Abstract
Thermophilic bacteria are regarded as attractive production organisms for cost-efficient conversion of renewable resources to green chemicals, but their genetic accessibility is a major bottleneck in developing them into versatile platform organisms. In this study, we aimed to isolate thermophilic, facultatively anaerobic bacilli that are genetically accessible and have potential as platform organisms. From compost, we isolated 267 strains that produced acids from C5 and C6 sugars at temperatures of 55°C or 65°C. Subsequently, 44 strains that showed the highest production of acids were screened for genetic accessibility by electroporation. Two Geobacillus thermodenitrificans isolates and one Bacillus smithii isolate were found to be transformable with plasmid pNW33n. Of these, B. smithii ET 138 was the best-performing strain in laboratory-scale fermentations and was capable of producing organic acids from glucose as well as from xylose. It is an acidotolerant strain able to produce organic acids until a lower limit of approximately pH 4.5. As genetic accessibility of B. smithii had not been described previously, six other B. smithii strains from the DSMZ culture collection were tested for electroporation efficiencies, and we found the type strain DSM 4216T and strain DSM 460 to be transformable. The transformation protocol for B. smithii isolate ET 138 was optimized to obtain approximately 5 × 103 colonies per μg plasmid pNW33n. Genetic accessibility combined with robust acid production capacities on C5 and C6 sugars at a relatively broad pH range make B. smithii ET 138 an attractive biocatalyst for the production of lactic acid and potentially other green chemicals.
INTRODUCTION
Green chemicals are sustainable bio-based alternatives for chemicals based on fossil resources. Biomass is considered an attractive renewable resource for the production of such green chemicals. Major challenges for using biomass are to develop cost-effective production processes and to convert substrates that go beyond first-generation pure sugars. From this perspective, the use of microbial fermentation processes that convert lignocellulosic sugars is gaining considerable attention. For particular products natural producers can be used, but often Escherichia coli and Saccharomyces cerevisiae are used as platform organisms because of their well-known physiology and ease of engineering for the production of many different products (1, 2).
In general, most organisms used for the production of green chemicals and fuels are mesophiles, and current processes still are based mainly on first-generation feedstocks, i.e., sucrose from sugar beet or sugarcane or glucose derived from starches from corn or tapioca. Although the engineering of platform organisms broadens the spectrum of possible substrates (3), the efficiency of the process could be increased by the use of organisms naturally capable of degrading lignocellulose-derived sugars (4). To further increase the efficiency and reduce the costs of the microbial production of green chemicals, moderately thermophilic hosts offer several advantages over mesophilic hosts, including (i) reduced cooling costs, (ii) lower contamination risk, (iii) increased substrate and product solubility, and (iv) temperature optima of these bacteria matching those of enzymes used for simultaneous saccharification and fermentation (SSF), lowering the enzyme load and costs (5). The use of anaerobic or facultatively anaerobic bacteria eliminates the need for expensive aerated industrial reactors. For SSF, facultative anaerobes have the advantage over strict anaerobes that reduction of the medium is not necessary, as reduced medium has been shown to inhibit saccharolytic enzymes (6).
Altogether, organisms of particular interest for the production of green chemicals from lignocellulose in an SSF setting are (facultatively) anaerobic thermophiles that can grow in minimal medium and ferment a wide range of substrates, including C5 and C6 sugars. They potentially can be used as platform organisms if they are genetically accessible. A number of facultatively anaerobic thermophilic hosts have been studied for green chemical and fuel production (reviewed in references 7 and 8), such as Bacillus coagulans for lactic acid (9, 10), Bacillus licheniformis for 2,3-butanediol (11, 12), and Geobacillus thermoglucosidans for ethanol production (13). Examples of strictly anaerobic thermophiles that have been studied for biofuel production are Clostridium thermocellum (14, 15), several Caldicellulosiruptor spp. (16–18), as well as Thermoanaerobacterium spp. and Thermoanaerobacter spp. (18–21). For each of these species, one or more strains have been shown to be genetically accessible, and engineering tools have been developed (7, 22). Still, it is desirable to further develop more efficient production organisms and improved genetic tools. On the one hand, this will enable a better understanding and exploitation of the diversity in metabolism found in thermophilic organisms, and on the other hand, this will expand the genetic toolbox for thermophiles, which at the moment is still rather limited compared to those available for mesophilic platform organisms.
In this study, we aimed to extend the collection of potential industrial platform organisms by isolating thermophilic, facultatively anaerobic bacilli and selecting a suitable production host by screening for C5 and C6 sugar utilization, organic acid production, fermentation performance, and genetic accessibility in order to select a new biocatalyst that has the potential to be developed into a platform organism.
MATERIALS AND METHODS
Media, cultivation methods, and strains.
Thermophile minimal medium (TMM; adjusted from reference 23) contained, per liter, 8.37 g morpholinepropanesulfonic acid (MOPS) and 100 ml 10× concentrated salt solution (TSS-10×, containing, per liter, 2.3 g K2HPO4, 5.1 g NH4Cl, 50 g NaCl, 14.7 g Na2SO4, 0.8 g NaHCO3, 2.5 g KCl, 18.7 g MgCl2 · 6H2O, 4.1 g CaCl2 · 2H2O, 0.08 g SrCl2 · 6H2O, 0.08 g H3BO3, 9.0 g NaNO3). The pH was set at 6.94 at room temperature, unless indicated otherwise. The medium was autoclaved for 20 min at 121°C and cooled to a maximum of 80°C, after which substrate and 10 ml per liter of 1 mM FeSO4 · 7H2O in 0.4 M tricine was added.
TMMY is TMM supplemented with 0.5 g/liter yeast extract (Roth, Germany).
Thermophile vitamin medium with yeast extract (TVMY) resembles TMMY, with the exception that SrCl2 · 6H2O, H3BO3, and NaNO3 are omitted and that metal and vitamin mixtures are added after sterilization. The metal mix (1,000× concentrated) contained, per liter, 16.0 g MnCl2 · 6H2O, 1.0 g ZnSO4, 2.0 g H3BO3, 0.1 g CuSO4 · 5H2O, 0.1 g Na2MoO4 · 2H2O, 1.0 g CoCl2 · 6H2O, 7.0 g FeSO4 · 7H2O. The vitamin mix (1,000× concentrated) contained, per liter, 0.1 g thiamine; 0.1 g riboflavin; 0.5 g nicotinic acid; 0.1 g pantothenic acid; 0.5 g pyridoxamine HCl; 0.5 g pyridoxal HCl; 0.1 g d-biotin; 0.1 g folic acid; 0.1 g p-aminobenzoic acid; 0.1 g cobalamin.
LB2 contained, per liter, 10 g tryptone (Oxoid, United Kingdom), 5 g yeast extract (Roth), 100 ml TSS-10×. The pH was set between 6.94 and 7.00 at room temperature.
TGP2 (adjusted from reference 24) contained, per liter, 17 g tryptone (Oxoid), 3 g neutralized soy peptone (Oxoid), 100 ml TSS-10×. The pH was set between 6.94 and 7.00 at room temperature. After autoclaving, 4 ml/liter sterile 60% (vol/vol) glycerol and 4 g/liter filter-sterilized 200 g/liter sodium pyruvate were added.
For all tube and plate cultures, carbon substrates were used at a concentration of 10 g/liter. For cultivation in reactors, carbon substrates were used at a concentration of 30 g/liter unless indicated otherwise. Substrates were added separately as 50% sterile solutions after autoclaving of the medium; xylose and arabinose were filter sterilized, and glucose and sucrose were autoclaved. For plates, 5 g/liter GelRite (Roth) was added. The acid indicator bromocresol purple was used to monitor acidification in plates and tubes. Anaerobic cultivation of plates was performed in an anaerobic jar (Oxoid) containing a GasPak (Oxoid). All strains were grown routinely at their isolation temperature (55°C or 65°C), unless stated otherwise. All strains obtained from DSMZ were routinely grown at 55°C.
To evaluate the optimum temperature of strain ET 138, cells were grown overnight from glycerol stock in 10 ml LB2 at 55°C. The next morning, they were diluted to an optical density at 600 nm (OD600) of 0.05 in 20 ml TMMY supplemented with 10 g/liter glucose in 50 ml Greiner tubes and incubated in water baths at different temperatures without shaking. A second experiment was performed in the same way, but with the overnight cultures grown at the same temperatures as those to which they were transferred the next morning. Tested temperatures were 37, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 57, 60, 63, 66, and 69°C. All of these experiments were performed in triplicate. The same experimental setup was used to evaluate the optimum pH of ET 138, but then the temperature was set at 55°C and the initial pH of the medium was varied from 3.0 to 7.0 with intervals of 0.5. For pH values from 3.0 to 5.5, morpholineethanesulfonic acid (MES) buffer was used instead of MOPS.
The following strains were obtained from DSMZ (Germany): G. thermoglucosidans DSM 2542T, B. coagulans DSM 1T, B. smithii DSM 4216T, B. smithii DSM 459, B. smithii DSM 460, B. smithii DSM 2319, B. smithii DSM 2320, and B. smithii DSM 2321.
Sampling, isolation, and initial selection.
Fresh compost samples were collected from two compost heaps at Recom Ede (The Netherlands) on 5 August 2010, both consisting solely of plant material. One heap was sieved through a 2- to 3-cm mesh and the other through a 1- to 2-cm mesh. The heaps were mixed every 2 weeks, resulting in an environment suitable for isolating facultatively anaerobic organisms. The temperature of the compost was 75°C at ∼30-cm depth, and sampling material was taken from the surface until ∼30-cm depth. Samples were taken by scraping compost into a plastic jar, transported to the laboratory (∼20 min) at ambient temperature, and stored at 4°C until use (1 to 4 weeks).
Five different conditions were applied during the isolation procedure: condition 1, sucrose at 55°C; condition 2, sucrose at 65°C supplemented with 5 g/liter CaCO3; condition 3, sucrose at 55°C supplemented with 5 g/liter CaCO3; condition 4, glucose at 65°C supplemented with 5 g/liter CaCO3; and condition 5, glucose at 55°C supplemented with 5 g/liter CaCO3. In all conditions, TMM was used, and it contained the indicated carbon source and was kept the same throughout the whole procedure for each condition. The isolation procedure was as follows. For each of the two compost types, 5 g compost was added to a 250-ml Erlenmeyer flask containing 50 ml TMM medium supplemented with the indicated carbon source and shaken for 6 h at 120 rpm at either 55°C or 65°C. After 6 h, dilution series were plated on the same medium containing 5 g/liter GelRite (Roth) and grown anaerobically for 24 to 48 h at either 55°C or 65°C. Subsequently, single colonies were picked and each colony was streaked onto 4 plates, containing TMM with either glucose, xylose, arabinose, or sucrose and incubated anaerobically for 24 to 48 h at the same temperature. Strains grown on 3 or more sugars, of which at least one was a C5 sugar and one was a C6 sugar, were inoculated into 8 ml of their isolation medium containing bromocresol purple in 15-ml screw-cap tubes and incubated without shaking for 24 to 48 h at 55 or 65°C. For the conditions containing CaCO3, this was added to the tubes as 500 μl autoclaved solution containing 80% (vol/vol) CaCO3 grains. For strains that grew and acidified the medium, glycerol stocks were made by the addition of 60% (vol/vol) glycerol to make 15% (vol/vol) and stored at −80°C.
In order to perform a production-based screening, all isolated strains were inoculated in duplicate from glycerol stocks into 8 ml TMMY supplemented with 10 g/liter glucose and 500 μl of 80% CaCO3 in 15-ml tubes, placed horizontally while shaking at 40 rpm, and incubated for 48 h at their isolation temperature (55°C or 65°C), after which the OD600 and pH were measured and high-pressure liquid chromatography (HPLC) analysis was performed. Based on HPLC results, pure cultures were made for selected strains by transferring single colonies twice on TMMY plates supplemented with 10 g/liter glucose before finally transferring them to the same medium in liquid form to make stocks after 24 to 30 h of growth. Pure cultures were subjected to 16S identification and used for further studies.
16S rRNA identification and phylogenetic analysis.
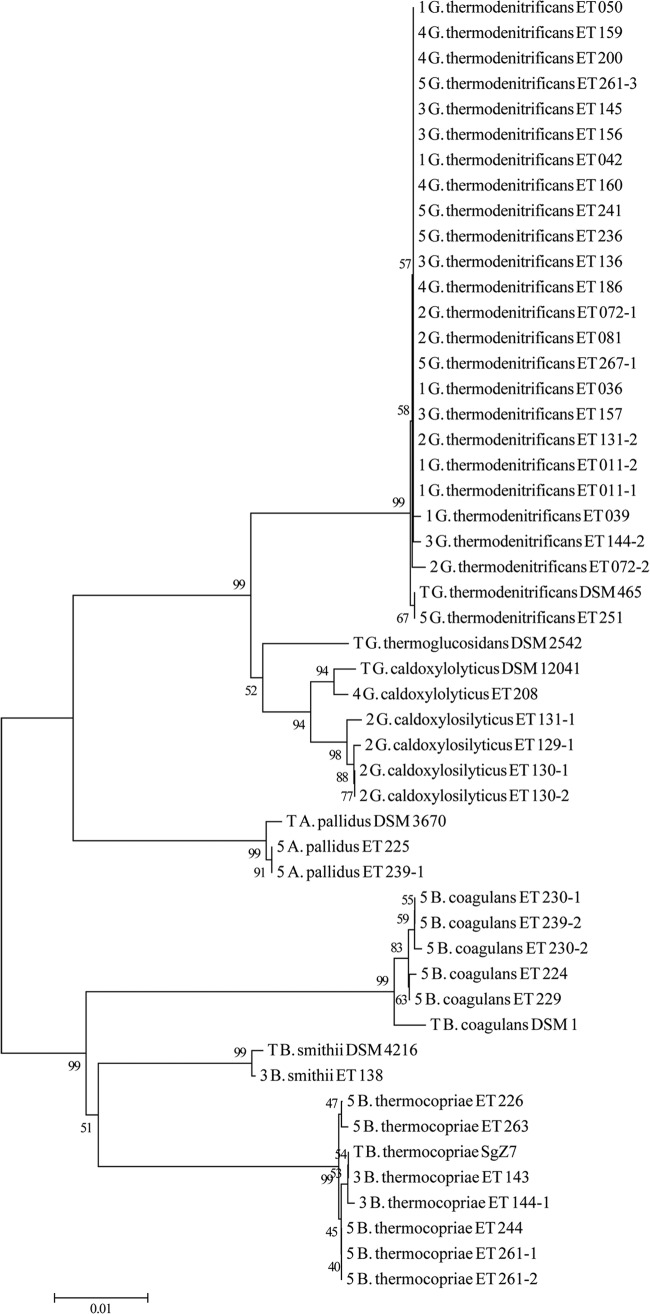
A little material from single colonies was transferred from plates to a PCR tube using a sterile toothpick and subsequently incubated in a microwave for 1.5 min at 800 W. PCR mix was added containing 2.5 U DreamTaq DNA polymerase (Fermentas, United Kingdom), DreamTaq buffer (Fermentas), 1 mM deoxynucleoside triphosphates (dNTPs) (Fermentas), 0.2 μM primers GM3 (AGAGTTTGATCATGGC) and GM4 (TACCTTGTTACGACTT), and MilliQ water to a total volume of 50 μl. PCR products were checked on agarose gel. Products were purified either by GeneJet PCR purification kit (Fermentas) or GeneJet gel extraction kit (Fermentas) or by a sequencing company, either BaseClear (The Netherlands) or GATC (Germany). GM3 and GM4 sequences were assembled to one sequence using CloneManager and manually curated and trimmed to 1,358 bp, after which BLASTn was used for identification against the 16S database. Mega6 software (25) was used for creating the alignment using Clustal (26), after which the neighbor-joining method (27) was used to create the phylogram. Bootstrap analysis (28) was performed using 1,000 replicate analyses.
Type strain sequences used for the phylogenetic tree were trimmed to the same 1,358 bp as the isolate sequences and were derived from the following GenBank accession numbers: NR_043021.1 (G. thermodenitrificans OHT-1 [=DSM 465]), NR_036987.1 (B. smithii DSM 4216), NR_109664.1 (B. thermocopriae SgZ7), NR_115727.1 (B. coagulans DSM 1), NR_026515.1 (A. pallidus DSM 3670), NR_043022.1 (G. thermoglucosidasius BGSC 95A1 [=G. thermoglucosidans DSM 2542]), and NR_028708.1 (G. caldoxylosilyticus DSM 12041).
Testing genetic accessibility.
Depending on their 16S rRNA gene-based genus identification, strains selected in the initial selection were subjected to a transformation protocol for either Geobacillus (24) or Bacillus (29), with slight modifications as follows. Strains were grown overnight at their isolation temperature in 10 ml medium in a 50-ml Greiner tube, and the next morning, cultures were diluted to an OD600 of 0.08 in 100 ml medium for Bacillus species and 50 ml medium for Geobacillus species. For strains belonging to the genus Bacillus, LB2 medium was used, and for strains belonging to the Geobacillus genus, TGP2 medium was used, with the exception of Geobacillus strains that grew better on LB2 (isolates ET 036, ET 050, ET 130, ET 144-2, and ET 261-3). Cultures were grown until they reached an OD600 of 0.45 to 0.65 for Bacillus species and around 1.00 for Geobacillus species. After the cultures were washed 3 times with SG buffer for Bacillus species (per liter, 171.2 g sucrose, 0.2 g MgCl2, 50 ml glycerol) or 4 times with electroporation buffer for Geobacillus species (per liter, 91 g sorbitol, 91 g mannitol, 100 ml glycerol), final pellets were resuspended in 240 μl SG buffer for Bacillus species and 900 μl electroporation buffer for Geobacillus species, after which cells were stored at −80°C or electroporation was performed directly. Electroporation settings were a voltage of 2.0 kV, a capacity of 25 μF, and a resistance of 200 Ω in a 2-mm cuvette (based on reference 30) for all strains. Additional settings were tested as 1.5 kV, 25 μF, and 600 Ω in a 1-mm cuvette (29) for Bacillus strains ET 138, ET 143, ET 224, ET 226, ET 229, ET 230-1, ET 239-2, ET 244, ET 261-1, and ET 263; 2.5 kV, 25 μF, and 600 Ω in a 1-mm cuvette (24) for Geobacillus strains ET 006, ET 011-1, ET 036, ET 042, ET 050, and ET 208; and 2.5 kV, 25 μF, and 600 Ω in a 2-mm cuvette (based on reference 24) for Geobacillus strains ET 006, ET 036, ET 042, ET 050, ET 072, ET 129, and ET 208. Recovery was performed at 3°C below the isolation temperatures for 3 h in RG2 medium (LB2 with 121 g/liter sucrose and 10 g/liter glucose) for Bacillus species and for 2 h in LB2 or TGP2 supplemented with 0.5% (wt/vol) glucose for Geobacillus species. G. thermoglucosidans DSM 2542T and B. coagulans DSM 1T were used as positive controls. All strains were tested with plasmid pNW33n (Bacillus Genetic Stock Centre), and 7 μg/ml chloramphenicol was added to the plates after transformation. Negative controls were performed by electroporating without adding any plasmid DNA.
Colonies appearing after transformation were streaked to a new plate and grown overnight, after which grown streaks were subjected to colony PCR as described above, using primers BG3464 (AACTCTCCGTCGCTATTGTAACCA) and BG3465 (TATGCGTGCAACGGAAGTGAC). To confirm transformation, positive colonies were inoculated into 10 ml LB2 or TGP2 supplemented with 7 μg/ml chloramphenicol in a 50-ml tube and grown overnight at their isolation temperature. From this culture, plasmid was extracted using the GeneJet plasmid miniprep kit (Fermentas) with 4 mg/ml lysozyme (Sigma, USA) added to the resuspension buffer, in which resuspended pellets were incubated for 30 min at 37°C. Extracted plasmid was used for PCR as described above and subjected to restriction analysis using 550 to 650 ng plasmid DNA with Fermentas enzymes StuI and HindIII for 2 h at 37°C. Plasmid extraction on nontransformed strains was used as a negative control. In the case of inconclusive band patterns, the isolated plasmid DNA was transformed to E. coli DH5α and subsequently reisolated and digested.
Fermentations.
Precultures of transformable isolates were grown overnight from glycerol stocks in 10 ml LB2 at 55°C in a 50-ml Greiner tube without shaking. The next morning, 10 ml culture was transferred to 40 ml TMMY medium supplemented with 30 g/liter glucose in a 250-ml Erlenmeyer flask and incubated at 55°C without shaking. When the cultures reached an OD600 of 0.3 to 0.7 (exponential growth phase), 20 ml was used to inoculate a reactor containing 1 liter TMMY medium without MOPS and supplemented with 30 g/liter glucose. Glass reactors of 2-liter volume were used (Applikon, The Netherlands) under the control of an ADI 1010 Bio-controller (Applikon) with an ADI 1025 Bio-console. Temperature was controlled at 55°C, stirring speed was 150 rpm, and pH was maintained at 6.5, unless stated otherwise, by addition of 3 M KOH. Growth was monitored off-line by absorbance at 600 nm (OD600), and sugar and fermentation products were measured by HPLC.
Fermentations of ET 138 at different pHs were performed in the same way as those described above, but overnight cultures were transferred to TMMY medium with the same pH as the reactor (either 4.5, 5.5, or 6.5), and pHs of the reactors were controlled at pH 4.5, 5.5, or 6.5.
For the comparison of ET 138 on glucose and xylose, precultures were inoculated from glycerol stock and grown overnight at 55°C in 10 ml TVMY medium supplemented with either 10 g/liter glucose or 10 g/liter xylose in a 50-ml tube at 150 rpm. The next morning, 1 ml culture was transferred to 50 ml of the same medium in a 250-ml Erlenmeyer flask at 55°C and 150 rpm. When this culture reached an OD600 between 0.5 and 0.7, 20 ml was used to inoculate a reactor. In the reactor, 1 liter TVMY medium without MOPS was used, supplemented with 20 or 25 g/liter xylose or glucose, as indicated.
Analytical techniques.
Sugars and fermentation products were quantified using an HPLC system from Thermo containing a P2000 pump, an AS3000 autosampler, a UV-visible (UV-Vis) 1000 detector, and an RI-150 refraction index detector. A Shodex RSpak KC-811 cation-exchange column was used with a mobile phase of 3 mM H2SO4 and was operated at 0.8 ml/min and 80°C. All samples were diluted 1:1 with 10 mM dimethylsulfoxide (DMSO) in 0.04 N H2SO4. d-Lactate and l-lactate were distinguished via enzymatic assay kits (MegaZyme D-LATE and K-DATE).
Unknown peaks observed in the HPLC spectrum in the xylose fermentation were collected from HPLC, freeze-dried, and analyzed by nuclear magnetic resonance (NMR) by Biqualys (The Netherlands). NMR identification was confirmed by running the corresponding standards on HPLC.
GenBank accession numbers.
All of the 16S rRNA gene sequences generated in this study have been submitted to the GenBank database under accession numbers KP010222 to KP010265.
RESULTS
Isolation, initial selection, and identification.
Thermophiles able to grow on minimal medium (TMM) at 55°C or 65°C were obtained from compost (Table 1). Single colonies (842) from the dilution series of compost samples were transferred to plates containing different carbon sources (glucose, sucrose, xylose, or arabinose). All colonies grew on glucose, and 584 colonies also grew on one or both of the C5 sugars xylose and arabinose at 55°C or 65°C. These 584 colonies then were inoculated into liquid TMM supplemented with a pH indicator under microaerobic conditions. Glycerol stocks were made of 267 strains that showed both growth and acidification in liquid culture on glucose (Table 1). To select the best acid-producing strains, all 267 isolates were inoculated in duplicate in liquid TMM supplemented with glucose, CaCO3, and 0.5 g/liter yeast extract and grown for 48 h under microaerobic conditions, after which HPLC analysis was performed. For each isolation condition, strains were found that either produced relatively high quantities of total products or showed products that were rarely found among strains isolated under the same conditions. In total, 35 strains were selected from the five different isolation conditions (Table 1; also see Table S1 in the supplemental material) to make pure cultures to screen for genetic accessibility. For 10 isolates (ET 011, ET 072, ET 129, ET 130, ET 131, ET 144, ET 230, ET 239, ET 261, and ET 267), purification revealed distinct morphologies and resulted in more than one pure culture from the same original colony (e.g., 144-1 and 144-2). The 44 pure cultures were identified by 16S rRNA gene sequence analysis (Fig. 1). The majority of strains were identified as Geobacillus thermodenitrificans, followed by Bacillus thermocopriae, Geobacillus caldoxylosilyticus, Bacillus coagulans, and Aeribacillus pallidus. One strain of Bacillus smithii was obtained. The highest species diversity was obtained with isolation condition 5, whereas it was the lowest under conditions 1 and 4, in which mainly G. thermodenitrificans strains were found. As expected, at 65°C, mainly Geobacillus species were obtained, whereas at 55°C, Bacillus species were also found and the species diversity was larger.
TABLE 1.
Overview of the number of isolates per step for each isolation condition
| Isolation condition (no.)a | Strain (ET) no. | No. of colonies selected from dilution series and plated on 4 sugars | No. of strains: |
||
|---|---|---|---|---|---|
| Growing on at least 3 sugarsb and inoculated into liquid medium | Acidified and stocked | Selected based on HPLC data and purifiedc | |||
| TMMscr, 65°C (1) | 001–054 | 257 | 217 | 53 | 5 |
| TMMscr + CaCO3, 65°C (2) | 055–131 | 260 | 151 | 77 | 5 |
| TMMscr + CaCO3, 55°C (3) | 132–157 | 65 | 45 | 26 | 7 |
| TMMglc + CaCO3, 65°C (4) | 158–213 | 130 | 77 | 56 | 5 |
| TMMglc + CaCO3, 55°C (5) | 214–267 | 130 | 94 | 55 | 13 |
| Total | 842 | 584 | 267 | 35 | |
Abbreviations: TMMscr, TMM containing sucrose; TMMglc, TMM containing glucose.
Tested sugars were d-glucose, d-xylose, l-arabinose, and sucrose. To be selected, strains should grow on at least one of the C5 sugars.
The final 35 selected strains were purified, resulting in 44 pure cultures, which subsequently were tested for genetic accessibility.
FIG 1.
Neighbor-joining tree of 16S rRNA gene sequences of selected isolates and type strains. The first number indicates the isolation round, whereas a T at that position indicates a type strain. Numbers at the nodes represent bootstrap values out of 1,000 replicates.
HPLC data for the tube test for the selected strains are shown in Table S1 in the supplemental material. Independent of the species, all strains produced mainly lactate, with acetate as the major by-product and succinate as a minor by-product. Gas production was not analyzed. All strains, except for Bacillus smithii, also produced small amounts of one or more other minor products, such as ethanol, 2,3-butanediol (2,3-BDO), formate, propionate, or malate. By-products varied not only between species but also between strains of the same species. For example, Geobacillus thermodenitrificans ET 236 and ET 251 produced malate, ethanol, formate, and 2,3-BDO, whereas G. thermodenitrificans ET 136 did not produce detectable amounts of malate and 2,3-BDO but did produce larger amounts of ethanol, and G. thermodenitrificans ET 241 did not produce any ethanol. Most B. thermocopriae strains produced propionate. The addition of CaCO3 in the tubes might have enhanced production of C4-dicarboxylic acids, but these were never observed as major products. The highest titer of these products was reached by strain G. caldoxylosilyticus ET 208, with 0.58 g/liter succinate and 0.18 g/liter malate, together constituting 15% of its total production, with its main products being lactate and acetate. The total amount of products varied strongly, from 1.27 g/liter to 7.61 g/liter. On average, strains isolated in condition 5 (glucose at 55°C with CaCO3) produced the most acids, while strains from condition 2 (sucrose at 65°C) produced the least. Strains isolated at 55°C produced, on average, more acid than strains isolated at 65°C. No difference was observed in average production between strains isolated on sucrose and glucose.
Screening for genetic accessibility by electrotransformation.
The 44 selected pure cultures were tested for their transformation efficiency by electroporation. Depending on the species, slightly modified versions of either a Bacillus (29) or a Geobacillus (24) protocol were used to prepare competent cells. Subsequently, the obtained cell suspensions were electroporated using E. coli-Geobacillus/Bacillus shuttle vector pNW33n. Strains that were successfully transformed with pNW33n were B. smithii strain ET 138 and G. thermodenitrificans strains ET 144-2 and ET 251. Transformation was confirmed by colony PCR and by isolating plasmid DNA from the transformed strains, which subsequently was confirmed to be pNW33n by restriction analysis of the isolated plasmid DNA from the transformants (G. thermodenitrificans strains ET 144-2 and ET 251) or from retransformed E. coli (B. smithii ET 138) (see Fig. S1 in the supplemental material). Transformation of G. thermodenitrificans strains ET 144-2 and ET 251 according to the Geobacillus protocol (with settings of 2.0 kV, 25 μF, and 200 Ω in a 2-mm cuvette) resulted in 6 and 4 colonies per μg DNA, respectively. B. smithii strain ET 138, made competent according to the Bacillus protocol and electroporation settings (29), yielded 3 colonies per μg DNA, whereas the Geobacillus electroporation settings yielded 33 colonies per μg DNA for this strain. Subsequent retesting of other Bacillus isolates with the Geobacillus settings did not result in any transformants. For strains ET 144-2 and ET 251, transformation was reproducible but not always successful, whereas transformation for ET 138 was highly reproducible despite the low efficiency. For 14 strains, occasionally colonies also appeared when no DNA was added during electroporation, while the strains did not grow on chloramphenicol prior to transformation (see Table S1 in the supplemental material). None of the colonies formed by these strains tested positive in the PCR on the pNW33n repB-cat gene combination. As transformation is crucial for the development of an organism into an efficient platform organism, only the three transformable strains were selected for further studies.
Fermentation and growth characteristics of transformable strains.
Transformable isolates B. smithii ET 138 and G. thermodenitrificans ET 144-2 and ET 251 were tested in 1-liter pH-controlled fermentations on glucose. Similar to the results of the acidifying tube tests, for all three strains, lactate was the major product and acetate the second most abundant product, with succinate being a minor by-product (Table 2). For all three strains, ethanol production was observed in one of the two duplicate fermentations, while this was not observed during the tube experiments for B. smithii ET 138 (see Table S1 in the supplemental material). G. thermodenitrificans ET 251 showed very low titers and productivity in the reactor, namely, a total of 6.58 ± 1.35 g/liter with 0.05 ± 0.01 g/liter/h (Table 2), even though it performed well in tubes (see Table S1). G. thermodenitrificans ET 144-2 produced a total of 14.80 ± 4.03 g/liter products but showed a rather low productivity of 0.08 ± 0.01 g/liter/h (Table 2). In contrast, B. smithii ET 138 outperformed the other strains both in product titer and productivity, with 19.35 ± 2.34 g/liter at 0.14 ± 0.01 g/liter/h (Table 2). As it also outperformed the other two isolates in transformation efficiency, B. smithii ET 138 was selected for further studies.
TABLE 2.
Fermentation performance of transformable isolatesa
| Strain | Time (h) | Amt of productb (g/liter) |
Avg productivityc (g/liter/h) | |||||
|---|---|---|---|---|---|---|---|---|
| Lac | Ace | Suc | Mal | Eth | Total | |||
| B. smithii ET 138 | 135 ± 8 | 18.15 ± 3.03 | 0.95 ± 0.45 | 0.09 ± 0.00 | ND | 0.16 ± 0.22 | 19.35 ± 2.34 | 0.14 ± 0.01 |
| G. thermodenitrificans ET 144-2 | 190 ± 70 | 13.22 ± 3.26 | 1.01 ± 0.16 | 0.29 ± 0.22 | 0.06 ± 0.08 | 0.21 ± 0.30 | 14.80 ± 4.03 | 0.08 ± 0.01 |
| G. thermodenitrificans ET 251 | 132 ± 11 | 5.70 ± 1.38 | 0.68 ± 0.05 | 0.15 ± 0.04 | ND | 0.04 ± 0.06 | 6.58 ± 1.35 | 0.05 ± 0.01 |
The results shown are the averages from duplicates ± standard deviations.
Abbreviations: Lac, lactate; Ace, acetate; Suc, succinate; Mal, malate; Eth, ethanol; ND, not detected.
Average productivity was calculated by dividing total products by fermentation time.
B. smithii ET 138 showed growth at the tested temperatures between 37 and 63°C, with optimal growth between 51 and 57°C and no growth at 66°C or higher. The optimum pH was found to be 6.5, with little difference between pH values of 6.0 to 7.0 (data not shown). In further experiments, growth conditions were kept at 55°C and pH 6.5. The lowest initial pH still supporting growth was 4.5. To further evaluate the ability of B. smithii ET 138 to ferment at low pH, fermentations in pH-controlled reactors were performed at pH 4.5, 5.5, and 6.5 (Table 3). During the first 43 h of fermentation, product titers and productivities at pH 5.5 and 6.5 were almost identical, with 10.3 g/liter at 0.24 g/liter/h and 10.8 g/liter at 0.25 g/liter/h, respectively. After 141 h, both titer and productivity were 1.5-fold lower at pH 5.5 than at pH 6.5 (Table 3). At pH 4.5, the strain still was able to grow and produce for 43 h, but titer and productivity were reduced to 3.6 g/liter and 0.08 g/liter/h. Hardly any acid was produced after 43 h, indicating severe acid stress (Table 3). At pH 5.5 and 6.5, the relative product distributions were equal, with 96.5% lactate, 3% acetate, and 0.5% succinate. At pH 4.5, this changed slightly to 91% lactate, 8% acetate, and 1% succinate.
TABLE 3.
Fermentation of B. smithii ET 138 at different pHs
| pH | Time (h) | Amt of product (g/liter)b |
Avg productivityc (g/liter/h) | |||
|---|---|---|---|---|---|---|
| Lac | Ace | Suc | Total | |||
| 4.5 | 43a | 3.3 | 0.3 | 0.0 | 3.6 | 0.08 |
| 5.5 | 43 | 9.9 | 0.3 | 0.1 | 10.3 | 0.24 |
| 141 | 14.0 | 0.6 | 0.1 | 14.7 | 0.10 | |
| 6.5 | 43 | 10.5 | 0.3 | 0.0 | 10.8 | 0.25 |
| 141 | 21.1 | 0.6 | 0.1 | 21.8 | 0.15 | |
At pH 4.5, no more acids were produced after 43 h, so no further time points are shown.
Abbreviations: Lac, lactate; Ace, acetate; Suc, succinate.
Average productivity was calculated by dividing total products by fermentation time.
We next evaluated the ability of B. smithii ET 138 to ferment xylose (Fig. 2; also see Fig. S2 in the supplemental material), which is the most abundant sugar in lignocellulose next to glucose. The medium was changed from TMMY to TVMY, and lower substrate concentrations of 20 g/liter were used to decrease browning of the medium with xylose, which was observed when using 30 or 25 g/liter xylose, probably due to a Maillard reaction. The growth curves on both substrates were highly similar (Fig. 2; also see Fig. S2). On both glucose and xylose, growth stopped after approximately 24 h, while acid production continued with only a small decrease in productivity, indicating that production is uncoupled from growth. The product profiles on both substrates were found to be similar, with lactate being the major product, i.e., 91% (13.9 g/liter) on xylose and 92% (17.46 g/liter) on glucose, with an optical purity of 99% l-lactate. On both sugars, acetate was the major by-product, followed by minor amounts of succinate and malate (Fig. 2). In the xylose fermentation, two unknown peaks were observed in the HPLC spectrum, and they were analyzed by NMR. One peak was identified as xylitol and was produced in amounts of approximately 1.5 g/liter. The other peak was likely to be an acetylated form of xylose but could not be further identified with certainty. Whereas glucose is completely consumed at the end of fermentation, xylose is not. This might be related to browning of the medium after approximately 70 h, which also was observed at 25 g/liter xylose concentrations (see Fig. S2) and was followed by a slight pyruvate accumulation.
FIG 2.
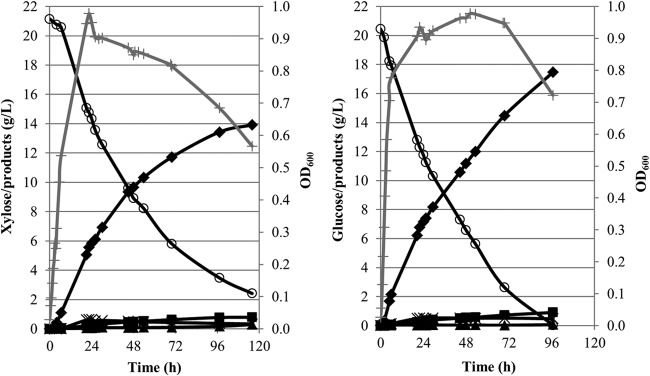
Fermentation of B. smithii ET 138 on xylose and glucose. Fermentation was carried out in 1 liter TVMY supplemented with 20 g/liter xylose or glucose at 55°C, pH 6.5, 150 rpm, and without any gas addition. Gray line with plus symbol, OD600; open circles, xylose or glucose; filled diamonds, lactate; closed squares, acetate; crosses, malate and succinate; closed triangles, pyruvate. During the xylose fermentation, the browning of the medium was observed after approximately 70 h.
Optimization of transformation for B. smithii ET 138 and other B. smithii strains.
The electroporation protocol for B. smithii ET 138 was optimized for increased transformation efficiencies. Out of 5 different tested electroporation settings (see Table S2 in the supplemental material), the best were found to be 2.0 kV, 25 μF, and 400 Ω in a 2-mm cuvette, resulting in 160 colonies per μg DNA (Table 4). The induction of antibiotic resistance by the addition of a sublethal (1,000× diluted) concentration of antibiotics after 2 h of recovery as described for B. coagulans (31) had no effect (see Table S2). In contrast, the recovery medium had a large impact. Changing the original RG2 recovery medium (LB2 with 121 g/liter sucrose and 10 g/liter glucose) to LB2 resulted in an increase in transformation frequencies to ∼1,000 colonies per μg DNA. The transformation efficiency could be further increased to ∼2,000 by growing the cells in larger flasks prior to making them competent to allow for more aeration and faster growth (Table 4). Substituting the SG buffer for the Geobacillus electroporation buffer strongly reduced efficiencies to 40 colonies per μg DNA (see Table S2). Lastly, we tested smaller amounts of added plasmid DNA. In all initial experiments, 1.0 to 2.5 μg DNA was added. Using only 20 ng DNA for transformation of B. smithii ET 138 resulted in a maximum efficiency of ∼5,000 colonies per μg DNA (Table 4). Storage of competent cells at −80°C was tested for periods of up to 1 year and did not affect the transformation efficiency.
TABLE 4.
Optimization of electrotransformation for B. smithii ET 138
| Parameter change | Final ODa | Period of growth (h)b | Electroporation setting |
Cuvette size (mm) | Amt (μg) of DNA | Recovery mediumc | CFUd | ||
|---|---|---|---|---|---|---|---|---|---|
| Voltage (kV) | μF | Ω | |||||||
| Settingse | 0.483 | 1.75 | 1.5 | 25 | 600 | 1 | 1 | RG2 | 3 |
| Settingsf | 0.483 | 1.75 | 2.0 | 25 | 200 | 2 | 1 | RG2 | 33 |
| Settings | 0.424 | 2.9 | 2.0 | 25 | 400 | 2 | 0.7 | RG2 | 157 |
| Recovery medium | 0.519 | 1.75 | 2.0 | 25 | 400 | 2 | 1 | LB2 | 960 |
| Recovery medium | 0.519 | 1.75 | 2.0 | 25 | 400 | 2 | 1 | RG2 | 5 |
| Fast growthg | 0.539 | 1 | 2.0 | 25 | 400 | 2 | 2.5 | LB2 | 1,900 |
| μg DNA addedg | 0.617 | 1.3 | 2.0 | 25 | 400 | 2 | 0.2 | LB2 | 2,409 |
| μg DNA addedg | 0.617 | 1.3 | 2.0 | 25 | 400 | 2 | 0.02 | LB2 | 5,118 |
Final OD600 after the indicated number of hours when growing cells prior to making them competent.
Number of hours cells had grown before making them competent.
RG2 is LB2 with 121 g/liter sucrose and 10 g/liter glucose (29).
CFU indicate CFU per μg DNA. Using fresh cells or cells stored at −80°C did not change CFU counts.
Settings from reference 29.
Settings are based on reference 30, the same settings as those used for screening of Geobacillus strains.
In these experiments, after overnight growth, cells were transferred into 500-ml Erlenmeyer flasks or 1-liter bottles (having a similar bottom surface) instead of into a 250-ml Erlenmeyer flask to allow for more aeration.
We also tested B. smithii strains from the DSMZ culture collection for transformation to evaluate their genetic accessibility. Using the optimized protocol for ET 138, a transformation efficiency of 10 to 100 colonies per μg plasmid DNA was obtained for type strain DSM 4216T, which is approximately 20 to 200 times lower than that for strain ET 138 under the same conditions. Also for DSM 4216T, different electroporation conditions and recovery media were tested (data not shown). For DSM 4216T, the difference between recovery medium LB2 and RG2 was less pronounced than that for ET 138. The best settings for DSM 4216T were 1.5 kV, 25 μF, and 600 Ω in a 1-mm cuvette with recovery for 3 h in LB2, yielding 10 to 200 colonies per μg plasmid DNA. Under the tested conditions, transformation for the type strain was found to be less reproducible than that for isolate ET 138. The optimal settings for type strain DSM 4216T and isolate ET 138 were used to evaluate the transformation of 5 more publically available B. smithii strains. Of those strains, only DSM 460 generated transformants (25 colonies per μg DNA), but no attempts for further optimization were made. The best and most reproducibly transformable B. smithii strain so far is our isolate ET 138, with a maximum of ∼5,000 colonies per μg DNA.
Similar attempts to increase transformation efficiencies by changing electroporation settings and buffers for ET 144-2 were unsuccessful (data not shown). For strain ET 251, no transformation optimization was performed, because this strain showed poor fermentation performance (Table 2).
DISCUSSION
In this study, we aimed at isolating thermophilic bacilli that are genetically accessible, and our isolation conditions were aimed at finding industrially relevant strains: minimal medium was used, the temperature was set at moderately thermophilic temperatures, both microaerobic and anaerobic conditions were applied during the isolation procedure to find facultative anaerobes, and utilization of both C5 and C6 sugars was evaluated. Using 5 different isolation conditions, we isolated 267 strains from compost; of these, 44 were found to belong to the family Bacillaceae by 16S rRNA gene sequencing. The observed abundance of the genera Bacillus and Geobacillus (notably G. thermodenitrificans) among the isolates from the thermophilic compost is in line with a recent report describing the microbial succession during thermophilic composting (32). In contrast to Li et al., we did not find any Ureibacillus species or B. licheniformis but isolated several B. coagulans strains and one B. smithii strain (32). These differences might be explained by the fact that for isolation, we used a minimal medium and compost that consisted solely of plant material, whereas the other study made use of rich LB medium and compost that contained both plant material and manure (32, 33).
Product formation on glucose was evaluated for all our isolates. In general, the product profiles are in line with other reports on thermophilic bacilli (23, 33, 34). Whereas these studies mainly focused on a particular species or strain, our data provide a comparison of several thermophilic Bacillus species and strains grown under the same conditions. This revealed homogeneity in the main products lactate and acetate, whereas a large diversity was observed in the formation of by-products between species as well as between strains of the same species. This diversity creates opportunities for the use of these organisms for the production of different chemicals and their use as platform organisms or sources of enzymes and pathways. G. thermoglucosidans DSM 2542T and B. coagulans DSM 1T were included as benchmarks, because these two organisms have been studied for production of ethanol (35) and lactate (31), respectively, and both are genetically accessible (24, 31, 36). B. smithii ET 138 was the only strain from our study that had genetic accessibility and product titers comparable to those of the benchmark organisms.
To the best of our knowledge, this is the first large-scale screen of genetic accessibility among different species of thermophilic bacilli. Although several thermophilic Bacillus species have been described to be genetically accessible (11, 24, 29, 31, 37–39), often laborious protocols, such as protoplast transformation, are required. Moreover, for each species, highly variable transformation efficiencies have been reported for a limited number of strains, suggesting that genetic accessibility is very strain specific. Strain specificity also is implicated by Patel et al., who reported only one of their B. coagulans isolates to be genetically accessible (34). This is supported by our findings: in the first isolation round, only 2 out of 25 tested G. thermodenitrificans isolates were genetically accessible, and no G. stearothermophilus or B. coagulans strains were transformable under the tested conditions. After testing the electrotransformation for all selected isolates, 3 strains were transformable: B. smithii ET 138 and G. thermodenitrificans ET 144-2 and ET 251, of which ET 138 was the most reproducible. A second isolation round, in which we enriched strains in reactors, resulted in four G. thermoglucosidans strains that could be reproducibly transformed and one G. toebii strain that was not reproducibly transformable (data not shown). Further optimization of the protocols per strain or the use of other transformation methods, such as protoplast transformation or conjugation, might increase the number of transformable strains, but for high-throughput engineering and use as a platform organism, electroporation is a fast and convenient method. As B. smithii ET 138 outperformed the other strains in both fermentation and transformation via simple electroporation, we focused optimization experiments on this strain. The optimized electroporation efficiency of B. smithii ET 138 of 5 × 103 using pNW33n is close to that found in B. coagulans P4-102B with 1 × 104 using pNW33n (29) and G. thermoglucosidans with 1 × 104 using E. coli-Geobacillus shuttle vector pUCG18 (24).
B. smithii originally was reclassified from B. coagulans (40), but contrary to B. coagulans, genetic accessibility has not been reported previously. B. smithii has been reported to produce several biotechnologically interesting enzymes, such as nitrile hydratase (41), endoinulinase (42), and alkaline lipase (43). Its biotechnological potential was further reported in an isolation study targeted at xylose-utilizing, ethanol-tolerant ethanol producers (44), as well as during lignocellulose-based composting, where B. smithii was among three bacterial compost isolates showing ligninolytic activity (45). Several B. smithii strains were isolated from a sugar beet factory and shown to harbor glycosylated S-layer proteins and to secrete lactic acid as their main product (46). The sugar beet factory isolates showed temperature curves ranging from 37°C to 65°C, although not all strains grew at 37°C and the type strain did not grow at 65°C. The sugar beet extraction plant that was used as the isolation source was around pH 4.5, indicating acid tolerance of the strains (46). In line with these findings, our isolated strain, ET 138, grows between 37°C and 65°C, with highly similar growth rates between 51 and 57°C, and it is still well able to grow and produce acids at pH 5.5 and even to some extent at pH 4.5. Such robustness against temperature and pH fluctuations is an important advantage in industrial applications to increase process stability. Another useful industrial property is its capacity to uncouple growth and acid production. Its robustness, combined with the ability to efficiently utilize C5 and C6 sugars present in lignocellulosic substrates and being genetically accessible, makes B. smithii an attractive biocatalyst for lactic acid production, having the potential to be developed into a novel platform organism. Genome sequencing currently is ongoing, as well as the development of genetic tools to allow its further exploitation by metabolic engineering.
Conclusion.
Thermophilic bacilli able to ferment glucose and xylose can readily be isolated from compost. All produce lactic acid and acetic acid under microaerobic conditions, while the production of other products, such as succinate, malate, ethanol, formate, 2,3-butanediol, and propionate, is variable. Genetic accessibility via electroporation is scarce and strain dependent. We have isolated B. smithii ET 138 and established an efficient transformation protocol. The combination of fermentation robustness and genetic accessibility makes this strain an attractive biocatalyst and candidate for further development into a platform organism for production of green chemicals from renewable resources.
Supplementary Material
ACKNOWLEDGMENTS
We thank Willem van Harten from Recom Ede for allowing us to sample their compost. We are grateful to Jacques Vervoort of Biqualis for discussions and NMR analyses and to Ron Winkler from Dutch Technology Foundation STW for his involvement in an earlier phase of this work.
This work was supported financially by Corbion.
We have no conflicts of interest to declare. R.V.K. is employed by the commercial company Corbion (Gorinchem, The Netherlands).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03640-14.
REFERENCES
- 1.Chen X, Zhou L, Kangming T, Kumar A, Singh S, Prior BA, Wang Z. 2013. Metabolic engineering of Escherichia coli: a sustainable industrial platform for bio-based chemical production. Biotechnol Adv 31:1200–1223. doi: 10.1016/j.biotechadv.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Hong K-K, Nielsen J. 2012. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci 69:2671–2690. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buschke N, Schäfer R, Becker J, Wittmann C. 2013. Metabolic engineering of industrial platform microorganisms for biorefinery applications–optimization of substrate spectrum and process robustness by rational and evolutive strategies. Bioresour Technol 135:544–554. doi: 10.1016/j.biortech.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 4.Blumer-Schuette SE, Brown SD, Sander KB, Bayer EA, Kataeva I, Zurawski JV, Conway JM, Adams MWW, Kelly RM. 2014. Thermophilic lignocellulose deconstruction. FEMS Microbiol Rev 38:393–448. doi: 10.1111/1574-6976.12044. [DOI] [PubMed] [Google Scholar]
- 5.Ou MS, Mohammed N, Ingram LO, Shanmugam KT. 2009. Thermophilic Bacillus coagulans requires less cellulases for simultaneous saccharification and fermentation of cellulose to products than mesophilic microbial biocatalysts. Appl Biochem Biotechnol 155:379–385. doi: 10.1007/s12010-008-8509-4. [DOI] [PubMed] [Google Scholar]
- 6.Podkaminer KK, Kenealy WR, Herring CD, Hogsett DA, Lynd LR. 2012. Ethanol and anaerobic conditions reversibly inhibit commercial cellulase activity in thermophilic simultaneous saccharification and fermentation (tSSF). Biotechnol Biofuels 5:43. doi: 10.1186/1754-6834-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosma EF, van der Oost J, de Vos WM, van Kranenburg R. 2013. Sustainable production of bio-based chemicals by extremophiles. Curr Biotechnol 2:360–379. doi: 10.2174/18722083113076660028. [DOI] [Google Scholar]
- 8.Taylor MP, Eley KL, Martin S, Tuffin MI, Burton SG, Cowan DA. 2009. Thermophilic ethanologenesis: future prospects for second-generation bioethanol production. Trends Biotechnol 27:398–405. doi: 10.1016/j.tibtech.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Ingram LO, Shanmugam KT. 2011. Evolution of D-lactate dehydrogenase activity from glycerol dehydrogenase and its utility for D-lactate production from lignocellulose. Proc Natl Acad Sci U S A 108:18920–18925. doi: 10.1073/pnas.1111085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma K, Maeda T, You H, Shirai Y. 2014. Open fermentative production of l-lactic acid with high optical purity by thermophilic Bacillus coagulans using excess sludge as nutrient. Bioresour Technol 151:28–35. doi: 10.1016/j.biortech.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Chen T, Zhao X, Chamu J. 2012. Metabolic engineering of thermophilic Bacillus licheniformis for chiral pure D-2,3-butanediol production. Biotechnol Bioeng 109:1610–1621. doi: 10.1002/bit.24427. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Zhang L, Li K, Wang Y, Gao C, Han B, Ma C, Xu P. 2013. A newly isolated Bacillus licheniformis strain thermophilically produces 2,3-butanediol, a platform and fuel bio-chemical. Biotechnol Biofuels 6:123. doi: 10.1186/1754-6834-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cripps RE, Eley K, Leak DJ, Rudd B, Taylor M, Todd M, Boakes S, Martin S, Atkinson T. 2009. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab Eng 11:398–408. doi: 10.1016/j.ymben.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Shao X, Jin M, Guseva A, Liu C, Balan V, Hogsett D, Dale BE, Lynd L. 2011. Conversion for Avicel and AFEX pretreated corn stover by Clostridium thermocellum and simultaneous saccharification and fermentation: insights into microbial conversion of pretreated cellulosic biomass. Bioresour Technol 102:8040–8045. doi: 10.1016/j.biortech.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 15.van der Veen D, Lo J, Brown S, Johnson C, Tschaplinski T, Martin M, Engle N, den Berg R, Argyros A, Caiazza N, Guss A, Lynd L. 2013. Characterization of Clostridium thermocellum strains with disrupted fermentation end-product pathways. J Ind Microbiol Biotechnol 40:725–734. doi: 10.1007/s10295-013-1275-5. [DOI] [PubMed] [Google Scholar]
- 16.Chung D, Cha M, Guss AM, Westpheling J. 2014. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc Natl Acad Sci U S A 111:8931–8936. doi: 10.1073/pnas.1402210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willquist K, van Niel EWJ. 2012. Growth and hydrogen production characteristics of Caldicellulosiruptor saccharolyticus on chemically defined minimal media. Int J Hydrogen Energy 37:4925–4929. doi: 10.1016/j.ijhydene.2011.12.055. [DOI] [Google Scholar]
- 18.Svetlitchnyi V, Kensch O, Falkenhan D, Korseska S, Lippert N, Prinz M, Sassi J, Schickor A, Curvers S. 2013. Single-step ethanol production from lignocellulose using novel extremely thermophilic bacteria. Biotechnol Biofuels 6:31. doi: 10.1186/1754-6834-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao S, Mikkelsen MJ. 2010. Metabolic engineering to improve ethanol production in Thermoanaerobacter mathranii. Appl Microbiol Biotechnol 88:199–208. doi: 10.1007/s00253-010-2703-3. [DOI] [PubMed] [Google Scholar]
- 20.Saripan AF, Reungsang A. 2013. Biohydrogen production by Thermoanaerobacterium thermosaccharolyticum KKU-ED1: culture conditions optimization using xylan as the substrate. Int J Hydrogen Energy 38:6167–6173. doi: 10.1016/j.ijhydene.2012.12.130. [DOI] [Google Scholar]
- 21.Bhandiwad A, Shaw AJ, Guss A, Guseva A, Bahl H, Lynd LR. 2014. Metabolic engineering of Thermoanaerobacterium saccharolyticum for n-butanol production. Metab Eng 21:17–25. doi: 10.1016/j.ymben.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Taylor M, van Zyl L, Tuffin I, Leak D, Cowan D. 2011. Genetic tool development underpins recent advances in thermophilic whole-cell biocatalysts. Microb Biotechnol 4:438–448. doi: 10.1111/j.1751-7915.2010.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fong J, Svenson C, Nakasugi K, Leong C, Bowman J, Chen B, Glenn D, Neilan B, Rogers P. 2006. Isolation and characterization of two novel ethanol-tolerant facultative-anaerobic thermophilic bacteria strains from waste compost. Extremophiles 10:363–372. doi: 10.1007/s00792-006-0507-2. [DOI] [PubMed] [Google Scholar]
- 24.Taylor MP, Esteban CD, Leak DJ. 2008. Development of a versatile shuttle vector for gene expression in Geobacillus spp. Plasmid 60:45–52. doi: 10.1016/j.plasmid.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 29.Rhee MS, Kim JW, Qian Y, Ingram LO, Shanmugam KT. 2007. Development of plasmid vector and electroporation condition for gene transfer in sporogenic lactic acid bacterium, Bacillus coagulans. Plasmid 58:13–22. doi: 10.1016/j.plasmid.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Narumi I, Sawakami K, Nakamoto S, Nakayama N, Yanagisawa T, Takahashi N, Kihara H. 1992. A newly isolated Bacillus stearothermophilus K1041 and its transformation by electroporation. Biotechnol Tech 6:83–86. doi: 10.1007/BF02438695. [DOI] [Google Scholar]
- 31.Van Kranenburg R, Van Hartskamp M, Heintz E, Anthonius J, Van Mullekom E, Snelders J. February 2007. Genetic modification of homolactic thermophilic Bacilli. WO patent WO/2007/085443.
- 32.Li R, Li L, Huang R, Sun Y, Mei X, Shen B, Shen Q. 2014. Variations of culturable thermophilic microbe numbers and bacterial communities during the thermophilic phase of composting. World J Microbiol Biotechnol 30:1737–1746. doi: 10.1007/s11274-013-1593-9. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Zhao X, Chamu J, Shanmugam KT. 2011. Isolation, characterization and evolution of a new thermophilic Bacillus licheniformis for lactic acid production in mineral salts medium. Bioresour Technol 102:8152–8158. doi: 10.1016/j.biortech.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Patel MA, Ou MS, Harbrucker R, Aldrich HC, Buszko ML, Ingram LO, Shanmugam KT. 2006. Isolation and characterization of acid-tolerant, thermophilic bacteria for effective fermentation of biomass-derived sugars to lactic acid. Appl Environ Microbiol 72:3228–3235. doi: 10.1128/AEM.72.5.3228-3235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atkinson A, Cripps R, Eley K, Rudd B, Todd M. September 2008. Thermophilic micro-organisms for ethanol production. WO patent WO2008/038019 A3.
- 36.Kovacs AT, van Hartskamp M, Kuipers OP, van Kranenburg R. 2010. Genetic tool development for a new host for biotechnology, the thermotolerant bacterium Bacillus coagulans. Appl Environ Microbiol 76:4085–4088. doi: 10.1128/AEM.03060-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanchard K, Robic S, Matsumura I. 2014. Transformable facultative thermophile Geobacillus stearothermophilus NUB3621 as a host strain for metabolic engineering. Appl Microbiol Biotechnol 98:6715–6723. doi: 10.1007/s00253-014-5746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao C, Xue Y, Ma Y. 2011. Protoplast transformation of recalcitrant alkaliphilic Bacillus sp. with methylated plasmid DNA and a developed hard agar regeneration medium. PLoS One 6:e28148. doi: 10.1371/journal.pone.0028148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki H, Yoshida KI. 2012. Genetic transformation of Geobacillus kaustophilus hta426 by conjugative transfer of host-mimicking plasmids. J Microbiol Biotechnol 22:1279–1287. doi: 10.4014/jmb.1203.03023. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura LK, Blumenstock I, Claus D. 1988. Taxonomic study of Bacillus coagulans Hammer 1915 with a proposal for Bacillus smithii sp. nov. Int J Syst Bacteriol 38:63–73. doi: 10.1099/00207713-38-1-63. [DOI] [Google Scholar]
- 41.Takashima Y, Kawabe T, Mitsuda S. 2000. Factors affecting the production of nitrile hydratase by thermophilic Bacillus smithii SC-J05-1. J Biosci Bioeng 89:282–284. doi: 10.1016/S1389-1723(00)88835-0. [DOI] [PubMed] [Google Scholar]
- 42.Gao W, Bao Y, Liu Y, Zhang X, Wang J, An L. 2009. Characterization of thermo-stable endoinulinase from a new strain Bacillus smithii T7. Appl Biochem Biotechnol 157:498–506. doi: 10.1007/s12010-008-8313-1. [DOI] [PubMed] [Google Scholar]
- 43.Lailaja VP, Chandrasekaran M. 2013. Detergent compatible alkaline lipase produced by marine Bacillus smithii BTMS 11. World J Microbiol Biotechnol 29:1349–1360. doi: 10.1007/s11274-013-1298-0. [DOI] [PubMed] [Google Scholar]
- 44.Qi X, Zhang Y, Tu R, Lin Y, Li X, Wang Q. 2011. High-throughput screening and characterization of xylose-utilizing, ethanol-tolerant thermophilic bacteria for bioethanol production. J Appl Microbiol 110:1584–1591. doi: 10.1111/j.1365-2672.2011.05014.x. [DOI] [PubMed] [Google Scholar]
- 45.Jurado M, López MJ, Suárez-Estrella F, Vargas-García MC, López-González JA, Moreno J. 2014. Exploiting composting biodiversity: study of the persistent and biotechnologically relevant microorganisms from lignocellulose-based composting. Bioresour Technol 162:283–293. doi: 10.1016/j.biortech.2014.03.145. [DOI] [PubMed] [Google Scholar]
- 46.Messner P, Scheberl A, Schweigkofler W, Hollaus F, Rainey FA, Burghardt J, Prillinger H. 1997. Taxonomic comparison of different thermophilic sugar beet isolates with glycosylated surface layer (S-layer) proteins and their affiliation to Bacillus smithii. Syst Appl Microbiol 20:559–565. doi: 10.1016/S0723-2020(97)80027-6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.