmH and fucK genes were indicative of enteric E. coli. The genes are responsible for encoding fimbrial-like adhesion proteins and sugar kinases for pentulose and hexalose, respectively (32). Both adhesion and sugar kinases may be important for colonization and resource acquisition, respectively, in the gut (32–34). Additionally, rfaI, a gene that encodes lipopolysaccharide biosynthesis protein, was identified as highly enriched in environmental E. coli (32). A mechanistic explanation for the latter association is not clear. However, LPS core biosynthesis genes have been linked to efficiency of E. coli adhesion to abiotic surfaces (35).
Phylogroup characterization of E. coli also provides insight into phenotypes. Within the E. coli species, there is a well-known genetic substructure that covaries with phenotypic traits (36). Seven phylogroups (A, B1, B2, C, D, E, and F) are used to describe the substructure, with strains from different phylogroups occupying different ecological niches (36, 37). For example, A and B1 strains appear to be generalists found within guts of a range of vertebrates and also are more prevalent in freshwater samples than other strains (36). Also, B1 strains are more persistent in the environment (i.e., soils and waters) than A, B2, or D strains (38, 39). Conversely, B2 strains are rarer in ectotherms and freshwater samples, and, when present, are associated with reduced within-host E. coli phylogroup diversity (36, 40). Phylogroup identification is also useful for identifying human health risks, as diarrheal disease-causing E. coli strains are more likely to be B1 and E strains and extraintestinal infections are more likely to be caused by B2 and D (36).
Recognizing the increasing interest in the role of environmental matrices (hands, soils, and surfaces) in enteric disease transmission in low-income countries, this study quantified the false-positive rate of the Colilert detection assay for E. coli detection in soils and on hands in Bangladesh. The false-positive rates for the environmental matrices were compared to false-positive rates from human, cattle, and chicken feces. One goal of the study was to identify whether or not β-glucuronidase-based assays are appropriate for detection of E. coli on hands and in soil. Using β-glucuronidase-positive E. coli isolates, we further investigated whether or not there were genotypic and phenotypic characteristics of the E. coli isolates that differed based on source. Specifically, we investigated whether or not there were differences in the results of the individual biochemical assays of the API-20E test, the Clermont phylogroup, and the presence of genes that indicate an environmental or enteric source (sfmH, rfaI, and fucK).
MATERIALS AND METHODS
Study site.
All samples (hand, soil, and fecal composite) were collected from rural households in the Mymensingh district of central Bangladesh. The main forms of income in this region are agriculture and raising livestock. The most common types of animals owned include poultry (chickens and ducks), cattle, and goats (41). The samples were collected during the months of November 2013 to March 2014. Institutional Review Board approval for the collection of human fecal samples for this project was obtained from the International Center for Diarrheal Diseases Research, Bangladesh.
Soil sampling.
Field staff collected 25 soil samples from 25 different households from an outdoor location within the compound, designated by the household as the most recent play area for young children in the household. All soil samples were collected from the top layer of a 30-cm by 30-cm square outlined by a metal stencil disinfected in 70% ethanol. The soil was collected into a sterile Whirl-Pak bag using a sterile scoop (Nasco, Fort Atkinson, WI, USA). The sample was stored on ice prior to and during transport to the laboratory and during processing within the laboratory. All samples were processed within 6 h of collection. Bacteria were eluted from approximately 20 g (19.95 to 20.05 g) of soil using 200 ml distilled water by mixing with an Interscience BagMixer for 1 min at speed 2 with a 10-mm gap (Interscience, Rockland, MA, USA). From the supernatant, 1 ml of homogenized solution was immediately collected prior to particle settling and added to a prefilled Whirl-Pak bag with 99 ml distilled water for a 102 dilution. A 104 dilution then was created by removing 1 ml from the 102 dilution and adding it to 99 ml distilled water. The 104 dilution was mixed and enumerated using the most-probable-number (MPN) IDEXX Quanti-Tray system with Colilert reagents according to the manufacturer's instructions. The samples were incubated for 18 h at 44.5°C, a modification of the Colilert protocol for detection of thermotolerant E. coli that is in line with previous studies investigating the presence of E. coli in environmental assays (16, 42).
Hand sampling.
Field staff collected 25 hand samples according to the hand rinse sampling method (43). Each hand sample was collected from a different child who was <5 years old, and each child was from a different home. In brief, children were asked to place first one hand, and then the other, into an 800-ml Whirl-Pak bag (Nasco Corp., Fort Atkinson, WI) containing 250 ml of sterile water. Each hand was placed in the bag for 30 s, which included 15 s of shaking and 15 s of the enumerator massaging the hand through the plastic bag. The samples were stored on ice and transported to the laboratory for processing within 6 h. A 50-ml aliquot of each sample was mixed with 50 ml of sterile distilled water. E. coli cells were enumerated using the MPN IDEXX Quanti-Tray system with Colilert reagents according to the manufacturer's instructions. The samples were incubated for 18 h at 44.5°C, a modification of the Colilert protocol for detection of E. coli intended to reduce the growth of nonspecific background colonies present on hands (44).
Fecal composite sampling.
Five fecal composite samples were collected from cattle, chicken, and humans for a total of 15 samples. Field staff trained in animal feces identification collected cattle and chicken feces. Although the staff targeted fresh cattle and chicken feces that appeared to be deposited on the same day, the exact age is unknown. After feces were identified, the field staff confirmed the presence of the appropriate animal (cattle or chicken) and then collected >2 g from the top or middle of the sample using a sterile collection spoon and 50-ml centrifuge tube. Care was taken to avoid collecting soil alongside the sample. Human fecal samples were collected by leaving a stool collection kit (collection container, aluminum foil, Whirl-Pak bag, and gloves) at enrolled households in the morning. The samples then were collected within 24 h. All samples were stored on ice and transported to the laboratory for processing within 12 h. Aliquots of three to four individual fecal specimens of the same animal type were combined to form a 2.0-g composite before being diluted and vortexed to form a 20-ml solution in distilled or molecular-grade water. Cattle composites were further diluted to 10−4, 10−6, and 10−8; chicken and human composites were diluted to 10−6, 10−8, and 10−10. Samples then were enumerated using Colilert with incubation for 18 h at 35.5°C.
Presumptive E. coli colony isolation and species-level identification.
Following incubation, quantitative estimates of E. coli were determined according to the manufacturer's instructions. IDEXX Quanti-Trays were stored at 4°C prior to E. coli isolation. The back of each IDEXX Quanti-Tray was disinfected by spraying with 70% ethanol and allowing to air dry. Four presumptive positive large wells from each IDEXX Quanti-Tray then were punctured using 5-ml pipettes, and the contents of the well were transferred into microcentrifuge tubes. The sample was streaked onto MacConkey agar and incubated at 35°C for 18 to 24 h. Characteristics of the colonies were recorded. A representative colony type was used to spike both 2 ml of LB broth and 2 ml of Colilert reagent by splitting the colony in half using a sterile loop. If more than one colony morphology was observed on the MacConkey agar, a representative colony of each morphology type consistent with E. coli (pink/red colonies on red agar) was spiked to LB broth and Colilert reagent. Up to four colony morphologies from each MacConkey plate were chosen. The LB broth and Colilert reagent were incubated at 35°C for 18 to 24 h. Visual evidence of turbidity and fluorescence were recorded for LB broth and Colilert, respectively.
The E. coli isolates exhibiting both fluorescence in Colilert and turbidity in LB broth again were streaked to MacConkey agar from the turbid LB broth. If more than one isolate from the same fluorescing large well met this criterion, only one was chosen for the second MacConkey agar isolation step. If no fluorescence was observed in any of the isolates from a large well, the predominant colony type was streaked from LB broth to MacConkey agar. The MacConkey agar was incubated at 35°C for 18 to 24 h. The MacConkey agar plates were observed for uniform colony morphology as an indication that only one isolate was present.
A single colony then was used for species-level identification via an oxidase test followed by an API 20-Etest kit (bioMérieux Inc., Marcy l'Etoile, France) according to the manufacturer's instructions. The API-20E assays use a series of 21 biochemical assays to identify enterobacteria and other nonfastidious Gram-negative bacteria. The results of the tests provide species-level identification along with a quantified level of confidence. Confidence that an isolate is E. coli is dependent on the presence of β-galactosidase; indole production; arabinose, glucose, mannitol, rhamnose, and sorbitol fermentation/oxidation, as well as the absence of arginine decarboxylase; citrate utilization; hydrogen sulfide production; urea hydrolysis; tryptophan deaminase; gelatinase; acetoin production via Voges-Proskauer test; myo-inositol fermentation/oxidation; and oxidase (45). The results of any individual biochemical test alone are insufficient to definitively identify E. coli. Therefore, E. coli isolates exhibiting reactions that do not align with the prototypical E. coli profile may still be characterized as E. coli, although the confidence level may decrease correspondingly.
The API-20E kits were incubated at 35.5°C for 18 to 24 h. Quality control of the kits was performed by inoculating kits with five quality-control microorganisms (Enterobacter cloacae, E. coli, Klebsiella pneumonia, Proteus mirabilis, and Stenotrophomonas maltophilia). Additionally, sterile water was processed alongside every 30 isolates as a negative control. The online analytical profile index (apiweb.bioMérieux.com) was used to identify both the presumptive bacterial species and confidence associated with the identification.
Phylogenetic group determination.
The phylogroups (A, B1, B2, C, D, E, and F) were determined for a subset of 87 presumptive E. coli isolates. Of these, 15, 12, and 17 isolates were from fecal composite samples of cattle, chicken, and humans, respectively. An additional 23 isolates were from hand samples, and 20 isolates were from soil samples. The subset of isolates was chosen such that the first presumptive E. coli isolate identified per sample (fecal composite, soil, or hand rinse sample) was included; additional presumptive E. coli isolates were randomly chosen to increase sample size. Isolates identified as presumptive E. coli were preserved in Primestore MTM (Longhorn Vaccines and Diagnostics, San Antonio, TX, USA) at room temperature for 2 weeks total during storage and subsequent transport to Johns Hopkins University (Maryland, USA), where we performed all molecular work. Phylogroups were determined for each isolate using the updated Clermont method (46). In brief, DNA was extracted from cell lysate using the QIAamp DNA minikit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Phylogroups were assigned based on the results of a multiplex PCR assay for detection of genes chuA, yjaA, and arpA as well as DNA fragment TspE4.Ce using a multiplex PCR kit (Qiagen) (46). If the E. coli isolate phylogroups remained undetermined, we then screened for the presence/absence of C-specific or E-specific primers, as described by Clermont et al. (46).
Detection of functional genes.
In the same subset of 87 E. coli isolates used for phylogroup determination, we investigated the presence/absence of sfmH and rfaI genes. Presence/absence of the fucK gene was investigated in a subset of 25 isolates (5 each soil, hand, and human fecal composite, 6 chicken fecal composite, and 4 cow fecal composite samples were randomly selected). Presence/absence was determined using endpoint PCR with the Quantitect PCR master mix (Qiagen) according to the manufacturer's instructions. Primers and probes were designed using E. coli sequences for E. coli UMN026 (accession number NC_011751.1) as a characteristic enteric strain and E. coli SMS-3-5 (accession number NC_010498.1) as a characteristic environmental strain (Table 1). The designed primers and probes then were adjusted to increase alignment with additional E. coli sequences containing the specific gene based on results of a nucleotide query to the National Center for Biotechnology Information database (blast.ncbi.nlm.nih.gov/Blast.cgi). Only in silico assessments of specificity and sensitivity were performed. To ensure specificity, the PCR products of 8 sfmH, 8 rfaI, and 5 fucK genes were sequenced and compared to published genes (gene identifiers 945407, 5586429, and 946022, respectively) to verify that PCR assays detected target genes. The sensitivity of the primers/probes was not assessed.
TABLE 1.
Primers and probes used for detection of functional genes in E. coli isolates
| Gene | Function | Primer/probe name | Sequence | Size (bp) |
|---|---|---|---|---|
| sfmH | Fimbrial-like adhesion protein | sfmH.F | 5′-TGCGTCTGGAAGCCAGTGCC-3′ | 220 |
| sfmH.R | 5′-GCGCTAAACGGCCCYTCGGT-3′ | |||
| sfmH.P | 5′-GTCTGGATGCAGCTGCGGCAG-3′ | |||
| rfaI | Lipopolysaccharide biosynthesis protein | rfaI.F | 5′-TGCTTGGCGCAATAACGAGCA-3′ | 142 |
| rfaI.R | 5′-TCGGCCCGACAAAACCCTGG-3′ | |||
| rfaI.P | 5′-TGCGCRCTTGGATAACCGGCCCAGTA-3′ | |||
| fucK | Sugar kinases for 5-C and 6-C sugars | fucK.F | 5′-ACAGAACAGCGCCGCAGCAA-3′ | 162 |
| fucK.R | 5′-ACCTGGGCGCTRGGACCAT-3′ | |||
| fucK.P | 5′-CGCCCTTTTTGGCGCTGGTGCCG-3′ |
Statistical analyses.
All statistical analyses were performed in R version 3.0.1 (R Foundation for Statistical Computing, Vienna, Austria). Fisher's exact tests were used to investigate relationships between the source of E. coli isolates and API-20E biochemical assay results, phylogroups, and presence/absence of functional genes. The models were run using three different categorizations for isolation source. The first categorization investigated enteric versus environmental isolates by including cattle, chicken, and human isolates in one category (“enteric”) and hands and soil (“environmental”) in the second. To investigate the distinction between soil E. coli isolates and enteric isolates, the second categorization removed hand samples, which may contain isolates from both soil and enteric sources, from the analysis by investigating enteric samples in one category and soil in the second. Because E. coli isolates from cattle, chicken, and human isolates may differ substantially from another, the third categorization investigated cattle isolates, chicken isolates, human isolates, hand isolates, and soil isolates as five separate categories. Statistical significance was defined at α = 0.05. Finally, we performed linear discriminant analysis (LDA) using the lda function in the MASS package to identify E. coli isolate groupings based on biochemical assay and phylogroup data and to identify relationships with enteric E. coli sources. LDA was used primarily to visualize clusters of similar E. coli isolates using a priori knowledge of categorization. Nonparametric LDA is frequently applied to discrimination of samples based on binary variables and has been reported to perform better than alternative methods (i.e., logistic regression) for data sets with small sample sizes (47).
RESULTS
False-positive rate of Colilert.
In total, 282 isolates were identified: 95 isolates from soil samples, 95 isolates from hand samples, 32 isolates from chicken fecal composite samples, 32 isolates from cattle fecal composite samples, and 28 isolates from human fecal composite samples. Presumptive E. coli was identified using the API-20E kit in 279 of the 282 (98.9%) IDEXX tray wells that fluoresced in Colilert reagents, corresponding to a false-positive rate of 1.1% (95% confidence interval, 0, 2.3). Three isolates were identified as not being E. coli: (i) one isolate from a chicken fecal composite sample was identified as oxidase positive and no further identification was attempted, (ii) one isolate from cattle fecal composite sample was identified as a member of the Kluyvera genus using the API-20E kit, although discrimination was poor, with only 29% identification confidence, and (iii) one isolate from a hand sample was identified as presumptive Escherichia vulneris, although the discrimination was low, with only 59% confidence.
For the fecal composite samples, the Colilert false-positive rate was 2.1% (0, 5.0). Splitting the fecal composites by source, the false-positive rates were similar. The false-positive rate for cattle was 3.1% (0, 9.2). The false-positive rate also was 3.1% (0, 9.2) for chicken samples. The false-positive rate was equal to the lower limit of detection, or <3.6% (0, <10.4), for human composite samples. The false-positive rate for Colilert for hand samples was 1.1% (0, 3.1). The false-positive rate was equal to the lower limit of detection of <1.1% (0, <3.1) for soil samples.
Confidence of E. coli identification.
Despite the low overall false-positive rates for E. coli isolated from Colilert, there was variability in the level of confidence assigned to the identification (Table 2). Most (88%) isolates were identified as E. coli with acceptable, good, very good, or excellent confidence. These classifications, specified by the API-20E identification kits, correspond to quantitative estimates of ≥80.0% likelihood that the isolates were E. coli based on their biochemical profiles. The remaining isolates were identified as not likely E. coli (1%), matching E. coli at the genus level (2%), or matching E. coli with low confidence (8%). Therefore, the rate of identification of isolates with less than acceptable confidence was 11.5% (7.9, 15.5). Although the rate was higher for fecal samples at 15.2% (7.9, 22.6) than it was for both hand and soil samples at 9.5% (3.6, 15.4) and 9.5% (3.6, 15.4), respectively, the difference was not statistically significant (P = 0.23 by two-tailed Z test).
TABLE 2.
Identification confidence level of isolates as determined by API-20E enterobacterial identification strips, stratified by isolate source
| Isolate source | No. of isolates | No. (%) of isolates at each confidence level |
No. (%) of isolates identified to genus level | No. (%) of isolates that were not E. coli | ||||
|---|---|---|---|---|---|---|---|---|
| Excellent (≥99.9%) | Very good (99.0–99.8%) | Good (90.0–98.9%) | Acceptable (80.0–89.9%) | Low | ||||
| Fecal composites | 92 | 10 (11) | 52 (57) | 16 (17) | 0 (0) | 5 (8) | 7 (5) | 2 (2) |
| Cattle | 32 | 6 (19) | 16 (50) | 5 (16) | 0 (0) | 4 (13) | 0 (0) | 1 (3) |
| Chicken | 32 | 1 (3) | 21 (66) | 6 (19) | 0 (0) | 3 (9) | 0 (0) | 1 (3) |
| Human | 28 | 3 (11) | 15 (54) | 5 (18) | 0 (0) | 0 (0) | 5 (18) | 0 (0) |
| Hand | 95 | 21 (22) | 48 (51) | 17 (18) | 0 (0) | 0 (8) | 8 (0) | 1 (1) |
| Soil | 95 | 12 (13) | 56 (59) | 17 (18) | 1 (1) | 7 (7) | 2 (2) | 0 (0) |
| Total | 282 | 43 (15) | 156 (55) | 50 (18) | 1 (0) | 7 (8) | 22 (2) | 4 (1) |
Biochemical assays.
For nine of the 21 biochemical assays tested as part of the API-20E kit (citrate, urease, tryptophanedeaminase, Voges-Proskauer reaction, gelatinase, glucose fermentation, mannitol fermentation, arabinose fermentation, or oxidase tests), there was no variability in the response of presumptive E. coli isolates. Of the other 12 assays, variability was limited in three. Specifically, only one isolate (soil) was negative for o-nitrophenyl-β-galactosidase; two isolates (hand and cattle feces) were positive for amygdalin fermentation, and one isolate (soil) was positive for hydrogen sulfide production. Of the nine remaining biochemical assays, two were correlated with a coefficient of >0.40: ornithine decarboxylase and sucrose fermentation (Spearman's ρ, 0.41; P < 0.001). Ornithine decarboxylase test results were removed from subsequent statistical analyses to limit effects of multicollinearity.
A logit model of enteric versus environmental samples using biochemical assay results for the remaining eight tests (arginine dihydrolase; lysine decarboxylase; indole production; and oxidation/fermentation of inositol, sorbitol, rhamnose, sucrose, and melibiose) explained only 8.5% of variability (Nagelkerke R2 = 0.085), with oxidation/fermentation of rhamnose and melibiose as the only statistically significant coefficients (Table 3). Both rhamnose (χ2 = 3.9, P = 0.048) and melibiose oxidation/fermentation (χ2 = 4.0, P = 0.046) were more frequently detected in enteric isolates than in environmental isolates. A logit model comparing biochemical assay results between enteric and soil samples explained only 10% of the variability (Nagelkerke R2 = 0.10) (Table 3). Oxidation/fermentation of sucrose and melibiose were statistically significant in the model but were not statistically different in the frequency of detection in enteric compared to soil samples (for sucrose, χ2 = 2.8 and P = 0.10; for melibiose, χ2 = 1.7 and P = 0.19).
TABLE 3.
Logit models examining relationship between E. coli isolate biochemical assay results and isolation source
| Model parameter | Value by isolate comparisona |
|||
|---|---|---|---|---|
| Enteric vs environmental |
Enteric vs soil |
|||
| Coefficient | Pr(>|Z|) | Coefficient | Pr(>|Z|) | |
| Intercept | −0.61 | 0.58 | −0.56 | 0.65 |
| Arginine dihydrolase | 6.2 | 0.76 | – | |
| Lysine decarboxylase | −0.12 | 0.77 | −0.28 | 0.56 |
| Indole production | −0.84 | 0.32 | −0.96 | 0.31 |
| Inositol | −1.2 | 0.36 | −7.2 | 0.78 |
| Sorbitol | 0.30 | 0.47 | 0.17 | 0.72 |
| Rhamnose | 0.90 | 0.03* | 0.59 | 0.23 |
| Sucrose | −0.52 | 0.08 | −0.77 | 0.02* |
| Melibiose | 1.6 | 0.01* | 1.7 | 0.03* |
| n | 281 | 186 | ||
| Pseudo-R2 | 0.085 | 0.104 | ||
| P | 0.024 | 0.034 | ||
An asterisk denotes results statistically significant at α < 0.05. The dash denotes the exclusion of the arginine dihydrolase parameter in the enteric versus soil model due to lack of variation. Pr(>|Z|) refers to the probability that the coefficient is not zero as calculated using the Z test.
Phylogroup determination.
Most of the E. coli isolates were identified as phylogroup A (40%) or B1 (36%). Phylogroups E (11%), C (6%), D (6%), and F (3%) also were detected (Table 4). Phylogroup frequency was not statistically significantly associated with enteric compared to environmental samples (χ2 = 4.8, P = 0.43) or enteric compared to soil samples alone (χ2 = 5.1, P = 0.40). However, isolation source (cattle feces, chicken feces, human feces, hand, or soil) was statistically significantly associated with phylogroups (χ2 = 38.3, P = 0.008).
TABLE 4.
Phylogroup of E. coli isolates by source type
| Isolate source | No. (%) of isolates in each phylogroupa |
Total | |||||
|---|---|---|---|---|---|---|---|
| A | B1 | C | D | E | F | ||
| Environmental | 20 (47) | 16 (37) | 2 (5) | 1 (2) | 4 (9) | 0 (0) | 43 |
| Hands | 10 (43) | 9 (39) | 2 (8) | 1 (4) | 1 (4) | 0 (0) | 23 |
| Soil | 10 (50) | 7 (35) | 0 (0) | 0 (0) | 3 (15) | 0 (0) | 20 |
| Enteric | 15 (34) | 15 (34) | 3 (7) | 4 (9) | 5 (11) | 2 (5) | 44 |
| Cattle | 4 (27) | 6 (40) | 0 (0) | 0 (0) | 4 (27) | 1 (7) | 15 |
| Chicken | 9 (75) | 3 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 12 |
| Human | 2 (12) | 6 (35) | 3 (18) | 4 (24) | 1 (6) | 1 (6) | 17 |
| Total | 35 (40) | 31 (36) | 5 (6) | 5 (6) | 9 (10) | 2 (2) | 87 |
Percentages report the proportion of a phylogroup within a source type.
Functional gene detection.
Because all of the 25 isolates we initially tested (100%) contained the fucK gene, we did not continue to test for the fucK gene in the remaining 65 samples. Of the five fucK samples sequenced, all aligned to the reference gene with a maximum identity value range of 93 to 97%. Almost all (97%) of the 90 isolates we tested contained the sfmH gene; the three isolates without the sfmH gene included two from soil samples and one from a human fecal sample. Of the eight sfmH genes sequenced, all aligned to the reference gene with a maximum identity value range of 95 to 99%. Additionally, most (87%) of the 90 isolates we tested contained the rfaI gene. The isolates without the rfaI gene included two from cow fecal composite samples, one from a human fecal composite sample, six from soil samples, and three from hand samples. Of the eight rfaI genes sequenced, all aligned to the reference gene with a maximum identity value range of 98 to 99%. The limited variability of the results for fucK and sfmH genes precluded subsequent statistical analysis of presence/absence of gene types by E. coli isolation source. Using Fisher's exact test, the presence of the rfaI gene was not statistically significantly different in enteric versus environmental isolates (P = 0.12), by isolation source (P = 0.27), or in enteric versus soil isolates (P = 0.06). Notably, in the latter comparison, rfaI was present in a greater fraction of enteric samples (93%) than soil samples (67%).
Linear discriminant analysis.
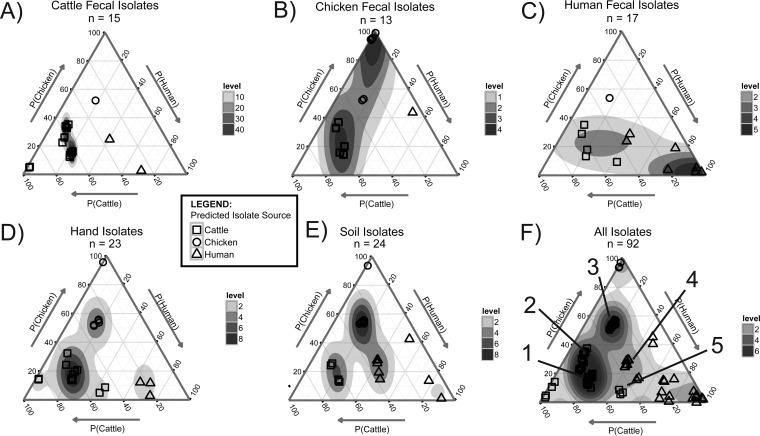
The linear discriminant analysis was performed on the data from 15 cattle, 13 chicken, and 17 human fecal E. coli isolates. The sample sizes were determined by sample availability. Applying the LDA function to the same isolates (i.e., the training data set) resulted in the identification of more than 10 clusters of isolates with similar biochemical and phylogroup profiles. The clusters can be visualized on ternary plots using the estimated probabilities that each isolate is from cattle, chicken, or human feces (Fig. 1). For example, some isolate clusters were isolated only from cattle, chicken, or human feces and appear in the respective corners of the ternary plots. The corners represent a high probability that the isolates are from cattle, chicken, or human feces. Examining the characteristics of these isolates highlights characteristics that are descriptive of the fecal source. The isolates identified in the LDA as very likely cattle isolates, for example, are characterized as unable to ferment/oxidize sorbitol. Similarly, the analysis identified as very likely chicken isolates those from phylogroup A (with yjaA gene) that are unable to ferment/oxidize rhamnose. Isolates identified as very likely human isolates are unable to ferment/oxidize melibiose. Other isolate clusters, however, appear in the interior of the ternary plots, representative of the uncertainty about the enteric source as predicted by LDA. Isolates within clusters 1 and 2 (Fig. 1F, annotations 1 and 2) are from phylogroups B1 (cluster 1) or D or F (cluster 2) with the ability to oxidize/ferment sucrose and were isolated from every source type. Cluster 3 (Fig. 1F, annotation 3) isolates are phylogroup A, sucrose oxidation/fermentation negative, and also are detected in every source type (Fig. 1A to E). Cluster 4 (Fig. 1F, annotation 4) isolates are from phylogroup B1, unable to ferment sucrose, and isolated from cattle feces, human feces, and soil samples. Cluster 5 (Fig. 1F, annotation 5) isolates are phylogroup A or C with the ability to oxidize/ferment sucrose and were isolated from hand samples and human fecal samples.
FIG 1.
Linear discriminant analysis results for estimating the probability that an E. coli isolate is from cattle feces, P(Cattle), chicken feces, P(Chicken), or human feces, P(Human), based on the isolates' biochemical assay results and Clermont phylogroup. The ternary plots are used to visualize the fecal source probabilities for E. coli isolates collected from cattle feces (A), chicken feces (B), human feces (C), hands (D), and soil (E). (F) Ternary plot overlays of all of the samples together, with annotations (1 to 5) indicating clusters of isolates with similar biochemical and phylogroup profiles. Each marker represents one isolate. The sample locations in the plot are shifted slightly by the addition of random noise for visualization to avoid sample overlap. Density gradients were estimated using linear interpolation to visualize relative sample densities at locations within the ternary plots, with levels used to describe the relative local sample density at a given spatial location.
Applying the LDA function to 23 E. coli isolates from hands and 24 isolates from soil samples identified clusters based on phylogroup and biochemical assays (Fig. 1D and E). Of the E. coli isolates from hands, one is similar to chicken fecal isolates (99% probability), two to cattle isolates (86% probability), and three to human isolates (>65% probability) (Fig. 1D). The remaining 17 isolates are similar to isolates in clusters 1, 2, 3, and 5. Likewise, E. coli isolates from soil samples identified one isolate similar to chicken isolates (86% probability) and two similar to human isolates (>65% probability). The rest of the isolates were from clusters 1 to 4 (Fig. 1E).
DISCUSSION
Colilert, a β-glucuronidase-based E. coli detection assay, is sufficiently specific for the detection of E. coli in diverse environmental matrices (soils and hands).
With the increased awareness of the role of environmental matrices in enteric disease transmission, especially in low-income countries, rapid and field-based methods for fecal indicator bacterium detection are needed. Currently, studies rely on existing E. coli detection assays for diverse matrices despite limited data available on assay specificity (12, 13). We demonstrate, using Colilert, that β-glucuronidase-based assays detect E. coli on hands and in soils with low false-positive rates when incubated at an elevated temperature. In fact, the fraction of E. coli isolates with at least an acceptable level of confidence, as defined by the API-20E kits, collected from soil and hand samples was not significantly different from isolates collected from feces. The low false-positive rates quantified here are comparable to rates reported in drinking and recreational waters (21 and http://www.idexx.fr/pdf/fr_fr/water/7537-01-colilert-18-report-eng2.pdf). Given the low false-positive rate, definitive identification (i.e., 16S rRNA gene sequencing) of the three presumptively different microorganisms responsible for the false positives was not performed. We conclude that Colilert is an appropriate method for detection of E. coli on hands and in soils when incubated at 44.5°C.
E. coli isolated from hand and soil samples in Bangladesh are genotypically diverse, representing five of the seven phylogroups.
Strains from phylogroups A and B1 were the most abundant, comprising 84% of isolates from environmental samples, consistent with the perception of A and B1 strains as generalists (36). Although B1 strains were identified as more persistent in studies of E. coli survival in both soil and water than A, B2, or D (36, 38, 39), our study did not show a substantial difference in relative abundance of B1 isolates in soil or on hands compared to feces. Phylogroups C, D, and E also were detected on hands and/or in soils, albeit infrequently. Strains from C and D phylogroups were detected only in human feces and on hands, providing some evidence that E. coli on hands is from contact with human feces. The detection of phylogroups B1 and E, which contain the majority of diarrheal disease-causing E. coli strains, is consistent with our previous reported detection of virulence genes in E. coli isolated from soils in Tanzania (1, 36). Further analysis for pathogenicity, however, would be warranted to differentiate pathogenic E. coli strains from nonpathogenic strains within the B1 and E phylogroups.
E. coli isolates' ability to ferment or oxidize specific sugars may provide some insight into fecal source.
The LDA identified that the inability to ferment/oxidize melibiose is a prototypical trait of human fecal E. coli isolates. Melibiose is a disaccharide composed of glucose and galactose. In our data set, 100%, 94%, and 81% of cattle, chicken, and human isolates were able to ferment/oxidize melibiose. These data align with previous studies; notably, Farmer et al., who reported 75% of isolates from human clinical samples were able to ferment/oxidize melibiose compared to the 98% and 97% reported for cattle and bird isolates by Godbout-DeLasalle and Higgins (45, 48). Using LDA on biochemical assay results highlights the potential to use a series of easily characterized phenotypic traits to distinguish the fecal source of environmental E. coli.
The presence/absence of the three functional genes fucK, sfmH, and rfaI provides no useful information for identifying the presence of environmental E. coli isolates in this setting.
The sfmH and fucK genes, which were identified as highly enriched in E. coli from enteric sources, encode fimbrial-like adhesion proteins and sugar kinases, respectively. Both genes were found in almost all of the E. coli isolates tested, including those isolated from hand and soil samples. Similarly, the rfaI gene, which was identified as highly enriched in E. coli from environmental sources, also was detected in most of the strains tested. Culturing E. coli prior to gene detection may have influenced our results; direct detection may provide more accurate gene prevalence data. Nevertheless, our findings suggest one or more of the following: (i) the fucK, sfmH, and rfaI genes are not differentially present in enteric, as opposed to environmental, isolates in Bangladeshi households; (ii) the genomic sequences used in the study by Luo et al. to identify differential presence of fucK, sfmH, and/or rfaI are not representative of E. coli isolates from Bangladeshi households (32); or (iii) all E. coli organisms detected are enteric source isolates (hence, the detection of fucK and sfmH) that also contain the rfaI gene. The last explanation may be the most probable, because the hand and soil samples were incubated at elevated temperature, selecting for thermotolerant E. coli that may be of enteric source.
We demonstrate that isolates from soils and hands are indistinguishable, using traditional biochemical and phylogrouping data, from fecal E. coli isolates.
Although the persistence and growth of E. coli in environmental samples, including soil samples, has been demonstrated previously (28–30), we have not yet identified a reliable method for distinguishing naturalized or autochthonous strains from enteric E. coli strains. The application of LDA to identify and visualize clusters of E. coli with similar biochemical assay and phylogroup profiles highlighted a subset of E. coli isolates that appear to be uniquely associated with cattle, chicken, or human feces. However, these isolates were the exception, not the rule. Most E. coli isolates shared profiles with isolates across multiple source types (cattle feces, chicken feces, human feces, hand, or soil). Our findings suggest E. coli isolates from different sources are largely indistinguishable based on biochemical and phylogroup data. One area that may provide an opportunity to differentiate E. coli isolates by source is the investigation of the presence/absence of functional genes that provide a competitive advantage within the enteric or environmental niches (39). However, we demonstrated that presumptive enteric genes fucK and sfmH and environmental gene rfaI are not sufficient, as they cooccur within our sample subset. Additional work is needed to identify and assess other candidate gene targets.
Study limitations.
First, the study relied on E. coli isolated using Colilert and only investigated phenotypic and genotypic characteristics of β-glucuronidase-positive E. coli. Therefore, E. coli isolates that do not possess β-glucuronidase activity, such as E. coli O157:H7, are excluded from analyses. Studies of false-negative rates of E. coli detection using β-glucuronidase activity in environmental samples report rates as high as 10 to 20% (49). Although future studies may consider using methods capable of isolating β-glucuronidase-negative E. coli isolates, our study relied on Colilert as a screening assay due to its widespread use in field studies (9). Second, incubation of the environmental samples (hand and soil) at 44.5°C selected for thermotolerant E. coli and may have impaired the growth of naturalized and/or autochthonous E. coli. The elevated temperatures for hand and soil samples were used because thermotolerant E. coli is perceived to be a better indicator of fecal contamination in environmental reservoirs than E. coli (16, 42, 44). In a study investigating incubation temperature impacts on wild-type E. coli growth in Colilert, Matthews et al. demonstrated little difference between 35°C and 45°C (50). Regardless, the lack of an observable difference in phenotypic or genotypic characteristics between environmental and enteric E. coli isolates in this study may be due to the relative selectivity of incubation temperatures. Future studies investigating E. coli as an indicator of fecal contamination on hands and in soils may choose to incubate Colilert at 44.5°C to ensure low false-positive rates, whereas studies investigating the presence of autochthonous and/or naturalized E. coli in environmental reservoirs may choose to incubate Colilert at the lower recommended temperature of 35.5°C. A third limitation is that it is possible that the functional gene assays we developed are not sufficiently specific or sensitive for use on environmental E. coli isolates. We confirmed the genes detected in our study align with published sequences for fucK, sfmH, and rfaI genes and are confident the false-positive rate is low. However, there may be a significant false-negative rate for the sfmH and rfaI genes due to sequence variability of the genes in primer and probe target regions.
The study findings may not necessarily be generalizable to other sites.
Bangladesh is a tropic environment. Furthermore, only 57% of the population have access to improved sanitation as defined by the Joint Monitoring Program (51). Latrines are the most common sanitation type (>70% of the population have access), although many latrines are shared (28% of the population shares sanitation facilities with more than one household) (51). Open defecation, practiced by 3% of the population, is relatively common (51). Although previous research demonstrated the presence of a slab on a pit latrine did not influence E. coli concentrations in soils in Tanzania, it remains unclear what influence more dramatic differences in sanitation technologies may have on E. coli concentrations (1). It remains unclear what influence sanitation conditions and/or animal ownership (e.g., chickens, ducks, cattle, and goats) have on the study findings; similar studies in other locations are warranted before generalizations can be made.
Substantially more research is needed on the impacts of high concentrations of E. coli on hands and in soils in low-income countries.
Multiple studies have reported concentrations of fecal indicator bacteria 10 to 100 times higher on surfaces and hands in low-income countries than in high-income countries (1, 3, 4). Also, a systematic review by Gruber et al. identifies E. coli in stored drinking water as a risk factor in diarrheal disease incidence (52). However, it remains uncertain what the health impacts are for chronic exposures to E. coli in the environment. Although enterotoxigenic E. coli, and its close relatives, Shigella spp., have been identified as two of the four etiological agents most frequently responsible for moderate-to-severe diarrheal disease globally, it is clear that only a subset of E. coli isolates in households are pathogenic (1, 8). The health impacts from chronic exposure to nonpathogenic E. coli are not well characterized. For example, Humphrey et al. hypothesizes that chronic exposures to fecal bacteria result in malnutrition and stunting mediated via environmental enteropathy (53). Therefore, tools are needed to better understand the level and extent of fecal indicator bacteria exposures in low-income countries. In this study, we demonstrate that Colilert, and likely other β-glucuronidase-based assays, can be readily used for detection of E. coli on hands and in soils with a low false-positive rate, especially in households in rural Bangladesh. Furthermore, using traditional biochemical assays and phylogrouping, we demonstrate that E. coli strains in these environments are diverse and indistinguishable from E. coli isolated from cattle, chicken, and human feces.
ACKNOWLEDGMENTS
We thank Rebecca Pinekenstein for providing laboratory support.
The work was supported by the Osprey Foundation of Maryland, Inc., and the Johns Hopkins Water Institute.
REFERENCES
- 1.Pickering AJ, Julian TR, Marks SJ, Mattioli MC, Boehm AB, Schwab KJ, Davis J. 2012. Fecal contamination and diarrheal pathogens on surfaces and soil show spatial heterogeneity within Tanzanian households and no association with improved sanitation. Environ Sci Technol 46:5736–5743. doi: 10.1021/es300022c. [DOI] [PubMed] [Google Scholar]
- 2.Julian TR, MacDonald LH, Guo Y, Marks SJ, Kosek M, Yori PP, Pinedo SR, Schwab KJ. 2013. Fecal indicator bacteria contamination of fomites and household demand for surface disinfection products: a case study from Peru. Am J Trop Med Hyg 89:869–872. doi: 10.4269/ajtmh.12-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinclair RG, Gerba CP. 2011. Microbial contamination in kitchens and bathrooms of rural Cambodian village households. Lett Appl Microbiol 52:144–149. doi: 10.1111/j.1472-765X.2010.02978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Julian TR, Pickering AJ, Leckie JO, Boehm AB. 2013. Enterococcus spp on fomites and hands indicate increased risk of respiratory illness in child care centers. Am J Infect Control 41:728–733. doi: 10.1016/j.ajic.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Pickering AJ, Julian TR, Mamuya S, Boehm AB, Davis J. 2011. Bacterial hand contamination among Tanzanian mothers varies temporally and following household activities. Trop Med Int Health 16:233–239. doi: 10.1111/j.1365-3156.2010.02677.x. [DOI] [PubMed] [Google Scholar]
- 6.Ram PK, Jahid I, Halder AK, Nygren B, Islam MS, Granger SP, Molyneaux JW, Luby SP. 2011. Variability in hand contamination based on serial measurements: implications for assessment of hand-cleansing behavior and disease risk. Am J Trop Med Hyg 84:510–516. doi: 10.4269/ajtmh.2011.10-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattioli MCM, Pickering AJ, Gilsdorf R, Davis J, Boehm AB. 2013. Hands and water as vectors of diarrheal pathogens in Bagamoyo, Tanzania. Environ Sci Technol 47:355–363. doi: 10.1021/es303878d. [DOI] [PubMed] [Google Scholar]
- 8.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 9.Chuang P, Trottier S, Murcott S. 2011. Comparison and verification of four field-based microbiological tests: H2S test, Easygel®, Colilert®, Petrifilm™. J Water Sanit Hyg Dev 1:68–85. doi: 10.2166/washdev.2011.026. [DOI] [Google Scholar]
- 10.Stauber C, Miller C, Cantrell B, Kroell K. 2014. Evaluation of the compartment bag test for the detection of Escherichia coli in water. J Microbiol Methods 99:66–70. doi: 10.1016/j.mimet.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Rice EW, Allen M, Edberg S. 1990. Efficacy of beta-glucuronidase assay for identification of Escherichia coli by the defined-substrate technology. Appl Environ Microbiol 56:1203–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stauber CE, Walters A, de Aceituno AMF, Sobsey MD. 2013. Bacterial contamination on household toys and association with water, sanitation and hygiene conditions in Honduras. Int J Environ Res Public Health 10:1586–1597. doi: 10.3390/ijerph10041586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omar KB, Potgieter N, Barnard TG. 2010. Development of a rapid screening method for the detection of pathogenic Escherichia coli using a combination of Colilert® Quanti-Trays/2000 and PCR. Water Sci Technol 10:7–13. doi: 10.2166/ws.2010.862. [DOI] [Google Scholar]
- 14.Eckner KF. 1998. Comparison of membrane filtration and multiple-tube fermentation by the Colilert and Enterolert methods for detection of waterborne coliform bacteria, Escherichia coli, and enterococci used in drinking and bathing water quality monitoring in southern Sweden. Appl Environ Microbiol 64:3079–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer CJ, Tsai Y-L, Lang AL, Sangermano LR. 1993. Evaluation of Colilert marine water for detection of total coliforms and Escherichia coli in the marine environment. Appl Environ Microbiol 59:786–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yakub GP, Castric DA, Stadterman-Knauer KL, Tobin MJ, Blazina M, Heineman TN, Yee GY, Frazier L. 2002. Evaluation of Colilert and Enterolert defined substrate methodology for wastewater applications. Water Environ Res 74:131–135. doi: 10.2175/106143002X139839. [DOI] [PubMed] [Google Scholar]
- 17.Edberg SC, Allen MJ, Smith DB. 1988. National field evaluation of a defined substrate method for the simultaneous enumeration of total coliforms and Escherichia coli from drinking water: comparison with the standard multiple tube fermentation method. Appl Environ Microbiol 54:1595–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landre J, Gavriel A, Lamb A. 1998. False-positive coliform reaction mediated by Aeromonas in the Colilert defined substrate technology system. Lett Appl Microbiol 26:352–354. doi: 10.1046/j.1472-765X.1998.00347.x. [DOI] [PubMed] [Google Scholar]
- 19.Pisciotta JM, Rath DF, Stanek PA, Flanery DM, Harwood VJ. 2002. Marine bacteria cause false-positive results in the Colilert-18 rapid identification test for Escherichia coli in Florida waters. Appl Environ Microbiol 68:539–544. doi: 10.1128/AEM.68.2.539-544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies CM, Apte SC, Peterson SM, Stauber JL. 1994. Plant and algal interference in bacterial β-d-galactosidase and β-d-glucuronidase assays. Appl Environ Microbiol 60:3959–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao K-K, Chao C-C, Chao W-L. 2004. Evaluation of Colilert-18 for detection of coliforms and Escherichia coli in subtropical freshwater. Appl Environ Microbiol 70:1242–1244. doi: 10.1128/AEM.70.2.1242-1244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez J, Berrocal CI, Berrocal L. 1986. Evaluation of a commercial β-glucuronidase test for the rapid and economical identification of Escherichia coli. J Appl Bacteriol 61:541–545. doi: 10.1111/j.1365-2672.1986.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 23.Rice E, Allen M, Brenner D, Edberg S. 1991. Assay for beta-glucuronidase in species of the genus Escherichia and its applications for drinking-water analysis. Appl Environ Microbiol 57:592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyle M, Schoeni J. 1984. Survival and growth characteristics of Escherichia coli associated with hemorrhagic colitis. Appl Environ Microbiol 48:855–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pao S, Davis CL, Friedrich LM, Parish ME. 2002. Utilization of fluorogenic assay for rapid detection of Escherichia coli in acidic fruit juice. J Food Prot 65:1943–1948. [DOI] [PubMed] [Google Scholar]
- 26.Fang W, Vikerpuur M, Sandholm M. 1995. A fluorometric β-glucuronidase assay for analysis of bacterial growth in milk. Vet Microbiol 46:361–367. doi: 10.1016/0378-1135(95)00044-B. [DOI] [PubMed] [Google Scholar]
- 27.Gerba CP. 2009. Indicator microorganisms, p 485–499. In Maier RM, Pepper IL, Gerba CP (ed), Environmental microbiology. Academic Press, San Diego, CA. [Google Scholar]
- 28.Byappanahalli M, Fujioka R. 2004. Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Sci Technol 50:27–32. [PubMed] [Google Scholar]
- 29.Hardina C, Fujioka R. 1991. Soil: The environmental source of Escherichia coli and enterococci in Hawaii's streams. Environ Toxic Water 6:185–195. doi: 10.1002/tox.2530060208. [DOI] [Google Scholar]
- 30.Vital M, Hammes F, Egli T. 2008. Escherichia coli O157 can grow in natural freshwater at low carbon concentrations. Environ Microbiol 10:2387–2396. doi: 10.1111/j.1462-2920.2008.01664.x. [DOI] [PubMed] [Google Scholar]
- 31.Berthe T, Ratajczak M, Clermont O, Denamur E, Petit F. 2013. Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl Environ Microbiol 79:4684–4693. doi: 10.1128/AEM.00698-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. 2011. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A 108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang D-E, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klemm P. 1985. Fimbrial adhesins of Escherichia coli. Rev Infect Dis 7:321–340. doi: 10.1093/clinids/7.3.321. [DOI] [PubMed] [Google Scholar]
- 35.Genevaux P, Bauda P, DuBow MS, Oudega B. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch Microbiol 172:1–8. doi: 10.1007/s002030050732. [DOI] [PubMed] [Google Scholar]
- 36.Donnenberg M. 2013. Escherichia coli: pathotypes and principles of pathogenesis. Academic Press, San Diego, CA. [Google Scholar]
- 37.Chaudhuri RR, Henderson IR. 2012. The evolution of the Escherichia coli phylogeny. Infect Genet Evol 12:214–226. doi: 10.1016/j.meegid.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Bergholz PW, Noar JD, Buckley DH. 2011. Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl Environ Microbiol 77:211–219. doi: 10.1128/AEM.01880-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White A, Sibley K, Sibley C, Wasmuth J, Schaefer R, Surette M, Edge T, Neumann N. 2011. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl Environ Microbiol 77:7620–7632. doi: 10.1128/AEM.05909-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno E, Johnson JR, Pérez T, Prats G, Kuskowski MA, Andreu A. 2009. Structure and urovirulence characteristics of the fecal Escherichia coli population among healthy women. Microbes Infect 11:274–280. doi: 10.1016/j.micinf.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Roess AA, Winch PJ, Ali NA, Akhter A, Afroz D, El Arifeen S, Darmstadt GL, Baqui AH. 2013. Animal husbandry practices in rural Bangladesh: potential risk factors for antimicrobial drug resistance and emerging diseases. Am J Trop Med Hyg 89:965–970. doi: 10.4269/ajtmh.12-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Characklis GW, Dilts MJ, Simmons OD III, Likirdopulos CA, Krometis L-AH, Sobsey MD. 2005. Microbial partitioning to settleable particles in stormwater. Water Res 39:1773–1782. doi: 10.1016/j.watres.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Pickering AJ, Davis J, Walters SP, Horak HM, Keymer DP, Mushi D, Strickfaden B, Chynoweth JS, Liu J, Blum A, Rogers K, Boehm AB. 2010. Hands, water, and health: fecal contamination in Tanzanian communities with improved, non-networked water supplies. Environ Sci Technol 44:3267–3272. doi: 10.1021/es903524m. [DOI] [PubMed] [Google Scholar]
- 44.Greene LE, Freeman MC, Akoko D, Saboori S, Moe C, Rheingans R. 2012. Impact of a school-based hygiene promotion and sanitation intervention on pupil hand contamination in western Kenya: a cluster randomized trial. Am J Trop Med Hyg 87:385–393. doi: 10.4269/ajtmh.2012.11-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farmer J, Davis BR, Hickman-Brenner F, McWhorter A, Huntley-Carter G, Asbury M, Riddle C, Wathen-Grady H, Elias C, Fanning G. 1985. Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol 21:46–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 47.Asparoukhov OK, Krzanowski WJ. 2001. A comparison of discriminant procedures for binary variables. Comput Stat Data Anal 38:139–160. doi: 10.1016/S0167-9473(01)00032-9. [DOI] [Google Scholar]
- 48.Godbout-DeLasalle F, Higgins R. 1986. Biotyping of clinical isolates of Escherichia coli of animal origin, using the Analytab API 20E system. Can J Vet Res 50:418. [PMC free article] [PubMed] [Google Scholar]
- 49.Chao W. 2006. Evaluation of Colilert-18 for the detection of coliforms and Escherichia coli in tropical fresh water. Lett Appl Microbiol 42:115–120. doi: 10.1111/j.1472-765X.2005.01814.x. [DOI] [PubMed] [Google Scholar]
- 50.Matthews RL, Tung R. 2014. Broader incubation temperature tolerances for microbial drinking water testing with enzyme substrate tests. J Water Health 12:113–121. doi: 10.2166/wh.2013.076. [DOI] [PubMed] [Google Scholar]
- 51.WHO. 2014. Progress on drinking water and sanitation: 2014 update. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 52.Gruber JS, Ercumen A, Colford JM Jr. 2014. Coliform bacteria as indicators of diarrheal risk in household drinking water: systematic review and meta-analysis. PLoS One 9:e107429. doi: 10.1371/journal.pone.0107429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humphrey JH. 2009. Child undernutrition, tropical enteropathy, toilets, and handwashing. Lancet 374:1032–1035. doi: 10.1016/S0140-6736(09)60950-8. [DOI] [PubMed] [Google Scholar]