Abstract
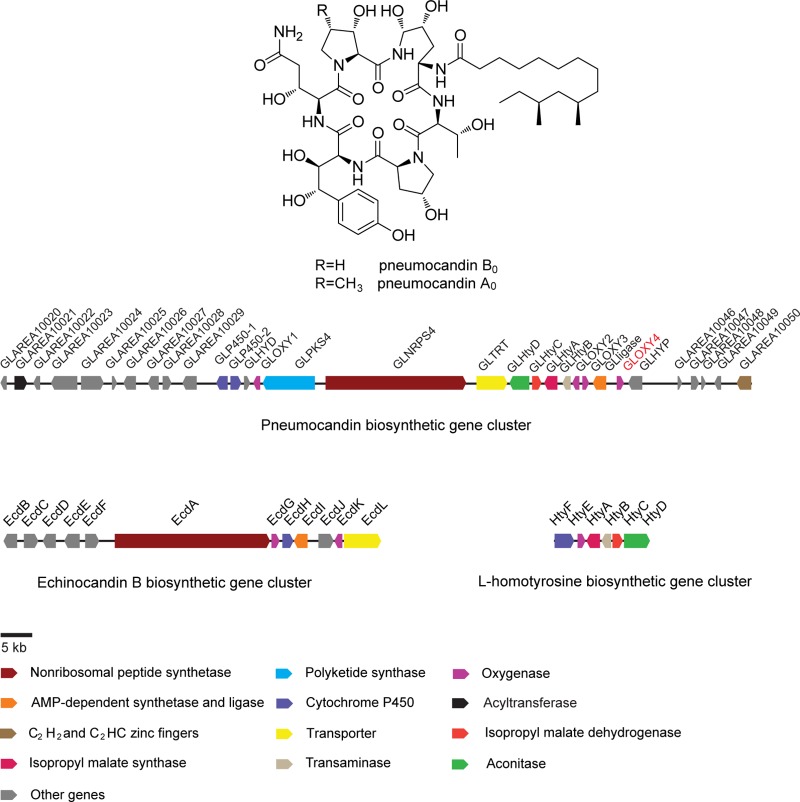
Pneumocandins produced by the fungus Glarea lozoyensis are acylated cyclic hexapeptides of the echinocandin family. Pneumocandin B0 is the starting molecule for the first semisynthetic echinocandin antifungal drug, caspofungin acetate. In the wild-type strain, pneumocandin B0 is a minor fermentation product, and its industrial production was achieved by a combination of extensive mutation and medium optimization. The pneumocandin biosynthetic gene cluster was previously elucidated by a whole-genome sequencing approach. Knowledge of the biosynthetic cluster suggested an alternative way to produce exclusively pneumocandin B0. Disruption of GLOXY4, encoding a nonheme, α-ketoglutarate-dependent oxygenase, confirmed its involvement in l-leucine cyclization to form 4S-methyl-l-proline. The absence of 4S-methyl-l-proline abolishes pneumocandin A0 production, and 3S-hydroxyl-l-proline occupies the hexapeptide core's position 6, resulting in exclusive production of pneumocandin B0. Retrospective analysis of the GLOXY4 gene in a previously isolated pneumocandin B0-exclusive mutant (ATCC 74030) indicated that chemical mutagenesis disrupted the GLOXY4 gene function by introducing two amino acid mutations in GLOXY4. This one-step genetic manipulation can rationally engineer a high-yield production strain.
INTRODUCTION
Pneumocandins are lipohexapeptides of the echinocandin family and potently prevent fungal cell wall formation by noncompetitive inhibition of β-1,3-glucan synthase. Due to their high efficacy and reduced toxicity compared to azoles and amphotericin B, echinocandin-type antibiotics have rapidly risen to use as first-line therapies for the treatment of invasive fungal infections (1, 2). Pneumocandins are differentiated from other echinocandins by their 3R-hydroxyl-l-glutamine in place of l-threonine at the fifth position of the core hexapeptide and by a 10R,12S-dimethylmyristoyl side chain (3) which renders the molecule less prone to cause red blood cell lysis than other naturally occurring echinocandins with palmitoyl or linoleoyl side chains (4). Pneumocandin A0 is the most prevalent pneumocandin in fermentations of wild-type (wt) Glarea lozoyensis and was the first to be isolated and structurally elucidated (5). Subsequently, other pneumocandins were isolated, including pneumocandin B0, which was chosen as the starting point for semisynthesis of the first echinocandin-type antifungal drug, caspofungin acetate (Cancidas) (2, 6, 7). Pneumocandin B0 was selected as the natural product for semisynthesis of clinical quantities of caspofungin because of its superior potency and pathogen spectrum (2, 8).
Interest in the biosynthesis of the pneumocandins stems not only from their potent and fungus-specific antifungal activity but also from the need to understand reactions peculiar to their biosynthesis (9). A 10R,12S-dimethylmyristic acid is acylated to a hexapeptide comprised of the nonproteinogenic amino acids 4R,5R-dihydroxyl-l-ornithine, 3S-hydroxyl-4S-methyl-l-proline, 3S-hydroxyl-l-proline, 4R-hydroxyl-l-proline, 3S,4S-dihydroxyl-l-homotyrosine, and 3R-hydroxyl-l-glutamine, which are cooperatively assembled by a polyketide synthase (PKS) and a nonribosomal peptide synthetase (NRPS).
In our previous study (10), the pneumocandin gene cluster was identified by whole-genome bioinformatic analysis and by gene homology comparison to the recently characterized echinocandin B gene cluster from Aspergillus rugulosus (11). Gene disruption of the polyketide synthase-encoding gene GLPKS4 or the nonribosomal peptide synthetase-encoding gene GLNRPS4 in the gene cluster abolished pneumocandin biosynthesis (Fig. 1) (10). The commonalities of the pneumocandin and echinocandin B pathways and other echinocandin-type pathways are striking, as most genes among these clusters appear to be orthologs despite significant organizational differences (3) (Fig. 1). However, unlike ecdA, GLNRPS4 sits downstream of and adjacent to a gene (GLPKS4) encoding a highly reducing polyketide synthase. These two core genes are centrally located in the pneumocandin biosynthetic gene cluster (Fig. 1) and are independently transcribed and translated. Instead of the echinocandin acyl side chain originating from cytosolic fatty acids as has been proposed for the echinocandin pathway, a dedicated PKS, encoded by GLPKS4, synthesizes the 10R,12S-dimethylmyristoyl side chain in pneumocandins.
FIG 1.
The pneumocandins and their biosynthetic gene cluster in Glarea lozoyensis and the gene clusters for echinocandin biosynthesis from Aspergillus rugulosus.
Purification of pneumocandin B0 was challenging due to its structural similarity to the more abundant pneumocandin A0 (A0:B0 ratio = 7:1) (2). Merck researchers invested considerable effort to alter the ratio through iterative rounds of mutagenesis and medium optimization, and eventually the A0:B0 ratio was shifted from 7:1 to 1:80 (12). Although the ratio was significantly improved, low levels of pneumocandin A0 and other minor products continued to complicate large-scale purification processes (13). Therefore, identification and manipulation of the gene(s) responsible for the shift from the naturally prevailing production of pneumocandin A0 to fermentations where pneumocandin B0 predominates may lead to more efficient large-scale production and downstream processing of pneumocandin B0. Furthermore, a full understanding of competing pathways and intervention in their reactions may optimize substrate flow into pneumocandin intermediates and lead to reductions in impurities and to high-yield production strains.
MATERIALS AND METHODS
Strains and plasmids.
The pneumocandin-producing strain was the wt G. lozoyensis strain ATCC 20868. Escherichia coli DH5α (TaKaRa, Japan) was used for DNA propagation and was cultured at 37°C and 220 rpm in Luria-Bertani (LB) broth with the appropriate antibiotics. Agrobacterium tumefaciens AGL-1 was used for transformation and was cultured at 28°C and 220 rpm in YEB broth (10) (5 g sucrose, 1 g yeast extract, 10 g peptone, 0.5 g MgSO4 · 7H2O, distilled H2O to 1 liter, with the pH adjusted to 7.0). Carbenicillin and kanamycin were added at a concentration of 50 μg/ml for culture of A. tumefaciens AGL-1 bearing the disruption vector. The plasmid pAg1-H3, which was described previously, was used for gene disruption vector construction and transformation (14). Genomic DNA of G. lozoyensis was extracted as previously described by Zhang et al. (14). A DNA fragment upstream of the 5′ region of GLOXY4 was amplified by employing genomic DNA of wt G. lozoyensis ATCC 20868 as the template and S1 and R1 as primers. The primers added PvuII and ApaI restriction sites. The product was cloned into the pMD18-T vector (TaKaRa, Japan), and the 1.2-kb fragment was released as a PvuII-ApaI fragment that was placed between the PvuII/ApaI sites of pAg1-H3 to produce pAg1-H3-GLOXY4L. Similarly, a 1.2-kb DNA fragment representing the 3′ region of GLOXY4 and part of the downstream intergenic region between GLOXY4 and GLHYP was also amplified by PCR, using genomic DNA of wt G. lozoyensis ATCC 20868 as the template and S2 and R2 as primers. These primers added AscI and SbfI restriction sites. Following cloning of the PCR product into the pMD18-T vector, it was released as an AscI-SbfI fragment and inserted into the AscI/SbfI sites of pAg1-H3-GLOXY4L to generate the completed disruption vector pAg1-H3-GLOXY4 (Fig. 2). The homologous fragment amplifications were carried out as follows. One microliter of the prepared genomic DNA template from wt G. lozoyensis ATCC 20868 was added to a 50-μl PCR amplification system using Phusion high-fidelity DNA polymerase following the manufacturer's instructions (NEB). All PCRs were carried out in a Veriti 96-well thermal cycler (PE Applied Biosystems). The amplification program consisted of predenaturation at 98°C for 30 s followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 57°C for 30 s, and elongation at 72°C for 40 s, with a final extension step at 72°C for 10 min. The primers and restriction enzymes used for disruption vector construction are listed in Table 1.
FIG 2.
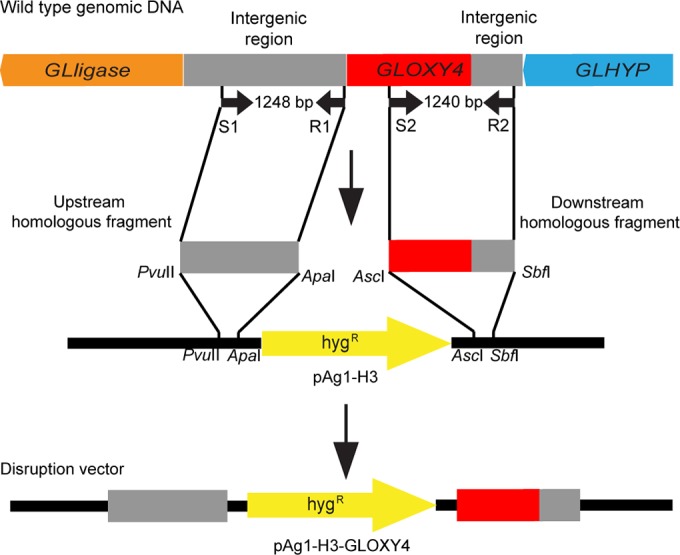
Construction of disruption vector pAg1-H3-GLOXY4. The upstream and downstream homologous fragments were first amplified from wt genomic DNA. The two fragments were then digested by restriction enzymes, followed by insertion into the upstream and downstream multiple-cloning sites of the hygromycin resistance gene (hygR) to give the disruption vector pAg1-H3-GLOXY4.
TABLE 1.
GLOXY4 primers used in this studya
| Primer name | Sequence (5′ to 3′) | Restriction enzyme | PCR fragment size (kb) |
|---|---|---|---|
| S1 | CAGCTGGAGAAGTTCAAGAGCAGGAT | PvuII | 1.2 |
| R1 | GGGCCCCAGGGCAGATATTTCAATTAG | ApaI | |
| S2 | GGCGCGCCGATGGTCCAAAACCTCAAAG | AscI | 1.2 |
| R2 | CCTGCAGGGTTGGGTTGGAGTACAAAAG | SbfI | |
| W | GGAGGCTTTCATCGTCGTC | 1.5 (wt) | |
| X | CGAGTCCTCCCACATCAT | 3.4 (KO) | |
| Y | TGACGGACGGCTTACATG | No product (wt) | |
| I | CGAGGGCAAAGGAATAGAGTAG | 2.7 (KO) |
KO, knockout. Restriction enzymes were used for the construction of disruption vectors. Underlined sequences identify the restriction sites.
Gene disruption and fungal transformation.
The protocol for gene disruption in G. lozoyensis was performed as described by Zhang et al. (14), with modifications. The conidia of G. lozoyensis were washed with 0.05% Tween 20, vortexed for 15 min, rinsed twice with distilled water, and suspended in 750 μl of distilled H2O. The cultivation of A. tumefaciens and the subsequent transformation method were described by Chen et al. (10). The conidial suspension (750 μl) was mixed with an equal volume of A. tumefaciens culture, vortexed for 2 min, and spread onto IMAS agar. IMAS agar consisted of the following (per liter of distilled water): 400 ml 2.5× salt solution, 1.8 g glucose, 5 ml glycerol, 40 ml 1 M MES (morpholineethanesulfonic acid), and 2 ml 100 mM acetosyringone (MES and acetosyringone were added after sterilization). The 2.5× salt solution consisted of the following (per liter of distilled water): 3.625 g K2HPO4, 5.125 g KH2PO4, 1.25 g MgSO4 · 7H2O, 0.375 g NaCl, 0.165 g CaCl2 · 2H2O, 0.0062 g FeSO4 · 7H2O, and 1.25 g (NH4)2SO4. The mixture of conidial suspension and A. tumefaciens culture was cocultivated at 28°C for 2 days. The coculture was then covered with M-100 medium supplemented with 300 μg/ml cefotaxime and 200 μg/ml hygromycin B. M-100 medium consisted of the following (per liter of distilled water): 62.5 ml M-100 salt solution, 10 g glucose, 3 g KNO3, and 15 g agar. M-100 salt solution consisted of the following (per liter of distilled water): 16 g KH2PO4, 4 g Na2SO4, 8 g KCl, 2 g MgSO4 · 7H2O, 1 g CaCl2, and 8 ml M-100 trace element solution. M-100 trace element solution consisted of the following (per liter of distilled water): 30 mg H3BO3, 70 mg MnCl2 · 4H2O, 200 mg ZnCl2, 20 mg Na2MoO4 · 2H2O, 50 mg FeCl3 · 6H2O, and 200 mg CuSO4 · 5H2O. The cultures were incubated at 25°C for 2 to 3 weeks before isolation of hygromycin B-resistant transformants. Transformants with the desired gene disruption were identified by PCR. Aliquots (0.5 μl) of genomic DNA template from hygromycin-resistant strains and the wt (control) strain were added to 25-μl PCR amplification mixtures along with EasyTaq polymerase (Transgen, China). PCRs were performed with the following program: predenaturation at 94°C for 180 s followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and elongation at 72°C for 3.5 min, with a final extension step at 72°C for 10 min. PCR primers used for screening are listed in Table 1.
Fermentation and compound extraction procedures.
The procedures and media for fermentation were described by Schwartz et al. (5). Briefly, conidia from oat bran agar (4% oat bran, 2% agar in tap water) were inoculated into 10 ml of seed medium (KF medium). The seed medium was incubated for 5 days with agitation at 220 rpm. For the production of pneumocandins, 0.4 ml of seed medium was inoculated into 10 ml of production medium (H medium) (15) in a tube. The production culture was agitated at 220 rpm at 25°C for 14 days. To extract the pneumocandins, an equal volume of methanol was added to each culture tube, and tubes were agitated at 220 rpm for 1 h at 25°C, followed by filtration to remove cells. Extracts were evaporated to dryness under a vacuum, followed by addition of 1 ml methanol to dissolve the samples.
HPLC and high-resolution mass spectrometry (MS) analysis.
The chromatographic profiles of crude extract samples were detected with an Agilent 1260 high-pressure liquid chromatograph (HPLC) equipped with a diode array detector (DAD), with wavelength scanning from 190 nm to 400 nm. Ten-microliter aliquots of crude extracts were injected for each run and eluted with a solvent gradient of 10 to 100% B for 28 min (solvent A, 0.1% formic acid in H2O; solvent B, 0.1% formic acid in acetonitrile) on a C18 reverse-phase column (Agilent Zorbax Eclipse Plus C18; 4.6 mm × 150 mm × 5 μm). The column effluent, eluting at a flow rate of 1 ml/min, was monitored at 210 nm. Titers were calculated from the peak area obtained by integration of the elution profiles. Pure pneumocandin B0 (Molcan, Canada) was used to determine the response factor. Titer values are the averages ± standard deviations for three replicate cultures.
High-resolution MS analysis of the samples was performed on an Agilent 6538 ultrahigh-definition accurate-mass quadrupole time of flight (Q-TOF) LC-MS system interfaced with an Agilent 1200 series HPLC-chip–MS system. The LC-chip system used a 40-nl enrichment column and a 75-μm by 43-mm analytical column packed with Zorbax 300SB-C18 (5 μm). The solvents were 0.1% formic acid in water (A) and 90% acetonitrile in water with 0.1% formic acid (B). The flow rate was 3.5 nl/min for loading the sample onto the enrichment column and 600 nl/min for the analytical column. Samples were loaded onto the enrichment column by use of 20% solvent B. The gradient for the analytical column was as follows: 20% solvent B at 0 min, 90% solvent B at 13 min, and 20% solvent B at 15 min. The Q-TOF system was operated in positive mode with a capillary voltage of 1,800 V and a drying gas flow rate of 4 liters/min at 340°C.
Sequence alignment and protein structure modeling.
Protein sequences of the oxygenases from G. lozoyensis ATCC 20868 (wt) and ATCC 74030 (mutant) and the adenylation (A) domains from GLNRPS4 and EcdA were aligned by using M-Coffee with the default parameters recommended by the website (16). Oxygenase protein structures were predicted by using SWISS-MODEL (17), with the putative 2-oxoglutarate-Fe(II) oxygenase family protein as the template (RCSB Protein Data Bank [PDB] ID 3oox). The A domain structures were generated via homology modeling using SWISS-MODEL, with the phenylalanine adenylation domain (RSCB PDB ID 1AMU) as the template.
RESULTS AND DISCUSSION
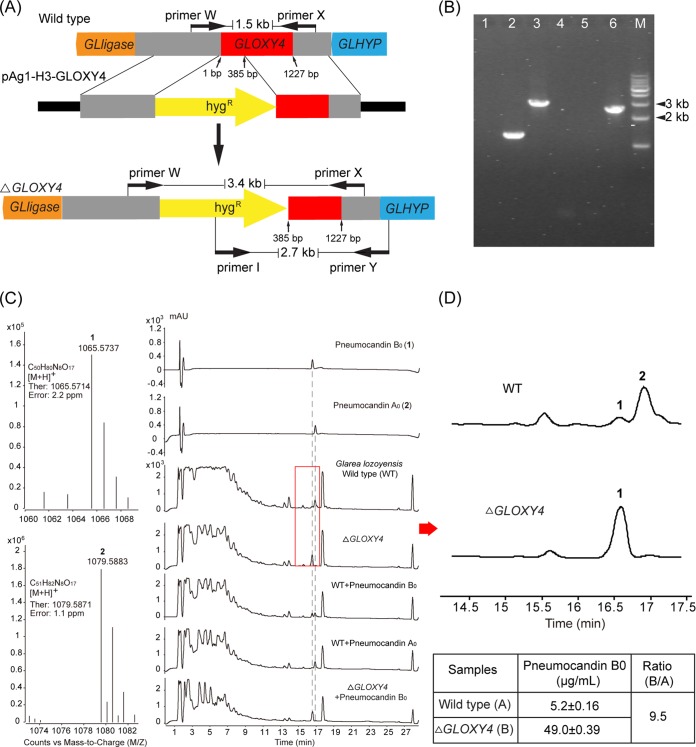
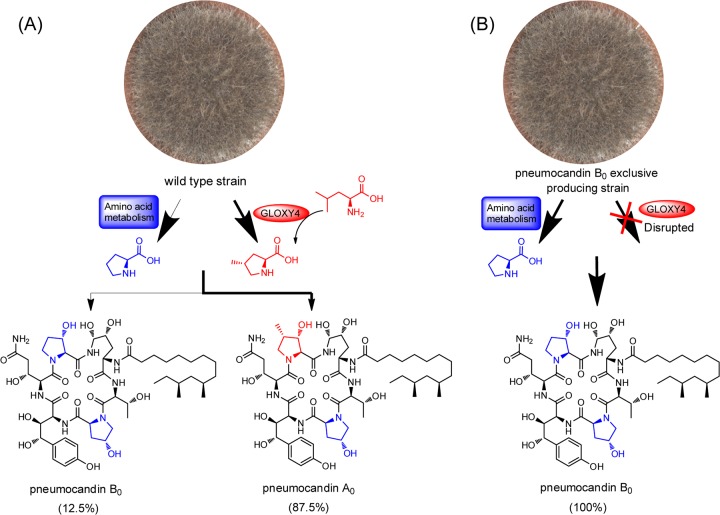
The pneumocandin scaffold has three l-proline derivatives: 3S-hydroxyl-4S-methyl-l-proline or 3S-hydroxyl-l-proline in the sixth position and 4R-hydroxyl-l-proline in the third. Feeding of labeled precursors established that these residues have different origins: 3S-hydroxyl-l-proline and 4R-hydroxyl-l-proline are derived from l-proline, while 3S-hydroxyl-4S-methyl-l-proline is derived from l-leucine (18). The biosynthesis of 4S-methyl-l-proline was proposed to occur in two steps, i.e., oxidation of a terminal methyl moiety of l-leucine followed by cyclization (18, 19). Four nonheme iron, α-ketoglutarate-dependent oxygenases are encoded in the pneumocandin biosynthetic gene cluster, and one of them (Fig. 1) was predicted to be involved in l-leucine cyclization. High amino acid similarity between the l-leucine cyclization enzyme EcdK of A. rugulosus, involved in echinocandin biosynthesis (20), and the oxygenase GLOXY4 of G. lozoyensis (62% amino acid identity) predicted that GLOXY4 catalyzes l-leucine cyclization in pneumocandin biosynthesis. The GLOXY4 gene was disrupted by A. tumefaciens-mediated transformation with a hygromycin resistance gene, which was used as the selection marker. The target gene GLOXY4 was inactivated by replacing its 5′ end (∼400 bp) with a hygromycin resistance gene. The hygromycin-resistant clones were rescued from the medium, and mutants with the desired disruption of the target gene were verified by PCR analysis. DNA fragments of the intact GLOXY4 locus in the wild-type strain and the GLOXY4 locus disrupted with the hygromycin resistance gene in the mutant strain were amplified by use of primers W and X and primers I and Y (Fig. 3A and B). A product of approximately 1.5 kb would be expected from an intact copy of the gene amplified by use of primers W and X, with no band expected from the intact copy of the gene amplified with primers I and Y. Products of about 3.4 kb with primers W and X and 2.7 kb with primers I and Y would reflect the presence of the disrupted gene (Fig. 3A and B). One positive mutant and wt G. lozoyensis were fermented for pneumocandin product analysis. The identities of pneumocandins B0 and A0 were confirmed by HPLC analysis of injection of authentic standards alone or coinjection with fermentation extracts and by high-resolution mass spectrum analysis (Fig. 3C). HPLC analysis of the fermentation extracts showed that the GLOXY4 disruption mutant strain failed to produce pneumocandin A0 but continued to produce pneumocandin B0 (Fig. 3C and D). Failure to produce 3S-hydroxyl-4S-methyl-l-proline led to its replacement by 3S-hydroxyl-l-proline in the sixth position of pneumocandin A0. Thus, GLOXY4 was confirmed to be responsible for 4S-methyl-l-proline biosynthesis (Fig. 4A). Quantitative analysis showed that the titer of pneumocandin B0 increased 9.5-fold in the GLOXY4 disruption mutant compared to the wt strain (Fig. 3D), suggesting that GLNRPS4 is able to incorporate 3S-hydroxyl-l-proline as the sole substrate at the sixth position, thus leading to the exclusive biosynthesis of pneumocandin B0 (Fig. 4B).
FIG 3.
Disruption of GLOXY4 abolishes pneumocandin A0 production. (A) Schematic of GLOXY4 disruption. The 5′ end of GLOXY4 was replaced by a hygromycin resistance gene. Primers W/X and I/Y were used for detection of the target disruption mutant. (B) PCR analysis of the ΔGLOXY4 strain. Lanes: 1, negative control with primers W and X; 2, amplification of genomic DNA from wt strain with primers W and X; 3, amplification of genomic DNA from ΔGLOXY4 strain with primers W and X; 4, negative control with primers I and Y; 5, amplification of genomic DNA from wild-type strain with primers I and Y; 6, amplification of genomic DNA from ΔGLOXY4 strain with primers I and Y; M, 1-kb DNA ladder. (C) High-resolution mass spectrum analysis of pneumocandins B0 (1) and A0 (2) from the wt strain and HPLC profiles of pneumocandin B0 and A0 standards and fermentation extracts of the ΔGLOXY4 and wild-type strains alone and coinjected with the standards. (D) Comparison of the ΔGLOXY4 and wild-type strains shows the absence of pneumocandin A0 production and increased pneumocandin B0 production in the ΔGLOXY4 strain.
FIG 4.
Biosynthetic scheme for exclusive production of pneumocandin B0. (A) Pneumocandins from the wt strain. The thinner arrow indicates the minor route for pneumocandin B0 biosynthesis, while the thick arrow indicates the predominant route for pneumocandin A0 biosynthesis. (B) Blockage of pneumocandin A0 biosynthesis in the GLOXY4 disruption mutant leads to exclusively pneumocandin B0 production. The hydroxylated proline residues originating from l-proline are shown in blue, and the hydroxylated 4-methyl-l-proline residue originating from l-leucine is shown in red.
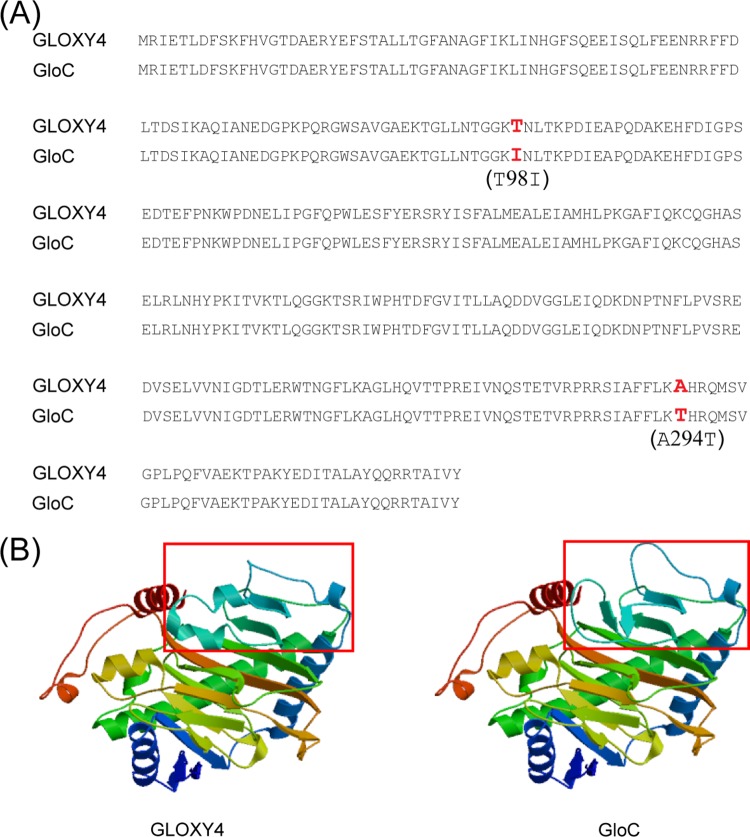
In the wt strain, pneumocandin B0 is the minor compound compared to the major compound pneumocandin A0 (A0:B0 ratio = 7:1). The selection of pneumocandin B0 as the eventual starting material for caspofungin required extensive classical mutagenesis and production medium optimization work to eliminate the production of pneumocandin A0 and other competing pneumocandins, eventually shifting the pneumocandin A0/pneumocandin B0 ratio to 1:80 (12). The genome of a pneumocandin B0-exclusive strain (G. lozoyensis ATCC 74030) has been sequenced and thus afforded the opportunity for retrospective analysis of the effects of mutagenesis (21). The amino acid sequences of wt GLOXY4 (ATCC 20868) and the equivalent protein in the mutant strain ATCC 74030, designated GloC (62% identity with EcdK), which was predicted to catalyze l-leucine cyclization (9), differed by two amino acids. The residues of wt GLOXY4 at positions 98 and 294 were l-threonine and l-alanine, respectively, while the equivalent residues of GloC were mutated to l-isoleucine and l-threonine (Fig. 5A). Substitution of a polar amino acid (Thr) for a nonpolar amino acid (Ile) at position 98 (T98I) and the opposite situation at position 294 (A294T) are predicted to modify the three-dimensional structure of mutated GLOXY4 (Fig. 5B). The failure to incorporate 3S-hydroxyl-4S-methyl-l-proline into the peptide core may have been an indirect result of this structural alteration compared to GLOXY4 and further suggests that these two amino acids may be essential for maintaining the proper structural conformation needed to catalyze the cyclization of l-leucine to form 4S-methyl-l-proline. Although the malfunction of GLOXY4 may indirectly increase pneumocandin B0 production, it is possible that chemical mutagenesis in ATCC 74030 affected other proteins that may have contributed to increased pneumocandin B0 production. Specific biochemical confirmation of the influence of the two mutated amino acids on the function of GLOXY4 will require further study. Recently, an enzyme identical to GLOXY2, GloF, was characterized for the mutant strain ATCC 74030 (9). GloF was found to be responsible for catalyzing the hydroxylation of l-proline to generate 3S-hydroxyl-4S-methyl-l-proline, 3S-hydroxyl-4S-methyl-l-proline, and 4S-hydroxyl-l-proline. Thus, GLOXY2 plays a critical role in providing the essential proline-derived building blocks for pneumocandin biosynthesis, including that of pneumocandin B0. In summary, we found that GLOXY4, a nonheme iron, α-ketoglutarate-dependent oxygenase, is essential for the cyclization of l-leucine to form 4S-methyl-l-proline of the pneumocandin A0 scaffold. The substantial homology between the pneumocandin GLOXY4 and the echinocandin B tailoring oxygenase EcdK suggested that GLOXY4 catalyzes two oxidation steps on C-5 of l-leucine, followed by a spontaneous dehydration to generate 3-methyl-pyrroline-5-carboxylic acid, and possibly that a reductase acts on the last step to generate 4S-methyl-l-proline as proposed for echinocandin B (20).
FIG 5.
Predicted linear and three-dimensional structures of wt GLOXY4 and GloC. (A) Amino acid sequence alignment of GLOXY4 and GloC. (B) Protein structure modeling of GLOXY4 and GloC. The predicted affected region is framed in red.
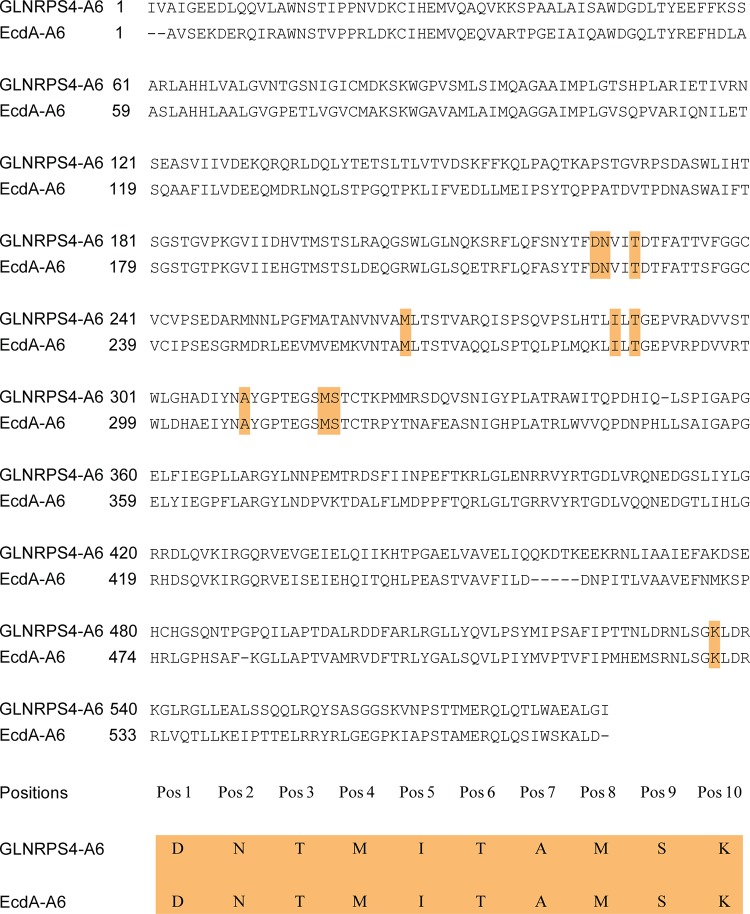
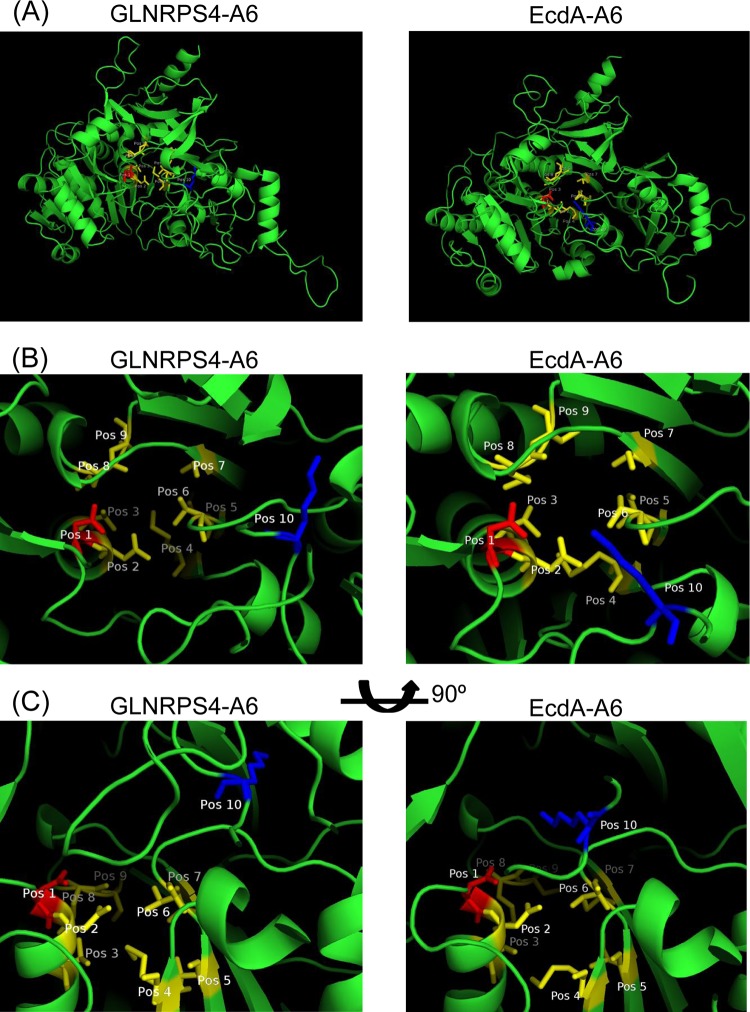
Unlike the ΔGLOXY4 mutation, disruption of ecdK of the echinocandin pathway apparently abolished echinocandin B production, because no echinocandin variant with 3S-hydroxyl-l-proline at position 6 was observed. Furthermore, to our knowledge, an echinocandin-producing strain simultaneously producing sixth-position methyl-proline and desmethyl-proline echinocandin variants in a manner such as that in G. lozoyensis has not been reported (22). In special cases, l-proline alone has been forced into the sixth position of pneumocandins by overfeeding l-proline in the medium (23). These results indicate a subtle difference in affinity for oxidized l-prolines between the two NRPS proteins, which may be due to substantial differences in amino acid composition (55% identity). To gain insight into the functional difference between the sixth A domains of GLNRPS4 and EcdA, we conducted an in silico analysis of their sequences. First, we aligned the A domain sequences by using the NCBI BLAST tool; the identity and similarity between them are 59% and 73%, respectively (Fig. 6). The 10-residue substrate specificity-determining sequence (10-amino-acid code) of the A domains is thought to be the structural basis of substrate recognition of the A domains of NRPS proteins. The 10-amino-acid codes of the sixth A domains of GLNRPS4 and EcdA were extracted by using the Web-based software NRPSpredictor2 (24), and the 10-amino-acid code of GLNRPS4 was DNTMITAMSK. EcdA has exactly the same 10-amino-acid code as GLNRPS4 (Fig. 6), indicating that these two A domains activate the same, or at least very similar, amino acids (3S-hydroxyl-4S-methyl-l-proline and 3S-hydroxyl-l-proline). Homology modeling and analysis of the putative substrate-binding pockets of the GLNRPS4 and EcdA A domains may provide clues that can rationalize the observed differences in substrate specificities (Fig. 7). The key difference in the binding pocket appears to be in the relative positions between amino acids at position 1 (Pos 1) and position 10 (Pos 10) (Fig. 7B and C), which are thought to stabilize the substrate amino acid (25). The predicted three-dimensional distance between Asp (Pos 1) and Lys (Pos 10) in EcdA was much closer than that in GLNRPS4. The shorter distance between Asp and Lys in EcdA seemed to form a more compact substrate-binding pocket than that in GLNRPS4 and may conceivably lead to stricter substrate recognition for only 3S-hydroxyl-4S-methyl-l-proline than the case for GLNRPS4, which recognizes both 3S-hydroxyl-4S-methyl-l-proline and 3S-hydroxyl-l-proline.
FIG 6.
Comparison of the whole sequences and 10-residue substrate specificity-determining sequences of the sixth adenylation domains of GLNRPS4 and EcdA.
FIG 7.
Comparison of the structures of the sixth adenylation domains of GLNRPS4 and EcdA, generated via homology modeling using SWISS-MODEL, with the RSCB PDB ID 1AMU structure as the template. (A) Three-dimensional structures of adenylation domains from GLNRPS4 and EcdA. (B) Top-down views of the modeled substrate-binding pockets. (C) Side-on views generated by rotating the structures 90° around the horizontal plane. Residues are colored and labeled according to their positions in the 10-amino-acid specificity code. Amino acids at position 1 and position 10 are shown in red and blue, respectively, whereas the amino acids in the rest of the positions are shown in yellow.
Disruption of GLOXY4 therefore makes possible the exclusive incorporation of 3S-hydoxy-l-proline into the sixth position and a shift from the dominant pneumocandin A0 to exclusively pneumocandin B0 production. Such a mutant would be a critical first step in the rational engineering of a high-yield producer of pneumocandin B0.
ACKNOWLEDGMENTS
We acknowledge Jan Tkacz for his critical review of the manuscript and for his pioneering research on the biosynthetic origins of the pneumocandins, which was critical for the postgenomic elucidation of the echinocandin-type pathways. We also thank two anonymous reviewers for their valuable input on the manuscript's final version.
This work was supported by National Natural Science Foundation of China (NSFC) grant 31328001 to Z.A., NSFC grant 30625001 to X.L., Welch Foundation grant AU00024 to Z.A., a University of Texas System Star award to Z.A., and University of Texas Health Science Center at Houston faculty startup funds to G.F.B.
REFERENCES
- 1.Denning DW. 2003. Echinocandin antifungal drugs. Lancet 362:1142–1151. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- 2.Balkovec JM, Hughes DL, Masurekar PS, Sable CA, Schwartz RE, Singh SB. 2014. Discovery and development of first in class antifungal caspofungin (CANCIDAS®)—a case study. Nat Prod Rep 31:15–34. doi: 10.1039/c3np70070d. [DOI] [PubMed] [Google Scholar]
- 3.Bills GF, Li Y, Chen L, Yue Q, Niu X, An Z. 2014. New insights into the echinocandins and other fungal non-ribosomal peptides and peptaibiotics. Nat Prod Rep 31:1348–1375. doi: 10.1039/C4NP00046C. [DOI] [PubMed] [Google Scholar]
- 4.Richards FM, Eisenberg DS, Kim PS, Scolnick EM. 2001. Drug discovery and design, vol 56 Academic Press, San Diego, CA. [Google Scholar]
- 5.Schwartz RE, Giacobbe RA, Bland JA, Monaghan RL. 1989. L-671,329, a new antifungal agent. I. Fermentation and isolation. J Antibiot 42:163–167. [DOI] [PubMed] [Google Scholar]
- 6.Kartsonis NA, Nielsen J, Douglas CM. 2003. Caspofungin: the first in a new class of antifungal agents. Drug Resist Updat 6:197–218. doi: 10.1016/S1368-7646(03)00064-5. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz RE, Sesin DF, Joshua H, Wilson KE, Kempf AJ, Goklen KA, Kuehner D, Gailliot P, Gleason C, White R, Inamine E, Bills GF, Salmon P, Zitano L. 1992. Pneumocandins from Zalerion arboricola. I. Discovery and isolation. J Antibiot 45:1853–1866. [DOI] [PubMed] [Google Scholar]
- 8.Schmatz D, Abruzzo G, Powles M, McFadden D, Balkovec J, Black R, Nollstadt K, Bartizal K. 1992. Pneumocandins from Zalerion arboricola. IV. Biological evaluation of natural and semisynthetic pneumocandins for activity against Pneumocystis carinii and Candida species. J Antibiot 45:1886–1891. [DOI] [PubMed] [Google Scholar]
- 9.Houwaart S, Youssar L, Hüttel W. 2014. Pneumocandin biosynthesis: involvement of a trans-selective proline hydroxylase. Chembiochem 15:2365–2369. doi: 10.1002/cbic.201402175. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Yue Q, Zhang X, Xiang M, Wang C, Li S, Che Y, Ortiz-López F, Bills GF, Liu X, An Z. 2013. Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis. BMC Genomics 14:339. doi: 10.1186/1471-2164-14-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cacho RA, Jiang W, Chooi YH, Walsh CT, Tang Y. 2012. Identification and characterization of the echinocandin B biosynthetic gene cluster from Emericella rugulosa NRRL 11440. J Am Chem Soc 134:16781–16790. doi: 10.1021/ja307220z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masurekar PS, Fountoulakis JM, Hallada TC, Sosa MS, Kaplan L. 1992. Pneumocandins from Zalerion arboricola. II. Modification of product spectrum by mutation and medium manipulation. J Antibiot 45:1867–1874. [DOI] [PubMed] [Google Scholar]
- 13.Connors N, Pollard D. 2005. Pneumocandin B0 production by fermentation of fungus Glarea lozoyensis: physiological and engineering factors affecting titer and structural analogue formation, p 515–536. In An Z. (ed), Handbook of industrial mycology, vol 22 Marcel Dekker, New York, NY. [Google Scholar]
- 14.Zhang A, Lu P, Dahl-Roshak A, Paress P, Kennedy S, Tkacz JS, An Z. 2003. Efficient disruption of a polyketide synthase gene (pks1) required for melanin synthesis through Agrobacterium-mediated transformation of Glarea lozoyensis. Mol Genet Genomics 268:645–655. [DOI] [PubMed] [Google Scholar]
- 15.Tkacz JS, Giacobbe RA, Monaghan RL. 1993. Improvement in the titer of echinocandin-type antibiotics—a magnesium-limited medium supporting the biphasic production of pneumocandin A0 and pneumocandin B0. J Ind Microbiol 11:95–103. doi: 10.1007/BF01583681. [DOI] [PubMed] [Google Scholar]
- 16.Wallace IM, O'Sullivan O, Higgins DG, Notredame C. 2006. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res 34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adefarati AA, Hensens OD, Jones E, Tkacz JS. 1992. Pneumocandins from Zalerion arboricola. V. Glutamic acid- and leucine-derived amino acids in pneumocandin A0 (L-671,329) and distinct origins of the substituted proline residues in pneumocandins A0 and B0. J Antibiot 45:1953–1957. [DOI] [PubMed] [Google Scholar]
- 19.Luesch H, Hoffmann D, Hevel JM, Becker JE, Golakoti T, Moore RE. 2003. Biosynthesis of 4-methylproline in cyanobacteria: cloning of nosE and nosF genes and biochemical characterization of the encoded dehydrogenase and reductase activities. J Org Chem 68:83–91. doi: 10.1021/jo026479q. [DOI] [PubMed] [Google Scholar]
- 20.Jiang W, Cacho RA, Chiou G, Garg NK, Tang Y, Walsh CT. 2013. EcdGHK are three tailoring iron oxygenases for amino acid building blocks of the echinocandin scaffold. J Am Chem Soc 135:4457–4466. doi: 10.1021/ja312572v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Youssar L, Gruning BA, Erxleben A, Gunther S, Huttel W. 2012. Genome sequence of the fungus Glarea lozoyensis: the first genome sequence of a species from the Helotiaceae family. Eukaryot Cell 11:250. doi: 10.1128/EC.05302-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emri T, Majoros L, Tóth V, Pócsi I. 2013. Echinocandins: production and applications. Appl Microbiol Biotechnol 97:3267–3284. doi: 10.1007/s00253-013-4761-9. [DOI] [PubMed] [Google Scholar]
- 23.Petersen LA, Hughes DL, Hughes R, DiMichele L, Salmon P, Connors N. 2001. Effects of amino acid and trace element supplementation on pneumocandin production by Glarea lozoyensis: impact on titer, analogue levels, and the identification of new analogues of pneumocandin B0. J Ind Microbiol Biotechnol 26:216–221. doi: 10.1038/sj.jim.7000115. [DOI] [PubMed] [Google Scholar]
- 24.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res 33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stachelhaus T, Mootz HD, Marahiel MA. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]