Abstract
Epigenetic events including chromatin remodeling and histone modifications have recently emerged as important contributors to a variety of neurodevelopmental disorders. This review focuses on CHARGE syndrome, a multiple anomaly condition caused by mutations in the gene encoding CHD7, an ATP-dependent chromatin remodeling protein. CHD7 exhibits pleiotropic effects during embryonic development, consistent with highly variable clinical features in CHARGE syndrome. In this review, a historical description of CHARGE is provided, followed by establishment of diagnostic criteria, gene discovery, and development of animal models. Current understanding of epigenetic CHD7 functions and interacting proteins in cells and tissues is also presented, and final emphasis is placed on challenges and major questions to be answered with ongoing research efforts.
Keywords: chromatin remodeling, CHARGE Syndrome, autism, intellectual disability
Introduction
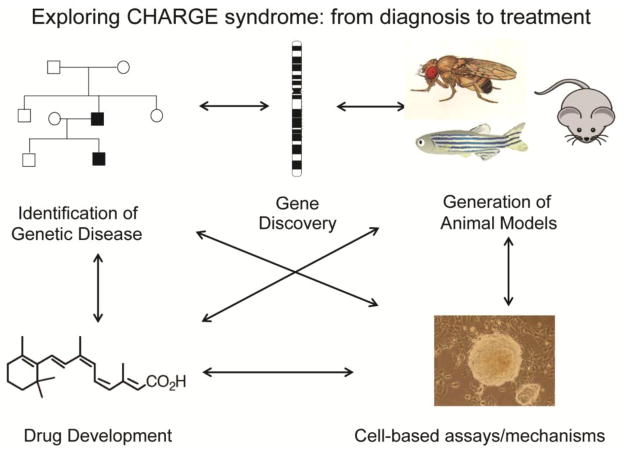
Genes that encode histone modifying enzymes are increasingly being identified as causative in a variety of developmental disorders. Included among such disorders is CHARGE syndrome, caused by heterozygous mutations in the gene encoding CHD7, a chromatin remodeling protein, is characterized by Coloboma, Heart defects, Atresia of the choanae, Retardation of growth and development, Genital hypoplasia, and Ear abnormalities including deafness and vestibular disorders [1]. CHARGE syndrome affects 1 in 10,000 to 1 in 8,000 newborns worldwide [2,3]. Individuals with CHARGE syndrome exhibit broad variability in clinical features, even between affected individuals from the same family who carry the same mutation. The mechanisms underlying this variability are not known, but are likely related to changes in gene expression or genetic modifiers. Roles for CHD7 in cells include formation of large protein complexes and regulation of movement of nucleosomes along DNA. In this review, the historical significance of CHARGE association (later renamed CHARGE syndrome) is presented, followed by the impact of CHD7 gene discovery on diagnosis and understanding of the phenotypic spectrum association with CHARGE. Finally, an exploration of recent discoveries and challenges is provided, highlighting the pleiotropic effects of CHD7 and interacting partners in developing cells and tissues. Figure 1 illustrates the general paradigm by which CHARGE syndrome and CHD7 research has evolved: from family studies and descriptions, to gene identification and generation/characterization of animal models, to development of cell and molecule based approaches and, ultimately, back to treatments for affected individuals.
Figure 1.
Illustration of the paradigm for CHARGE clinical description, gene discovery, development of animal models, and pursuit of molecular and cell based therapies.
CHARGE: from association to syndrome
The first reports of individuals who were later found to have CHARGE occurred simultaneously in 1979. “Choanal atresia and associated multiple anomalies” was described by Dr. Bryan Hall in a series of 17 patients ascertained primarily by bilateral or unilateral posterior choanal atresia [4]. The same year, Hittner and colleagues published a series of 9 children and one adult, including a mother-daughter, with colobomatous microphthalmia, heart disease, abnormalities of the external ear with associated hearing loss, and mental retardation [5]. The phrase “CHARGE association” was coined two years later by Pagon and colleagues, when an additional 21 individuals were described as having a similar constellation of features, including choanal atresia or ocular coloboma or both, and at least four of the seven most common findings (coloboma, congenital heart disease, choanal atresia, postnatal growth deficiency, mental retardation and/or CNS anomalies, hypogonadism, and ear anomalies/deafness) [6]. Interestingly, Pagon et al remarked that “At least two separate, but perhaps indistinguishable, genetic forms may exist: autosomal dominant inheritance is suggested by one family in which a mother and daughter appear to be affected; autosomal recessive inheritance seems likely in another family in which an affected sister and brother have normal parents”. This prescient observation, written 23 years before the gene for CHARGE was identified, was correct in concluding that autosomal dominant transmission can occur, since it is now known that heterozygous mutations in CHD7 cause CHARGE; however, autosomal recessive inheritance has not been reported, probably because loss of both CHD7 alleles is embryonic lethal. An alternative explanation for the family with two affected children and unaffected parents is germline mosaicism for a de novo mutation.
These early descriptive studies of affected individuals and their family members established a solid foundation for the variety of clinical features observed in CHARGE, and led to the first set of proposed diagnostic criteria by Blake in 1998 [7]. Additional input by Verloes elevated the importance of semicircular canal dysgenesis, revised the diagnostic criteria to include typical and atypical cases, and provided impetus to change the official name to “CHARGE syndrome” [8]. It is highly noteworthy (though perhaps not surprising to the dysmorphologists and clinicians involved) that both diagnostic criteria by Blake (1998) and Verloes (2005) have survived beyond discovery of the CHD7 gene, suggesting high specificity and sensitivity for these early astute observations.
As mentioned above, CHARGE syndrome was considered early on to be a potential genetic disorder, based upon reports of familial transmission, similarity of features between affected individuals, and chromosomal abnormalities in isolated cases. However, early searches yielded no major causative genes [9]. This situation changed dramatically in 2004, when Vissers et al reported identification of CHD7 as the causative gene for CHARGE [1]. Their report built upon earlier studies of a child with a large deletion of chromosome 8q12 and another child with a balanced translocation of chromosomes 6 and 8, and was the first successful identification of a single gene disorder using array comparative genomic hybridization (cGH). In retrospect, it is ironic that the technology (array cGH) used to discover the CHD7 gene in CHARGE does not typically reveal the genetic mutation, since the vast majority of individuals with CHARGE have single base pair mutations or very small deletions/duplications that do not affect the entire gene [10].
Over 500 different human pathogenic mutations in CHD7 have been identified thus far (www.chd7.org), in all but one of the 37 coding exons and in some intronic sequences. These mutations affect known protein domains, including the N-terminal chromodomains, helicase domains, and c-terminal SANT/BRK domains. However, only one study has systematically tested the effects of CHD7 mutations in biochemical assays, showing that mutations disrupt ATP-depending nucleosome sliding along DNA and accessibility of chromatin to restriction enzymes [11]. Although not yet available, a rapid, high-throughput, cell or animal-based assay would be highly informative not only for confirming the pathogenicity of these variants for clinical diagnosis and counseling, but for providing critical information about novel and known protein functions.
Variability in CHARGE clinical features
In the 10 years since CHD7 was discovered there have been 222 publications indexed in www.pubmed.org using “CHD7” as a keyword. Early reports included extensive genotype-phenotype analyses of large cohorts of individuals [12–15]. Results of these studies suggest that individuals with atypical CHARGE who do not meet established criteria tend to have missense mutations, whereas individuals with typical CHARGE more often have loss-of-function or deletion mutations. In another series of studies, investigators asked whether individuals with isolated CHARGE features also have CHD7 mutations. To date, such studies indicate that CHD7 mutations are rarely associated with isolated hypogonadotropic hypogonadism and congenital heart disease but not with isolated semicircular canal dysplasia and clefting [16–20]. Other reports point to rare individuals with developmental delay, autism spectrum disorder, or intellectual disability and CHD7 mutations [21–23]. Further biochemical analyses of these newly reported mutations are necessary to establish their effects on CHD7 protein function.
Individuals with CHARGE face a variety of health challenges that change with age. In the first few months of life, major difficulties include feeding, respiratory dysfunction, and cardiac disease. CHARGE is also the second leading cause of deaf-blindness (after Usher syndrome), although most individuals with CHARGE have some degree of hearing and visual abilities. CHARGE is also a major cause of balance disturbance, facial palsy, impaired pain sensation, and hyposmia/anosmia. These sensory impairments are often identified shortly after birth while families are adjusting to medical, life-threatening illnesses in their baby. In later childhood, developmental delays, speech and hearing impairments, and growth delays become intense areas of focus for physicians and families. Children with CHARGE syndrome often require hormonal supplementation, attributable to hypogonadotropic hypogonadism, hypothyroidism, growth hormone deficiency, or a combination of each. Children and young adults with CHARGE may also have developmental challenges that require special accommodations for sensory impairments, learning disabilities, and attention disorders, and may benefit from supportive services to help them transition to adulthood.
Several recent studies have summarized the prevalence of specific disorders associated with CHARGE. The most commonly reported condition is dysplasia or aplasia of the semicircular canals, which is considered almost pathognomonic for CHARGE since it occurs in 94–98% of affected individuals regardless of CHD7 mutation status [24,25]. Ocular coloboma is also highly penetrant in CHARGE, occurring in 75–81% of individuals with CHARGE and CHD7 mutations [24,25]. Cardiac abnormalities, cranial nerve dysfunction, choanal atresia and clefting are less common, varying from 30–75% [24,25]. The high degree of variability in penetrance of clinical features by organ system suggests that certain cells or tissues are more susceptible to altered CHD7 function than others. Identification of these tissue specific CHD7 requirements will certainly influence the design of rationale therapies toward promoting organ and tissue regeneration.
CHD7 functions, animal models and interacting partners
The clinical features of CHARGE are consistent with emerging information about the functions of CHD7 protein. CHD7 forms large complexes with other proteins, and acts in the cell nucleus to move nucleosomes along, or away from, DNA [11]. This function of CHD7 endows it with a critical role in co-regulation of gene expression at thousands of sites in the human genome. Like other CHD family member proteins, CHD7 binds preferentially to methylated histones at specific Lysine residues, and has no a priori preference for binding directly to specific DNA sequences [26–28]. CHD7 has been reported to be enriched at methylated histones in enhancer regions and near transcription start sites, and loss of Chd7 in cells leads to misregulation of thousands of other genes [28]. A major challenge, therefore, is to determine which downstream target genes share responsibility for the phenotypes observed in CHARGE, and whether these target genes might be sensitive to treatment.
Several key areas of research have been pursued since the CHD7 gene was discovered in CHARGE. Mouse models have proven invaluable for recapitulating the phenotypes observed in humans, and for testing responses to altered vitamin A signaling pathways [29–31]. Kismet, the Drosophila orthologue of Chd7, also exhibits abnormalities reminiscent of CHARGE, including disrupted neural development [32–34]. In kismet mutant flies, RNA polymerase II transcriptional elongation is abnormal, suggesting another potential avenue for intervention [33]. Work in zebrafish has also uncovered deficits in ribosomal RNA dysgenesis, and overlapping functions between kismet/CHD7 and other genes that contribute to other craniofacial conditions, including Treacher-Collins syndrome [35,36]. Studies on Xenopus laevis also showed that neural crest derivatives are sensitive to changes in Chd7 dosage and exhibit CHARGE-like features, providing yet another potential route for early developmental intervention [37]. There is also emerging data that CHD7 binds to and down-regulates the tumor suppressor p53, and may act to control programmed cell death [38].
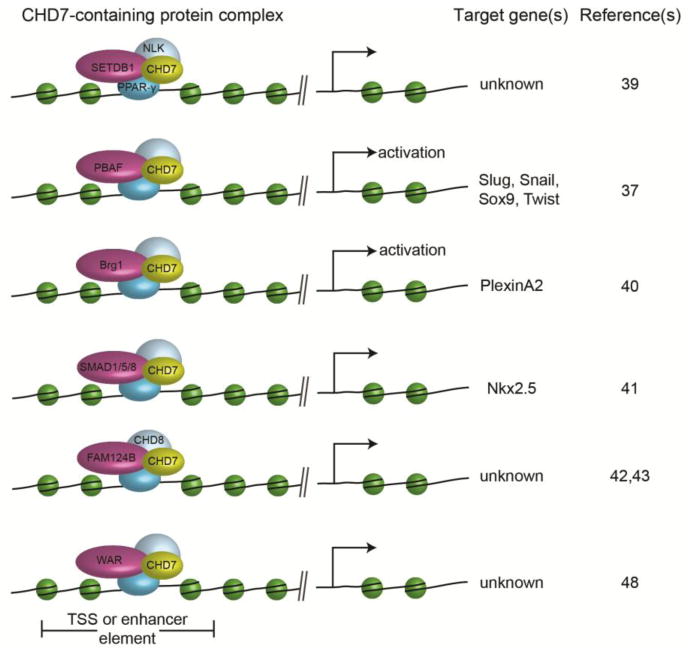
The epigenetic effects of CHD7 on chromatin and gene regulation appear to depend largely upon the precise composition(s) of large protein complexes with which it interacts (Fig. 2). In mesenchymal stem cells, CHD7 forms a complex with SETDB1, a histone methyltransferase, and Nemo-Like Kinase (NLK) in response to Wnt5a-mediated signaling [39]. This complex then translocates to the nucleus where it binds to and regulates PPAR-γ transactivation of target genes to promote cell fate decisions [39]. In developing neural crest cells, CHD7 interacts with members of the PBAF complex to regulate expression of Twist, Slug, Snail, and Sox9 and with Brg1 specifically to promote PlexinA2 expression [37,40]. Cardiac myocytes, some of which are likely to have their developmental origins in neural crest, also express CHD7 and form complexes with R-SMAD proteins SMAD1/5/8, (components of the BMP signaling pathway) which then regulate expression of the cardiac transcription factor gene Nkx2.5 [41]. These examples illustrate the variety of protein complexes and cell types/tissues involved in CHD7 functions.
Figure 2.
Cartoon showing known CHD7 binding partners and target genes. Additional details and references are provided in the text.
In addition to these known tissue-specific factors, CHD7 also binds to other proteins based on in vitro assays in heterologous cells. CHD8, a member of the third subfamily of CHD chromatin remodeling proteins, and FAM124B were recently shown to form a large protein complex, but its function in cells and tissues has not yet been defined [42,43]. In humans, mutations in CHD8 were recently reported in individuals with autism, macrocephaly and craniofacial dysmorphisms, but no other features suggestive of CHARGE syndrome [44–46]. Interestingly, mutations in two other CHD family genes, CHD2 and CHD4, were recently associated with seizures and developmental delay, raising the possibility that mutations in additional family member genes in other human neurological disorders remain to be identified. It is not yet known whether CHD2 or CHD4 form protein complexes with CHD7.
Critical questions for CHARGE and CHD7 research
Future research will need to address several key questions. To date, little is known about the factors that regulate expression of CHD7. In mice, the Chd7 gene is highly expressed in embryonic stem cells and in the early (e7.5-e8.5) embryo, and becomes progressively downregulated [47]. By e12.5, Chd7 expression is restricted to neural progenitors in the brain, cranial ganglia, and enteric nervous system, and to a variety of neural crest derivatives [30]. It is also expressed in heart, kidney, limb bud, and craniofacial mesenchyme, among other tissues. This gradual restriction of Chd7 gene expression argues for the presence of active factors that bind to the Chd7 gene promoter and restrict its expression. Identification of these factors will be useful because such factors could be targets for interventions aimed at correction or de-repression of mutant alleles.
An open question that requires intensive study is the genetic cause(s) of CHARGE in individuals who meet clinical criteria yet have no identifiable mutation in CHD7. Recent studies suggest that roughly 10–20% of individuals with CHARGE do not have CHD7 gene mutations or gene dosage abnormalities as a genetic explanation [10]. Notably, there is significant overlap in clinical features between CHARGE and other single gene or oligo-genic disorders including Kabuki syndrome, Renal-coloboma syndrome, Branchio-oto-renal (BOR) and Branchio-otic (BO) syndromes, Kleefstra syndrome, Mowat-Wilson syndrome, and Mandibulofacial dysostosis [48–53]. The overlap in features between CHARGE and Kabuki syndromes may be explained in part by common interacting partners. Like KMT2D, the H3K4 histone methyltransferase mutated in some individuals with Kabuki syndrome, CHD7 was recently shown to interact with members of the WAR complex of proteins (WDR5, ASH2L, and RbBP5), suggesting that regulation of target genes by this complex may be similar in cells and tissues affected in CHARGE and Kabuki [48].
Application of next-generation sequencing techniques, including whole exome sequencing and whole genome sequencing, should help uncover new mutations in CHD7 or in novel genes associated with CHARGE. Whole genome sequencing, with deep coverage, may also help identify somatic mutations (those occurring later than the zygotic stage), mutations in genes with low coverage on whole exome sequencing, and mutations in noncoding DNA regions. A very recent and promising study of 1500 individuals with nonsyndromic intellectual disability used a step-wise approach with array CGH (to detect genomic deletions or duplications), followed by whole exome sequencing (to identify mutations in exons), then whole genome sequencing (to identify noncoding mutations or mutations in genes that were poorly covered by exome sequencing) [54]. This approach led to a genetic diagnosis in 60% of cumulative cases, suggesting that next generation sequencing technologies may also uncover novel genetic causes of CHARGE [54].
Finally, advances in small molecule or chemical based treatments will require the availability of human cell lines that harbor mutations in CHD7. Use of induced pluripotent stem cells from affected individuals, compared to cells corrected using CRISPR/Cas9 gene editing, will be very powerful for determining effects of CHD7 dysfunction in a wide variety of cells and tissues, and will be useful for studying cellular responses to medications that require functional CHD7.
Conclusion
CHARGE syndrome is a relatively common, multiple birth defect condition that is being increasingly recognized and diagnosed worldwide. Identification of CHD7 mutations in 2004 as the primary cause of CHARGE has led to astonishing advances in understanding of how disrupted chromatin remodeling can impact human health, and has posed new questions about how to harness this information to develop targeted therapies. Treatment and prevention of CHD7 mutation-related effects on developing cells and tissues are goals that will require intense study of epigenetic changes in chromatin across developmental space and time, using newly developed mouse, fly, fish, and frog models. Design and implementation of these therapies will also require close collaboration among affected individuals, their physicians, and researchers. Fortunately, the time is now, the field is well poised for such advances, and affected individuals and their families are waiting with bated breath.
Acknowledgments
DM Martin is supported by NIH R01-DC009410 and The University of Michigan Donita B. Sullivan MD Research Professorship Funds. She also serves as Chair of the Scientific Advisory Board of the CHARGE Syndrome Foundation. She receives reimbursement for travel to board meetings 1–2 times per year. She also received funds to support a local research symposium on chromatin and development.
Footnotes
Conflict of Interest
DM Martin declares no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Vissers LE, van Ravenswaaij CM, Admiraal R, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004 Sep;36(9):955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 2.Issekutz KA, Graham JM, Jr, Prasad C, Smith IM, Blake KD. An epidemiological analysis of CHARGE syndrome: preliminary results from a Canadian study. Am J Med Genet A. 2005 Mar 15;133(3):309–317. doi: 10.1002/ajmg.a.30560. [DOI] [PubMed] [Google Scholar]
- 3.Kallen K, Robert E, Mastroiacovo P, Castilla EE, Kallen B. CHARGE Association in newborns: a registry-based study. Teratology. 1999 Dec;60(6):334–343. doi: 10.1002/(SICI)1096-9926(199912)60:6<334::AID-TERA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 4.Hall BD. Choanal atresia and associated multiple anomalies. J Pediatr. 1979;95(3):395–398. doi: 10.1016/s0022-3476(79)80513-2. [DOI] [PubMed] [Google Scholar]
- 5.Hittner HM, Hirsch NJ, Kreh GM, Rudolph AJ. Colobomatous microphthalmia, heart disease, hearing loss, and mental retardation--a syndrome. Journal of Pediatric Ophthalmology & Strabismus. 1979;16(2):122–128. doi: 10.3928/0191-3913-19790301-10. [DOI] [PubMed] [Google Scholar]
- 6.Pagon RA, Graham JM, Jr, Zonana J, Yong SL. Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J Pediatr-Us. 1981;99(2):223–227. doi: 10.1016/s0022-3476(81)80454-4. [DOI] [PubMed] [Google Scholar]
- 7.Blake KD, Davenport SL, Hall BD, et al. CHARGE association: an update and review for the primary pediatrician. Clinical pediatrics. 1998 Mar;37(3):159–173. doi: 10.1177/000992289803700302. [DOI] [PubMed] [Google Scholar]
- 8.Verloes A. Updated diagnostic criteria for CHARGE syndrome: a proposal. Am J Med Genet A. 2005 Mar 15;133(3):306–308. doi: 10.1002/ajmg.a.30559. [DOI] [PubMed] [Google Scholar]
- 9.Martin DM, Probst FJ, Fox SE, et al. Exclusion of PITX2 mutations as a major cause of CHARGE association. Am J Med Genet. 2002 Jul 22;111(1):27–30. doi: 10.1002/ajmg.10473. [DOI] [PubMed] [Google Scholar]
- 10••.Janssen N, Bergman JE, Swertz MA, et al. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum Mutat. 2012 Aug;33(8):1149–1160. doi: 10.1002/humu.22086. This paper summarizes recent genotype-phenotype information about individuals with CHARGE syndrome. It also highlights a publicly available database, www.chd7.org, for CHD7 mutations and their classification. [DOI] [PubMed] [Google Scholar]
- 11••.Bouazoune K, Kingston RE. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc Natl Acad Sci U S A. 2012 Nov 20;109(47):19238–19243. doi: 10.1073/pnas.1213825109. Bouazoune and Kingston provide the first evidence that CHD7 in an ATP-dependent chromatin remodeling protein, with roles in regulation of nucleosome position and restriction enzyme accessibility to chromatin and DNA. They also show that mutations in CHD7 association with CHARGE syndrome disrupt these nuclear functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalani SR, Safiullah AM, Fernbach SD, et al. Spectrum of CHD7 Mutations in 110 Individuals with CHARGE Syndrome and Genotype-Phenotype Correlation. Am J Hum Genet. 2006 Feb;78(2):303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jongmans MC, Admiraal RJ, van der Donk KP, et al. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet. 2006 Apr;43(4):306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aramaki M, Udaka T, Kosaki R, et al. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J Pediatr. 2006 Mar;148(3):410–414. doi: 10.1016/j.jpeds.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 15.Sanlaville D, Etchevers HC, Gonzales M, et al. Phenotypic spectrum of CHARGE syndrome in fetuses with CHD7 truncating mutations correlates with expression during human development. J Med Genet. 2005 Sep 16; doi: 10.1136/jmg.2005.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HG, Kurth I, Lan F, et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008 Oct;83(4):511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felix TM, Hanshaw BC, Mueller R, Bitoun P, Murray JC. CHD7 gene and non-syndromic cleft lip and palate. Am J Med Genet A. 2006 Oct 1;140(19):2110–2114. doi: 10.1002/ajmg.a.31308. [DOI] [PubMed] [Google Scholar]
- 18.Green GE, Huq FS, Emery SB, Mukherji SK, Martin DM. CHD7 mutations and CHARGE syndrome in semicircular canal dysplasia. Otology & neurotology : official publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology. 2014 Sep;35(8):1466–1470. doi: 10.1097/MAO.0000000000000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corsten-Janssen N, du Marchie Sarvaas GJ, Kerstjens-Frederikse WS, et al. CHD7 mutations are not a major cause of atrioventricular septal and conotruncal heart defects. Am J Med Genet A. 2014 Sep 24; doi: 10.1002/ajmg.a.36747. [DOI] [PubMed] [Google Scholar]
- 20.Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013 Jun 13;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tucker T, Zahir FR, Griffith M, et al. Single exon-resolution targeted chromosomal microarray analysis of known and candidate intellectual disability genes. Eur J Hum Genet. 2014 Jun;22(6):792–800. doi: 10.1038/ejhg.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang YH, Yuen RK, Jin X, et al. Detection of Clinically Relevant Genetic Variants in Autism Spectrum Disorder by Whole-Genome Sequencing. Am J Hum Genet. 2013 Jul 10; doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012 May 10;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zentner GE, Layman WS, Martin DM, Scacheri PC. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A. 2010 Mar;152A(3):674–686. doi: 10.1002/ajmg.a.33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergman JE, Janssen N, Hoefsloot LH, Jongmans MC, Hofstra RM, van Ravenswaaij-Arts CM. CHD7 mutations and CHARGE syndrome: the clinical implications of an expanding phenotype. J Med Genet. 2011 May;48(5):334–342. doi: 10.1136/jmg.2010.087106. [DOI] [PubMed] [Google Scholar]
- 26.Schnetz MP, Bartels CF, Shastri K, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009 Apr;19(4):590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnetz MP, Handoko L, Akhtar-Zaidi B, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010 Jul;6(7):e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011 Aug;21(8):1273–1283. doi: 10.1101/gr.122382.111. Zentner and colleagues demonstrate that CHD7 binds to “poised” enhancers, which show preference for regulation of ectodermal derivatives. This study, combined with their earlier reports, confirm that CHD7 has a broad repertoire of binding sites in the mammalian genome, including transcription start sites and enhancer regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Micucci JA, Layman WS, Hurd EA, et al. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum Mol Genet. 2014 Jan 15;23(2):434–448. doi: 10.1093/hmg/ddt435. The authors of this study provide the first evidence that changes in retinoic acid signaling can influence the developmental effects of CHD7 on inner ear development. Using a combination of mouse genetics and in vitro neural stem cell assays, they show that CHD7 is a key regulator of neural stem cell proliferation and that CHD7 deficiency leads to impaired neuronal development in the brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurd EA, Capers PL, Blauwkamp MN, et al. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome. 2007 Feb;18(2):94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- 31.Bosman EA, Penn AC, Ambrose JC, Kettleborough R, Stemple DL, Steel KP. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 2005 Nov 15;14(22):3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 32.Melicharek DJ, Ramirez LC, Singh S, Thompson R, Marenda DR. Kismet/CHD7 regulates axon morphology, memory and locomotion in a Drosophila model of CHARGE syndrome. Hum Mol Genet. 2010 Nov 1;19(21):4253–4264. doi: 10.1093/hmg/ddq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005 Apr;132(7):1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- 34.Daubresse G, Deuring R, Moore L, et al. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development. 1999 Mar;126(6):1175–1187. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- 35.Patten SA, Jacobs-McDaniels NL, Zaouter C, Drapeau P, Albertson RC, Moldovan F. Role of Chd7 in zebrafish: a model for CHARGE syndrome. PLoS One. 2012;7(2):e31650. doi: 10.1371/journal.pone.0031650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balow SA, Pierce LX, Zentner GE, et al. Knockdown of fbxl10/kdm2bb rescues chd7 morphant phenotype in a zebrafish model of CHARGE syndrome. Dev Biol. 2013 Oct 1;382(1):57–69. doi: 10.1016/j.ydbio.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bajpai R, Chen DA, Rada-Iglesias A, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010 Feb 18;463(7283):958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Nostrand JL, Brady CA, Jung H, et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature. 2014 Aug 3; doi: 10.1038/nature13585. This manuscript shows that the activation of the tumor suppressor p53 is associated with some features of CHARGE syndrome in mice. The authors also present evidence that CHD7 binds to and suppresses p53, suggesting that CHD7 is also involved in regulation of cell survival. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takada I, Mihara M, Suzawa M, et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007 Nov;9(11):1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Xiong Y, Shang C, et al. Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1738–1743. doi: 10.1073/pnas.1218072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Harmelink C, Peng Y, Chen Y, Wang Q, Jiao K. CHD7 interacts with BMP R-SMADs to epigenetically regulate cardiogenesis in mice. Hum Mol Genet. 2014 Apr 15;23(8):2145–2156. doi: 10.1093/hmg/ddt610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batsukh T, Pieper L, Koszucka AM, et al. CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Hum Mol Genet. 2010 Jul 15;19(14):2858–2866. doi: 10.1093/hmg/ddq189. [DOI] [PubMed] [Google Scholar]
- 43.Batsukh T, Schulz Y, Wolf S, et al. Identification and Characterization of FAM124B as a Novel Component of a CHD7 and CHD8 Containing Complex. PLoS One. 2012;7(12):e52640. doi: 10.1371/journal.pone.0052640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernier R, Golzio C, Xiong B, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014 Jul 17;158(2):263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014 Feb;37(2):95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012 Dec 21;338(6114):1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Randall V, McCue K, Roberts C, et al. Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J Clin Invest. 2009 Nov;119(11):3301–3310. doi: 10.1172/JCI37561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Schulz Y, Freese L, Manz J, et al. CHARGE and Kabuki syndromes: a phenotypic and molecular link. Hum Mol Genet. 2014 Aug 15;23(16):4396–4405. doi: 10.1093/hmg/ddu156. In this paper, the authors highlight the significant phenotypic similarity between CHARGE and Kabuki syndrome. Kabuki syndrome is caused by mutations in the KMT2D gene, which encodes a H3K4 histone methyltransferase. The authors further demonstrate that CHD7 and KMT2D also share interacting protein partners. [DOI] [PubMed] [Google Scholar]
- 49.Schimmenti LA. Renal coloboma syndrome. Eur J Hum Genet. 2011 Dec;19(12):1207–1212. doi: 10.1038/ejhg.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song MH, Kwon TJ, Kim HR, et al. Mutational analysis of EYA1, SIX1 and SIX5 genes and strategies for management of hearing loss in patients with BOR/BO syndrome. PLoS One. 2013;8(6):e67236. doi: 10.1371/journal.pone.0067236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleefstra T, Kramer JM, Neveling K, et al. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet. 2012 Jul 13;91(1):73–82. doi: 10.1016/j.ajhg.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lines MA, Huang L, Schwartzentruber J, et al. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet. 2012 Feb 10;90(2):369–377. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenger TL, Harr M, Ricciardi S, et al. CHARGE-like presentation, craniosynostosis and mild Mowat-Wilson Syndrome diagnosed by recognition of the distinctive facial gestalt in a cohort of 28 new cases. Am J Med Genet A. 2014 Oct;164(10):2557–2566. doi: 10.1002/ajmg.a.36696. [DOI] [PubMed] [Google Scholar]
- 54.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014 Jul 17;511(7509):344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]