Summary
In placental mammals, balanced expression of X-linked genes is accomplished by X chromosome inactivation (XCI) in female cells. In humans, random XCI is initiated early during embryonic development. To investigate whether reprogramming of female human fibroblasts into induced pluripotent stem cells (iPSCs) leads to reactivation of the inactive X chromosome (Xi), we have generated iPSC lines from fibroblasts heterozygous for large X-chromosomal deletions. These fibroblasts show completely skewed XCI of the mutated X chromosome, enabling monitoring of X chromosome reactivation (XCR) and XCI using allele-specific single-cell expression analysis. This approach revealed that XCR is robust under standard culture conditions, but does not prevent reinitiation of XCI, resulting in a mixed population of cells with either two active X chromosomes (Xas) or one Xa and one Xi. This mixed population of XaXa and XaXi cells is stabilized in naive human stem cell medium, allowing expansion of clones with two Xas.
Graphical Abstract

Highlights
-
•
Robust X chromosome reactivation in human iPSCs with large X-chromosomal deletions
-
•
Female human iPSCs with two active X chromosomes
-
•
Expansion of human iPSCs with two active X chromosomes in naive human stem cell medium
In this article, Gribnau and colleagues used fibroblasts, obtained from female carriers heterozygous for large X-chromosomal deletions, to generate human induced pluripotent stem cells (hiPSCs) and show that reprogramming leads to robust reactivation of the inactive X chromosome.
Introduction
Inactivation of one of the two X chromosomes in eutherian female cells by X chromosome inactivation (XCI) is an epigenetic process, which compensates for potential dosage differences of X-linked genes between female XX and male XY cells (Lyon, 1961). Mechanistic and regulatory aspects of XCI have been extensively studied during mouse development and for mouse embryonic stem cells (mESCs). These mESCs are derived from the inner cell mass (ICM) of the blastocyst and contain two active X chromosomes (Xa), but will undergo XCI upon in vitro differentiation. The noncoding Xist RNA is crucial for XCI and becomes upregulated upon differentiation of mESCs. Xist coats the future Xi, attracting chromatin remodeling enzymes that infer the transcriptional shutdown of the Xi (reviewed in Barakat and Gribnau, 2012; Pollex and Heard, 2012). Several components of the regulatory network driving XCI are conserved between mice and humans, but many questions regarding human XCI remain unanswered. In contrast to undifferentiated mESCs, most human ESC lines (hESCs) are in a post-XCI state and are prone to epigenetic fluidity (Silva et al., 2008). This variation in regulation and stability of the XCI state between these eutherian species might reflect suboptimal culture conditions for the human cells, resulting in a gradual progression toward a more differentiated state, including initiation of XCI. Alternatively, the XCI process itself may have reached a more advanced state in the human ICM compared with the mouse so that XCI in the hESCs derived from the ICM has occurred already prior to or during ESC derivation.
The derivation of human induced pluripotent stem cells (hiPSCs) from fibroblasts (Takahashi et al., 2007) offers new opportunities to study XCI in human cells. For mouse fibroblasts, it has been shown that the Xi becomes reactivated during the reprogramming process, followed by random XCI (rXCI) upon differentiation of these miPSCs (Maherali et al., 2007; Stadtfeld et al., 2008). Similar to studies involving hESC lines, previous studies of XCI in hiPSCs have provided varying results. Systematic analysis of multiple female hiPSC lines derived from several fibroblast populations under different reprogramming strategies indicated that all hiPSC lines retained the Xi inherited from the starting fibroblasts (Amenduni et al., 2011; Ananiev et al., 2011; Cheung et al., 2011; Tchieu et al., 2010). In another study, it was found that in all hiPSC lines derived from one fibroblast population with established rXCI, one and the same X chromosome had become the Xi in all lines, indicating involvement of cell selection processes (Pomp et al., 2011). In contrast, other studies showed reactivation of the Xi, an apparent reversal of XCI that is herein referred to as X chromosome reactivation (XCR), in all or a limited number of hiPSC lines, but XCI was reinitiated upon differentiation of these hiPSC lines (Bruck and Benvenisty, 2011; Kim et al., 2011; Marchetto et al., 2010). XCR followed by reinitiation of XCI and stable establishment of the Xi upon hiPSC differentiation is a crucial step that needs to take place for hiPSCs to be applied for various purposes. If hiPSC lines do not pass through this series of events, they show signs of stochastic reactivation of the Xi inherited from the founder fibroblasts (Mekhoubad et al., 2012). This erosion of XCI is detrimental for studies involving cell types generated from female hiPSCs, as it can be expected that many of these cell types will be prone to gene dosage inequalities. Therefore, the availability of such hiPSC lines with stable XCR, having two active X chromosomes as in mESCs, would greatly advance research on modeling of X-linked human diseases and studies on regulatory mechanisms of human XCI.
The varying results regarding XCR and XCI obtained for hiPSCs may be explained by different reprogramming techniques and the growth conditions in which hiPSCs are generated and maintained. In a recent study, it was found that growth of hESCs and hiPSCs in defined conditions (naive human stem cell medium [NHSM]) results in more naive iPSCs and leads to efficient loss of Xi specific markers, including XIST RNA and Xi-specific histone modifications, which are re-established upon differentiation (Gafni et al., 2013). Although these NHSM-cultured hESCs and hiPSCs resemble mESCs and miPSCs in several aspects, it is unclear whether the described loss of Xi-specific markers under these growth conditions has resulted in XCR, as expression of X-linked loci was not assessed (Gafni et al., 2013).
Skewed rXCI is obtained in cells that carry a large X chromosomal deletion on either one of the two X chromosomes outside the X inactivation center (XIC), based on selection against cells that inactivate the intact X chromosome. Here, we have generated and analyzed hiPSC lines derived from female carriers with such heterozygous large X chromosomal deletions, which were expanded under different growth conditions (standard versus NHSM). These hiPSCs provide a powerful model system to study XCR and XCI, facilitated by allele-specific expression analysis. Single-cell expression analysis of these hiPSC lines, when generated under standard culture conditions, revealed robust XCR, which was followed by XCI. Hence, the presence of two active X chromosomes in these hiPSCs is not a stable condition, as it is in undifferentiated mESCs. However, growth of the present hiPSC lines in NHSM conditions did stabilize the equilibrium between XCR and XCI over several passages and allowed rapid expansion of hiPSC lines with two active X chromosomes.
Results
Cell Lines with Non-rXCI for hiPSC Formation
To generate novel hiPSC lines to investigate XCR and XCI during reprogramming at a single-cell level, we screened cell repositories for female cell lines harboring large deletions on one of the X chromosomes. Because of secondary selection, an X chromosome with a large deletion is preferentially inactivated in female carriers, providing a sensitive assay to identify hiPSC lines that have reactivated the Xi inherited from the founder fibroblasts, which results in biallelic expression of polymorphic informative genes.
Cell lines were collected through different sources, and the gene content was further characterized by multiplex ligation-dependent probe amplification, single nucleotide polymorphism (SNP) array, and DNA-FISH analysis (Figures 1A, 1B and S1; Table S1). The fibroblast lines X12, X14, and X15 containing informative large deletions of Xq were selected for further studies examining XCR and XCI. The respective deletions concern regions outside the XIC. Deletions including the XIC would block XCI of the X chromosome carrying the deletion, whereas large deletions that do not include the XIC will lead to clonal selection of the cells that have inactivated the mutant X chromosome, keeping the intact X chromosome as the Xa. In addition, 46,XX fibroblasts not carrying an X-chromosomal deletion and 47,XXX fibroblasts derived from a patient with triple X syndrome were included as controls (Brosens et al., 2014). RNA-FISH analysis indicated that in fibroblast cell lines X12, X14, and X15, 99% of the cells showed one XIST cloud per nucleus (Figure 1C; n = 200 cells analyzed per cell line). In the 47,XXX fibroblasts, two XIST clouds marking two inactive X chromosomes were found in almost every cell (Figure 1C), in agreement with the rule that all but one X chromosome becomes inactivated per diploid genome (Harnden, 1961). HUMARA analysis amplifies a polymorphic region in the androgen receptor (AR) gene after digestion with a methylation sensitive enzyme and is used to detect the methylated Ar allele marking the Xi (Allen et al., 1994). This analysis indicated that rXCI is completely skewed toward inactivation of one of the two X chromosomes, for all three X12, X14, and X15 fibroblast cell lines (Table S1). Both in the 46,XX and the 47,XXX fibroblast cell lines, rXCI without such skewing was observed (Table S1) (Brosens et al., 2014). Preferential inactivation of one specific X chromosome in fibroblast line X12 was confirmed by RFLP RT-PCR analysis of a SNP in XIST (Figure 1D). Although no informative SNPs were available for XIST in the X14 and X15 fibroblast cell lines, analysis of X-linked genes showed completely skewed XCI in both cell lines (Figure S1D). We conclude that the X12, X14, and X15 fibroblast cell lines show highly skewed rXCI, most likely silencing the X carrying the deletion.
Figure 1.

Fibroblast Cell Lines with Skewed rXCI
(A) Schematic representation of the human X chromosome. The location of genes analyzed in this study is indicated. The dashed lines indicate the deletions in the X12, X14, and X15 fibroblast cell lines. Also DNA-FISH probes are indicated.
(B) DNA-FISH on metaphase chromosomes of X12 and X15 fibroblasts, using probes 1, 2, 3, and 4. Magnification of X chromosomes is shown in the insets.
(C) RNA-FISH detecting XIST RNA (FITC) in X12, X14, X15, and 47,XXX fibroblasts. DNA is counterstained with DAPI (blue, scale bar represents 10 μm).
(D) PCR and RT-PCR analysis with DNA and cDNA from X12, X14, and X15 fibroblast cell lines, amplifying an RFLP in XIST. PCR products were digested with MfeI to discriminate between both alleles.
Generation of hiPSC Lines
To generate hiPSC lines, X12, X14, X15, 46,XX, and 47,XXX fibroblasts were transduced with a polycistronic lentiviral vector expressing OCT4, SOX2, KLF4, and MYC, and a dTomato reporter, under the control of a retroviral promoter (SFFV) that is rapidly silenced during the reprogramming process (Warlich et al., 2011) (Figures 2A and 2B). These fibroblasts were plated on mouse embryonic fibroblasts and cultured in the presence of standard hESC medium. After approximately 10 days, small clusters of cells appeared that started to develop an hESC morphology. These clusters gradually lost the expression of the dTomato reporter (Figures 2B and S2A), which indicated proper silencing of the lentiviral transgene, required to establish fully reprogrammed hiPSCs. For each one of the different genotypes, we obtained several iPSC lines, of which the X12, X15, 46,XX, and 47,XXX iPSC lines were subjected to further analysis. All of these iPSC lines showed morphology resembling hESCs, with a stable 46,XX karyotype (or 47,XXX in case of lines derived from the 47,XXX fibroblasts) and expression of key endogenous pluripotency factors, including NANOG and REX1, at different passages (p) after establishment (Figures 2C–2E, S2C, and S2D; data not shown). Also, SNP arrays of various hiPSC lines confirmed an identical genome content compared with the founder fibroblasts (Figure S2D). Upon embryoid body (EB) differentiation, qPCR analysis indicated that most hiPSC lines showed expression of endodermal (FLK1, AFP), mesodermal (GATA2), and ectodermal (PAX6) marker genes (Figure 2G, only X12 clones are shown). This was confirmed by immunocytochemistry analysis on EB-differentiated iPSCs plated on slides, revealing GFAP (ectoderm), Vimentin (mesoderm), and AFP-positive cells for all iPSC lines, which supports our conclusion that all iPSC clones were completely reprogrammed and acquired full differentiation potential (Figure S2B; results of one representative X12 clone are shown).
Figure 2.
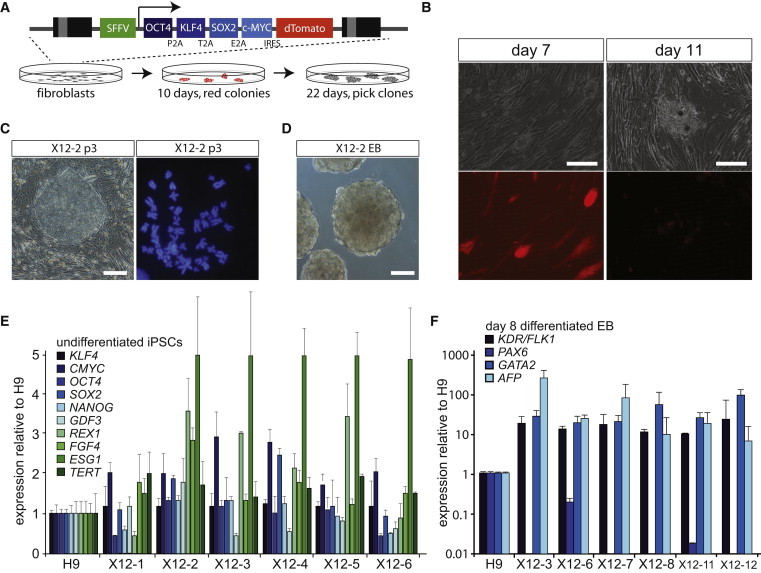
Generation of hiPSC Lines from Fibroblasts
(A) Schematic overview of the lentiviral reprogramming cassette (Warlich et al., 2011) and the procedure at 10 and 22 days after transduction, to establish the present hiPSC lines.
(B) Transduced fibroblasts express dTomato (left), which is silenced upon proper reprogramming (right, scale bar represents 100 μm).
(C) Representative picture of a hiPSC colony (p3 denotes passage 3) from iPSC line X12-2 (left, scale bar represents 100 μm), and karyogram of iPSC X12-2, revealing a 46,XX karyotype (right).
(D) Representative picture of EBs derived from iPSC line X12-2 after 8 days of differentiation (scale bar represents 100 μm).
(E) qRT-PCR analysis of pluripotency factors in X12 hiPSC lines (1–6). Expression of the same factors in an hESC line (H9) served as a control and was set at 100%.
(F) qRT-PCR analysis of differentiation associated markers in day 8 (d8) EBs from X12 hiPSC lines. Gene expression was normalized to a differentiated hESC line H9.
Early Passage Female hiPSCs Show Reactivation of the Xi
The X12, X15, and 47,XXX iPSCs were subjected to immuno-RNA-FISH analysis at passages 3–5 to investigate the XCI status of these cell lines. This analysis showed that in several iPSC lines generated from the X12 and X15 fibroblasts only 30% of the nuclei displayed XIST-coated X chromosomes (Figures 3A and 3D). In hiPSC lines generated from the 47,XXX fibroblasts, many cells did not show Xi markers or showed signs of only one Xi (Figures 3A and 3E). In all cell lines, analysis of heterochromatin markers associated with the Xi, including enrichment of H3K27me3, indicated variable staining, with many cells not displaying all characteristic Xi features (Figure 3A; data not shown). Also, Barr bodies (e.g., DAPI-dense, heterochromatic regions covering the Xi) were detected in a minority of cells. Strikingly, in many colonies, cells with all Xi hallmarks, including depletion of H3K9ac, were most often found at the edges or in the center of the colonies, where most of the differentiated cells are found. In contrast, cells without XIST and associated Xi markers were found in a donut-shaped region surrounding the middle of the colonies (Figures 3B and 3C).These results could be explained by (1) loss of X chromosome(s), (2) loss of XIST expression and Xi markers during hiPSC cell culture and colony formation, similar to findings for hESC lines (Bruck and Benvenisty, 2011), or (3) XCR in a pluripotent subpopulation of hiPSCs, possibly followed by XCI. Option 1 was excluded, as karyotyping and SNP array analysis did not reveal a significant population of cells with X chromosome aneuploidies at different passages after establishment of the iPSC lines. To distinguish between options 2 and 3, we first analyzed the methylation pattern of the Xi by HUMARA analysis, focusing on the hiPSC lines derived from the X12 and X15 fibroblasts. If the Xi in the hiPSCs is the same X chromosome as the Xi inherited from the founder fibroblasts, HUMARA analysis will detect completely skewed methylation of the Ar gene. Furthermore, using this analysis, XCR in all cells would lead to an absence of methylation at the Ar gene, whereas XCR followed by rXCI, which might also involve the intact X chromosome would result in methylation of the previously unmethylated AR allele. HUMARA analysis for genomic DNA of undifferentiated X15 hiPSC lines showed the initial 100:0 skewing ratio (Figure 3F), which was also observed for the founder fibroblast cell line, compatible with either an absence of XCR, or XCR in a small subpopulation of cells that cannot be detected by this method. In undifferentiated X12 p3 hiPSC lines, we observed methylation, in some clones up to 12%, also of the second Ar allele located on the intact X chromosome (Figure 3F). This finding indicates that XCR in the X12 p3 hiPSCs is unstable and that these cells have started rXCI also on the intact X chromosome in a subpopulation of cells. We next performed allele-specific expression analysis of XIST using RFLP RT-PCR analysis on RNA from X12 hiPSCs, the only hiPSC line with an informative XIST SNP. On the whole cell population level, we detected XIST expression from both X chromosomes (Figure 3G). This supported the findings with the HUMARA analysis, indicating that XIST upregulation and initiation of XCI have occurred following XCR, in X12 hiPSC lines. In the day 8 differentiated progeny of X12 hiPSC lines, we detected monoallelic XIST expression (Figure 3H) comparable to that in the original X12 fibroblasts. This is explained by survival, at later passages, of cells that have silenced the mutant X chromosome and loss of cells that have initiated XCI on the intact X chromosome.
Figure 3.
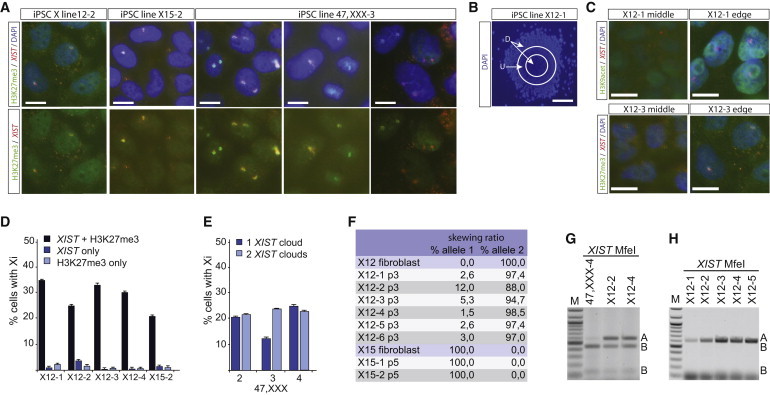
XCI Analysis of hiPSCs
(A) Immuno-RNA-FISH analysis on hiPSC lines X12-2, X15-2, and 47,XXX-3 (all cells in A–E and G, passage 3), detecting XIST (Rhodamine Red) and H3K27me3 (FITC). In the upper panels, the nuclei are counterstained with DAPI (scale bar represents 10 μm).
(B) Example of an X12-1 hiPSC clone, at lower magnification. The center part and edge of colony are indicated with D, the undifferentiated middle part of the colony with an U (scale bar represents 100 μm).
(C) Immuno-RNA-FISH detecting XIST (Rhodamine Red) and H3K9ac (FITC, top) and detecting XIST (Rhodamine Red) and H3K27me3 (FITC, bottom) on cells found in U (left) and in D (right) (scale bar represents 10 μm).
(D) Quantification of immuno-RNA-FISH analysis of representative hiPSC lines X12 (1–4) and X15-2. Percentages of cells with both an XIST cloud and H3K27me3 accumulation, only an XIST cloud, or only H3K27me3 accumulation are shown (mean ± SD, n = 3 experiments, >100 cells were counted per experiment).
(E) Quantification of immuno-RNA-FISH analysis of representative hiPSC lines 47,XXX (2–4). Percentage of cells with either one or two XIST clouds is plotted (mean ± SD, n = 3 experiments, >100 cells were counted per experiment).
(F) Quantification of all HUMARA analyses performed with different fibroblast and hiPSC lines, passages 3 and 5 (p3 and p5).
(G) Allele-specific RT-PCR analysis of XIST. PCR products were digested using MfeI to distinguish between both parental alleles. The hiPSC lines X12-2 and X12-4 show biallelic XIST expression, which was not found in the original fibroblast line (compare to Figure 1D).
(H) As for (G), but here day 8 differentiated X12 hiPSC lines are assessed.
Biallelic Expression of X-Linked Loci and Reversal of XIST Expression
The observed presence of XIST-negative cells within colonies at early passages of the X12 hiPSC lines is in agreement with the present findings for the whole-cell populations with the HUMARA assay and XIST expression analysis. Taken together, the results provide evidence that the hiPSCs, cultured under the above standard conditions, engage in both XCR and subsequent initiation of XCI.
To precisely evaluate the dynamics of XCR, we performed allele-specific single-cell RT-PCR analysis, amplifying RFLPs in the X-linked genes XIST, SUVAR39H1, and G6PD at different time points after reprogramming. X12 iPSC and 47,XXX iPSC lines were sorted via fluorescence-activated cell sorting (FACS) in 96-well plates using antibodies against the pluripotency-associated surface markers SSEA4 and Tra1-81 at p0, p3/4, and p25 after establishment. All sorted cells were dTomato negative. Passage 0 cells were isolated 20 days after the start of the reprogramming procedure, prior to picking. For all time points, the SSEA4 and Tra1-81 double-positive fraction represented more than 40% of the total viable cell fraction (Figure 4A; data not shown), and qPCR analysis showed increased expression of the pluripotency markers in pooled double-positive SSEA4+/Tra1-81+ and GCTM2+/CD9+ sorted cells (Figure 4B). SSEA4+/Tra1-81+ double-positive (dTomato−) cells were sorted in lysis buffer, and single-cell nested RT-PCR was performed, followed by restriction digestion to distinguish between expression from the different alleles. Analysis of XIST in X12 hiPSCs revealed a high percentage of cells not expressing XIST and cells displaying expression of XIST emanating from the Xa (allele B) at all stages after reprogramming (Figures 4D and 4E). Also, allele-specific expression analysis of X-linked SUVAR39H1 revealed a high percentage of cells displaying expression of the previously inactive allele B (Figure S3). Interestingly, only a small percentage of cells displayed XIST and SUVAR39H1 expression both exclusively from the alleles A, as was found for the founder fibroblasts used to generate the iPSCs (Figures 4C–4E and S3). In addition, the percentage of cells expressing only the XIST and SUVAR39H1 alleles A did not markedly drop after establishment of the iPSC line. Similar results were obtained with iPSCs generated from 47,XXX fibroblasts analyzed at p0 and p25, revealing a considerable amount of cells that showed biallelic expression of G6PD, indicating reactivation of the Xi (Figure 4E). These results indicate robust reactivation of the Xi during the early stages of the reprogramming process. In addition, the results of the RNA-FISH analysis on iPSC clones X12-2 and X12-4 suggest that in most cells expressing XIST this RNA is detected only at very low levels (Figures 3D and 4E). To test whether XCR involved multiple genes on the X chromosome, we performed allele-specific expression analysis of three additional informative genes located at different positions on the X. RNA was isolated from pooled cells of different X12 clones. In two of the three tested iPSC clones (X12-23 and X12-24), we found biallelic expression of HUWE1, ATP7A, and NROB1, whereas in the iPSC line X12-19 only NROB1 expression was biallelic (Figure S4), indicating that the robustness of XCR is variable between clones.
Figure 4.

Single-Cell RT-PCR Analysis of Sorted hiPSCs
(A) FACS analysis of hiPSCs stained for the pluripotency-associated surface markers SSEA4 and TRA1-81 (gate used to sort hiPSCs is shown).
(B) qRT-PCR analysis of NANOG in living sorted H9 ESCs, and X12-2 and 47,XXX-4 hiPSCs, comparing unsorted cells (U), SSEA4−/TRA1-81− double-negative cells, CD9+/GCTM2+ double-positive cells, and SSEA4+/TRA1-81+ double-positive cells. Results are normalized for GAPDH. NANOG expression in unsorted H9, the positive control, was set at 100.
(C) Allele-specific expression analysis of SUVAR39H1 on gDNA and cDNA isolated from X12 fibroblasts.
(D) Single-cell RT-PCR analysis of SSEA1+/TRA1-81+ sorted cells from hiPSC line X12-2 (passage 25, p25). Shown are the cells numbered 49–60. Allele-specific expression of XIST and SUVAR39H1 was assessed by digestion with MfeI and MspI, distinguishing the parental alleles (indicated with A and B).
(E) Quantification of (D) for XIST (left graph) and SUVAR39H1 (middle graph) on X12 iPSC lines (X12-2 up to X12-24, and the pooled cells) at different passages (p0 up to p25) after reprogramming. The right graph shows the quantification of allele-specific expression analysis of G6PD on SSEA4+/Tra1-81+ sorted 47,XXX iPSCs (for all experiments n = 96 cells per cell line).
(F) Dome-shaped morphology of NHSM cultured hiPSCs (scale bar represents 100 μm).
(G–K) TRA1-81 (G), ID3 (H), and STAT3 (I) expression is increased, and DNMT3A (J) and DNMT3B (K) expression is decreased in NHSM-cultured iPSC lines. Shown is the mean intensity of TRA1-81 determined by FACS analysis. For ID3, STAT3, DNMT3A, and DNMT3B expression per iPSC line is shown relative to H9 hESCs cultured in human ES medium.
(L) XIST expression analysis as in (H)–(K).
(M) XIST RNA-FISH analysis on X12-23 iPSCs grown in NHSM, passage 6 (>100 cells were counted per experiment, scale bar represents 10 μm).
(N) Quantification of cells with XIST-coated X chromosomes in different X12 iPSC lines (p6).
(O) Allele-specific expression analysis of XIST and SUVAR39H1 at different passages (p0–p4) of X12 hiPSC lines growing in NHSM (alleles A and B, as in Figure 4D).
Recently, hESCs and hiPSCs have been generated with more naive characteristics closely resembling the mouse mESCs (Gafni et al., 2013). Naive hiPSC lines were established by reprogramming human fibroblasts in NHSM medium, but could also be generated by culturing hiPSCs established in standard hESC medium in NHSM. To test whether NHSM medium would enhance XCR in our iPSCs, we passaged (p3/4) four different hiPSC lines in NHSM medium, which already at passage one resulted in morphologically mouse-like hiPSCs that could be passaged through trypsinization (Figure 4F). FACS and qRT-PCR analysis indicated that expression of TRA1-81, ID3, and STAT3 was upregulated and that expression of DNMT3A and DNMT3B was downregulated in NHSM culture conditions as described (Gafni et al., 2013) (Figures 4G–4K). Other tested markers, including TEAD4 and DUSP10, did not show consistent changes. XIST RNA-FISH and qRT-PCR analysis on these hiPSC lines, fixed after six passages in NHSM, indicated a near loss of XIST clouds and XIST expression in most lines (Figures 4L–4N), consistent with reported findings (Gafni et al., 2013). Allele-specific RT-PCR analysis indicated no change in the allelic expression ratio of XIST after culturing the hiPSCs in NHSM, with different iPSC lines showing either biallelic or monoallelic expression of XIST (Figure 4O). For most clones, the allele-specific expression ratio of SUVAR39H1 was stable throughout several passages in NHSM. Also, allele-specific expression analysis of HUWE1, ATP7A, and NROB1, on three different X12 iPSC lines, indicated no change in the expression status before and after transfer to NHSM. These findings indicate that culture of the hiPSC lines in NHSM does not have an effect on XCR, but reduces XIST expression. This indicates that indeed a further shift toward stable XCR in hiPSCs can be obtained by culturing iPSCs in NHSM.
Discussion
Here, we have investigated the dynamics of XCI in hiPSCs derived from human female fibroblast cultures with completely skewed rXCI. This approach allowed us to include analysis of XCR and XCI at the single-cell level. The present results show that, at the single cell level, reprogramming of human cells into hiPSCs results in XCR, even at early passages. Biallelic expression of different X-linked loci indicates that the Xi is reactivated in a large part of the cell population of our hiPSC lines, but is variable between iPSC lines. In several hiPSC lines, we also detected XIST expression and expression of several X-linked genes from the allele that was not active in the starting fibroblast culture. Human iPSC lines share a clonal origin, and therefore, this switch in expression from one allele to the other, or biallelic expression, indicates that during the reprogramming process XCR must have occurred in a high proportion of cells. Differentiation of hiPSCs was found to result in completely skewed rXCI, similar to that of the starting fibroblast population, indicating a high selective pressure against cells that have inactivated the intact X chromosome.
A recent study indicated that XIST expression can be lost upon prolonged passaging of female hiPSCs, referred to as “erosion of XCI” (Mekhoubad et al., 2012). This epigenetic erosion was found to be irreversible and correlated with a loss of differentiation characteristics and is very relevant for disease-modeling procedures. Erosion of XCI in hiPSC lines can be effectively prevented by XCR followed by XCI upon differentiation. Several studies have been published assessing XCR during the reprogramming process. Derepression attributed to XCR of genes located on the Xi has been observed in hESC lines (Lengner et al., 2010) and has been reported in studies involving gene expression profile comparison of multiple female hESC and hiPSC lines (Bruck and Benvenisty, 2011). Very recently, XCR was shown to happen efficiently early during reprogramming of human cells, but that this was quickly followed by initiation of XCI upon generation of nascent hiPSC lines, leading to XaXi cells only (Kim et al., 2014). These authors concluded that XCR was the result of overexpression of the exogenous reprogramming factors and that shutdown of the reprogramming cassette leads to XCI initiation. In contrast, our study indicates that in the present reprogramming and culture conditions, hiPSCs maintain the XaXa state in a high percentage of cells in the absence of ectopic expression of the reprogramming factors. The present single-cell allele-specific expression analysis revealed expression of the SUVAR39H1, HUWE1, ATP7A, NROB1, and G6PD alleles located at different positions on the X chromosome that was the Xi in the starting fibroblasts, in a high (>50%) proportion of cells of X12 and 47,XXX hiPSC lines. This effect was already present 20 days after transduction, arguing against erosion of XCI in our hiPSC lines and favoring XCR of the silenced Xi upon reprogramming. Our studies also showed that XaXa cells represent the major population of cells at all stages after reprogramming, indicating that the present experimental conditions prevented robust precocious initiation of XCI, but allowed cells to maintain either the XaXa or the XaXi situation. The present findings would be in agreement with the suggestion that culturing hiPSCs in naive growth conditions facilitates XCR, which was not addressed in this initial report (Gafni et al., 2013). Indeed, our studies indicate that NHSM growth conditions effectuate a reduction in the percentage of cells with XIST-coated X chromosomes. However, this does not lead to changes in the allele-specific expression ratio of SUVAR39H1, HUWE1, ATP7A, and NROB1 in most hiPSC lines, and although we only analyzed our cells at an early stage after changing to the NHSM condition (passage 4), these findings suggest that XCR and XCI characteristics are hiPSC line specific and established during reprogramming and cannot be changed afterward. Recently, two other protocols have been described to revert primed into naive hESCs (Takashima et al., 2014; Theunissen et al., 2014). Surprisingly, one study reports initiation of XCI after induction of the naive state (Theunissen et al., 2014), whereas another study indicates loss of XIST and H3K27me3 accumulation after a reset of primed to naive ESCs (Takashima et al., 2014). These findings emphasize that knowledge about the transcriptional status of the X chromosomes in the ICM of the female preimplantation human embryo will be crucial to conclude which of these conditions result in hESCs that resemble ICM cells most.
The results presented herein strongly suggest that XCR of the Xi is an important first step in reprogramming of human female fibroblasts and demonstrate that rXCI is often de novo initiated during the culture of the generated hiPSC lines, most likely triggered by partial differentiation of the hiPSCs. In addition, we found that XCR appears to occur remarkably efficient under standard reprogramming conditions and that subsequent NHSM culture conditions stabilize this XaXa state. This facilitates rapid expansion and genetic manipulation of established female hiPSC lines with two active X chromosomes, to be used for various applications, in particular disease modeling, offering differentiated cell types that have newly established an Xi, thereby avoiding effects related to erosion of XCI. Furthermore, this provides us with a powerful model system to study human XCI, allowing us to examine the effects of a wide spectrum of X-linked mutations and deletions on XCI.
Experimental Procedures
Cell Lines
GM07148 (X12), GM03923 (X14), and GM03827 (X15) fibroblasts were obtained from the Corriel cell repository, and 47,XXX fibroblasts were established from a skin biopsy of a triple X patient (Brosens et al., 2014). All fibroblast were cultured in standard fibroblast medium.
hiPSC Generation and Culture
To generate iPSCs, human fibroblasts from lines X12 (p12), X15 (p4), and 47,XXX (p14) were reprogrammed according to Warlich et al. (2011), with minor modifications. NHSM medium and culture conditions were as described by Gafni et al. (2013). Detailed information is provided in the Supplemental Experimental Procedures.
Immuno-RNA-FISH Analysis
Detailed protocols and probes for RNA-FISH and immuno-RNA-FISH have been described (Barakat and Gribnau, 2014; Barakat et al., 2011; Jonkers et al., 2009). For immunostainings, the following antibodies were used: anti-Nanog (1:100, Abcam), anti-KLF4 (1:250, Abcam), anti-H3K27me3 (1:500, Diagenode), anti-H3K4me3 (1:1000, Upstate), anti-H3K9ac (1:1000, Sigma), anti-CD9 (Invitrogen), anti-GCTM2 (BD), anti-SSEA4 (BD), and anti-TRA1-80 (BD).
RT-PCR and Single-Cell RT-PCR
All primers used are described in Table S2. For single-cell RT-PCR, SSEA1+/TRA1-81+ double-positive cells were sorted via FACS in 96-well plates containing 9 μl lysis buffer using a BD FACSAria apparatus. Lysis buffer consisted of 8 μl 2xReaction mix (SuperScript One-Step RT-PCR kit; Invitrogen), 10 U RNaseOut (Invitrogen), and 0.15% IGEPAL CA-630 (Sigma). cDNA was prepared with gene-specific outer primers and further processed as described in the Supplemental Experimental Procedures. PCR products were precipitated and digested with the indicated restriction enzymes (New England Biolabs) to distinguish expression from the different alleles. RFLPs in XIST, SUVAR39H1, G6PD, and SNPs in HUWE1, NROB1, and ATP7A were identified by PCR amplification and Sanger sequencing, followed by allele-specific expression analysis using cDNA-specific primer sets described in Table S2.
SNP Array and HUMARA Analysis
To map the deletions in our fibroblast cell lines, SNP array was performed using Human CYTO SNP 12 version 1 arrays (Illumina), aligned to human genome build 18. A detailed description of the HUMARA assay applied to determine skewing of XCI is provided in the Supplemental Experimental Procedures.
DNA-FISH Analysis
To visualize the deletion in cell lines, DNA-FISH was performed according to standard procedures. Probes used were BAC CTD-3076O23 (BAC1FITC, Xq23, 108,9 Mb, HG19), RP11-799O20 (BAC2, A595, Xq25, 123 Mb), RP11-75N13 (BAC3, FITC, Xq21.1, 84,5 Mb), and RP1-279N11 (BAC4, A595, Xq13.3, 75,8 Mb).
Statistics and qRT-PCR Analysis
For qRT-PCR expression analysis, the average and SD of three biological replicates are shown.
Author Contributions
T.S.B., M.G., A.K., R.-J.G., J.A.G., J.L., and J.G. conceived the experiments, and T.S.B. and J.G. wrote the manuscript. T.S.B., M.G., B.H., T.L., B.E., H.D., R.L., N.S., and M.B. performed the experiments.
Acknowledgments
We thank Dr. Axel Schambach for providing the reprogramming vector and Dr. Dorota Kurek and Dr. Derk ten Berge for providing anti-GCTM2 antibody. Dr. Sabrina Roth is acknowledged for advice on single-cell RT-PCR. J.G. was supported by NWO VICI and ERC grants.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- Allen R.C., Nachtman R.G., Rosenblatt H.M., Belmont J.W. Application of carrier testing to genetic counseling for X-linked agammaglobulinemia. Am. J. Hum. Genet. 1994;54:25–35. [PMC free article] [PubMed] [Google Scholar]
- Amenduni M., De Filippis R., Cheung A.Y., Disciglio V., Epistolato M.C., Ariani F., Mari F., Mencarelli M.A., Hayek Y., Renieri A. iPS cells to model CDKL5-related disorders. Eur. J. Hum. Genet. 2011;19:1246–1255. doi: 10.1038/ejhg.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev G., Williams E.C., Li H., Chang Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS ONE. 2011;6:e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat T.S., Gribnau J. X chromosome inactivation in the cycle of life. Development. 2012;139:2085–2089. doi: 10.1242/dev.069328. [DOI] [PubMed] [Google Scholar]
- Barakat T.S., Gribnau J. Combined DNA-RNA fluorescent in situ hybridization (FISH) to study X chromosome inactivation in differentiated female mouse embryonic stem cells. J. Vis. Exp. 2014 doi: 10.3791/51628. Published online June 14, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat T.S., Gunhanlar N., Pardo C.G., Achame E.M., Ghazvini M., Boers R., Kenter A., Rentmeester E., Grootegoed J.A., Gribnau J. RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genet. 2011;7:e1002001. doi: 10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosens E., de Jong E.M., Barakat T.S., Eussen B.H., D’haene B., De Baere E., Verdin H., Poddighe P.J., Galjaard R.J., Gribnau J. Structural and numerical changes of chromosome X in patients with esophageal atresia. Eur. J. Hum. Genet. 2014;22:1077–1084. doi: 10.1038/ejhg.2013.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck T., Benvenisty N. Meta-analysis of the heterogeneity of X chromosome inactivation in human pluripotent stem cells. Stem Cell Res. (Amst.) 2011;6:187–193. doi: 10.1016/j.scr.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Horvath L.M., Grafodatskaya D., Pasceri P., Weksberg R., Hotta A., Carrel L., Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum. Mol. Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Harnden D.G. Nuclear sex in triploid XXY human cells. Lancet. 1961;2:488. doi: 10.1016/s0140-6736(61)92457-6. [DOI] [PubMed] [Google Scholar]
- Jonkers I., Barakat T.S., Achame E.M., Monkhorst K., Kenter A., Rentmeester E., Grosveld F., Grootegoed J.A., Gribnau J. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- Kim K.Y., Hysolli E., Park I.H. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc. Natl. Acad. Sci. USA. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.Y., Hysolli E., Tanaka Y., Wang B., Jung Y.W., Pan X., Weissman S.M., Park I.H. X chromosome of female cells shows dynamic changes in status during human somatic cell reprogramming. Stem Cell Rep. 2014;2:896–909. doi: 10.1016/j.stemcr.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner C.J., Gimelbrant A.A., Erwin J.A., Cheng A.W., Guenther M.G., Welstead G.G., Alagappan R., Frampton G.M., Xu P., Muffat J. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollex T., Heard E. Recent advances in X-chromosome inactivation research. Curr. Opin. Cell Biol. 2012;24:825–832. doi: 10.1016/j.ceb.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Pomp O., Dreesen O., Leong D.F., Meller-Pomp O., Tan T.T., Zhou F., Colman A. Unexpected X chromosome skewing during culture and reprogramming of human somatic cells can be alleviated by exogenous telomerase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Silva S.S., Rowntree R.K., Mekhoubad S., Lee J.T. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D.T., Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J., Kuoy E., Chin M.H., Trinh H., Patterson M., Sherman S.P., Aimiuwu O., Lindgren A., Hakimian S., Zack J.A. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warlich E., Kuehle J., Cantz T., Brugman M.H., Maetzig T., Galla M., Filipczyk A.A., Halle S., Klump H., Schöler H.R. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol. Ther. 2011;19:782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


