Abstract
Cancer of unknown primary (CUP) represents a common and important clinical problem. There is evidence that most CUPs are metastases of carcinomas whose primary site cannot be recognized. Driven by the hypothesis that the knowledge of primary cancer could improve patient’s prognosis, we investigated microRNA expression profiling as a tool for identifying the tissue of origin of metastases. We assessed microRNA expression from 101 formalin-fixed, paraffin-embedded (FFPE) samples from primary cancers and metastasis samples by using a microarray platform. Forty samples representing ten different cancer types were used for defining a cancer-type-specific microRNA signature, which was used for predicting primary sites of metastatic cancers. A 47-miRNA signature was identified and used to estimate tissue-of-origin probabilities for each sample. Overall, accuracy reached 100% for primary cancers and 78% for metastases in our cohort of samples. When the signature was applied to an independent published dataset of 170 samples, accuracy remained high: correct prediction was found within the first two options in 86% of the metastasis cases (first prediction was correct in 68% of cases). This signature was also applied to predict 16 CUPs. In this group, first predictions exhibited probabilities higher than 90% in most of the cases. These results establish that FFPE samples can be used to reveal the tissue of origin of metastatic cancers by using microRNA expression profiling and suggest that the approach, if applied, could provide strong indications for CUPs, whose correct diagnosis is presently undefined.
Keywords: microRNA, cancer with unknown primary, metastasis, microarray
Introduction
Ten to fifteen per cent of cancer cases are first diagnosed as metastases. In spite of efforts, the primary cancer site is never identified in about one-third of these cases, even after extensive clinical, advanced imaging, and immunohistochemical (IHC) analyses [1–5]. If no primary is identified, the tumour is defined as cancer of unknown primary (CUP), a diagnosis that is associated with 3–5% of new cancers [3]. Hence, CUP ranks among the ten most frequent cancer diagnoses and because of its poor prognosis, it ranks fourth as the cause of cancer-associated deaths in western countries [6,7], thus representing a common and important clinical problem.
Because of their aggressive clinical behaviour, it has been hypothesized that CUPs may constitute a distinct cancer entity [8]. However, no specific genetic factors or mutations that uniquely characterize CUP have yet been described, while the identification of a primary site can be achieved in 70–80% of patients based on post-mortem autopsies. From these data, it was demonstrated that most CUPs originate from carcinomas: 85–90% adenocarcinomas and poorly differentiated carcinomas, 5–10% squamous carcinomas, and 5% neuroendocrine carcinomas. The lungs represent the most common primary site of CUPs, followed by various gastrointestinal cancers (pancreas, colon, stomach, liver). Frequently occurring cancers, such as breast and prostate, have rarely been identified as primary sites of CUPs [6,9–11].
As therapy and clinical management largely rely on tumour type and extent of disease, in the case of CUP treatment, options have been defined according to the most likely primary [1]. For CUPs that do not belong to a defined subcategory, standardized chemotherapeutic regimens have been proposed [12]. This approach, however, did not seem to be very effective. In fact, the prognosis of patients with CUPs remains poor, with median survival ranging from 6 to 10 months [13]. Although all patients with CUPs have advanced, metastatic disease, those for whom the primary source of cancer is identified have longer survival [14].
Driven by the hypothesis that the knowledge of primary cancer can establish a more rational therapeutic approach and potentially improve patient’s prognosis and quality of life, there have been important efforts in the last decade to find markers and methods able to improve the diagnosis of CUPs. In particular, protocols involving a combination of thorough physical examination, advanced imaging techniques, and IHC markers have been developed to improve the rate of primary identification [1]. To this end, multigene expression profiling seems to be a potentially excellent approach for tissue-of-origin identification, and microRNAs seem to be very important markers in cancer [15,16]. Indeed, also if specific microRNAs modulated during the metastatic process have been identified [17], both mRNA and microRNA expression assays proved their ability to reveal the tissue of origin by using cancer-specific genes retained by metastasis, which led to the recent development of molecular tests based on microarray or quantitative PCR methods (see Monzon and Koen for a review [18]).
Here, we performed a primary site prediction of 101 biopsies, comprising 16 CUPs, based on a 47-miRNA classifier. The study demonstrated high sensitivity and specificity, and CUP prediction was largely in agreement with autopsy statistics. Last, but not least, it confirmed the possibility of using FFPE specimens in routine microarray-based analyses without any loss of samples.
Materials and methods
Patients and tumour samples
Eighty-five patients who had primary or metastatic carcinomas and who were diagnosed and treated at the University Hospital of Ferrara between 2005 and 2008 were included in this study. The study was approved by the local Institutional Ethical Committee. For 16 patients, both the primary and the corresponding metastatic tissues were available, giving a total of 101 specimens. Tumour classes comprised ten different tumour types (Supporting information, Supplementary Tables 1 and 2). All samples were from formalin-fixed, paraffin-embedded (FFPE) specimens only. To exclude the possibility of inaccurate prediction due to contamination of tumour material with normal tissue, two expert pathologists examined all the cases and microdissected the samples.
RNA extraction
RNA was isolated from 20 µm thick FFPE tumour sections using the Recover All™ Total Nucleic Acid Isolation Kit for FFPE from Ambion (Austin, TX, USA) (#AM1975) according to the manufacturer’s instructions. Sample quality was assessed by Nanodrop (Thermo Scientific, Waltham, MA, USA).
Microarray analysis
MiRNA expression was investigated using the Agilent Human miRNA microarray v.2 (#G4470B; Agilent Technologies, Santa Clara, CA, USA). This microarray consists of 60-mer DNA probes synthesized in situ and contains 15 000 features which represent 723 human miRNAs, sourced from the Sanger miRBASE database (Release 10.1). RNA labelling and hybridization were performed in accordance with the manufacturer’s instructions. An Agilent scanner and Feature Extraction 10.5 software (Agilent Technologies) were used to obtain the microarray raw data. Microarray results were analysed by using GeneSpring GX 11 software (Agilent Technologies). Data transformation was applied to set all the negative raw values at 1.0, followed by a quantile normalization and a log2 transformation. Filters on gene expression were used to keep only the miRNAs expressed in at least one sample (flagged as P). The list of 47 predictors was identified by comparing the miRNA expression levels across ten different tumour types. A 1.5 fold-change filter and ANOVA (analysis of variance) statistical test were applied. Differentially expressed genes were employed in cluster analysis, using the Manhattan correlation as a measure of similarity. For cluster image generation, an additional step of normalization on the gene median across all samples was added. All microarray data have been submitted to ArrayExpress, accession number E-TABM-1135.
Tumour prediction
MiRNAs for tissue-of-origin prediction were determined by using the GeneSpring software ANOVA test. The Prediction Analysis of Microarray (PAM) algorithm was then applied, without feature selection (threshold = 0), using 40 primary carcinomas from ten different tissue types as the training set. The test sets included 45 metastases originating from nine specific sites and 16 metastases for which the tissue of origin remained unknown. The same approach was used to predict samples from the ten tissue types present in the dataset published by Rosenfeld et al [19].
Results
Patient characteristics
RNA extraction and microarray hybridization were successfully performed for all 101 FFPE samples archived in the course of 4 years (2005–2008). Characteristics of the patients enrolled in this study are described in the Supporting information, Supplementary Tables 1 and 2. Selected primary tumour types comprised tumours with a high incidence in the general population, such as lung, stomach, colorectal, and breast cancer [20], and frequently identified as primary tumours of CUPs at autopsy [9]. The metastatic samples were from the most common sites of metastasis from solid tumours (lung, bone, liver, lymph node, and brain) as well as from other sites. Histological information about the tumours employed in this study is available in the Supporting information, Supplementary Table 2. Sixteen patients diagnosed with cancer with unknown primary (CUP) were also selected [1,5,21].
Before initiating the study, we tested and confirmed that miRNAs from FFPE samples could maintain a tissue-specific expression pattern (data not shown). The choice of this type of sample has the advantage that it is the most common type of specimen used in routine histo-pathological work-up. Importantly, no sample was excluded from analyses because of technical reasons.
Primary tumours display a distinct miRNA expression profile and metastases retain a large part of their primary tumour profile
To identify a panel of miRNAs able to reveal the tissue of origin of metastases, we analysed a training set of 40 primary tumours, representative of ten different cancer types (breast, colon, endometrium, stomach/gastric, kidney, liver, lung, pancreas, prostate, and skin melanoma). We assessed the expression levels of 723 human miRNAs in all samples by using an Agilent miRNA microarray platform. To select microRNAs whose expression profile could characterize each type of tumour, we performed an ANOVA test across the primary tumours. After filtering out the low-expressed and low-variant probes, the analysis identified 47 miR-NAs (listed in Table 1).
Table 1.
List of the 47 miRNAs used for cancer molecular classification
| No | MicroRNA | ANOVA p value |
Gene family |
miRNA cluster | Cytoband | Chromosome coordinates (GRCh37) |
|---|---|---|---|---|---|---|
| 1 | hsa-miR-10a | 8.39E–05 | mir-10 | – | 17q21.33 | 17:46657200–46657309 [−] |
| 2 | hsa-miR-10a* | 4.01E–03 | mir-10 | – | 17q21.33 | 17:46657200–46657309 [−] |
| 3 | hsa-miR-122 | 5.13E–18 | mir-122 | – | 18q21.32 | 18: 56118306–56118390 [+] |
| 4 | hsa-miR-126* | 1.30E–07 | mir-126 | – | 9q34.3 | 9:139565054–139565138 [+] |
| 5 | hsa-miR-135b | 1.86E–05 | mir-135 | – | 1q32.1 | 1:205417430–205417526 [−] |
| 6 | hsa-miR-141 | 4.55E–12 | mir-8 | mir-200c/mir-141 | 12p13.31 | 12:7073260–7073354 [+] |
| 7 | hsa-miR-145 | 5.98E–09 | mir-145 | mir-143/mir-145 | 5q33.1 | 5:148810209–148810296 [+] |
| 8 | hsa-miR-146a | 3.25E–06 | mir-146 | – | 5q33.3 | 5:159912359–159912457 [+] |
| 9 | hsa-miR-149 | 4.82E–07 | mir-149 | – | 2q37.3 | 2: 241395418–241395506 [+] |
| 10 | hsa-miR-181a-2* | 4.26E–07 | mir-181 | mir-181a-2/mir-181b-2 | 9q33.3 | 9:127454721–127454830 [+] |
| 11 | hsa-miR-182 | 1.06E–07 | mir-182 | mir-183/mir-96/mir-182 | 7q32.2 | 7:129410223–129410332 [−] |
| 12 | hsa-miR-183 | 5.89E–07 | mir-183 | mir-183/mir-96/mir-182 | 7q32.2 | 7:129414745–129414854 [−] |
| 13 | hsa-miR-187* | 1.05E–03 | mir-187 | – | 18q12.2 | 18:33484781–33484889 [−] |
| 14 | hsa-miR-192 | 2.18E–13 | mir-192 | mir-194-2/mir-192 | 11q13.1 | 11:64658609–64658718 [−] |
| 15 | hsa-miR-193a-3p | 3.24E–06 | mir-193 | – | 17q12 | 17:29887015–29887102 [+] |
| 16 | hsa-miR-194 | 1.38E–11 | mir-194 | mir-194-1/mir-215 | 1q41 | 1:220291499–220291583 [−] |
| 17 | hsa-miR-194* | 3.71E–12 | mir-194 | mir-194-1/mir-215 | 1q41 | 1:220291499–220291583 [−] |
| 18 | hsa-miR-200a | 5.15E–09 | mir-8 | mir-200b/mir-200a/mir-429 | 1p36.2 | 1:1103243–1103332 [+] |
| 19 | hsa-miR-200a* | 1.80E–06 | mir-8 | mir-200b/mir-200a/mir-429 | 1p36.2 | 1:1103243–1103332 [+] |
| 20 | hsa-miR-200b | 7.07E–11 | mir-8 | mir-200b/mir-200a/mir-429 | 1p36.2 | 1:1102484–1102578 [+] |
| 21 | hsa-miR-200b* | 1.80E–05 | mir-8 | mir-200b/mir-200a/mir-429 | 1p36.2 | 1:1102484–1102578 [+] |
| 22 | hsa-miR-200c | 9.29E–17 | mir-8 | mir-200c/mir-141 | 12p13.31 | 12:7072862–7072929 [+] |
| 23 | hsa-miR-204 | 1.28E–08 | mir-204 | – | 9q21.13 | 9: 73424891–73425000 [−] |
| 24 | hsa-miR-205 | 8.35E–07 | mir-205 | – | 1q32.2 | 1:209605478–209605587 [+] |
| 25 | hsa-miR-210 | 3.09E–04 | mir-210 | – | 11p15.4 | 11:568089–568198[−] |
| 26 | hsa-miR-211 | 9.88E–27 | mir-204 | – | 15q13.3 | 15:31357235–31357344 [−] |
| 27 | hsa-miR-215 | 4.64E–12 | mir-192 | mir-194-1/mir-215 | 1q41 | 1:220291195–220291304 [−] |
| 28 | hsa-miR-30a | 5.82E–05 | mir-30 | – | 6q13 | 6:72113254–72113324 [−] |
| 29 | hsa-miR-30a* | 7.62E–07 | mir-30 | – | 6q13 | 6:72113254–72113324 [−] |
| 30 | hsa-miR-30c | 1.24E–08 | mir-30 | – | 1 p34.2 | 1:41222956–41223044 [+] |
| 31 | hsa-miR-31 | 8.96E–04 | mir-31 | – | 9p21.3 | 9: 21512114–21512184 [−] |
| 32 | hsa-miR-31* | 6.40E–04 | mir-31 | – | 9p21.3 | 9: 21512114–21512184 [−] |
| 33 | hsa-miR-340* | 6.16E–10 | mir-340 | – | 5q35.3 | 5:179442303–179442397 [−] |
| 34 | hsa-miR-342-3p | 2.97E–06 | mir-342 | – | 14q32.3 | 14:100575992–100576090 [+] |
| 35 | hsa-miR-361-5p | 4.51E–06 | mir-361 | – | Xq21.2 | X:85158641–85158712 [−] |
| 36 | hsa-miR-363 | 3.21E–07 | mir-363 | mir-106a/mir-18b/mir-20b/mir-19b-2/ mir-92a-2/mir-363 |
Xq26.2 | X: 133303408–133303482 [−] |
| 37 | hsa-miR-375 | 2.10E–07 | mir-375 | – | 2q35 | 2: 219866367–219866430 [−] |
| 38 | hsa-miR-485-5p | 2.14E–02 | mir-485 | 17 miRNA cluster | 14q32.3 | 14:101521756–101521828 [+] |
| 39 | hsa-miR-506 | 1.72E–19 | mir-506 | mir-508/mir-507/mir-506/mir-513a-2 | Xq27.3 | X:146312238–146312361 [−] |
| 40 | hsa-miR-508-3p | 5.36E–13 | mir-506 | mir-508/mir-507/mir-506/mir-513a-2 | Xq27.3 | X:146318431–146318545 [−] |
| 41 | hsa-miR-509-3p | 5.48E–26 | mir-506 | mir-509-1/mir-509-2/mir-509-3 | Xq27.3 | X:146342050–146342143 [−] |
| 42 | hsa-miR-510 | 5.75E–09 | mir-506 | mir-510/mir-514-1/mir-514-2/mir-514-3 | Xq27.3 | X:146353853–146353926 [−] |
| 43 | hsa-miR-514 | 5.76E–23 | mir-506 | mir-510/mir-514-1/mir-514-2/mir-514-3 | Xq27.3 | X:146360765–146360862 [−] |
| 44 | hsa-miR-552 | 2.20E–10 | mir-552 | – | 1 p34.3 | 1:35135200–35135295 [−] |
| 45 | hsa-miR-650 | 1.03E–06 | mir-650 | – | 22q11.23 | 22:23165270–23165365 [+] |
| 46 | hsa-miR-873 | 1.02E–04 | mir-873 | – | 9p21.1 | 9:28888877–28888953 [−] |
| 47 | hsa-miR-96 | 4.87E–10 | mir-96 | mir-183/mir-96/mir-182 | 7q32.2 | 7:129414532–129414609 [−] |
The ability of this list of miRNAs to classify the ten types of primary tumours was assessed by a clusterization algorithm (Figure 1). Every tumour type displayed an exclusive pattern of miRNA expression, with the exception of breast and lung cancers which exhibited very similar profiles, as best shown by a graphical representation of tissue miRNA average expression (Supporting information, Supplementary Figure 1). Breast and lung carcinomas were indeed sometimes mixed up in the cluster analysis of Figure 1. Gastric and pancreatic cancers were two other classes of tumours exhibiting overlapping expression patterns and sometimes mixed up in the cluster analysis. Although surprising, these findings were not entirely unexpected, as they were also detected in a previous report [22]. Overall, this list of miR-NAs exhibited a tissue-specific expression pattern able to distinguish different tumour types and could constitute a good candidate for the classification of metastases.
Figure 1.
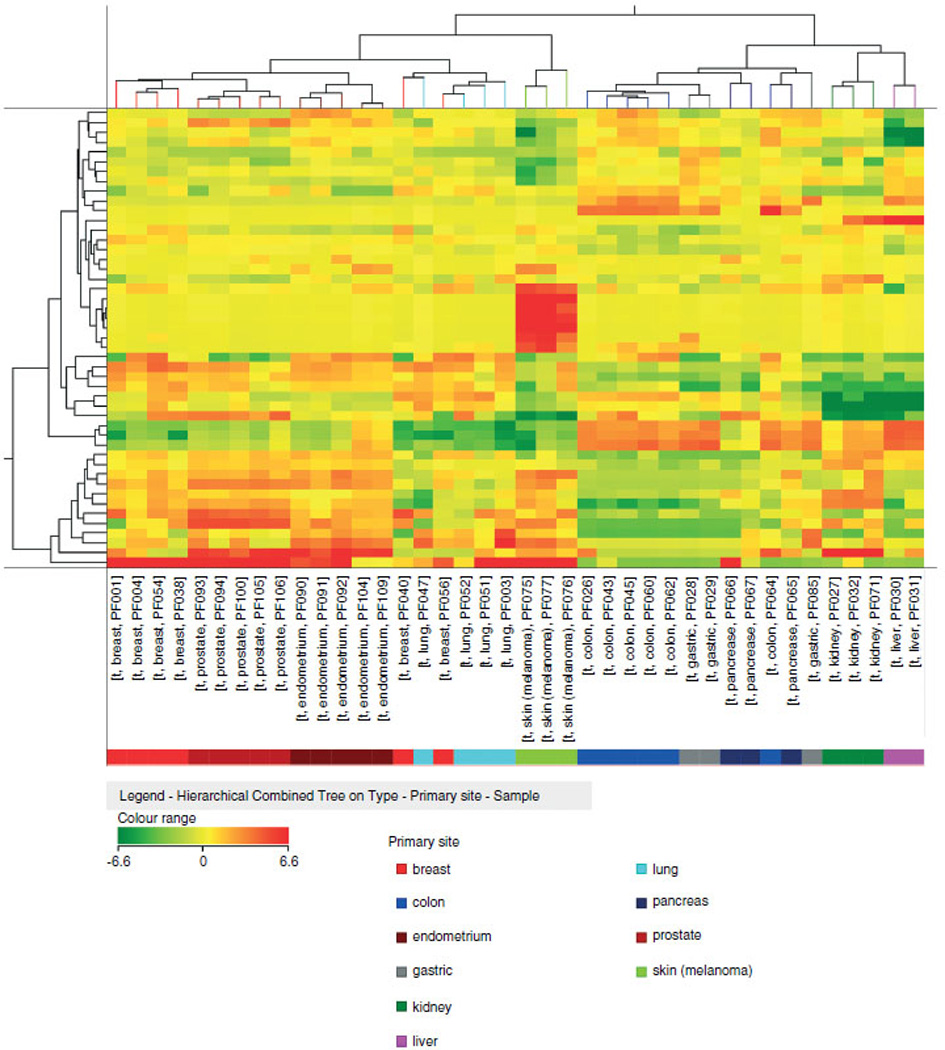
Classification of 40 carcinomas using the 47-miRNA signature. Cluster analysis of 40 primary carcinomas in accordance to the expression of the 47-miRNA classifier. Samples are grouped according to their tissue of origin. The colours of the genes represented on the heat map correspond to the expression values normalized on miRNA mean expression across all samples: green indicates down-regulated; red indicates up-regulated in the sample.
To assess similarities between primary cancers and metastases, we used the panel of 47 cancer-specific miRNAs to classify primary tumours and metastases together. Graphical representation of the similarities between primaries and metastases by cluster analysis revealed a good separation of samples according to the tissue of origin (Supporting information, Supplementary Figure 2), which is highlighted by displaying their average expression (Figure 2). Overall, the primary origin appears to be the main determinant of the metastasis miRNA profile.
Figure 2.
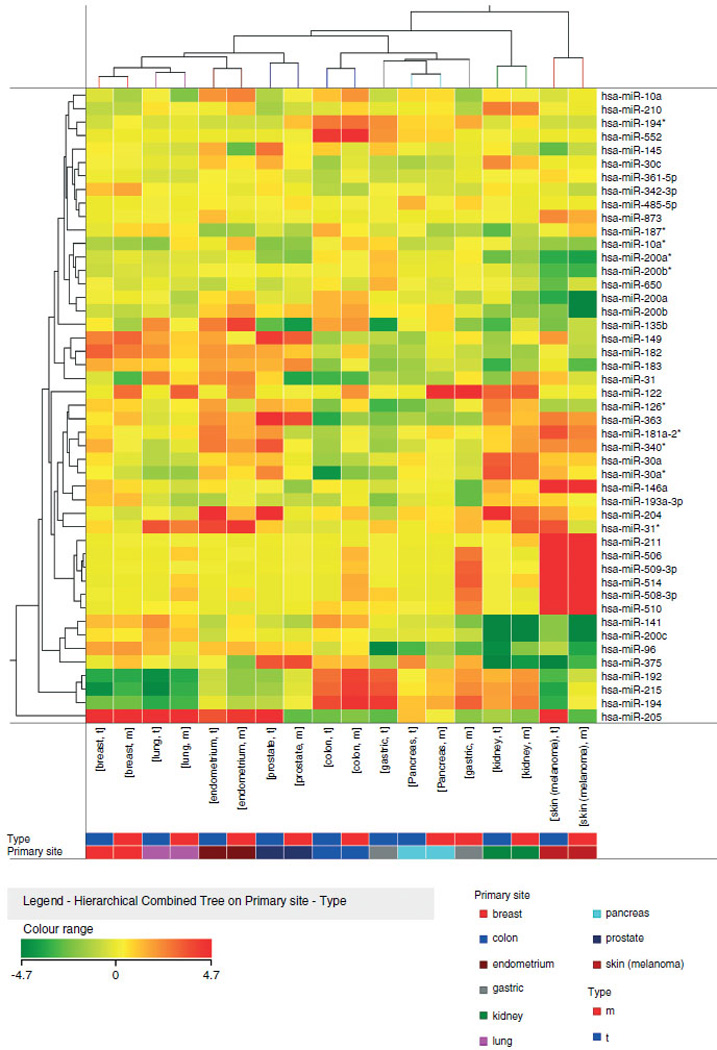
Average expression of 47 miRNAs in primary and metastatic tumours. Heat-map representation of the average expression of 47 miRNAs in metastatic and primary carcinomas from ten different tissues. Metastases and primaries from the same origin exhibit highly similar miRNA expression and are grouped together. The colours of the genes represented on the heat map correspond to the expression values normalized on miRNA mean expression across all samples: green indicates down-regulated; red indicates up-regulated in the tissue.
Tissue classifier development
To assess the predictive potential of the list of 47 differentially expressed miRNAs, a tissue classifier was developed by applying the supervised principal component approach [23]. All 47 miRNAs were retained by the predictive algorithm to achieve the highest accuracy in primary and metastasis prediction. The PAM centroids used for tumour prediction are shown in the Supporting information, Supplementary Figure 3. The panel of centroids (values based on miRNA expression) identified by the PAM algorithm highlights the tissue-specific miRNA expression and points out the best discriminating miRNAs. As examples, miR-211, miR-146a, and miR-506/miR-508/miR-509/miR-510/ miR-514 miRNAs are highly expressed in melanoma; miR-122 in liver and kidney cancers; miR-192/miR-194/miR-215 in colon and gastric carcinomas; and miR-96 and miR-182 in breast, lung, prostate and endometrium cancers.
We used primary tumours as a training set for both cross validation and test set prediction; metastases were only used as a test set. The predictive algorithm assigned a prediction probability for each sample of belonging to every possible class, thereby producing a list of possible primaries for every sample. In test set prediction, 100% of examined samples from primary tumours were assigned to the correct class, with a probability higher than 90% in each case (Supporting information, Supplementary Figure 4A and Supplementary Table 3). For metastases, the accuracy of prediction was 73% and at probability >0.1, the correct class was detected in 78% of cases (Supplementary Figure 4B and Supplementary Table 4). The results, summarized in Table 2, establish that the correct class of primary tumour is most often found as the first class. If not first, it can be found within the first two or three predictions.
Table 2.
Summary of prediction results in primary tumours and metastases
| Correct Predictions at Probability |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Correct first prediction |
Correct first prediction |
>0.9 |
>0.5 |
>0.1 |
||||||||
| Training set (primaries) |
tot | % | Test set (metastases) |
tot | % | tot | % | tot | % | tot | % | |
| Breast | 6 | 6 | 100.0 | 7 | 6 | 85.7 | 5 | 71.4 | 6 | 85.7 | 6 | 85.7 |
| Colon | 6 | 6 | 100.0 | 10 | 9 | 90.0 | 9 | 90.0 | 9 | 90.0 | 9 | 90.0 |
| Endometrium | 5 | 5 | 100.0 | 4 | 2 | 50.0 | 1 | 25.0 | 2 | 50.0 | 2 | 50.0 |
| Gastric | 3 | 3 | 100.0 | 5 | 3 | 60.0 | 2 | 40.0 | 3 | 60.0 | 3 | 60.0 |
| Kidney | 3 | 3 | 100.0 | 5 | 4 | 80.0 | 4 | 80.0 | 4 | 80.0 | 4 | 80.0 |
| Liver | 2 | 2 | 100.0 | 0 | 0 | – | – | – | – | – | – | – |
| Lung | 4 | 4 | 100.0 | 6 | 4 | 66.7 | 2 | 33.3 | 4 | 66.7 | 5 | 83.3 |
| Melanoma | 3 | 3 | 100.0 | 3 | 3 | 100.0 | 3 | 100.0 | 3 | 100.0 | 3 | 100.0 |
| Pancreas | 3 | 3 | 100.0 | 3 | 2 | 66.7 | 2 | 66.7 | 2 | 66.7 | 2 | 66.7 |
| Prostate | 5 | 5 | 100.0 | 2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 50.0 |
| Tot Correct predictions | 40 | 40 | 100.0 | 45 | 33 | 73.3 | 28 | 62.2 | 33 | 73.3 | 35 | 77.8 |
| Specificity* | 100.0 | 75.6 | 84.4 | 75.6 | 66.7 | |||||||
Specificity is calculated on the basis of the incorrect predictions at the specified probability (>90%, >50%, >10%). See Supplementary Table 4 for details.
Performance of the 47-miR predictor on a published dataset
To verify the reliability of the 47-miRNA classifier in an independent set of samples, a publicly available dataset of tumours and metastasis microRNA profile [19] were re-analysed by using the predictive method and the 47-miRNA signature described herein (with the exclusion of six miRNAs whose expression was not available in the published dataset). Expression data of the published dataset were obtained using a custom array based on Agilent technology developed by Rosenfeld et al [19].
Of the whole dataset, we only investigated primaries/metastases belonging to the ten cancer site categories for which our 47-miRNA classifier was developed. To this end, we employed the same training set and test set used by Rosenfeld et al: 135 samples (primaries and metastases) in the training set and 35 samples in the test set. In this experimental condition, the classifier reached an 87% accuracy (correct class predicted as first choice) in training set prediction and a 69% accuracy in test set prediction, frequencies that did not differ significantly from published results. However, differently from published results, the method that we employed for prediction assigns a probability for each sample of belonging to each tumour class. Hence, the correct class could still be predicted with a significant probability. Indeed, by considering all the classes predicted with a probability greater than 0.1, the correct class of primary cancers was predicted in 95% of the cases and 85% for metastases (Table 3), frequencies that were surprisingly even higher than those of our own test set as well as of published results. These data proved that the 47-miRNA classifier was reliable and capable of predicting an independent set of samples whose results were produced using a different platform (although produced using the same Agilent technology).
Table 3.
Prediction of Rosenfeld et al. dataset by using our 47-miRNAs signature
| Correct predictions at probability |
Correct predictions at probability |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Correct first predictions |
>0.9 |
>0.5 |
>0.1 |
Correct first predictions |
>0.9 |
>0.5 |
>0.1 |
||||||||||||
| Tumour class | p/m1 | Training set | tot | % | tot | % | tot | % | tot | % | Test set | tot | % | tot | % | tot | % | tot | % |
| Breast | p | 2 | 1 | 50.0 | 0 | 0.0 | 1 | 50.0 | 2 | 100.0 | 1 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 100.0 |
| m | 18 | 14 | 77.8 | 5 | 27.8 | 14 | 77.8 | 15 | 83.3 | 4 | 2 | 50.0 | 0 | 0.0 | 2 | 50.0 | 4 | 100.0 | |
| Colon | p | 2 | 2 | 100.0 | 2 | 100.0 | 2 | 100.0 | 2 | 100.0 | 2 | 1 | 50.0 | 0 | 0.0 | 1 | 50.0 | 1 | 50.0 |
| m | 12 | 11 | 91.7 | 5 | 41.7 | 11 | 91.7 | 12 | 100.0 | 3 | 3 | 100.0 | 0 | 0.0 | 2 | 66.7 | 3 | 100.0 | |
| Endometrium | p | 5 | 4 | 80.0 | 0 | 0.0 | 4 | 80.0 | 5 | 100.0 | 2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 50.0 |
| m | 2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 50.0 | 1 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 100.0 | |
| Gastric | p | 3 | 2 | 66.7 | 0 | 0.0 | 2 | 66.7 | 3 | 100.0 | 2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 50.0 |
| m | 1 | 1 | 100.0 | 0 | 0.0 | 1 | 100.0 | 1 | 100.0 | 0 | 0 | – | 0 | – | 0 | – | 0 | – | |
| Kidney | p | 3 | 3 | 100.0 | 3 | 100.0 | 3 | 100.0 | 3 | 100.0 | 3 | 3 | 100.0 | 2 | 66.7 | 3 | 100.0 | 3 | 100.0 |
| m | 10 | 9 | 90.0 | 4 | 40.0 | 9 | 90.0 | 10 | 100.0 | 2 | 2 | 100.0 | 2 | 100.0 | 2 | 100.0 | 2 | 100.0 | |
| Liver | p | 4 | 4 | 100.0 | 4 | 100.0 | 4 | 100.0 | 4 | 100.0 | 2 | 1 | 50.0 | 1 | 50.0 | 1 | 50.0 | 1 | 50.0 |
| m | 0 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | 0 | – | 0 | – | 0 | – | 0 | – | |
| Lung | p | 29 | 29 | 100.0 | 14 | 48.3 | 26 | 89.7 | 29 | 100.0 | 4 | 4 | 100.0 | 1 | 25.0 | 4 | 100.0 | 4 | 100.0 |
| m | 13 | 11 | 84.6 | 2 | 15.4 | 8 | 61.5 | 12 | 92.3 | 1 | 1 | 100.0 | 1 | 100.0 | 1 | 100.0 | 1 | 100.0 | |
| Melanoma | p | 2 | 2 | 100.0 | 2 | 100.0 | 2 | 100.0 | 2 | 100.0 | 1 | 1 | 100.0 | 1 | 100.0 | 1 | 100.0 | 1 | 100.0 |
| m | 19 | 17 | 89.5 | 16 | 84.2 | 17 | 89.5 | 18 | 94.7 | 4 | 3 | 75.0 | 2 | 50.0 | 3 | 75.0 | 3 | 75.0 | |
| Pancreas | p | 5 | 4 | 80.0 | 3 | 60.0 | 3 | 60.0 | 5 | 100.0 | 2 | 2 | 100.0 | 0 | 0.0 | 2 | 100.0 | 2 | 100.0 |
| m | 1 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 100.0 | 0 | 0 | – | 0 | – | 0 | – | 0 | – | |
| Prostate | p | 3 | 2 | 66.7 | 1 | 33.3 | 2 | 66.7 | 3 | 100.0 | 1 | 1 | 100.0 | 0 | 0.0 | 1 | 100.0 | 1 | 100.0 |
| m | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 | 1 | 100.0 | 0 | 0 | – | 0 | – | 1 | – | 0 | – | |
| Tot Correct predictions | 135 | 117 | 86.7 | 61 | 45.2 | 109 | 80.7 | 129 | 95.6 | 35 | 24 | 68.6 | 10 | 28.6 | 24 | 68.6 | 30 | 85.7 | |
| Specificity* | 97.0 | 89.6 | 49.6 | 97.1 | 82.9 | 42.9 | |||||||||||||
p = primaries; m = metastases
Specificity is calculated on the basis of the incorrect predictions at the specified probability (>90%, >50%, >10%).
Molecular prediction of CUP origin
Finally, we applied the 47-miRNA diagnostic classifier to 16 patients diagnosed as CUP. The patients’ characteristics are described in the Supporting information, Supplementary Table 2. As previously described, the classifier assigns a probability for each sample of belonging to each tumour class. In our case, the choice is among ten different tissues of origin, which include the most frequently detected sites at autopsy [9]. Using the 47-miRNA expression, we were able to predict for every CUP patient the most probable primary, or couple of primaries, that could be investigated further with IHC examinations (Table 4). Our CUP predictions revealed 31% gastric origin, 25% lung, 19% pancreas, 12% liver, 6% colon, and 6% kidney. Importantly, most predictions (12 of 16) achieved a probability greater than 0.9. At this level of probability, the error rate is low (only 3% in Rosenfeld et al’s dataset), suggesting that these predictions are highly reliable. In addition, molecular predictions were also consistent with the hypotheses of primary sites proposed, but not proven, by pathologists or clinicians for our set of CUPs (see Table 4).
Table 4.
Clinico-pathological information and molecular prediction of cancers with unknown primary
| Molecular prediction at probability |
Class probability for each sample |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Type | Biopsy site |
Pathological and/or clinical hypothesis* |
Molecular prediction |
>90% | >50% | >10% | >10% | Breast | Colon | Endometrium | Gastric | Kidney | Liver | Lung | Melanoma | Pancreas | Prostate |
| PF007 | CUP | Liver | Gastroenteric | Colon | Colon | Colon | Colon | Colon > gastric |
3.07 E–19 | 9.18E–01• | 1.40E–19 | 8.17E–02 | 6.76E–21 | 1.04E–14 | 1.30E–16 | 1.30E–59 | 1.24E–04 | 7.48E–24 |
| PF012 | CUP | Liver | Gastric | Gastric | Gastric | Gastric | Gastric | 2.14E–09 | 8.76E–03 | 1.33E–07 | 9.87E–01 | 5.38E–16 | 2.39E–13 | 2.82E–10 | 1.68E–52 | 4.17E–03 | 5.35E–09 | |
| PF017 | CUP | Liver | Colecyst | Gastric | Gastric | Gastric | Gastric | Gastric > liver |
1.36E–18 | 4.04E–07 | 6.78E–15 | 9.86E–01 | 1.89E–10 | 1.42 E–02 | 2.13E–16 | 8.10E–37 | 6.94E–05 | 3.03E–22 |
| PF018 | CUP | Liver | Gastric/pancreas | Gastric | Gastric | Gastric | Gastric | Gastric > pancreas |
5.63E–13 | 6.28E–04 | 4.01 E–11 | 9.88E–01 | 1.45E–13 | 8.15E–12 | 6.03E–11 | 6.72E–49 | 1.12E–02 | 3.01 E—18 |
| PF020 | CUP | Pleura | Lung | Gastric | Gastric | Gastric | Gastric | Gastric | 8.51E–14 | 1.06E–04 | 6.14E–16 | 9.99E–01 | 1.06E–21 | 3.34E–25 | 6.10E–10 | 6.08E–35 | 1.22E–03 | 7.85E–21 |
| PF022 | CUP | Pleura | Digestive tract | Gastric | Gastric | Gastric > pancreas > colon |
Gastric > pancreas > colon |
1.34E–10 | 1.10E–02• | 1.39E–12 | 7.33E–01 | 5.61 E–20 | 8.37E–28 | 3.19E–08 | 1.60E–45 | 2.56E–01 | 8.19E–17 | |
| PF006 | CUP | Brain | Kidney/lung | Kidney | Kidney | Kidney | Kidney | Kidney | 2.95E–07 | 4.14E–14 | 1.21 E—10 | 3.36E–04 | 9.83E–01 | 5.93E–09 | 3.03E–07 | 1.10E–22 | 1.64E–02 | 3.54E–12 |
| PF008 | CUP | Liver | Liver | Liver | Liver | Liver | Liver | 1.62E–33 | 1.72E–27 | 8.70E–32 | 3.81 E—18 | 6.84E–10 | 1.00E+00 | 7.30E–37 | 3.43E–63 | 1.03E–20 | 1.75E–35 | |
| PF013 | CUP | Liver | Oesophagous | Liver | Liver | Liver | Liver | Liver | 2.10E–38 | 9.63E–37 | 1.14E–35 | 4.68E–25 | 1.03E–14 | 1.00E+00 | 5.80E–41 | 5.80E–58 | 2.79E–27 | 5.93E–38 |
| PF005 | CUP | Lung | Lung | Lung | Lung | Lung > breast > pancreas |
9.71 E–02 | 7.97E–13 | 9.37E–10 | 4.53E–08 | 1.62E–15 | 1.38E–26 | 8.68E–01 | 1.98E–35 | 3.44E–02 | 2.48E–09 | ||
| PF011 | CUP | Lung | Lung | Lung | Lung > breast |
Lung > breast |
1.02E–01 | 1.17E–12 | 5.70E–09 | 2.57E–12 | 2.62E–23 | 2.47E–42 | 8.98E–01 | 1.33E–41 | 1.16E–04 | 2.23E–09 | ||
| PF024 | CUP | Pleura | Lung | Lung | Lung | Lung | Lung | Lung > breast |
4.75E–02 | 3.36E–19 | 1.16E–06 | 6.11 E—19 | 3.29E–29 | 2.48E–51 | 9.52E–01 | 7.05E–44 | 1.58E–09 | 2.19E–06 |
| PF025 | CUP | Pleura | Lung | Lung | Lung | Lung | Lung | Lung | 3.11 E–04 | 4.73E–11 | 7.16E–10 | 1.19E–09 | 4.97E–24 | 2.34E–39 | 1.00E+00 | 1.17E–39 | 1.31 E–04 | 1.23E–11 |
| PF019 | CUP | Liver | Pancreas | Pancreas | Pancreas | Pancreas | Pancreas | Pancreas > lung |
4.44E–03 | 1.00E–06 | 3.07 E–06 | 9.52E–04 | 3.52E–11 | 7.28E–12 | 3.72E–02 | 3.18E–33 I | 9.57E–01 | 4.14E–09 |
| PF021 | CUP | Pleura | Lung | Pancreas | Pancreas | Pancreas | Pancreas | Pancreas > lung |
5.34E–05 | 7.57E–08 | 6.13E–11 | 7.74E–04 | 4.50E–17 | 6.21 E–28 | 1.12E–02 | 2.55E–39 | 9.88E–01 | 4.39E–12 |
| PF023 | CUP | Pleura | Lung | Pancreas | Pancreas | Pancreas > lung |
Pancreas > lung > breast |
1.38E–02 | 1.40E–10 | 6.94E–09 | 4.47E–06 | 2.04E–12 | 5.41 E—23 | 3.93E–01 | 9.05E–31 | 5.93E–01 | 1.68E–08 | |
Blank cells indicate that a hypothesis was not formulated due to clinical/pathological uncertainty.
Shaded areas highlight the highest probability for each sample.
Bold numbers highlight probabilities higher than 0.01.
Discussion
Several molecular tests have been recently described for the identification of the tissue of origin of metastatic carcinomas (see Monzon and Koen for a review [18]). Traditional gene expression microarrays cannot be properly performed on all FFPE samples, because RNA from this type of sample is not always suitable for labelling and hybridization, thereby constituting a hindrance in translating some very powerful gene expression profiles to daily clinical practice [18]. This is the case of the Pathwork Tissue of Origin Test (Pathwork Diagnostics) [24,25] and CupPrint (Agendia) [26–28] tests, based respectively on 1550 and 1900 genes. On the other hand, microRNA microarray profiles could be faster to generate—but equally accurate—than a panel of RT-qPCR-based genes or miRNAs, as described and patented up to now (Theros Cancer-TYPE ID [28] by bioTheranostics, and miRview mets [19,29] by Rosetta Genomics). The only molecular test that evaluates the expression of a small number of genes (10) by using the RT-qPCR technique is the CUP assay (Veridex) described by Talantov et al in 2006 and tested in CUPs in 2008 [30–32]. The main limitation of the CUP assay is the small number (six) of tissues of origin that can be identified by that test.
In our study, we identified a 47-miRNA classifier; we demonstrated the usefulness of miRNA microarray technology in predicting the tissue of origin of metastatic cancers from FFPE biopsy tissue; and we tested the classifier on an independent published dataset and on 16 CUPs. First, we confirmed the maintenance of a tumour-of-origin specific gene expression profile in metastatic carcinomas by using a molecular classification such as the clusterization algorithm. We then used a predictive approach (PAM) able to provide a well-defined probability of possible primaries for each tumour. With a probability cut-off set at 0.1, the prediction test showed an overall accuracy of about 90% (100% in primaries and 78% for metastases) for samples with known primary tumours (Table 2). The correct class of primary tumour was found as the first class or within the first two predictions in 78% of the cases, indicating that the 47-miRNA molecular profile is able to identify one or two possible tumours almost surely constituting the true tumour primary site for every blind metastatic tumour. If tumour sites predicted with a probability greater than 0.01 are included, the true primary site can almost surely be found among the first two or three tissues suggested (sensitivity is higher than 95%). Although this is a sub-optimal situation in the classification of some tumour types, probably because of the lack of very tissue-specific miRNAs, this information could still significantly help pathologists to establish a diagnosis and influence the CUP patient’s management, without the cost of investigating every possible tumour site [27,33,34], in particular in the challenging situation of moderately or poorly differentiated CUPs.
To validate the panel of 47 miRNAs, we assessed its performance on a publicly available dataset of miRNA profiles of human cancers and metastases, generated by Rosenfeld et al [19]. These authors produced their data for the prediction of the primary tumour sites of metastases and ended up identifying a 48-miRNA signature that achieved 71% prediction accuracy (although they used the combination from two different predictive methods to obtain the best accuracy level). The published 48-miRNA signature shares 15 miRNAs with our 47-miRNA signature (miR-122, miR-141, miR-146a, hsa-miR-182, miR-192, miR-193a-3p, miR-194, miR-200a, miR-200c, miR-205, miR-210, miR-31, miR-363, miR-375, miR-509-3p). When applied to their microarray expression data, the 47-miRNA signature revealed an accuracy in the prediction of the ten tumour classes (both primaries and metastases) equivalent to or even higher than the published accuracy. These results support the usefulness of the 47-miRNA signature for the identification of tumour tissue of origin, since the signature also worked very well on an independent set of samples whose miRNA expression was quantified with a different microarray platform.
When applied to CUPs, 75% (12 of 16) of samples were predicted at a probability greater than 0.9. At this level of probability, specificity is very high: 85% in our dataset and 98% in Roselfeld et al’s dataset. In support of the consistency of the results, within the limit of the small number of samples investigated, the detected classes were largely in agreement with the incidences of primary tumours reported by Pentheroudakis et al [9], in a review of post-mortem examinations of 644 cases of CUP patients, which revealed lung (27%), pancreas (24%), liver/bile duct (8%), kidney/adrenals (8%), bowel (7%), genital system (7%), and stomach (6%) as the most frequent sites of CUP origin.
The management of cancer patients is moving towards personalized procedures. In the case of CUPs, the application of microarray-based tests could help in better focusing on certain primary sites, thereby reducing patient’s burden and overall procedure costs. Whether these tests could also improve patient’s outcome and CUP morbidity remains to be established.
Supplementary Material
Acknowledgment
We wish to thank Dr Ranit Aharonov for providing data from their published dataset and the Microarray Facility at the University of Ferrara for performing the miRNA expression profiling. This work was supported by funding from the Ministry of Health Special Project Oncology, Regione Emilia-Romagna and by Associazione Italiana per la Ricerca sul Cancro.
Footnotes
No conflicts of interest were declared.
Author contribution statement
MN, MF, PQ, GL, IN, and CMC conceived and designed experiments. MF and MP performed experiments and analysed data. AV, BZ, RG, and EM carried out experiments. ML, GM, GQ, IM, and LU contributed materials and samples. MF, MN, PQ, and GL wrote the paper. All authors had final approval of the submitted and published versions.
SUPPORTING INFORMATION ON THE INTERNET
The following supporting information may be found in the online version of this article.
Figure S1. Cluster analysis of the average 47-miRNA expression in each tumour type.
Figure S2. Primary tumours and metastases cluster according to their tissue of origin.
Figure S3. Centroids used for tissue prediction.
Figure S4. Results of tissue prediction for primaries and metastases.
Table S1. Summary of samples and patients enrolled in the study.
Table S2. Detailed characteristics of the patients and tumour samples.
Table S3. Predicted classes and probabilities of primary tumours.
Table S4. Predicted classes and probabilities of metastases with known primary cancer.
References
- 1.Briasoulis E, Tolis C, Bergh J, et al. ESMO Minimum Clinical Recommendations for diagnosis, treatment and follow-up of cancers of unknown primary site (CUP) Ann Oncol. 2005;16(Suppl 1):i75–i76. doi: 10.1093/annonc/mdi804. [DOI] [PubMed] [Google Scholar]
- 2.Oien KA. Pathologic evaluation of unknown primary cancer. Sem Oncol. 2009;36:8–37. doi: 10.1053/j.seminoncol.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Pavlidis N, Fizazi K. Cancer of unknown primary (CUP) Crit Rev Oncol Hematol. 2005;54:243–250. doi: 10.1016/j.critrevonc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Freudenberg LS, Rosenbaum-Krumme SJ, Bockisch A, et al. Cancer of unknown primary. Recent Results Cancer Res. 2008;170:193–202. doi: 10.1007/978-3-540-31203-1_15. [DOI] [PubMed] [Google Scholar]
- 5.Varadhachary GR, Abbruzzese JL, Lenzi R. Diagnostic strategies for unknown primary cancer. Cancer. 2004;100:1776–1785. doi: 10.1002/cncr.20202. [DOI] [PubMed] [Google Scholar]
- 6.Pavlidis N, Briasoulis E, Hainsworth J, et al. Diagnostic and therapeutic management of cancer of an unknown primary. Eur J Cancer. 2003;39:1990–2005. doi: 10.1016/s0959-8049(03)00547-1. [DOI] [PubMed] [Google Scholar]
- 7.Levi F, Te VC, Erler G, et al. Epidemiology of unknown primary tumours. Eur J Cancer. 2002;38:1810–1812. doi: 10.1016/s0959-8049(02)00135-1. [DOI] [PubMed] [Google Scholar]
- 8.Pentheroudakis G, Briasoulis E, Pavlidis N. Cancer of unknown primary site: missing primary or missing biology? Oncologist. 2007;12:418–425. doi: 10.1634/theoncologist.12-4-418. [DOI] [PubMed] [Google Scholar]
- 9.Pentheroudakis G, Golfinopoulos V, Pavlidis N. Switching benchmarks in cancer of unknown primary: from autopsy to microarray. Eur J Cancer. 2007;43:2026–2036. doi: 10.1016/j.ejca.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Ayoub JP, Hess KR, Abbruzzese MC, et al. Unknown primary tumors metastatic to liver. J Clin Oncol. 1998;16:2105–2112. doi: 10.1200/JCO.1998.16.6.2105. [DOI] [PubMed] [Google Scholar]
- 11.van de Wouw AJ, Janssen-Heijnen ML, Coebergh JW, et al. Epidemiology of unknown primary tumours; incidence and population-based survival of 1285 patients in Southeast Netherlands, 1984–1992. Eur J Cancer. 2002;38:409–413. doi: 10.1016/s0959-8049(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 12.Huebner G, Link H, Kohne CH, et al. Paclitaxel and carboplatin vs gemcitabine and vinorelbine in patients with adeno- or undifferentiated carcinoma of unknown primary: a randomised prospective phase II trial. Br J Cancer. 2009;100:44–49. doi: 10.1038/sj.bjc.6604818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbruzzese JL, Abbruzzese MC, Lenzi R, et al. Analysis of a diagnostic strategy for patients with suspected tumors of unknown origin. J Clin Oncol. 1995;13:2094–2103. doi: 10.1200/JCO.1995.13.8.2094. [DOI] [PubMed] [Google Scholar]
- 14.Bishop JF, Tracey E, Glass P, et al. Prognosis of sub-types of cancer of unknown primary (CUP) compared to metastatic cancer. J Clin Oncol. 2007;25:21010. [Google Scholar]
- 15.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 16.Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 18.Monzon FA, Koen TJ. Diagnosis of metastatic neoplasms: molecular approaches for identification of tissue of origin. Arch Pathol Lab Med. 2010;134:216–224. doi: 10.5858/134.2.216. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nature Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 20.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 21.Dennis JL, Hvidsten TR, Wit EC, et al. Markers of adenocarcinoma characteristic of the site of origin: development of a diagnostic algorithm. Clin Cancer Res. 2005;11:3766–3772. doi: 10.1158/1078-0432.CCR-04-2236. [DOI] [PubMed] [Google Scholar]
- 22.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibshirani R, Hastie T, Narasimhan B, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumur CI, Lyons-Weiler M, Sciulli C, et al. Interlaboratory performance of a microarray-based gene expression test to determine tissue of origin in poorly differentiated and undifferentiated cancers. J Mol Diagn. 2008;10:67–77. doi: 10.2353/jmoldx.2008.070099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monzon FA, Lyons-Weiler M, Buturovic LJ, et al. Multicenter validation of a 1,550-gene expression profile for identification of tumor tissue of origin. J Clin Oncol. 2009;27:2503–2508. doi: 10.1200/JCO.2008.17.9762. [DOI] [PubMed] [Google Scholar]
- 26.Bridgewater J, van Laar R, Floore A, et al. Gene expression profiling may improve diagnosis in patients with carcinoma of unknown primary. Br J Cancer. 2008;98:1425–1430. doi: 10.1038/sj.bjc.6604315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horlings HM, van Laar RK, Kerst JM, et al. Gene expression profiling to identify the histogenetic origin of metastatic adenocarcinomas of unknown primary. J Clin Oncol. 2008;26:4435–4441. doi: 10.1200/JCO.2007.14.6969. [DOI] [PubMed] [Google Scholar]
- 28.Ma XJ, Patel R, Wang X, et al. Molecular classification of human cancers using a 92-gene real-time quantitative polymerase chain reaction assay. Arch Pathol Lab Med. 2006;130:465–473. doi: 10.5858/2006-130-465-MCOHCU. [DOI] [PubMed] [Google Scholar]
- 29.Rosenwald S, Gilad S, Benjamin S, et al. Validation of a microRNA-based qRT-PCR test for accurate identification of tumor tissue origin. Mod Pathol. 2010;23:814–823. doi: 10.1038/modpathol.2010.57. [DOI] [PubMed] [Google Scholar]
- 30.Talantov D, Baden J, Jatkoe T, et al. A quantitative reverse transcriptase-polymerase chain reaction assay to identify metastatic carcinoma tissue of origin. J Mol Diagn. 2006;8:320–329. doi: 10.2353/jmoldx.2006.050136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varadhachary GR, Raber MN, Matamoros A, et al. Carcinoma of unknown primary with a colon-cancer profile-changing paradigm and emerging definitions. Lancet Oncol. 2008;9:596–599. doi: 10.1016/S1470-2045(08)70151-7. [DOI] [PubMed] [Google Scholar]
- 32.Varadhachary GR, Talantov D, Raber MN, et al. Molecular profiling of carcinoma of unknown primary and correlation with clinical evaluation. J Clin Oncol. 2008;26:4442–4448. doi: 10.1200/JCO.2007.14.4378. [DOI] [PubMed] [Google Scholar]
- 33.Pimiento JM, Teso D, Malkan A, et al. Cancer of unknown primary origin: a decade of experience in a community-based hospital. Am J Surg. 2007;194:833–837. doi: 10.1016/j.amjsurg.2007.08.039. discussion 837–838. [DOI] [PubMed] [Google Scholar]
- 34.Varadhachary GR, Greco FA. Overview of patient management and future directions in unknown primary carcinoma. Semin Oncol. 2009;36:75–80. doi: 10.1053/j.seminoncol.2008.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


