Abstract
Chronic infection caused by the hepatitis B virus (HBV), is strongly associated with hepatitis, fatty liver and hepatocellular carcinoma. To investigate the underlying mechanisms, we characterize the metabolic features of host cells infected with the virus using systems biological approach. The results show that HBV replication induces systematic metabolic alterations in host cells. HBV infection up-regulates the biosynthesis of hexosamine and phosphatidylcholine by activating glutamine-fructose-6-phosphate amidotransferase 1 (GFAT1) and choline kinase alpha (CHKA) respectively, which were reported for the first time for HBV infection. Importantly suppressing hexosamine biosynthesis and phosphatidylcholine biosynthesis can inhibit HBV replication and expression. In addition, HBV induces oxidative stress and stimulates central carbon metabolism and nucleotide synthesis. Our results also indicate that HBV associated hepatocellular carcinoma could be attributed to GFAT1 activated hexosamine biosynthesis and CHKA activated phosphatidylcholine biosynthesis. This study provides further insights into the pathogenesis of HBV-induced diseases, and sheds new light on drug target for treating HBV infection.
Hepatitis B virus (HBV) causes hepatitis, an infectious illness of the liver, and a health problem. It is estimated that approximately 350 million people are chronically infected with HBV worldwide1. Persistent HBV infection can cause hepatofibrosis, and finally develop into hepatocellular carcinoma (HCC)2. About sixty percent of the liver cancer cases are associated with chronic HBV infection3. However, the underlying mechanisms of HBV-induced diseases are still unclear, and there is currently no cure for chronic HBV infection. It is therefore necessary to investigate the complex host cellular responses to HBV replication, which in turn could provide a deeper understanding of the pathogenic mechanisms and potential new targets for effective drug discovery.
HBV infection involves complicated interactions between the virus and host cell. Previous studies have provided some clues into the metabolic alterations caused by HBV infection, including the up-regulation of glutamate dehydrogenase 1 and isocitrate dehydrogenase4, and the transcriptional up-regulation of genes involved in lipid biosynthesis (e.g. ATP citrate lyase, fatty acid synthase)5. The lipid metabolic alterations induced by HBV infection were also found in cell models and HBV transgenic mice6,7. These reports suggest that there is an association between HBV infection and metabolic alterations in host cells. While target analysis is restrained for detection small numbers of metabolites, metabolomics techniques enable quantitative analysis of a larger range of metabolites, hence should provide a holistic view on the interactions between HBV and the host cell.
Metabolomics analysis method combining multivariate data analysis and spectral profile techniques, such as magnetic resonance spectroscopy (NMR), provides metabolic information and has been successfully applied in the evaluation of the systematic responses to exogenous stimuli8,9. Importantly, the results from metabolomics studies provide comprehensive information, enabling the generation of credible hypotheses, which can then be further validated by molecular biological assays.
In this study, we employ a HepG2.2.15 cell model, which is derived from the human hepatoblastoma cell line HepG2. HepG2.2.15 cells are well characterized by stable expression and replication of HBV and are widely used in studies on HBV infection10,11. We utilize metabolomics, proteomics and molecular biological assays to investigate the associations between HBV and the host cellular metabolism, with the objective of understanding the pathogenesis of HBV in greater depth.
Results
Metabolic characterization of HepG2.2.15 and HepG2 cells
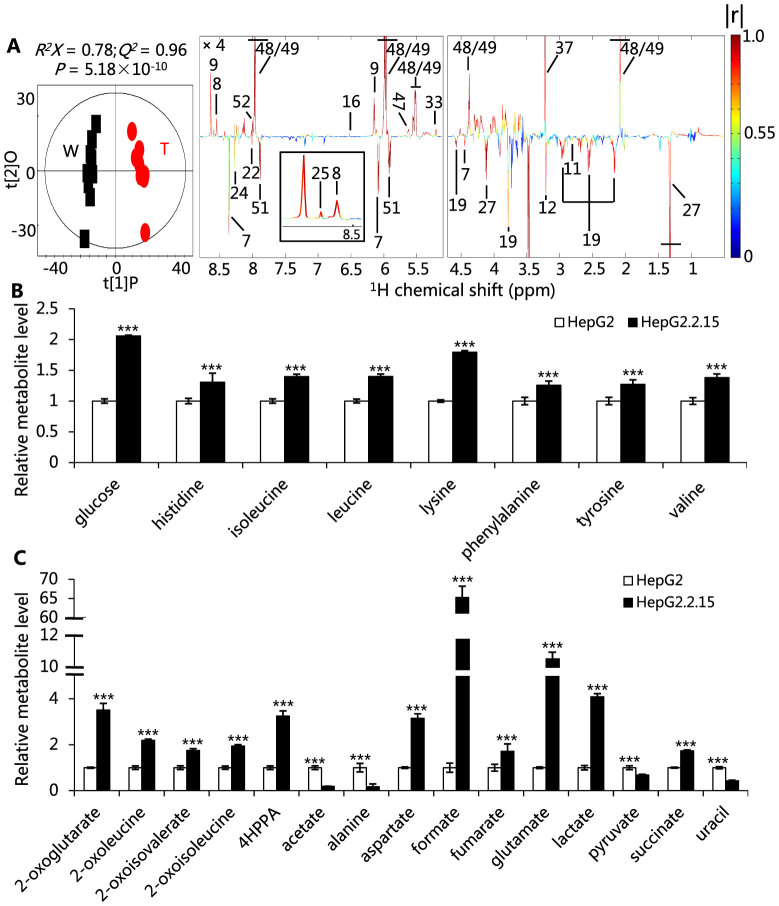
The metabolic profiles of HepG2.2.15 cells are notably different as compared with HepG2 cells. More than fifty metabolites, representing the major metabolic pathways, are identified (Fig. S1, Table S1). HepG2.2.15 cells, compared with HepG2, are characterized by higher levels of intracellular nucleotides and nucleotide sugars and lower levels of reduced glutathione and nucleosides (Fig. 1A, Table S2). These altered metabolites are involved in the metabolism of hexosamine, choline, carbohydrates, amino acids and nucleotides. The profiles of consumed and secreted metabolites by the cells are also significantly different (Fig. 1B, C). In addition, the compositions of fatty acids are altered by HBV replication (Table S3).
Figure 1. The alterations of metabolic profiles between HepG2.2.15 and HepG2 cells.
(A) Validated OPLS-DA scores and coefficient plots showing the discriminations of metabolic profiles of extracts of HepG2.2.15 (T) from HepG2 (W) cells. Here the resonance peaks pointing upward indicate an increase of metabolites in HepG2.2.15 cells and downwards a decrease. The color of the peaks represents correlation coefficient of a metabolite with the cutoff value of |r| is 0.602 (n = 10, P < 0.05). Detailed data are shown in Table S2. Keys: 7, adenosine; 8, adenosine 5′-diphosphate; 9, adenosine 5′-monophosphate; 11, Aspartate; 12, choline; 16, fumarate; 19, GSH; 22, guanosine; 24, inosine; 25, inosine 5′-monophosphate; 27, lactate; 33, N-acetyl-glucosamine; 37, phosphocholine; 47, UDP-glucuronate; 48, UDP-N-acetyl galactosamine; 49, UDP-N-acetyl glucosamine; 51, uridine; 52, uridine 5′-diphosphate. (B) Metabolites consumed by HepG2.2.15 and HepG2 cells, which are derived by subtracting levels of metabolites in the spent medium from fresh medium. Hence, higher levels of metabolites indicate more of the respective metabolites being consumed. (C) Metabolites secreted to cell medium. 4HPPA, 4-hydroxyphenylpyruvate. Data are shown as mean ± standard deviation (s.d.), n = 10, t-test, ***P < 0.001. The details of the metabolite assignment are shown in Figure S1 and Table S1.
In order to further confirm the metabolomic results, the levels of some enzymes are quantified. The mass spectrometer (MS) spectrum of one-peptide-identified protein NAGK, is checked manually (Fig. S2). The other enzymes used here are quantified with at least three peptides. The enzymatic alterations caused by HBV replication are shown in Table 1. The quantitation details are summarized in Table S4. These enzymes are mainly involved in central carbon metabolism, hexosamine metabolism and phenylalanine degradation, and their alterations are in accordance with the metabolomic results.
Table 1. Enzymes significantly changed between HepG2.2.15 and HepG2 cells.
| No. | Enzyme description (name) | Gene name | Metabolic pathway | Ratio (HepG2.2.15/HepG2) | P value |
|---|---|---|---|---|---|
| 1 | Fructose-bisphosphatealdolase (ALDOC) | ALDOC | Glycolysis | 1.27 | 0.045 |
| 2 | Glucose-6-phosphate isomerase (GPI) | GPI | Glycolysis | 1.31 | 2.50E-7 |
| 3 | Alpha-enolase (ENO1) | ENO1 | Glycolysis | 1.35 | 2.15E-5 |
| 4 | Phosphoglycerate kinase 1 (PGK1) | PGK1 | Glycolysis | 1.41 | 9.04E-6 |
| 5 | Triosephosphate isomerase (TPI) | TPI1 | Glycolysis | 1.22 | 0.008 |
| 6 | Succinate dehydrogenase (SDHB) | SDHB | TCA cycle | 2.32 | 0.009 |
| 7 | Malate dehydrogenase (MDH2) | MDH2 | TCA cycle | 1.66 | 5.54E-4 |
| 8 | Citrate synthase, mitochondrial (CS) | CS | TCA cycle | 1.67 | 3.70E-7 |
| 9 | Phosphoglucomutase-1 (PGM1) | PGM1 | Nucleotide-sugar biosynthesis | 1.61 | 0.016 |
| 10 | 6-Phosphogluconate dehydrogenase (PGD) | PGD | Pentose phosphate pathway | 1.16 | 0.046 |
| 11* | N-acetyl glucosamine kinase (NAGK) | NAGK | Hexosamine metabolism | 10.50 | - |
| 12 | 4-Hydroxyphenylpyruvate dioxygenase (HPD) | HPD | Phenylalanine degradation | 0.45 | 0.003 |
| 13 | Fumarylacetoacetase (FAA) | FAH | Phenylalanine degradation | 0.62 | 0.023 |
Note: *Fold change is greater than 10 without P value, as just one high quality peptide is quantified. More information can be found in Table S4.
HBV replication promotes central carbon metabolism
HepG2.2.15 cells consume more glucose and other nutrients (Fig. 1B). The consumed glucose is distributed into central carbon metabolism, providing energy and biomass for HBV replication. There is a significant increase in levels of lactate released by HepG2.2.15 cells into the medium (Fig. 1C), as a result of glucose consumption. Enzymes involved in the glycolysis pathway, including glucose-6-phosphate isomerase (GPI), fructose-bisphosphate aldolase (ALDOC), alpha-enolase (ENO1), phosphoglycerate kinase 1 (PGK1) and triosephosphate isomerase (TPI) are also up-regulated significantly in HepG2.2.15 cells (Table 1). These data suggest that glycolysis is being stimulated in HepG2.2.15 cells. Additionally, enzymes that regulate the tricarboxylic acid (TCA) cycle, such as citrate synthase (CS), succinate dehydrogenase (SDHB) and malate dehydrogenase (MDH2), are up-regulated in HepG2.2.15 cells (Table 1), which subsequently lead to the elevated levels of TCA cycle intermediates, such as 2-oxoglutarate (2-OG), succinate and fumarate (Fig. 1A, C). Furthermore, the elevated levels of adenosine 5′-monophosphate (AMP) and adenosine 5′-diphosphate (ADP) (Fig. 1A) can activate many enzymes in the regulation of glycolysis and the TCA cycle12. These observations suggest that HBV replication promotes glycolysis and the TCA cycle in host cells.
Glucose that participates in the pentose phosphate pathway is also up-regulated in HepG2.2.15 cells, which is manifested in the elevated levels of phosphogluconate dehydrogenase (PGD) (Table 1). This observation is consistent with the elevations in the levels of nucleotides, such as inosine-5′-monophosphate (IMP), AMP, ADP and uridine 5′-diphosphate (UDP), and depletions in the levels of nucleosides, such as guanosine, inosine, adenosine, and uridine, as well as uracil (Fig. 1A, C). The de novo biosynthesis of nucleotides requires the folate cycle to provide one carbon unit for the production of purine and pyrimidine. Formate is the major substrate in the folate cycle in human cells13. Here, the levels of formate in HepG2.2.15 cells are remarkably elevated (Fig. 1C). Taken together, these results show that the pentose phosphate pathway is promoted to provide ribose for nucleotide synthesis for HBV replication.
The promotion of hexosamine biosynthesis pathway is required for HBV replication
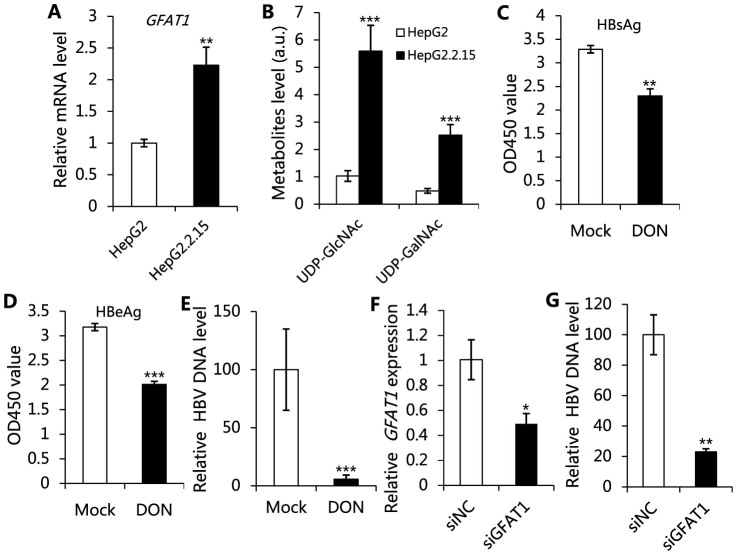
Hexosamine biosynthesis pathway (HBP) plays an important role in the nutrient signaling in mammalian cells. Glucose flux into this pathway is regulated by the rate-limiting enzyme, glutamine-fructose-6-phosphate amidotransferase (GFAT1), which is encoded by GFAT1 gene in humans14. We find that the transcriptional level of GFAT1 is much more up-regulated in HepG2.2.15 as compared with HepG2 (Fig. 2A), and, correspondingly, the levels of UDP-N-acetyl glucosamine (UDP-GlcNAc) and UDP-N-acetyl galactosamine (UDP-GalNAc), the most abundant end products of HBP, are markedly increased in HepG2.2.15 cells (Fig. 2B). In addition, elevated levels of both UDP-GlcNAc and UDP-GalNAc are also found in HepG2 transfected by HBV replication-competent plasmids (pHBV1.3) (Fig. S3A). Furthermore, N-acetyl glucosamine (GlcNAc) is significantly elevated in HepG2.2.15 (Fig. 1A). Consistent with this, GlcNAc kinase (NAGK) that converts GlcNAc to UDP-GlcNAc15 is up-regulated more than 10 fold in HepG2.2.15 cells (Table 1). Moreover, the levels of UDP-glucuronate (UDP-GlcA), another nucleotide sugar involved in polysaccharide biosynthesis, are markedly increased in HepG2.2.15 (Fig. 1A). UDP-GlcA biosynthesis is regulated by phosphoglucomutase-1 (PGM1), which is the first enzyme regulating the glucose flux into nucleotide sugar. Here, the expression of PGM1 is enhanced in HepG2.2.15 (Table 1). Taken together, these observations strongly indicate that HBP is promoted by HBV replication in cells.
Figure 2. Promotion of hexosamine biosynthesis is necessary for HBV replication.
(A) mRNA levels of GFAT1 in HepG2.2.15 and HepG2 cells. (B) Relative levels of UDP-N-acetyl glucosamine (UDP-GlcNAc) and UDP-N-acetyl galactosamine (UDP-GalNAc) in HepG2.2.15 and HepG2 cells. (C-E) The effects of inhibiting GFAT1 by culturing HepG2.2.15 cells with 2 mM DON. (C) Relative levels of HBsAg expression; (D) Relative levels of HBeAg expression; (E) Relative core-associated HBV DNA levels. (F and G) HepG2.2.15 cells were transfected by siRNA targeting GFAT1 (siGFAT1) or no sense control (siNC). (F) The transcriptional levels of GFAT1, data was normalized to actin; (G) Relative core-associated HBV DNA levels. For all graphs, data are shown as mean ± s.d., n = 3, t-test, *P < 0.05, **P < 0.01, ***P < 0.001.
In order to validate the essential role of HBP in HBV replication, we incubated HepG2.2.15 cells with 6-diazo-5-oxo-L-norleucine (DON), a glutamine analog that is capable of inhibiting the activity of GFAT1. The inhibition of GFAT1 leads to a significant reduction in the expression of hepatitis B surface antigen (HBsAg) (Fig. 2C). The DNA copy number of intracellular HBV decreases, which is accompanied by the reduced levels of HBV e antigen (HBeAg), an indicator of HBV DNA replication (Fig. 2D, E). We further employ RNA interference to down-regulate the expression of GFAT1 in order to validate the essential role of GFAT1 in HBV replication (Fig. 2F). The results show that down-regulation of GFAT1 can inhibit HBV DNA replication (Fig. 2G). These evidences show that stimulated HBP is essential for HBV replication.
Promotion of phosphatidylcholine biosynthesis is required for HBV replication
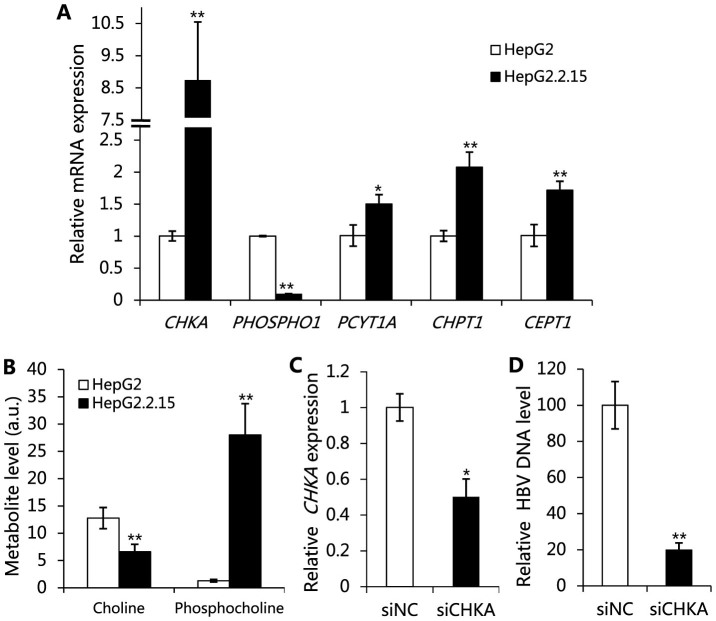
Phosphatidylcholine (PC) is a major phospholipid component of the biological membranes16 and a precursor for the biosynthesis of lipid signaling molecules17. Choline, phosphocholine (PCho) and fatty acids are the substrates for the biosynthesis of PC. We find that choline kinase α (CHKA), encoded by gene CHKA, is up-regulated at the transcriptional level. The transcriptional level of PHOSPHO1 encoding phosphocholine phosphatase (PHOSPHO1) is down-regulated in HepG2.2.15 as compared with HepG2 (Fig. 3A). CHKA catalyzes the phosphorylation of choline to form PCho, which is the first reaction in the CDP-choline pathway for PC synthesis, and PHOSPHO1 catalyzes the reverse reaction18. Hence, the up regulation of CHKA and down regulation of PHOSPHO1 are consistent with the reduced levels of choline and elevated levels of PCho found in HepG2.2.15 (Fig. 3B). We further find that the levels of choline-phosphate cytidylyltransferase A (PCYT1A), choline phosphotransferase 1 (CHPT1) and choline/ethanolamine phosphotransferase 1 (CEPT1) are up-regulated at the transcriptional level (Fig. 3A). PCYT1A catalyzes the rate-limiting step in PC synthesis (from PCho to CDP-choline), and both CHPT1 and CEPT1 catalyze the reaction from CDP-choline to PC19. In addition, the levels of total fatty acids, major components of PC, are distinctly increased in HepG2.2.15, while the levels of essential fatty acids are decreased (Fig. 4), suggesting the enhancement of fatty acid biosynthesis. This observation is confirmed by the up-regulation of fatty acid synthase (FASN), acetyl-CoA carboxylase (ACC), and stearoyl-CoA desaturase (Δ9 desaturase) in HBV infected cells reported previously7. These results strongly suggest that HBV can regulate the transcription of enzymes participating in PC synthesis, leading to the promotion of the biosynthesis of PC in HepG2.2.15 cells.
Figure 3. The relationship between HBV replication and phosphatidylcholine synthesis.
(A) The mRNA levels of the enzymes involved in the phosphatidylcholine synthesis. (B) The levels of choline and phosphocholine in HepG2.2.15 and HepG2 cells. (C and D) HepG2.2.15 cells were transfected by siRNA targeting CHKA (siCHKA) or no sense control (siNC). (C) Relative mRNA levels of CHKA. (D) Relative core-associated HBV DNA levels. For all graphs, data are shown as mean ± s.d., n = 3, t-test, *P < 0.05, **P < 0.01.
Figure 4. The alterations of fatty acid composition between HepG2.2.15 and HepG2 cells.
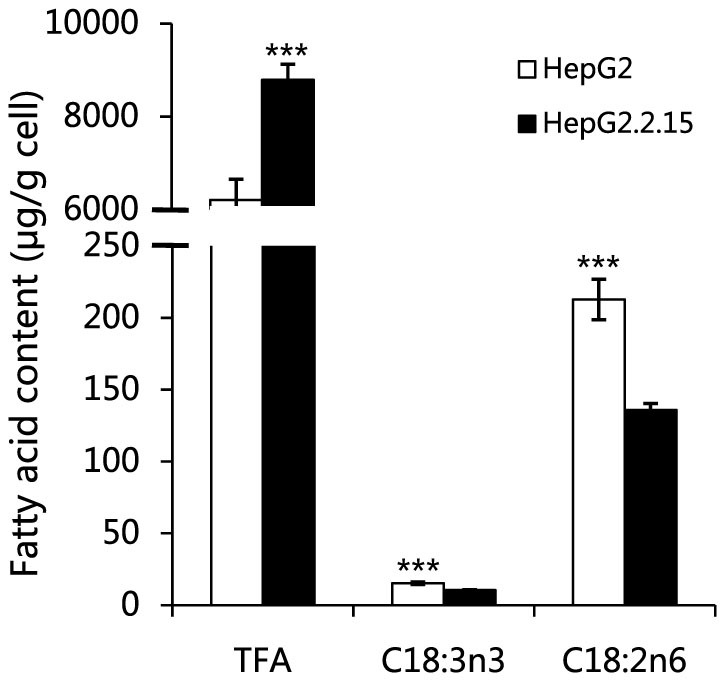
TFA, total fatty acids. Data are shown as mean ± s.d., n = 10, t-test, ***P < 0.001. More information can be found in Table S3.
To investigate the function of PC biosynthesis in HBV replication, we knocked down the expression of CHKA in HepG2.2.15 cells using targeted RNA interference (Fig. 3C). The results show that down-regulation of CHKA can drastically reduce the levels of HBV DNA (Fig. 3D). These results confirm that PC biosynthesis is essential for HBV replication.
Oxidative stress is triggered by HBV replication
Reduced glutathione (GSH) is an important antioxidant in living cells. The levels of GSH in both HepG2.2.15 cells and HepG2 cells transfected with pHBV1.3 are significantly lower than in HepG2 cells without transfection (Fig. 5A, Fig. S3B). This observation is consistent with previous studies on the chronic hepatitis B patients20. We further investigate enzymes involved in the biosynthesis of GSH encoded by genes GSS, GCLC and GCLM, at transcriptional level, and they are increased in HepG2.2.15 cells (Fig. 5B). The intracellular levels of glutamate and glycine, which are substrates for GSH biosynthesis, however, did not differ significantly between HepG2.2.15 and HepG2 cells (Fig. 5C). These results indicate that GSH biosynthesis is not inhibited and hence the reduced levels of GSH in HepG2.2.15 is likely due to the utilization of GSH in response to oxidative stress, induced by HBV replication. This notion is supported by the observation that the oxidization of branched chain amino acids in HepG2.2.15 cells result in the release of more α-ketoacids, including 2-oxoleucine (2-O-Leu), 2-oxoisovalerate (2-O-Val) and 2-oxoisoleucine (2-O-Ile) (Fig. 1C). α-ketoacids have been reported as efficient antioxidants21,22,23, therefore, elevated levels of α-ketoacids are likely used as alternative antioxidants in the case of GSH deficiency.
Figure 5. HBV replication induces oxidative stress as manifested by consumption of GSH.
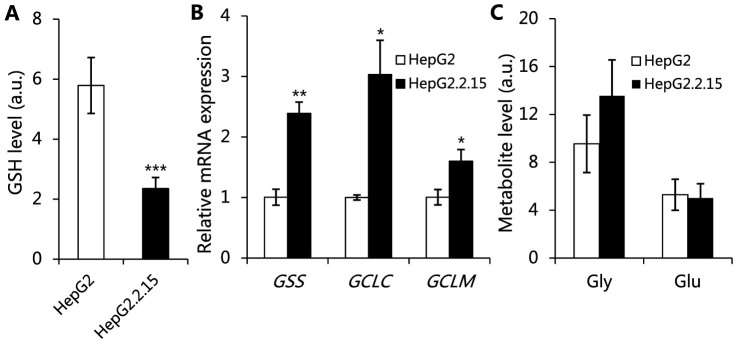
(A) The levels of GSH in HepG2.2.15 and HepG2 cells, data are shown as mean ± s.d., n = 10, t-test, ***P < 0.001. (B) The mRNA levels of enzymes involved in GSH biosynthesis in HepG2.2.15 and HepG2 cells, Data are shown as mean ± s.d., n = 3, t-test, *P < 0.05, **P < 0.01. (C) The relative levels of glycine and glutamate in the cell detected by 1H NMR, data are shown as mean ± s.d., n = 10, t-test, P > 0.05.
Discussion
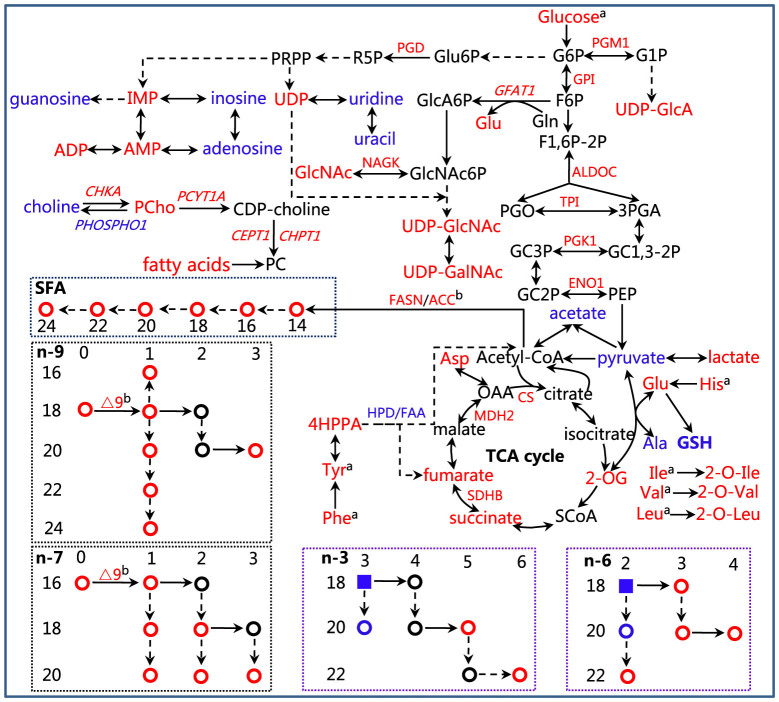
In this study, we identify the metabolic features associated with HBV infection using a metabolomics approach, molecular biology techniques, and MS-based enzyme expression analyses (Fig. 6). The results demonstrate that HBV can modify the metabolism of host cells to support its replication and expression. Therefore deciphering the metabolic pathways associated with HBV infection could shed new light on the pathological mechanisms and provide new targets for disease treatment.
Figure 6. Summary of the changes of metabolic pathways caused by HBV replication.
Metabolites, enzymes and mRNAs of enzymes are shown in red when their levels are higher in HepG2.2.15 or in blue when lower. The enzymes are display in roman type and mRNAs of the genes are shown in italic type. The full names of the enzymes are shown in Table 1. The dashed line means that the reactions involved in the transformation between metabolites need more than one step. Notes: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6-2P, fructose-1,6-diphosphate; R5P, Ribulose-5P; Glu6P, Gluconate-6 phosphate; GC1,3-2P, Glyceric acid 1,3-biphosphate; GC3P, 3-Phosphoglyceric acid; GlcA6P, glucosamine-6-phosphate; PEP, phosphoenolpyruvate; PGO, glycerone phosphate; 3PGA, glyceraldehyde-3-phosphate; PRPP, phosphoribosyl pyrophosphate; OAA, oxaloacetate; SCoA, succinyl CoA. aThe consumed metabolites; bResults from references.
Metabolic alterations induced by HBV are associated with carcinogenesis. Promoted HBP could contribute to the HBV-associated carcinogenesis. UDP-GlcNAc and UDP-GalNAc can form N-glycans to regulate the levels of surface glycoprotein to modify cell proliferation and differentiation24,25. UDP-GlcNAc is an important substrate that modifies proteins through O-linked GlcNAc (O-GlcNAc) in order to regulate the function of proteins26. The positive association between O-GlcNAc of proteins and the concentration of UDP-GlcNAc has been widely documented27,28. The O-GlcNAc is capable of modifying the products of tumor related genes, such as p53 and MYC29,30. O-GlcNAc of insulin-like growth factor proteins promote tumor progression and this leads to HCC in HBV infection31. In addition, hexosamine is also essential for the transcription and expression of tumor growth factor32,33. Taken together with our results we propose that HBP could play an important role in the development of HCC through O-GlcNAc modifying oncoproteins and signaling proteins. We also find that HBV replication promotes the biosynthesis of PC through up-regulation of relevant enzymes, such as CHKA and PCYT1A, with consistent elevations in the levels of PCho, a substrate for PC biosynthesis. The gene CHKA has been considered as a novel oncogene that mediates human cell transformation and tumorigenesis34, and the inhibitors of CHKA are efficient antitumor drugs both in vitro and in vivo35,36. The expression of CHKA is controlled by transcription factor MYC and HIF37,38, which are important in oncogenesis. Additionally, the RAS pathway can activate CHKA and PCYT1A through phosphatidylinositol 3-kinase (PI3K) and the mitogen-activated protein kinase (MAPK) signaling pathway during tumorigenesis, respectively39. Reversely, CHKA can regulate MAPK and PI3K-AKT signaling17. The results from these studies and our observations suggest that up-regulation of CHKA and PCYT1A in HBV replicating cells could contribute to the HCC induced by chronic HBV infection. This notion is reinforced by the close association between CHKA overexpression and increased levels of PCho and PC, with oncogenesis, in cells, animals and patients39,40,41. In addition, HBV replication could induce oxidative stress through producing more reactive oxygen species (ROS)6,42. This has been confirmed by our results showing that a reduced level of GSH was associated with HBV infection. ROS may activate signal pathways, such as those mediated by MAPK, PI3K, p53 and nuclear factor-κB (NF-κB), which are involved in hepatic carcinogenesis43,44.
Metabolic changes caused by HBV are associated with fatty liver and inflammation. In this study, we found that the total levels of fatty acids increased, which suggests that HBV chronic infection could induce a fatty liver. HBx is reported to play an important role in this pathological process45. However, the essential fatty acids, α-linolenic acid and linoleic acid, are present in lower levels in the HepG2.2.15 cell line compared with HepG2 cell line, and this indicates that HBV infection could also induce essential fatty acid deficiency-related diseases. The overexpression of GFAT1 in HBP has been documented to cause lipid accumulation and activation of inflammatory pathway in HepG2 cells46, implying that HBV could lead to hepatitis and fatty liver through the promotion of HBP. Hence, GFAT1-activated HBP contributes to multiple pathological processes associated with HBV infection and numerous benefits could be generated when GFAT1 is inhibited.
Metabolic profile analysis provides new information for antiviral treatment. Glycosylation is an important post-translational modification of proteins47, which is essential for HBV replication and secretion48. The disruption and inhibition of glycosylation in HBV replicating cells can reduce viral replication and secretion49,50. We confirm that the inhibition of HBP through GFAT1 can reduce HBV replication and expression. This result suggests that reduced viral replication could be due to the disruption of glycosylation of proteins through the inhibition of GFAT1-activated HBP. Most importantly, our data provides direct evidence that the inhibition of HBP could be a potential target for the therapy of HBV infection. Since PC accounts for more than 80% of total membrane lipids of HBsAg51, the up-regulation of PC biosynthesis in HepG2.2.15 cells can support the formation of the viral integrate membrane. A previous study has shown that silencing the expression of FASN, a regulator for lipid synthesis, can inhibit HBV replication52. Here, we demonstrate that silencing the expression of CHKA can inhibit HBV replication, indicating CHKA could be another potential target to treat HBV infection.
In summary, we have systematically reported the cellular metabolic responses to HBV replication. Metabolic pathways associated with HBV replication are identified. The promotion of both hexosamine and phosphatidylcholine biosynthesis, as two novel metabolic features caused by HBV replication, are documented. Importantly, these two metabolic pathways contribute to the pathological processes of HBV infection and are targets for reducing HBV replication. Our study provides new insights into the pathogenesis associated with chronic HBV infection and potential new drug therapy targets for combating HBV infection.
Methods
Cell culture for metabolomic analysis
Both HepG2.2.15 and HepG2 cells were cultured in DMEM medium with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. The cells were harvested by trypsinization at ~100% confluence and washed with cold PBS 3 times, then stored at −80°C until extraction.
Extraction of the intracellular metabolites
The cell pellets were resuspended in 1 mL water/methanol solution (1:2, v/v) and subjected to three freeze-thaw cycles, then sonicated in a wet ice bath for 15 min (1-min power following 1-min stop). The supernatant was collected after the centrifugation at 3200 × g at 4°C for 10 min. The remaining pellets were further extracted twice with the same procedure. The supernatants collected were pooled together and centrifuged at 12000 × g at 4°C for 10 min. The supernatant was lyophilized after removal of methanol by SpeedVac and stored at −80°C until NMR analysis.
Extraction of the extracellular metabolites
A total of 2 mL media from cultured cells was mixed with 4 mL methanol and placed in the ice for 30 min, followed by centrifugation at 1600 × g for 10 min at 4°C. 3 mL of supernatant was lyophilized and stored at −80°C until NMR analysis.
NMR spectroscopy
The extracts of the intracellular metabolites were dissolved in 600 μL 0.1 M phosphate buffer (pH 7.4, 99.9% D2O) containing 0.001% sodium 3-trimethylsilyl-1-[2,2,3,3-2H4] propionate (TSP)53. The extracts of the media were dissolved in 600 μL 0.08 M phosphate buffer (pH 7.4, 20% D2O) containing 0.02% TSP. All the samples were centrifuged at 16000 × g at 4°C for 10 min before transferred into the NMR tubes for the detection.
All 1D 1H NMR spectra of the cell extracts and media were acquired on a Bruker AVIII 600 MHz NMR spectrometer equipped with a cryogenic probe (Bruker Biospin, Germany) at 298 K. The first increment of NOESY pulse sequence with continuous wave irradiation on water peak during recycle delay and mixing time for water suppression was employed.
NMR spectral data analysis
The free induction decays were multiplied by an exponential window function with the line-broadening factor of 1 Hz prior to Fourier Transformation. Each spectrum was corrected for phase and baseline deformation manually using Topspin 3.0 (Bruker Biospin) and the chemical shift of TSP was calibrated at δ 0.00. The spectral region between δ 0.5 to δ 9.5 was integrated into bins with a width of 0.004 ppm using AMIX package (v3.8.3, Bruker Biospin), and the chemical shift region between δ 4.64 and δ 5.06 in the spectra of the cell extracts was removed because of imperfect water suppression. The areas of all bins were then normalized to the weight of respective cell pellets. The normalized data were used for multivariate analysis, and the model was constructed using the orthogonal projection to latent structure-discriminant analysis (OPLS-DA) with Pareto variance (Par) scaling and validated with a 7-fold cross validation method using SIMCA-P+ (v 12.0, Umetrics, Sweden). The parameter R2X is indication of the X variables being explained by the model and Q2 represents the predictability of the model. The significance of the model was also validated by another validation method, CV-ANOVA (P < 0.05)54. To assist the biological interpretation of the loadings generated from the model, the loadings were firstly back-transformed and then plotted with color-coded OPLS-DA coefficients in MATLAB 7.1 using an in-house script. The color code corresponds to the absolute value of the OPLS-DA correlation coefficients (|r|), which indicated the contribution of the corresponding variable to the group separation. The value of |r|, greater than 0.602, was considered to be significant (n = 10, P < 0.05).
A spectrum of the fresh medium was also acquired. Metabolites resonance peaks were integrated (MestReNova, Version 8.0.0, Mestrelab Research S. L., Spanish). The integration data was processed as described previously55 by taking different growth speed of HepG2 and HepG2.2.15 cells into consideration. The relative concentrations of metabolites, including those consumed and secreted into medium were then calculated using the difference between the fresh medium and the spent medium. We firstly normalized metabolites by the area under the growth curve, followed by doubling time correction.
Enzyme expression analysis
The levels of enzymes were detected using SILAC coupled with LC-MS/MS. The SILAC labeling, subcellular fractionation, and SCX-coupled RPLC-MS/MS were all carried out as described previously56. Tandem mass spectra were extracted by Analyst version 2.0. The raw MS spectra analysis was performed with ProteinPilot 4.0 (AB SCIEX, USA) using the Paragon algorithm57. Protein ratio P-value less than 0.05 indicated significant change.
Quantitative RT-PCR analysis
Total RNA was isolated using Trizol. Quantitative RT-PCR was performed using QuantiTect SYBR Green RT-PCR kit (QIAGEN). The primers sequences were given in Table S5. Gene transcription data were normalized to β-actin.
Enzyme-linked immunosorbent assay
The levels of HBsAg and HBeAg in culture supernatants of HepG2.2.15 cells were detected by the use of commercial routine diagnostic assay kits (Kehua, Shanghai) according to the manufacturer's instructions.
Transfection of small interfering RNA (siRNA)
The siRNAs target GFAT1/CHKA and negative control siRNA used in the experiment were obtained from GenePharma, Shanghai. The siRNA sequences for GFAT1 and CHKA are the same as described previously58,59. HepG2.2.15 cells were seeded at approximately 30% confluence and transfected with siRNAs by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendation.
Detection of the intracellular HBV DNA replication
HBV DNA were purified from intracellular core particles as described previously60 and quantified by real-time PCR using Sybr green reaction mix (Roche).
Author Contributions
H.Li., X.C., Y.W. and H.T. designed research; H.Li., W.Z., H.Lei., and L.Z. performed research; H.Li., X.W., L.G., H.T. and Y.W. analyzed the data; H.Li., Y.W. and H.T. wrote the paper.
Supplementary Material
Supplementary information
Table S4
Acknowledgments
We acknowledge financial supports from the Ministry of Science and Technology of China (2012CB934004, 2010CB912501, 2013CB911102), the National Natural Science Foundation of China (21175149, 21221064 and 21375144) and Chinese Academy of Sciences (KJCX2-YW-W13).
References
- Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat 11, 97–107 (2004). [DOI] [PubMed] [Google Scholar]
- Ganem D. & Prince A. M. Mechanisms of disease: Hepatitis B virus infection - Natural history and clinical consequences. N Engl J Med 350, 1118–1129 (2004). [DOI] [PubMed] [Google Scholar]
- Lai C. L. et al. Viral hepatitis B. Lancet 362, 2089–2094 (2003). [DOI] [PubMed] [Google Scholar]
- Tong A. P. et al. Proteomic analysis of cellular protein alterations using a hepatitis B virus-producing cellular model. Proteomics 8, 2012–2023 (2008). [DOI] [PubMed] [Google Scholar]
- Hajjou M. et al. cDNA microarray analysis of HBV transgenic mouse liver identifies genes in lipid biosynthetic and growth control pathways affected by HBV. J Med Virol 77, 57–65 (2005). [DOI] [PubMed] [Google Scholar]
- Yang F. et al. Expression of hepatitis B virus proteins in transgenic mice alters lipid metabolism and induces oxidative stress in the liver. J Hepatol 48, 12–19 (2008). [DOI] [PubMed] [Google Scholar]
- Kim K. H. et al. Hepatitis B virus X protein induces hepatic steatosis via transcriptional activation of SREBP1 and PPARγ. Gastroenterology 132, 1955–1967 (2007). [DOI] [PubMed] [Google Scholar]
- Li H. D. et al. Enhanced green fluorescent protein transgenic expression in vivo is not biologically inert. J Proteome Res 12, 3801–3808 (2013). [DOI] [PubMed] [Google Scholar]
- Claus S. P. et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol 4, 219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells M. A. et al. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA 84, 1005–1009 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. H. et al. Proteome responses to stable hepatitis B virus transfection and following interferon alpha treatment in human liver cell line HepG2. Proteomics 9, 1672–1682 (2009). [DOI] [PubMed] [Google Scholar]
- Williamson J. R. & Cooper R. H. Regulation of the citric acid cycle in mammalian systems. FEBS Lett 117, K73–K85 (1980). [DOI] [PubMed] [Google Scholar]
- Gregory J. F. et al. Primed, constant infusion with [2H3] serine allows in vivo kinetic measurement of serine turnover, homocysteine remethylation, and transsulfuration processes in human one-carbon metabolism. Am J Clin Nutr 72, 1535–1541 (2000). [DOI] [PubMed] [Google Scholar]
- Kornfeld R. Studies on L-glutamine D-fructose 6-phosphate aminotransferase. J Biol Chem 242, 3135–3141 (1967). [PubMed] [Google Scholar]
- Sasai K. et al. UDP-GlcNAc concentration is an important factor in the biosynthesis of beta 1,6-branched oligosaccharides: regulation based on the kinetic properties of N-acetylglucosaminyltransferase V. Glycobiology 12, 119–127 (2002). [DOI] [PubMed] [Google Scholar]
- Esko J. D. et al. Animal cells dependent on exogenous phosphatidylcholine for membrane biogenesis. Proc Natl Acad Sci USA 79, 1698–1702 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin A. et al. Selective inhibition of choline kinase simultaneously attenuates MAPK and PI3K/AKT signaling. Oncogene 29, 139–149 (2010). [DOI] [PubMed] [Google Scholar]
- Roberts S. J. et al. Human PHOSPHO1 exhibits high specific phosphoethanolamine and phosphocholine phosphatase activities. Biochem J 382, 59–65 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent C. CTP:phosphocholine cytidylyltransferase. Biochim Biophys Acta 1348, 79–90 (1997). [DOI] [PubMed] [Google Scholar]
- Fisgin N. T. et al. Oxidative stress and antioxidant defense in patients with chronic hepatitis B. Clin Lab 58, 273–280 (2012). [PubMed] [Google Scholar]
- Bunton C. A. Oxidation of α-diketones and α-keto-acids by hydrogen peroxide. Nature 163, 444–444 (1949).18115097 [Google Scholar]
- Andrae U. et al. Pyruvate and related α-ketoacids protect mammalian cells in culture against hydrogen peroxide-induced cytotoxitcity. Toxicol Lett 28, 93–98 (1985). [DOI] [PubMed] [Google Scholar]
- Salahudeen A. K. et al. Hydrogen peroxide-induced renal injury- A protective role for pyruvate in vitro and in vivo. J Clin Invest 88, 1886–1893 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K. S. et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 (2007). [DOI] [PubMed] [Google Scholar]
- Wellen K. E. et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev 24, 2784–2799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer F. I. & Hart G. W. O-glycosylation of nuclear and cytosolic proteins - Dynamic interplay between O-GlcNAc and O-phosphate. J Biol Chem 275, 29179–29182 (2000). [DOI] [PubMed] [Google Scholar]
- Robinson K. A. et al. Effects of diabetes and hyperglycemia on the hexosamine synthesis pathway in rat muscle and liver. Diabetes 44, 1438–1446 (1995). [DOI] [PubMed] [Google Scholar]
- McClain D. A. & Crook E. D. Hexosamines and insulin resistance. Diabetes 45, 1003–1009 (1996). [DOI] [PubMed] [Google Scholar]
- Shaw P. et al. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene 12, 921–930 (1996). [PubMed] [Google Scholar]
- Chou T. Y. et al. Glycosylation of the c-Myc transactivation domain. Proc Natl Acad Sci USA 92, 4417–4421 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad W. et al. Serine 204 phosphorylation and O-β-GlcNAC interplay of IGFBP-6 as therapeutic indicator to regulate IGF-II functions in viral mediated hepatocellular carcinoma. Virol J 8, 208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert C. et al. Overexpression of glutamine: fructose-6-phosphate-amidotransferase induces transforming growth factor-β1 synthesis in NIH-3T3 fibroblasts. FEBS Lett 488, 95–99 (2001). [DOI] [PubMed] [Google Scholar]
- Burt D. J. et al. P38 mitogen-activated protein kinase mediates hexosamine-induced TGFβ1 mRNA expression in human mesangial cells. Diabetologia 46, 531–537 (2003). [DOI] [PubMed] [Google Scholar]
- de Molina A. R. et al. Choline kinase is a novel oncogene that potentiates RhoA-induced carcinogenesis. Cancer Res 65, 5647–5653 (2005). [DOI] [PubMed] [Google Scholar]
- Hernando E. et al. A critical role for choline kinase-α in the aggressiveness of bladder carcinomas. Oncogene 28, 2425–2435 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Alcoceba R. et al. In vivo antitumor activity of choline kinase inhibitors: a novel target for anticancer drug discovery. Cancer Res 59, 3112–3118 (1999). [PubMed] [Google Scholar]
- Morrish F. et al. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene 28, 2485–2491 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glunde K. et al. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1 α signaling in a human prostate cancer model. Cancer Res 68, 172–180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glunde K. et al. Choline metabolism in malignant transformation. Nat Rev Cancer 11, 835–848 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerstaff E. et al. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer Res 61, 3599–3603 (2001). [PubMed] [Google Scholar]
- Bell J. D. & Bhakoo K. K. Metabolic changes underlying 31P MR spectral alterations in human hepatic tumours. NMR Biomed 11, 354–359 (1998). [DOI] [PubMed] [Google Scholar]
- Hagen T. M. et al. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc Natl Acad Sci USA 91, 12808–12812 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemin I. & Zoulim F. Hepatitis B virus induced hepatocellular carcinoma. Cancer Lett 286, 52–59 (2009). [DOI] [PubMed] [Google Scholar]
- Kuo M. T. & Savaraj N. Roles of reactive oxygen species in hepatocarcinogenesis and drug resistance gene expression in liver cancers. Mol Carcinog 45, 701–709 (2006). [DOI] [PubMed] [Google Scholar]
- Kim K. et al. Hepatitis B virus X protein induces lipogenic transcription factor SREBP1 and fatty acid synthase through the activation of nuclear receptor LXR alpha. Biochem J 416, 219–230 (2008). [DOI] [PubMed] [Google Scholar]
- Sage A. T. et al. Hexosamine biosynthesis pathway flux promotes endoplasmic reticulum stress, lipid accumulation, and inflammatory gene expression in hepatic cells. Am J Physiol Endocrinol Metabol 298, E499–E511 (2010). [DOI] [PubMed] [Google Scholar]
- Slawson C. et al. O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J Cell Biochem 97, 71–83 (2006). [DOI] [PubMed] [Google Scholar]
- Ito K. et al. Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J Virol 84, 12850–12861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. K. et al. Elevated expression of bisecting N-acetylglucosaminyltransferase-III gene in a human fetal hepatocyte cell line by hepatitis B virus. J Gastroenterol Hepatol 19, 1374–1387 (2004). [DOI] [PubMed] [Google Scholar]
- Block T. M. et al. Treatment of chronic hepadnavirus infection in a woodchuck animal model with an inhibitor of protein folding and trafficking. Nat Med 4, 610–614 (1998). [DOI] [PubMed] [Google Scholar]
- Satoh O. et al. Lipid composition of hepatitis B virus surface antigen particles and the particle-producing human hepatoma cell lines. J Lipid Res 31, 1293–1300 (1990). [PubMed] [Google Scholar]
- Zhang H. et al. Differential regulation of host genes including hepatic fatty acid synthase in HBV-transgenic mice. J Proteome Res 12, 2967–2979 (2013). [DOI] [PubMed] [Google Scholar]
- Xiao C. et al. An optimized buffer system for NMR-based urinary metabonomics with effective pH control, chemical shift consistency and dilution minimization. Analyst 134, 916–925 (2009). [DOI] [PubMed] [Google Scholar]
- Eriksson L. et al. CV-ANOVA for significance testing of PLS and OPLS (R) models. J Chemom 22, 594–600 (2008). [Google Scholar]
- Jain M. et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.-K. et al. Identification of host proteins involved in Japanese encephalitis virus infection by quantitative proteomics analysis. J Proteome Res 12, 2666–2678 (2013). [DOI] [PubMed] [Google Scholar]
- Shilov I. V. et al. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics 6, 1638–1655 (2007). [DOI] [PubMed] [Google Scholar]
- Li M.-D. et al. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab 17, 303–310 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glunde K. et al. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res 65, 11034–11043 (2005). [DOI] [PubMed] [Google Scholar]
- Meng Z. J. et al. Inhibition of hepatitis B virus gene expression and replication by endoribonuclease-prepared siRNA. J Virol Methods 150, 27–33 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Table S4