Abstract
Human T-lymphotropic virus type 1 (HTLV-1) is found in indigenous peoples of the Pacific Islands and the Americas, whereas type 2 (HTLV-2) is widely distributed among the indigenous peoples of the Americas, where it appears to be more prevalent than HTLV-1, and in some tribes of Central Africa. HTLV-2 is considered ancestral in the Americas and is transmitted to the general population and injection drug users from the indigenous population. In the Americas, HTLV-1 has more than one origin, being brought by immigrants in the Paleolithic period through the Bering Strait, through slave trade during the colonial period, and through Japanese immigration from the early 20th century, whereas HTLV-2 was only brought by immigrants through the Bering Strait. The endemicity of HTLV-2 among the indigenous people of Brazil makes the Brazilian Amazon the largest endemic area in the world for its occurrence. A review of HTLV-1 in all Brazilian tribes supports the African origin of HTLV-1 in Brazil. The risk of hyperendemicity in these epidemiologically closed populations and transmission to other populations reinforces the importance of public health interventions for HTLV control, including the recognition of the infection among reportable diseases and events.
Keywords: HTLV-1, HTLV-2, Indians, Origin, Americas
Abstract
O vírus linfotrópico de células T humanas do tipo 1 (HTLV-1) é encontrado em populações indígenas de países do Pacífico e Américas enquanto o tipo 2 (HTLV-2) é amplamente distribuído entre as populações indígenas das Américas, nas quais aparenta ser mais prevalente que o HTLV-1, e em algumas tribos da África Central, sendo considerado ancestral nas Américas e transmitido à população geral e de usuários de drogas injetáveis a partir da população indígena. No continente americano o HTLV-1 teria mais de uma origem, sendo trazido na era paleolítica pelos imigrantes através do estreito de Bering, através do tráfico de escravos no período colonial e com a imigração japonesa a partir do início do século XX, enquanto para o HTLV-2 teria sido trazido pelos imigrantes através do estreito de Bering. A endemicidade do HTLV-2 entre os indígenas do Brasil tornam a região amazônica brasileira a maior área endêmica do mundo para sua ocorrência e a revisão da infecção pelo HTLV-1 em todas as tribos brasileiras apoiam a origem africana do HTLV-1 no Brasil. O risco de hiperendemicidade nestas populações epidemiologicamente fechadas e de transmissão a outras populações reforçam a importância de medidas no âmbito da saúde pública para seu controle, incluindo o reconhecimento da infecção entre os agravos e eventos de notificação compulsória.
INTRODUCTION
The human T-lymphotropic virus type 1 (HTLV-1) and type 2 (HTLV-2), although closely related, have different geographical distributions8,21,23,35,59,63,72,93,110,119, pathogenesis10,95,97,98, and clinical manifestations8,10,21,23,35,59,63,97,110. Although types 3 (HTLV-3) and 4 (HTLV-4) have been found in populations of central Africa, they have not yet been associated with disease in humans19,133.
HTLV-1, the causative agent of HTLV-associated myelopathy/tropical spastic paraparesis (HAM/TSP), uveitis, infective dermatitis, and other inflammatory disorders21,35,59,110, is endemic in many parts of the world, including southwestern Japan, some of the Caribbean islands, South America, and foci in western and central Africa and Australo-Melanesia59. In turn, HTLV-2, despite there being no clear indications associating it with well-defined clinical manifestations, has been associated with sporadic cases of neurological disorders similar to HAM/TSP8,63, and has been observed as prevalent in native populations, such as the indigenous peoples of the Americas, certain tribes of pygmies in Africa23,63,72,93,119, and injection drug users (IDUs) in urban areas of the United States, Europe, and Latin America27,42,45,82,89,114,124,138.
In Brazil, approximately 517,000 Indians live in officially recognized indigenous lands, with an estimated additional 380,000 living outside these areas (78% of whom live in urban areas), for an estimated total population of 896,917 counted as indigenous by the 2010 census29. Both HTLV-1 and HTLV-2 are prevalent among Brazilian indigenous populations, with HTLV-2 being the most predominant among these individuals14,51,52,69,72,91,93,100,102,119,126.
The search strategy adopted in this review was intentionally broad so as to ensure the identification of all relevant studies published between 1980 and 2014 relating to infection by HTLV in indigenous populations in Latin America. Searches were made via the PubMed, Lilacs and Google Scholar electronic databases using the following terms: “HTLV”, “Indians”, “natives”, “Americas”. Lastly, multiple relevant articles were used to carry out the ‘Snowball’ method to supplement the review. However, the results of most of the studies published on this thematic should be taken with caution since the majority of them include rather small populations and not all the studies were carried out using stringent criteria of positivity, Western blot or PCR. In addition, HTLV prevalence tends to increase with age and is higher in women, but frequently information pertaining to age and sex of the studied populations has not been specified.
ORIGIN OF HTLV-1 AND HTLV-2 IN THE AMERICAS
HTLV-1 is endemic in South America, present in all 13 countries and prevailing in all ethnic populations22,26,59. In Brazil, for example, HTLV-1 is found in immigrants from other endemic foci, such as Africa and Japan53,79, and in those who are descended from Amerindians who have inhabited South America for thousands of years46,9,72,100,119.
There are two hypotheses about the origin of HTLV-1 in the Americas. The first considers the prehistoric migration of infected populations about 11,000 to 13,000 years ago across the Bering Strait into North America. These people migrated through Central America and spread into South America, where they settled mainly in the Andes and along the Amazon. The other hypothesis suggests that HTLV-1 originated in Africa and was brought to the Americas (including the Caribbean islands, United States, and South America) through the slave trade between the 16th and 19th centuries.
It is estimated that the separation of African and non-African human populations occurred around 75,000 to 287,000 years ago28,112, with gene flow occurring from pygmies to neighboring populations28. HTLV-1 and HTLV-2 infections among pygmies are the oldest, and although frequent transmission of simian T-lymphotropic virus type 1 (STLV-1) from apes to humans in Africa could suggest that these infections were the result of interspecies transmissions over the years87,90,115,130, the absence of non-human primates in Melanesia and Australia suggests that HTLV-1 existed among the Australoid people who first populated Australo-Melanesia around 60,000 years ago56,135.
The transcontinental migration of HTLV-1 is supported by studies showing that two strains of human leukocyte antigen (HLA)-A alleles are associated with HTLV-1 in endemic regions around the world. Both strains (HLA-A*26 and HLA-A*36) originally evolved in Africa and dispersed through Asian populations and indigenous North and South Americans120. This transcontinental dispersal of HTLV-1 partially overlapped the migration pattern of southeastern Asian mongoloids120,123. Interestingly, these alleles are associated with adult T-cell lymphoma (ATL), and phylogenetic analysis revealed that the ancestral genes of these alleles came from primate major histocompatibility complex genes, including those of gorillas, chimpanzees, and monkeys113. Taking this into consideration, it is possible that the HLA-A*26 and HLA-A*36 evolved > 50 million years ago and that carriers have become natural hosts of HTLV-1 due to a low immune responsiveness to the virus120.
Phylogenetic analysis of bone marrow samples from the femur of a mummified specimen revealed that the isolated clones were similar to those of the Indians of South America and belonged to a transcontinental subgroup closely related to HTLV-1 carriers, the Ainu of northern Japan, and some mongoloid Asian subgroups. These observations suggest that the Andean mummy's HTLV-1 could have originated from Asian paleo-mongoloids86.
The debate over the origin of HTLV-1 has primarily focused on empirical-level molecular phylogenetic analysis, particularly on how to better explain the phylogenetic origin of the cosmopolitan subtype of HTLV-1, to which the indigenous strains found in South America belong and which is the predominant subtype among the other isolates from various endemic and non-endemic areas of the Americas22. The subtypes, which do not appear to differ in pathogenicity, likely reflect the geographical origins and migrations of ancient populations77,80,118. Analysis of genetic sequencing has divided HTLV-1 into subtypes 1a (cosmopolitan), 1b (Central African), 1c (Melanesian), and 1d, 1e, 1f, and 1g (found in Central Africa). The cosmopolitan subtype is divided into five subgroups, depending on geographical location: transcontinental (A), to which most strains of HTLV-1 isolated from South America belong; Japanese (B); West African (C); North African (D)55,94; and Afro-Peruvian (E)128.
The transcontinental subgroup (A) has been characterized both in North America (United States and Canada) and South America (Argentina, Brazil, Chile, Colombia, French Guiana and Peru)23,53,59,77,118,122,127. The Japanese subgroup (B) has been found in the north, northeast, and southeast regions of Brazil; Canada; Colombia; and Peru and can be explained by the thousands of years of migration from Asia to the Americas and by recent Japanese immigration53,59,118,127,129. The West African subgroup (C) was identified in the Caribbean and French Guiana but not in Brazil, despite its introduction being associated with slave trafficking from West Africa and the high rate of infection among black South Americans23,53,59,122. The Afro-Peruvian subgroup (E) was characterized in two black individuals presenting a type of mitochondrial DNA identical to that found in some populations of West Africa55,128.
The migration of African people to the Americas through slave trade took place from mainly western and central Africa, and 40% of the approximately 10 million Africans arrived at Brazilian ports, making it intriguing that subgroup C is not found in Brazil53. Aiming to clarify the origin of HTLV-1 in Brazil, GALVÃO-CASTRO et al. 53 analyzed 243 sequences of the long terminal repeat region of isolates from descendants of various ethnic groups and geographical regions of the country, all of which were classified as the cosmopolitan subtype. Of these, 98% were in the transcontinental subgroup (A) and 3.3% in the Japanese subgroup (B), which is discordant with historical data indicating that the majority of Africans who came through Salvador were the original carriers of the West African subgroup (C).
The migration of African populations comprised several cycles: Guinea Cycle during the second half of the 16th century; Angola and Congo Cycle in the 17th century; Mina Coast Cycle during the first three-quarters of the 17th century; and Bay of Benin Cycle in the 18th and 19th centuries. It is possible that the occurrence of multiple introductions of some sequences of HTLV-1 in the post-Columbian era are clustered in Latin American groups with sequences of southern African ancestry that were segregated from the same ancestor of another group with a central African string. Thus, this relationship of ancestry suggests that this group was first introduced in South Africa due to the migration of the Bantu people of Central Africa to South Africa about 3,000 years ago and then to Brazil during the slave trade period between the 16th and 17th centuries53. Analysis of the distribution of haplotypes linked to the group of ß-globin genes demonstrated that 29.4% of the Bantu haplotype could explain why the majority of HTLV-1 isolates are grouped with those from southern Africa2. It is known that Africans of Bantu ethnicity were brought to Bahia between 1678 and 1810 and that approximately 2,400 African Bantu (with 100 coming from Angola and 2,300 from Madagascar) were brought between 1817 and 184337. During the colonization of South Africa by the British in the 17th and 18th centuries, many Africans migrated to neighboring regions currently known as Angola, Madagascar, and Mozambique, where they were captured and transported to Salvador37. In addition, there is evidence that the ports of departure of African slave ships often were not related to the ethnic and geographic origins of African transportees37. Thus, taken together, these results corroborate the hypothesis of multiple introductions of post-Columbian subgroup A in Brazil.
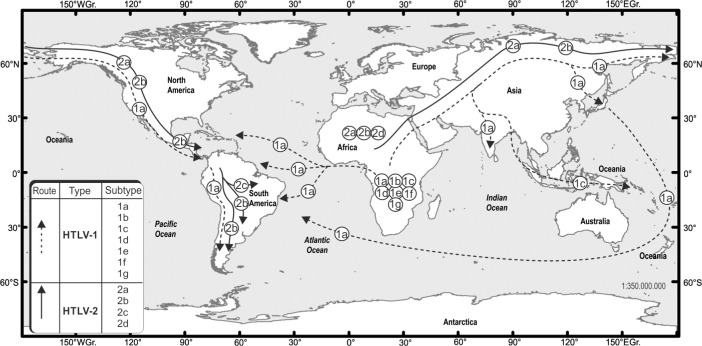
Most of non-indigenous persons infected by HTLV-1 in the Americas have probably been infected by virus strains originating from the African slave trade during the post-Columbian period2,37,44,53,57,59,75,122,128,130 and, it is today accepted that HTLV-1 in the Americas has more than one source: through migration from Asia through the Bering Strait86,94,104,134 during the Paleolithic era; the trafficking of slaves during the colonial period2,53,122,128,130; and more recently, the Japanese immigration in the early 20th century (Fig. 1)79,127,128.
Fig. 1. Origin of HTLV-1 and HTLV-2 in the Americas. Based on the references 53, 116, 120 and 128.
HTLV-1 and HTLV-2 are similar, with approximately 60% of their structures based on the same sequence of nucleic acids and 70% on the same sequence of amino acids20. Both viruses are very old and have evolved independently through transmission of STLV from nonhuman primates to the first humans62. HTLV-2 is divided into four subtypes: 2a, 2b, 2c, and 2d. Molecular studies confirm the presence of HTLV-2c in Brazil, the only country to identify this subtype43. The Brazilian variant of HTLV-2 has a tax region similar to that of subtype 2b and the env-like subtype 2a genomic long terminal repeat region33,43,72. Only the 2c variant was identified in Brazilian tribes, and this subtype of HTLV-2 is prevalent in IDU and non-IDU populations in Brazil3,43,69,70,72,119,125,126. HTLV-2 is considered an ancestral virus in the Americas because it is endemic among isolated indigenous groups, having been inherited by the general population and transmitted to IDUs from indigenous populations (Fig. 1)3,43,70,72,125,126.
The endemicity of HTLV-2 among various indigenous peoples of the Americas and the lack of evidence of infection with STLV-2 in New World monkeys have led to the conclusion that HTLV-2 has been present on the American continents since ancient times11,66,81,101,105. The hypothesis of independent development of HTLV-1 and HTLV-2 from a common ancestor after human migration to the Americas has a very low possibility of support due to the fact that HTLV-2 has been identified in isolates from pygmies in northwestern Zaire and Cameroon58,61, where HTLV-1 has not been identified, as well as the above mentioned absence of STLV-2 in New World monkeys75 and the slow evolutionary potential of HTLV-2116.
HTLV IN INDIGENOUS GROUPS OF OTHER COUNTRIES OF THE AMERICAS
Among native populations, HTLV-1 is found in the Pacific countries, including Australia and Melanesia, as well as in North, Central, and South America10, whereas HTLV-2 is endemic and widely distributed among the indigenous peoples of North, Central, and South America, where it appears to be more prevalent than HTLV-1, and in some tribes of Central Africa.
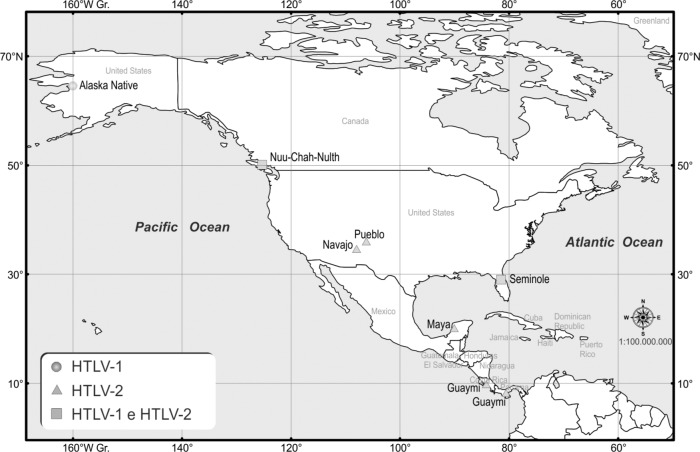
The population of American Indians and Alaska Natives is estimated at 5.2 million or 2% of the general US population, of which approximately 49% are not mixed with other races5. There are 566 tribes and 325 Indian reservations recognized by the US federal government5. In Canada, about 1.4 million Indians make up 4.3% of the total population, with the most populous group being the First Nations (61%) followed by Métis (32.3%) and Inuit (4.2%)1. In North America, there are few large, well-sampled studies of HTLV infection among Native Americans, and some have additional feature limitations, as shown by earlier serologic screening and confirmatory assays, or lack precise HTLV typing and molecular characterization of the virus23. A database of 2,047,740 blood donors was examined in North America for the period of 2000 to 2009 for both viruses, suggesting a prevalence of HTLV in the general population of 0.1% to 0.2% and showed a higher prevalence of HTLV-2 (14.7/100,000) than HTLV-1 (5.1/100,000) in the western and southwestern United States31, which could be attributed to endemic foci among American Indians30.
A previous study in blood donors had already demonstrated a rate of HTLV infection of 0.72/1,000 in New Mexico (most cases of which were attributed to HTLV-2) and, in turn, a higher prevalence among American Indian blood donors (1.0%-1.6%) than among non-Hispanic white donors (0.009%-0.06%)65. In the United States, HTLV is endemic among the Navajo and Pueblo people of New Mexico and Seminole people of Florida, with HTLV-2 presenting a higher prevalence85,88 (Table 1 and Fig. 2). In Alaska, the prevalence of HTLV among native peoples is 0.5% (two out of 380)34. This was confirmed as HTLV-1 in a subsequent study involving five other native individuals seropositive for HTLV from various geographical areas of Alaska, one being a blood donor with ATL, one with HAM/TSP, one with a neurological disease characterized by gait disturbance and urinary incontinence, and one with Hodgkin disease; all subjects with HTLV-1 had no risk factors for infection78.
Table 1. Prevalence of positivity for HTLV in indigenous groups of other countries of the Americas.
| Country | Author * | Population | N | HTLV-1 (%) | HTLV-2 (%) | ||
|---|---|---|---|---|---|---|---|
| United States | Levine et al. 199385 | Seminole | 106 | 14 | 13.2 | ||
| Lowis et al. 199988 | Seminole | 46 | 1 | 2.17 | 11 | 23.9 | |
| Davidson et al. 199034 | Alaska Natives | 380 | 2 | 0.5 | |||
| Canada | Peters et al. 2000 107 | Nuu-Chah-Nulth | 494 | 14 | 2.8 | 8 | 1.6 |
| Mexico | Gongora-Bianchi et al. 199760 | Maya | 440 | 1 | 0.23 | ||
| Panama | Lairmore et al. 199081 | Guaymi | 8 | 1 | 12.5 | ||
| Reeves et al. 1988111 | Guaymi | 317 | 25 | 7.9 | |||
| Pardi et al. 1993105 | Guaymi | 317 | 25 | 7.9 | |||
| Feigenbaum et al. 199446 | Guaymi | 109 | 9 | 8.3 | |||
| Vitek et al. 1995132 | Guaymi | 3686 | 352 | 9.5 | |||
| Costa Rica | Visoná et al. 1997131 | Guaymi | 8.0 | ||||
| Chile | Inostroza et al. 199167 | Mapuche | 405 | 3 | 0.7 | ||
| Cartier et al. 199324 | Huilliches/Mapuche | 199 | 1 | 0.5 | 2 | 1.0 | |
| Fujiyoshi et al. 199951 | Atacama | 217 | 9 | 4.1 | |||
| Alacalf | 23 | 8 | 34.8 | ||||
| Yahgan | 22 | 2 | 9.1 | ||||
| Rapa Nui | 132 | 1 | 0.8 | ||||
| Bolivia | Fujiyoshi et al. 199951 | Aymara | 151 | 8 | 5.3 | ||
| Quechua | 96 | 6 | 6.2 | ||||
| Colombia | Zamora et al. 1990136 | Paez | 32 | 2 | 6.3 | ||
| Dueñas-Barajas et al. 199239 | Wayuu | 523 | 1 | 1.6 | 3 | 4.8 | |
| Fujiyama et al. 199349 | Guahibo | 92 | 29 | 31.5 | |||
| Duenas-Barajas et al. 199340 | Waunana/Noanama | 143 | 3 | 2.1 | |||
| Tunebo | 40 | 2 | 5.0 | ||||
| Zaninovic et al. 1994137 | Inga | 62 | 1 | 1.6 | |||
| Kamsa | 59 | 5 | 8.5 | ||||
| Wayuu | 123 | 5 | 4.1 | ||||
| Arango et al. 19997 | Embera | 1014 | 10 | 1.0 | 7 | 0.7 | |
| Inga | 155 | 2 | 1.2 | 1 | 0.7 | ||
| Egea41 | Wayuu | 157 | 11 | 7.0 | |||
| Argentina | Ferrer et al. 199447 | Toba and Wichi | 175 | 24 | 13.7 | ||
| Bouzas et al. 199416 | Toba | 222 | 1 | 0.45 | 22 | 9.91 | |
| Biglione et al. 199912 | Wichi | 205 | 1 | 62 | 3.0 | ||
| Toba | 105 | 23 | 21.9 | ||||
| Medeot et al. 199992 | Toba | 72 | 2 | 2.78 | 2 | 2.78 | |
| Dipierre et al. 38 | Indians in Puna Jujeña | 86 | 2 | 2.32 | |||
| Ferrer et al. 199648 | Mapuche | 94 | 2 | 2.1 | |||
| Chorote | 171 | 61 | 35.6 | ||||
| Toba | 21 | 5 | 23.8 | ||||
| Wichi | 204 | 26 | 12.7 | ||||
| Chorote/Wichi | 14 | 4 | 28.5 | ||||
| Chorote/Chulupi | 10 | 8 | 80 | ||||
| Eirin et al. 201044 | Kolla | 112 | 11 | 9.8 | |||
| Fujiyoshi et al. 199951 | Puna | 88 | 2 | 2.3 | |||
Fig. 2. HTLV-1 and HTLV-2 among indigenous populations of North America and Central America. Based on the references from Table 1.
Table 1. Prevalence of positivity for HTLV in indigenous groups of other countries of the Americas (cont,).
| Country | Author * | Population | N | HTLV-1 (%) | HTLV-2 (%) | ||
|---|---|---|---|---|---|---|---|
| Paraguay | Ferrer et al. 199648 | Chulupi | 94 | 32 | 34 | ||
| Ayoreo | 51 | 2 | 3.9 | ||||
| Lengua | 49 | 5 | 10.2 | ||||
| Cabral et al. 199818 | Sanapaná | 30 | 2 | 6.7 | |||
| Angaité | 21 | 1 | 4.8 | ||||
| Fujiyoshi et al. 199951 | Chaco | 146 | 24 | 16.4 | |||
| Peru | Medeot et al. 199992 | Quechua | 40 | 1 | 2.5 | ||
| Shipibo-Konibo | 70 | 1 | 1.43 | ||||
| Harakmbet | 22 | 1 | 4.54 | ||||
| Huambisa | 42 | 1 | 2.38 | ||||
| Fujiyoshi et al. 199951 | Aymara | 62 | 1 | 1.6 | |||
| Sanchez-Palacios et al. 2003117 | Quechua women | 198 | 5 | 2.5 | |||
| Alva et al. 20104 | Shipibo-Konibo | 290 | 12 | 4.1 | 6 | 2.1 | |
| Ita et al. 201373 | Quechua | 389 | 11 | 2.82 | |||
| Blas et al. 201315 | Shipibo-Konibo women | 1253 | 74 | 5.9 | 47 | 3.8 | |
| Venezuela | Perez et al. 1993106 | Pume (Yaruro) | 210 | 12 | 5.7 | ||
| Leon-Ponte et al. 199683 | Guahibo | 166 | 41 | 24.7 | |||
| Leon-Ponte et al. 199884 | Yaruro/Guahibo | 41 | 25 | 61 | |||
| French Guiana | Talarmin et al. 1999122 | Arawack | 54 | 1 | 1.8 | ||
| Palikur | 78 | 2 | 2.6 | ||||
| Wayampi | 138 | 2 | 1.4 | ||||
Numbering as given in the references.
In Canada, studies in indigenous populations also have been limited. PICARD et al. (1995)108 observed that phylogenetic analysis of HTLV-1 strains recovered from indigenous people living on the coast of British Columbia in western Canada suggested multiple origins for the virus. Subsequently, ANDONOV et al. (2012)6 performed a phylogenetic analysis on various strains of HTLV-1 from the Nunavut population of Canada, of which 85% are Native Americans of British Columbia's Pacific coast, and Canadian non-indigenous people. The study demonstrated that strains in Nunavut (the cosmopolitan subtype) are related to those from East Asia and consistent with the presence of HTLV-1 in ancient indigenous peoples of the Canadian Arctic, noting the diversity of other strains of HTLV-1 analyzed from other Indians and reinforcing previous evidence of multiple incursions of this virus in indigenous populations of coastal British Columbia.
PETERS et al. (2000)107 found a prevalence of 2.8% for HTLV-1 and 1.6% for HTLV-2 among 494 serum samples from the Nuu-Chah-Nulth tribe of Vancouver Island in British Columbia, which is known for a high incidence of rheumatic disease. Although they found no association between arthropathy and HTLV-1, diseases such as ATL and HAM/TSP have been reported in this region36,54,103,107,109.
In Latin America, there are more than 400 indigenous groups, ranging from 45 million to 48 million individuals97. Most live in Bolivia, Guatemala, Peru, Ecuador, and Mexico9, comprising the majority of the population in Bolivia (62%) and Guatemala (60%)68, and less than 3% live in Paraguay, Venezuela, and Brazil29,68,96 (0.4% of the population)29. In South America, only Uruguay has no remaining indigenous population. In Mexico, the only Latin American country in North America, a 0.23% prevalence rate of HTLV-2 was found among 440 native Maya Indians and Mayans living in six communities on the Yucatan Peninsula60.
In the seven countries of Central America (Belize, Costa Rica, El Salvador, Guatemala, Honduras, Nicaragua, and Panama), some of which with strong commercial and cultural ties to the Caribbean islands, GESSAIN & CASSAR59 reviewed the high endemicity for HTLV-1 infection and associated diseases and demonstrated a very low HTLV-1 seroprevalence, with significant differences observed among the populations tested. In indigenous populations, HTLV-2 was endemic only among Guaymi people, with prevalence rates of 7.9%-8.5% in Panama46,81,105,111,132 and 8.0% in Costa Rica131 (Table 1 and Fig. 2).
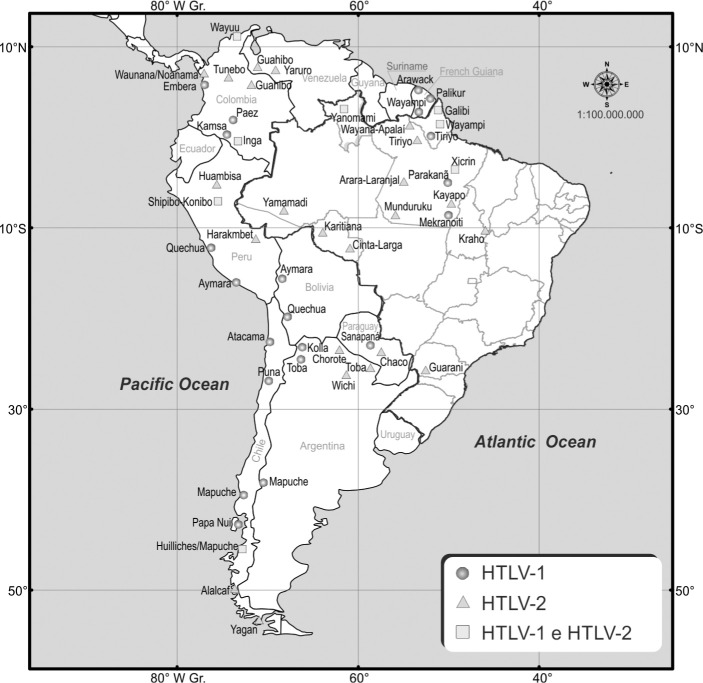
In South America, HTLV-2 predominates among indigenous groups (Table 1), with subtype 2b clearly prevailing in Amerindian populations121, except in Brazil, where it is characterized by subtype 2c23,43,69,72,119,126 (Fig. 1). HTLV-1 and HTLV-2 also differ in their geographical distribution among the various indigenous groups (Fig. 3). FUJIYOSHI et al. (1999)51 conducted a seroepidemiological study on indigenous peoples of the Andes of Colombia, Peru, Bolivia, Argentina, and Chile; Chiloé Island (Chilean coast); Easter Island (Chilean province of Polynesia); and the plains along the Atlantic coast from Colombia to the Orinoco, Amazon, and Patagonia, demonstrating an ethnic and geographically independent distribution between HTLV-1 and HTLV-2, with foci of HTLV-1 prevalent mainly in the Andean highlands and that of HTLV-2 in the coastal plains. FUJIYOSHI et al. (1995)50 also observed that HLA haplotypes in indigenous Andean groups with HTLV-1 and indigenous groups with HTLV-2 of the lower Orinoco (Venezuelan Amazon) were mutually exclusive. HLA haplotypes associated with HTLV-1 are commonly found in the known HTLV-1 endemic Indian and Japanese populations, whereas the haplotypes associated with HTLV-2 are specifically found among indigenous Orinoco and North American groups, suggesting that ethnic HLA haplotypes are separate from those native to South America and may be involved in the susceptibility to infection by HTLV-1 or HTLV-2. In southern Colombia, for example, HTLV-1 was first detected among natives belonging to the Paez people of the Andes136. Subsequently, HTLV-2 has been identified among the Wayuu, Guahibo, and Tunebo groups in the Guajira Peninsula in extreme northeastern Colombia (Caribbean Sea), with prevalence rates between 4.1% and 31.5%39,50,121,136. Other foci of HTLV-1 are found in various isolated indigenous populations (Wayuu, Waunana/Noanama, Inga, Kamsa, Embera), with prevalence rates from 1.0% to 8.5%7,39,40,137. Both viruses have been detected within some of these groups7,39, although in most, HTLV-1 and HTLV-2 appear to be mutually exclusive40,41,49,136,137, including a 31.5% HTLV-2 rate among Guahibo natives49 (Table 1). These observations suggest that the natives of South America could be divided into two major ethnic groups by an HTLV-1 and HTLV-2 carrier state that evolved among mongoloid populations and transmitted independently as two different strains among the indigenous peoples of the Andes highlands and coastal Atlantic plains41,50,51,137.
Fig. 3. HTLV-1 and HTLV-2 among indigenous populations of South America. Based on the references from Table 2.
In Venezuela, a high prevalence was found for HTLV-2, reaching 61% among Guahibo and Yaruro83,84,106, whereas in central and southern Bolivia, only HTLV-1 was detected among the Quechua and Aymara indigenous peoples, with a prevalence of 6.8% and 5.3%, respectively51. In the Peruvian Andes, Quechua and Aymara, the most populous indigenous groups in the region, also present solely HTLV-1, with prevalence rates from 1.6% to 2.82%51,73,92,117, whereas in the Amazon, the Peruvian Shipibo-Konibo notably carry both viruses, with high prevalence rates ranging from 1.43% to 5.9%4,15,92 and 2.1% to 3.8%4,15 for HTLV-1 and HTLV-2, respectively. A few cases of only HTLV-2 have also been reported in two other indigenous groups of the Peruvian Amazon92 (Table 1).
In Chile, where HTLV-1 seems to be endemic among groups of isolated indigenous peoples living in the Andes or in the southernmost region of the country59, prevalence ranges from 0.5% to 0.8% among the Mapuche and Rapa Nui24,51,67 and up to 4.1% among the Atacama were found51. Foci of HTLV-2 were reported among the Alacalf (34.8%)51, Yaghan (9.1%)51, and Huilliches/Mapuche (1.0%) peoples24.
In Argentina, ethnic and geographic restriction are also seen for both viruses13, with HTLV-1 prevalent among the Qulla and Puná of northwestern Argentina38,44,51 and HTLV-2 variably, but generally more prevalent among the Chorote, Wichi, Chulupi, and Toba peoples of the Chaco region, covering parts of Bolivia, Argentina, Brazil, and Paraguay12,13,16,47,48,51,92 (Table 1).
HTLV-2 is found almost exclusively among the indigenous population of Paraguay, who mostly live in the Gran Chaco region in the northwest, with a prevalence ranging from 3.9% to 34%18,48,51 (Table 1). In French Guiana (not included in Latin America), TALARMIN et al. (1993)122 did not detect HTLV-2 among the 847 indigenous peoples of the Arawak, Palikur, Wayampi, Galibi, Emerillon, and Wayana, but HTLV-1 was detected in 1.81% (5 out of 270) of the Arawak, Palikur, and Wayampi, with prevalence ranging from 1.4% to 2.6% (Table 1), with the virus probably acquired through contact with people of African descent during slave trade, according to phylogenetic analyses128.
HTLV IN INDIGENOUS POPULATIONS OF BRAZIL
The first description of HTLV infection among indigenous people in Brazil dates back to 1990 when, while assessing the prevalence of the virus in some human populations at risk, NAKAUCHI et al. 99 reported a 39% positivity for HTLV-1 in the serum of 82 Mekranoiti subjects and 20% in 55 Tiriyo subjects from Pará. Confirmatory tests of the final results of this study were positive in 3.63% (two out of 55), 12.19% (10 out of 82), and 13.88% (10 out of 72) for HTLV-1 among Tiriyo, Mekranoiti, and Xicrin subjects, respectively99,100.
Subsequent studies showed that HTLV-2 is predominant among Brazilian indigenous groups14,51,52,69,91,93,102,119,126, with an area of high endemicity in the Amazon region. HTLV-1, on the other hand, is present in isolated clusters69,119 (Table 2 and Fig. 3).
Table 2. Prevalence of positivity for HTLV in indigenous populations of Brazil.
| State | Tribe/Nation | Author * | N | HTLV-1 (%) | HTLV-2 (%) | ||
|---|---|---|---|---|---|---|---|
| Amazonas | Yamamadi | Ishak et al. 199569 | 36 | 2 | 5.6 | ||
| Amapá | Galibi | Ishak et al. 199569 | 148 | 1 | 0.67 | 3 | 2.0 |
| Wayampi | Ishak et al. 199569 | 71 | 1 | 1.4 | |||
| Shindo et al. 2002119 | 321 | 2 | 0.62 | ||||
| Pará | Kayapo | Maloney et al. 199291 | 264 | 88 | 33.3 | ||
| Black et al. 199414 | 703 | 28.0 | |||||
| Ishak et al. 199569 | 207 | 1 | 0.48 | 67 | 32.2 | ||
| Fujiyoshi et al. 199951 | 19 | 11 | 57.9 | ||||
| Vallinoto et al. 2002126 | 27 | 6 | 22.2 | ||||
| Kayapo ** | Novoa et al. 1997102 | 141 | 59 | 41.8 | |||
| Tiriyo | Nakauchi et al. 1992100 | 55 | 2 | 3.6 | |||
| Ishak et al. 199569 | 26 | 4 | 15.4 | ||||
| Vallinoto et al. 2002126 | 150 | 3 | 2.0 | ||||
| Shindo et al. 2002119 | 683 | 3 | 0.44 | ||||
| Xicrin | Nakauchi et al. 1992100 | 72 | 10 | 13.9 | |||
| Gabbai et al. 199352 | 206 | 31 | 15.0 | ||||
| Mekranoiti | Nakauchi et al. 1992100 | 82 | 10 | 12.2 | |||
| Parakanã | Gabbai et al. 199352 | 89 | 2 | 2.25 | |||
| Ishak et al. 199569 | 52 | 1 | 1.92 | ||||
| Arara Laranjal | Ishak et al. 199569 | 44 | 5 | 11.4 | |||
| Munduruku | Ishak et al. 199569 | 161 | 13 | 8.1 | |||
| Wayana-Apalaí | Ishak et al. 199569 | 50 | 1 | 2.0 | |||
| Roraima | Yanomami | Ishak et al. 199569 | 102 | 3 | 2.94 | 4 | 3.9 |
| Rondônia | Karitiana | Ishak et al. 199569 | 50 | 2 | 4.0 | ||
| Cinta-Larga | Ishak et al. 199569 | 50 | 1 | 2.0 | |||
| Tocantins | Kraho | Maloney et al. 199291 | 172 | 21 | 12.2 | ||
| Paraná | Guarani | Menna-Barreto et al. 200593 | 52 | 3 | 5.8 | ||
Numbering as given in the references.
Children born to HTLV-2-positive mothers.
ISHAK et al. (1995)69 investigated sera collected from 1,382 individuals belonging to 26 indigenous communities within endemic regions of HTLV-2. These communities are distributed through the states of Maranhão (Urubu-Kaapór), Amapá (Galibi, Palikur, Wayampi), Amazonas (Yamamadi), Roraima (Yanomami), Rondônia (Cinta Larga, Surui, Karitiana), and Pará (Wayana-Apalai, Tiriyo, Assurini Kuatinemo, Assurini Trocará, Zoé, Arara Laranjal, Arara Kurambê, Arara, Iriri, Araweté, Parakanã, Munduruku, and six different Kayapo tribes). The authors found HTLV-2 to be present in 17 of the 26 communities, providing evidence that the Amazon region of Brazil is the most endemic area in the world for HTLV-269,72. This study also found the presence of antibodies to HTLV-1 limited to five individuals: one Galibi (Amapá), three Yanomami (Roraima), and one Kayapo from the village of Aukre (Pará) (Table 2). The highest prevalence for HTLV-2 was observed among the Kayapo (32.2%), followed by Tiriyo (15.4%), Mundukuru (8.1%), and Arara Laranjal (11.4%) peoples69. Several other studies have also shown a very high prevalence of HTLV-2 among the Kayapo, at rates ranging from 28% to 57.9%14,51,91,102,126. Moreover, MALONEY et al. (1992)91 reported the presence of HTLV-2 in 12.2% (21 out of 172) of Krahos people in Tocantins.
Tiriyo and Wayampi indigenous peoples live at the border between Brazil, Suriname and French Guiana, respectively (approximately 1,700 Tiriyo, 750 of whom live in Brazil and are spread over 15 villages, and 1,200 Wayampis, 450 of whom live in Brazil in the state of Amapá)119. The presence of both HTLV-1100,119 and HTLV-269,119,126 was confirmed among Wayampi and Tiriyo from Brazil in whom HTLV-2 was predominant69,119,126, in contrast to what is observed among the indigenous population of the Amazon region of French Guiana, where only HTLV-1 is present and probably brought from Africa during the post-Columbian slave trading period74,122.
The south of Brazil, on the other hand, is directly related both geographically and ethnically to northern Argentina and southern Paraguay, areas known to be endemic for HTLV-212,16,18,47,48,51,92. A study of the Guarani Indians in southern Brazil reported a prevalence of 5.76% for HTLV-2 among 52 individuals examined, suggesting that the Guarani is another endemic indigenous group for this retrovirus and the need for molecular and phylogenetic studies with larger numbers of samples93.
The prevalence of infection reported in Brazil for the various ethnic groups ranges from 0.48% to 13.9% for HTLV-1 (Xicrin, Mekranoiti, Tiriyo, Yanomami, Galibi, Wayampi, and Kayapo)69,100,119 and 0.44% to 57.9% for HTLV-2 (Kayapo, Tiriyo, Xicrin, Kraho, Arara Laranjal, Mundukuru, Guarani, Yamamadi, Karitiana, Yanomami, Parakanã, Galibi, Wayana-Apalai, Cinta Larga, and Wayampi)14,51,52,69,91,93,102,119,126 (Table 2).
In addition to approximately two dozen cases initially reported in 1992 by NAKAUCHI et al. 100 among the Xicrin, Mekranoiti, and Tiriyo peoples of Brazil, only seven cases were confirmed as HTLV-1 infection among Brazilian indigenous69,119 (Table 2). The possibility of post-transfusion infection cannot be ruled out as a means of introduction of the virus to these ethnic groups with a low prevalence for retrovirus and in whom malaria is frequent76. These data support the African origin of HTLV-1 in Brazil introduced in the post-Columbian period through slave trade, similar to what would have occurred in French Guiana and the Caribbean basin2,37,53,57,74,122.
The importance of maintaining the endemicity of HTLV in these epidemiologically closed populations by transmission through sexual relations and vertical pathways is supported by evidence from the Kayapo Indians, who are experiencing a gradual increase in antibody prevalence with age and constant and continuous transmission between couples14,72. Molecular biology has confirmed a high prevalence (42%) of HTLV-2 among children born to HTLV-2-positive mothers71,102. Breastfeeding (and cross-feeding) is a primary source of nutrition for indigenous children, which increases the spread of the virus72. If transmission is not stopped, HTLV-2 will become hyperendemic14,69, and transmission to non-indigenous populations is increasingly possible despite the relative geographical isolation of the Kayapo. For example, some villages of Kayapo (Kubenkokre, Kokraimoro, Aukre, Kararaô, Gorotire, and Kikretum) are located near small towns maintained by commercial industries, agriculture, and mining, and it is common practice of indigenous Brazilian men to visit these towns to have sexual relations with non-indigenous women72.
Ignorance about the virus greatly increases the risk of transmission; thus, preventive measures to reduce the spread and transmission of retroviruses in indigenous populations initially depend on the identification of cases and educational programs17,23,32,139,140. The indigenous community should be adequately informed about the modes of HTLV transmission and the risks associated with prolonged breastfeeding and cross-feeding. Pregnant women should be routinely screened for HTLV infection and, if positive, should be given access to other alternatives to breast milk, such as formulas17,23,32,64. Woman who are likely to breast feed other children should be also screened for HTLV.
Regarding the prevention of sexual transmission of HTLV, programs should emphasize the importance of condom use, systematic screening for infection, and individual counseling17,23,32,139,140. Screening should also include the parents of the infected individual and siblings if the mother tests positive; children born to mothers with the virus should receive appropriate follow-up17,32,64.
In Brazil, HTLV is not considered a public health problem and, thus, has been widely neglected25,32,139. Like disease, injury, and public health events, cases of HTLV should be reportable to better approximate its incidence among indigenous peoples and to track its spread to the general population.
Revista do Instituto de Medicina Tropical de São Paulo on line.
Publications from 1984 to the present data are now available on: http://www.scielo.br/rimtsp
PAST ISSUES FROM 1959 ON (PDF)
SciELO – The Scientific Electronic Library OnLine - SciELO is an electronic virtual covering a selected collection of Brazilian scientific journals.
The library is an integral part of a project being developed by FAPESP – Fundação de Amparo à Pesquisa do Estado de São Paulo, in partnership with BIREME – the Latin American and Caribbean Center on Health Sciences Information.
SciELO interface provides access to its serials collection via an alphabetic list of titles or a subject index or a search by word of serial titles, publisher names, city of publication and subject.
The interface also provides access to the full text of articles via author index or subject index or a search form on article elements such as author names, words from title, subject and words from full text.
FAPESP/BIREME Project on Scientific Electronic Publications
Latin American and Caribbean Center on Health Sciences Information
Rua Botucatu 862 – 04023-901 São Paulo, SP – Brazil
Tel. (011) 5576-9863
scielo@bireme.br
ACKNOWLEDGMENTS
The authors would like to thank Dr. Augusto Cesar Penalva de Oliveira and Jerusa Smid for helpful discussions in the last years. They also thank Prof. Melchior Carlos do Nascimento (Universidade Federal de Alagoas), who prepared the maps used in this manuscript. FAPESP: 2012/23397-0; 2008/56427-4.
REFERENCES
- 1.Aboriginal Peoples in Canada: First Nations People, Métis and Inuit . Canada: Ministry of Industry; 2013. The National Household Survey, 2011. Available from: http://www12.statcan.gc.ca/nhs-enm/2011/as-sa/99-011-x/99-011-x2011001-eng.pdf. [Google Scholar]
- 2.Alcantara LCJ, Van Dooren S, Gonçalves MS, Kashima S, Costa MC, Santos FL, et al. Globin haplotypes of human T-cell lymphotropic virus type I-infected individuals in Salvador, Bahia, Brazil, suggest a post-Columbian African origin of this virus. J Acquir Immune Defic Syndr. 2003;33:536–42. doi: 10.1097/00126334-200308010-00016. [DOI] [PubMed] [Google Scholar]
- 3.Alcantara LCJ, Shindo N, Van Dooren S, Salemi M, Costa MCR, Kashima S, et al. Brazilian HTLV type 2a strains from Intravenous drug users (IDUs) appear to have originated from two sources: Brazilian Amerindians and European/North American IDUs. AIDS Res Hum Retroviruses. 2003;19:519–23. doi: 10.1089/088922203766774577. [DOI] [PubMed] [Google Scholar]
- 4.Alva IE, Orellana ER, Blas MM, Bernabe-Ortiz A, Cotrina A, Chiappe M, et al. HTLV-1 and -2 Infections among 10 Indigenous Groups in the Peruvian Amazon. Am J Trop Med Hyg. 2012;87:954–6. doi: 10.4269/ajtmh.2012.12-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Indians by the Numbers From the U.S. Census Bureau. Available from: http://www.infoplease.com/spot/aihmcensus1.html.
- 6.Andonov A, Coulthart MB, Pérez-Losada M, Crandall KA, Posada K, Posada D, et al. Insights into origins of human T-cell Lymphotropic Virus Type 1 based on new strains from aboriginal people of Canada. Infect Genet Evol. 2012;12:1822–30. doi: 10.1016/j.meegid.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Arango C, Maloney E, Rugeles MT, Bernal E, Bernal C, Borrero I, et al. HTLV-I and HTLV-II coexist among the Embera and Inga Amerindians of Colombia. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:102–3. doi: 10.1097/00042560-199901010-00021. [DOI] [PubMed] [Google Scholar]
- 8.Araujo A, Hall WW. Human T-lymphotropic virus type II and neurological disease. Ann Neurol. 2004;56:10–9. doi: 10.1002/ana.20126. [DOI] [PubMed] [Google Scholar]
- 9.Barreto SM, Miranda JJ, Figueroa P, Schmidt MI, Munoz S, Kuri-Morales PP, et al. Epidemiology in Latin America and the Caribbean: current situation and challenges. Int J Epidemiol. 2012;41:557–71. doi: 10.1093/ije/dys017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beilke MA, Theall KP, O'Brien M, Clayton JL, Benjamin SM, Winsor EL, et al. Clinical outcomes and disease progression among patients coinfected with HIV and human T lymphotropic virus types 1 and 2. Clin Infect Dis. 2004;39:253–63. doi: 10.1086/422146. [DOI] [PubMed] [Google Scholar]
- 11.Biggar RJ, Taylor ME, Neel JV, Hjelle B, Levine PH, Black FL, et al. Genetic variants of human T-lymphotropic virus type II in American Indian groups. Virology. 1996;216:165–73. doi: 10.1006/viro.1996.0043. [DOI] [PubMed] [Google Scholar]
- 12.Biglione M, Vidan O, Mahieux R, de Colombo M, de los Angeles de Basualdo M, Bonnet M, et al. Seroepidemiological and molecular studies of human T cell lymphotropic virus type II, subtype b, in isolated groups of Mataco and Toba Indians of northern Argentina. AIDS Res Hum Retroviruses. 1999;15:407–17. doi: 10.1089/088922299311150. [DOI] [PubMed] [Google Scholar]
- 13.Biglione MM, Berini CA. Aportes y consideraciones sobre la infección por los vírus linfotrópicos-T humanos tipo 1 y 2 en Argentina. Atualizaciones SIDA Infectol. 2013;21:84–94. [Google Scholar]
- 14.Black FL, Biggar RJ, Neel JV, Maloney EM, Waters DJ. Endemic transmission of HTLV type II among Kayapo Indians of Brazil. AIDS Res Hum Retroviruses. 1994;10:1165–71. doi: 10.1089/aid.1994.10.1165. [DOI] [PubMed] [Google Scholar]
- 15.Blas MM, Alva IE, García PJ, Cárcamo C, Montano SM, Mori N, et al. High prevalence of human T-lymphotropic virus Infection in Indigenous Women from the Peruvian Amazon. PLOS One. 2013;8: doi: 10.1371/journal.pone.0073978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouzas MB, Zapiola I, Quiruelas S, Gorvein D, Panzita A, Rey J, et al. HTLV type I and HTLV type II infection among Indians and natives from Argentina. AIDS Res Hum Retroviruses. 1994;10:1567–71. doi: 10.1089/aid.1994.10.1567. [DOI] [PubMed] [Google Scholar]
- 17.Brasil . Guia do manejo clínico do HTLV. Brasília: Ministério da Saúde; 2004. Ministério da Saúde. Secretaria de Vigilância em Saúde. Programa Nacional de DST e AIDS. [Google Scholar]
- 18.Cabral MB, Vera ME, Samudio M, Arias AR, Cabello A, Moreno R, et al. HTLV-I/II antibodies among three different groups from Paraguay. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:548–49. doi: 10.1097/00042560-199812150-00018. [DOI] [PubMed] [Google Scholar]
- 19.Callatini S, Chevalier SA, Duprez R, Bassot S, Froment A, Mahieux R, et al. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in Central Africa. Retrovirology. 2005;2:30. doi: 10.1186/1742-4690-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cann AJ, Chen ISY. Human T-cell leukemia virus types I and II. In: Fields BN, Knipe DM, Howley PM, Melnick JL, Roizman B, Straus SE, editors. Fields Virology. Philadelphia: Raven Publishers; 1996. pp. 1849–80. [Google Scholar]
- 21.Carneiro-Proietti AB, Ribas JG, Catalan-Soares BC, Martins ML, Brito-Melo GE, Martins OA, Filho, et al. Infeccção e doença pelos vírus linfotrópicos humanos de células T (HTLV-I/II) no Brasil. Rev Soc Bras Med Trop. 2002;35:499–508. doi: 10.1590/s0037-86822002000500013. [DOI] [PubMed] [Google Scholar]
- 22.Carneiro-Proietti ABF, Catalan-Soares B, Proietti FA. Human T cell lymphotropic viruses (HTLV-I/II) in South America: should it be a public health concern? J BiomedSci. 2002;9:587–95. doi: 10.1159/000067286. [DOI] [PubMed] [Google Scholar]
- 23.Carneiro-Proietti ABF, Catalan-Soares BC, Castro-Costa CM, Murphy EL, Sabino EC, Hisada M, et al. HTLV in the Americas: challenges and perspectives. Rev Panam Salud Publica. 2006;19:44–53. doi: 10.1590/s1020-49892006000100007. [DOI] [PubMed] [Google Scholar]
- 24.Cartier L, Araya F, Catillo JL, Zaninovic V, Hayami M, Miura T, et al. Southernmost carriers of HTLVI/II in the world. Jpn J Cancer Res. 1993;84:1–3. doi: 10.1111/j.1349-7006.1993.tb02774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casseb J. Is human T cell lymphotropic type 1 (HTLV-1)-associated myelopathy/tropical spastic paraparesis (HAM/TSP) syndrome a neglected disease? PLOS Negl Trop Dis. 2009;3:e487. doi: 10.1371/journal.pntd.0000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catalan-Soares BC, Proietti FA, Carneiro-Proietti ABF. Os vírus linfotrópicos de células T humanos (HTLV) na última década (1990-2000): aspectos epidemiológicos. Rev Bras Epidemiol. 2001;4:81–95. [Google Scholar]
- 27.Caterino-de-Araujo A, Casseb JS, Neitzert E, de Souza ML, Mammano F, Del Mistro A, et al. HTLV-I and HTLV-II infections among HIV-1 seropositive patients in Sao Paulo, Brazil. Eur J Epidemiol. 1994;10:165–71. doi: 10.1007/BF01730366. [DOI] [PubMed] [Google Scholar]
- 28.Cavalli-Sforza LL, Menozziand P, Piazza A. The history and geography of human genes. New Jersey: Princeton University Press; 1994. [Google Scholar]
- 29.Censo demográfico 2010: características gerais dos indígenas: resultados do universo. Rio de Janeiro: IBGE; 2012. [Google Scholar]
- 30.Chang YB, Kaidarova Z, Hindes D, Bravo M, Kiely N, Kamel H, et al. Seroprevalence and demographic determinants of human T-lymphotropic virus type-1 and -2 Infections among First-time blood donors, U.S. 2000-2009. J Infect Dis. 2014;209:523–31. doi: 10.1093/infdis/jit497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook LB, Taylor GP. HTLV-1 and HTLV-2 prevalence in the United States. J Infect Dis. 2014;209:486–7. doi: 10.1093/infdis/jit558. [DOI] [PubMed] [Google Scholar]
- 32.Costa CA, Furtado KCYO, Ferreira LSC, Almeida DS, Linhares AC, Ishak R, et al. Familial transmission of human T-cell lymphotrophic virus: silent dissemination of an emerging but neglected infection. PLOS Negl Trop Dis. 2013;7:e2272. doi: 10.1371/journal.pntd.0002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Covas DM, Kashima S. Complete nucleotide sequences of the genomes of two Brazilian specimens of human T lymphotropic virus type 2 (HTLV-2) AIDS Res Hum Retroviruses. 2003;19:689–97. doi: 10.1089/088922203322280919. [DOI] [PubMed] [Google Scholar]
- 34.Davidson M, Kaplan JE, Hartley TM, Lairmore MD, Lanier AP. Prevalence of HTLV-I in Alaska Natives. J Infect Dis. 1990;161:359–60. doi: 10.1093/infdis/161.2.359. [DOI] [PubMed] [Google Scholar]
- 35.de Thé GD, Bomford R. An HTLV-I vaccine: why, how, for whom? AIDS Res Hum Retroviruses. 1993;9:3816. doi: 10.1089/aid.1993.9.381. [DOI] [PubMed] [Google Scholar]
- 36.Dekaban GA, Oger JJ, Foti D, King EE, Waters DJ, Picard FJ, et al. HTLV-I infection associated with disease in aboriginal Indians from British Columbia: a serological and PCR analysis. Clin Diagn Virol. 1994;2:67–78. doi: 10.1016/0928-0197(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 37.Desrames A, Cassar O, Afonso PV, Gout O, Hermine O, Gessain A. Molecular epidemiology of HTLV-1 infection in the Caribbean area as compared to West Africa: relationship with the slave trade. Retrovirology. 2011.8:A90. [Google Scholar]
- 38.Dipierri JE, Tajima K, Cartier Robirosa L, Sonoda S. A seroepidemiological survey of HTLV-I/II carriers in the Puna Jujeña. Medicina (B Aires) 1999;59:717–20. [PubMed] [Google Scholar]
- 39.Dueñas-Barajas E, Bernal JE, Vaught DR, Briceño I, Durán C, Yanagihara R, et al. Coexistence of human T-lymphotropic virus types I and II among the Wayuu Indians from the Guajira Region of Colombia. AIDS Res Hum Retroviruses. 1992;8:1851–5. doi: 10.1089/aid.1992.8.1851. [DOI] [PubMed] [Google Scholar]
- 40.Dueñas-Barajas E, Bernal JE, Vaught DR, Nerurkar VR, Sarmiento P, Yanagihara R, et al. Human retroviruses in Amerindians of Colombia: high prevalence of human T cell lymphotropic virus type II infection among the Tunebo Indians. Am J Trop Med Hyg. 1993;49:657–63. doi: 10.4269/ajtmh.1993.49.657. [DOI] [PubMed] [Google Scholar]
- 41.Egea E. Polimorfismo genético del MHC y su asociación con la infección HTLV-II: una herramienta de epidemiología molecular en el análisis de subpoblaciones del Caribe colombiano. Rev Acad Colomb Cienc. 2002;26((99)):181–96. [Google Scholar]
- 42.Ehrlich GD, Glaser JB, LaVigne K, Quan D, Mildvan D, Sninsky JJ, et al. Prevalence of human T-cell leukemia/lymphoma virus (HTLV) type II infection among high-risk individuals: type- specific identification of HTLVs by polymerase chain reaction. Blood. 1989;74:1658–64. [PubMed] [Google Scholar]
- 43.Eiraku N, Novoa P, da Costa Ferreira M, Monken C, Ishak R, da Costa Ferreira O, et al. Identification and characterization of a new and distinct molecular subtype of human T cell lymphotropic virus type 2. J Virol. 1996;70:1481–92. doi: 10.1128/jvi.70.3.1481-1492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eirin ME, Berini CA, Jones LR, Dilernia DA, Puca AA, Biglione MM. Stable human T-cell lymphotropic virus type 1 (HTLV-1) subtype a/subgroup a endemicity in Amerindians from Northwest Argentina: a health problem to be resolved. J Med Virol. 2010;82:2116–22. doi: 10.1002/jmv.21834. [DOI] [PubMed] [Google Scholar]
- 45.Etzel A, Shibata GY, Rozman M, Jorge ML, Damas CD, Segurado AA. HTLV-1 and HTLV-2 infections in HIV-infected individuals from Santos, Brazil: seroprevalence and risk factors. J Acquir Immune Defic Syndr. 2001;26:185–90. doi: 10.1097/00042560-200102010-00015. [DOI] [PubMed] [Google Scholar]
- 46.Feigenbaum F, Fang C, Sandler SG. Human T-lymphotropic virus type II in Panamanian Guaymi Indians. Transfusion. 1994;34:158–61. doi: 10.1046/j.1537-2995.1994.34294143946.x. [DOI] [PubMed] [Google Scholar]
- 47.Ferrer JF, Del Pino N, Esteban E, Sherman MP, Dube S, Dube DK, et al. High rate of infection with the human T-cell leukemia retrovirus type II in four Indian populations of Argentina. Virology. 1993;197:576–84. doi: 10.1006/viro.1993.1631. [DOI] [PubMed] [Google Scholar]
- 48.Ferrer JF, Esteban E, Dube S, Basombrio MA, Segovia A, Peralta-Ramos M, et al. Endemic infection with human T cell leukemia/lymphoma virus type IIB in Argentinean and Paraguayan Indians: epidemiology and molecular characterization. J Infect Dis. 1996;174:944–53. doi: 10.1093/infdis/174.5.944. [DOI] [PubMed] [Google Scholar]
- 49.Fujiyama C, Fujiyoshi T, Miura T, Yashiki S, Matsumoto D, Zaninovic V, et al. A new endemic focus of human T lymphotropic virus type II carriers among Orinoco natives in Colombia. J Infect Dis. 1993;168:1075–7. doi: 10.1093/infdis/168.4.1075. [DOI] [PubMed] [Google Scholar]
- 50.Fujiyoshi T, Yashiki S, Fujiyama C, Kuwayama M, Miyashita H, Ohnishi H, et al. Ethnic segregation of HTLV-I and HTLV-II carriers among South American native Indians. Int J Cancer. 1995;63:510–5. doi: 10.1002/ijc.2910630409. [DOI] [PubMed] [Google Scholar]
- 51.Fujiyoshi T, Li HC, Lou H, YashIki S, Karino S, Zaninovic V, et al. Characteristic distribuition of HTLV type I and HTLV type II carriers among native ethnic groups in South America. AIDS Res Hum Retroviruses. 1999;15:1235–9. doi: 10.1089/088922299310124. [DOI] [PubMed] [Google Scholar]
- 52.Gabbai AA, Bordin JO, Vieira PB, Filho, Kuroda A, Oliveira ASB, Cruz MV, et al. Selectivity of human T lymphotropic virus type-1 (HTLV-1) and HTLV-2 infection among different populations in Brazil. Am J Trop Med Hyg. 1993;49:664–71. doi: 10.4269/ajtmh.1993.49.664. [DOI] [PubMed] [Google Scholar]
- 53.Galvão-Castro B, Alcântara LCJ, Grassi MFR, Mota-Miranda ACA, Queiroz ATL, Rego FFA, et al. Epidemiologia e origem do HTLV-1 em Salvador Estado da Bahia: a cidade com a mais elevada prevalência desta infecção no Brasil. Gaz Med Bahia. 2009;79:3–10. [Google Scholar]
- 54.Gascoyne RD, Kim SM, Oger JJ, Melosky BL, Dekaban GA. HTLV-I associated adult T cell leukemia/lymphoma: report of two cases from an Amerindian population in coastal northwest British Columbia. Leukemia. 1996;10:552–7. [PubMed] [Google Scholar]
- 55.Gasmi M, Farouqi B, d'Incan M, Desgranges C. Long terminal repeat sequence analysis of HTLV type I molecular variants identified in four north African patients. AIDS Res Hum Retroviruses. 1994;10:1313–5. doi: 10.1089/aid.1994.10.1313. [DOI] [PubMed] [Google Scholar]
- 56.Gessain A, Boeri E, Yanagihara R, Gallo RC, Francini G. Complete nucleotide sequence of a highly divergent human T-cell leukemia (lymphotropic) virus type I (HTLV-I) variant from Melanesia: genetic and phylogenetic relationship to HTLV-I strains from other geographical regions. J Virol. 1993;67:1015–23. doi: 10.1128/jvi.67.2.1015-1023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gessain A, Koralnik IJ, Füllen J, Boeri E, Mora C, Blank A, et al. Phylogenetic study of ten new HTLV-I strains from the Americas. AIDS Res Hum Retroviruses. 1994;10:103–6. doi: 10.1089/aid.1994.10.103. [DOI] [PubMed] [Google Scholar]
- 58.Gessain A, Mauclere P, Fromenti A, Biglione M, Hesran JYL, Tekaiat F, et al. Isolation and molecular characterization of a human T-cell lymphotropic virus type II (HTLV-II), subtype B, from a healthy Pygmy living in a remote area of Cameroon: an ancient origin for HTLV-II in Africa. Proc Natl Acad Sci USA. 1995;92:4041–5. doi: 10.1073/pnas.92.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gessain A, Cassar O. Epidemiological aspects and world distribution of HTLV-1 infection. Front Microbiol. 2012;3:388. doi: 10.3389/fmicb.2012.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gongora-Bianchi RA, Lal RB, Rudolph DL, Castro-Sansores C, Gonzalez-Martinez P, Pavia-Ruz N. Low prevalence of HTLV-II in Mayan Indians in the Yucatan Peninsula, Mexico. Arch Med Res. 1997;28:555–8. [PubMed] [Google Scholar]
- 61.Goubau P, Desmyter J, Ghesquiere J, Kasereka B. HTLV-II among pygmies. Nature. 1992;359:201. doi: 10.1038/359201a0. [DOI] [PubMed] [Google Scholar]
- 62.Goubau P, Vandamme AM, Desmyter J. Questions on the evolution of primate T-lymphotropic viruses raised by molecular and epidemiological studies of divergent strains. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13((Suppl 1)):S242–7. doi: 10.1097/00042560-199600001-00036. [DOI] [PubMed] [Google Scholar]
- 63.Hall WW, Ishak R, Zhu SW, Novoa P, Eiraku N, Takahashi H, et al. Human T lymphotropic virus type II (HTLV-II): epidemiology, molecular properties, and clinical features of infection. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:204–14. doi: 10.1097/00042560-199600001-00031. [DOI] [PubMed] [Google Scholar]
- 64.Hino S. Establishment of the milk-borne transmission as a key factor for the peculiar endemicity of human T-lymphotropic virus type 1 (HTLV-1): the ATL Prevention Program Nagasaki. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87:152–66. doi: 10.2183/pjab.87.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hjelle B, Mills R, Swenson S, Mertz G, Key C, Allen S. Incidence of hairy cell leukemia, mycosis fungoides, and chronic lymphocytic leukemia in first known HTLV-II-endemic population. J Infect Dis. 1991;163:435–40. doi: 10.1093/infdis/163.3.435. [DOI] [PubMed] [Google Scholar]
- 66.Ijichi S, Tajima K, Zaninovic V, Leon FE, Katahira Y, Sonoda S, et al. Identification of human T-cell leukemia virus type IIb infection in the Wayu, an aboriginal population of Colombia. Jpn J Cancer Res. 1993;84:1215–8. doi: 10.1111/j.1349-7006.1993.tb02824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Inostroza J, Diaz P, Saunier C. Prevalence of antibodies to HTLV-1 in South American Indians (Mapuches) from Chile. Scand J Infect Dis. 1991;23:507–8. doi: 10.3109/00365549109075102. [DOI] [PubMed] [Google Scholar]
- 68.International Work Group for Indigenous Affairs . Denmark: IWGIA; 2013. The indigenous World 2013. [Google Scholar]
- 69.Ishak R, Harrington WJ, Jr, Azevedo VN, Eiraku N, Ishak MO, Guerreiro JF, et al. Identification of human T cell lymphotropic virus type IIa infection in the Kayapo, an indigenous population of Brazil. AIDS Res Hum Retroviruses. 1995;11:813–21. doi: 10.1089/aid.1995.11.813. [DOI] [PubMed] [Google Scholar]
- 70.Ishak R, Ishak MOG, Azevedo VN, Santos DEM, Vallinoto ACR, Saraiva JCP, et al. Detection of HTLV-IIa in blood donors in an urban area of the Amazon Region of Brazil (Belém, Pará) Rev Soc Bras Med Trop. 1998;31:193–7. doi: 10.1590/s0037-86821998000200005. [DOI] [PubMed] [Google Scholar]
- 71.Ishak R, Vallinoto ACR, Azevedo VN, Lewis M, Hall WW, Guimarães Ishak MO. Molecular evidence of mother-to-child transmission of HTLV-IIc in the Kararao Village (Kayapo) in the Amazon Region of Brazil. Rev Soc Bras Med Trop. 2001;34:519–25. doi: 10.1590/s0037-86822001000600004. [DOI] [PubMed] [Google Scholar]
- 72.Ishak R, Vallinoto ACR, Azevedo VN, Ishak M de O. Epidemiological aspects of retrovirus (HTLV) infection among Indian populations in the Amazon Region of Brazil. Cad Saúde Pública. 2003;19:901–14. doi: 10.1590/s0102-311x2003000400013. [DOI] [PubMed] [Google Scholar]
- 73.Ita F, Mayer EF, Verdonck K, Gonzalez E, Clark D, Gotuzzo E. Human T-lymphotropic virus type 1 infection is frequent in rural communities of the southern Andes of Peru. Int J Infect Dis. 2014;19:46–52. doi: 10.1016/j.ijid.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 74.Kazanji M, Gessain A. Human T-cell lymphotropic virus types I and II (HTLV-I/II) in French Guiana: clinical and molecular epidemiology. Cad Saúde Pública. 2003;19:1227–40. doi: 10.1590/s0102-311x2003000500002. [DOI] [PubMed] [Google Scholar]
- 75.Kanzaki LIB, Casseb J. Human T-Lymphotropic viruses evolution possibly explained by primate Delta retrovirus geographical segregation. Retrovirology. 2008;1:15–20. [Google Scholar]
- 76.Kanzaki LIB, Casseb J. Unusual finding of HTLV-I infection among Amazonian Amerindians. Arch Med Res. 2007;38:897–900. doi: 10.1016/j.arcmed.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 77.Kashima S, Alcantara LC, Takayanagui OM, Cunha MA, Castro BG, Pombo-de- Oliveira MS, et al. Distribution of human T cell lymphotropic virus type 1 (HTLV-1) subtypes in Brazil: genetic characterization of LTR and tax region. AIDS Res Hum Retroviruses. 2006;22:953–9. doi: 10.1089/aid.2006.22.953. [DOI] [PubMed] [Google Scholar]
- 78.Kaplan JE, Lal RB, Davidson M, Lanier AP, Lairmore MD. HTLV-I in Alaska Natives. J Acquir Immune Defic Syndr. 1993;6:327–8. [PubMed] [Google Scholar]
- 79.Kitagawa T, Fujishita M, Taguachi H, Miyoshi T, Tadokoro M. Antibodies to HTLV-I in Japanese immigrants in Brazil. JAMA. 1986;256:2342. [PubMed] [Google Scholar]
- 80.Komurian F, Pelloquin F, de Thé G. In vivo genomic variability of human T-cell leukemia virus type I depends more upon geography than upon pathologies. J Virol. 1991;65:3770–8. doi: 10.1128/jvi.65.7.3770-3778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lairmore MD, Jacobson S, Gracia F, De BK, Castillo S, Larreategui M, et al. Isolation of human T-cell lymphotropic virus type 2 from Guaymi Indians in Panama. Proc Natl Acad Sci USA. 1990;87:8840–4. doi: 10.1073/pnas.87.22.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee H, Swanson P, Shorty VS, Zack JA, Rosenblatt JD, Chen IS. High rate of HTLV-II infection in seropositive i.v. drug abusers in New Orleans. Science. 1989;244:471–5. doi: 10.1126/science.2655084. [DOI] [PubMed] [Google Scholar]
- 83.Leon-Ponte M, Noya O, Bianco N, Echeverría de Perez G. Highly endemic human T- lymphotropic virus type II (HTLV-II) infection in a Venezuelan Guahibo Amerindian group. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:281–6. doi: 10.1097/00042560-199611010-00011. [DOI] [PubMed] [Google Scholar]
- 84.Leon-Ponte M, Echeverria de Perez G, Bianco N, Hengst J, Dube S, Love J, et al. Endemic infection with HTLV-IIB in Venezuelan Indians: molecular characterization. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17:458–64. doi: 10.1097/00042560-199804150-00011. [DOI] [PubMed] [Google Scholar]
- 85.Levine PH, Jacobson S, Elliott R, Cavallero A, Colclough G, Dorry C, et al. HTLV-II infection in Florida Indians. AIDS Res Hum Retroviruses. 1993;9:123–7. doi: 10.1089/aid.1993.9.123. [DOI] [PubMed] [Google Scholar]
- 86.Li HC, Fujiyoshi T, Lou H, Yashiki S, Sonoda S, Cartier L, et al. The presence of ancient human T-cell lymphotropic virus type I provirus DNA in an Andean mummy. Nat Med. 1999;5:1428–32. doi: 10.1038/71006. [DOI] [PubMed] [Google Scholar]
- 87.Liu HF, Goubau P, Van Brussel M, Van Laethem K, Chen YC, Desmyter J, et al. The three human T-lymphotropic virus type I subtypes arose from three geographically distinct simian reservoirs. J Gen Virol. 1996;77:359–68. doi: 10.1099/0022-1317-77-2-359. [DOI] [PubMed] [Google Scholar]
- 88.Lowis GW, Sheremata WA, Wickman PR, Dube S, Dube DK, Poiesz BJ. HTLV-II risk factors in Native Americans in Florida. Neuroepidemiology. 1999;18:37–47. doi: 10.1159/000026194. [DOI] [PubMed] [Google Scholar]
- 89.Lowis GW, Sheremata WA, Minagar A. Epidemiologic features of HTLV-II: serologic and molecular evidence. Ann Epidemiol. 2002;12:46–66. doi: 10.1016/s1047-2797(01)00231-9. [DOI] [PubMed] [Google Scholar]
- 90.Mahieux R, Chappey C, Georges-Courbot MC, Dubreuil G, Mauclere P, Georges A, et al. Simian T-cell lymphotropic virus type I from Mandrillus sphinx as a simian counterpart of human T-cell lymphotropic virus type I subtype D. J Virol. 1998;72:10316–22. doi: 10.1128/jvi.72.12.10316-10322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maloney EM, Biggar RJ, Neel JV, Taylor ME, Hahn BH, Shaw GM, et al. Endemic human T cell lymphotropic virus type II infection among isolated Brazilian Amerindians. J Infect Dis. 1992;166:100–7. doi: 10.1093/infdis/166.1.100. [DOI] [PubMed] [Google Scholar]
- 92.Medeot S, Nates S, Recalde A, Gallego S, Maturano E, Giordano M, et al. Prevalence of antibody to human T cell lymphotropic virus types 1/2 among aboriginal groups inhabiting northern Argentina and the Amazon region of Peru. Am J Trop Med Hyg. 1999;60:623–9. doi: 10.4269/ajtmh.1999.60.623. [DOI] [PubMed] [Google Scholar]
- 93.Menna-Barreto M, Bender AL, Bonatto SL, Freitas LB, Salzano FM, Tsuneto LT, et al. Human T-cell lymphotropic virus type II in Guaraní Indians, Southern Brazil. Cad Saúde Pública. 2005;21:1947–51. doi: 10.1590/s0102-311x2005000600045. [DOI] [PubMed] [Google Scholar]
- 94.Miura T, Fukunaga T, Igarashi T, Yamashita M, Ido E, Funahashi S, et al. Phylogenetic subtypes of human T-lymphotropic virus type I and their relations to the anthropological background. Proc Natl Acad Sci USA. 1994;91:1124–7. doi: 10.1073/pnas.91.3.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montanheiro P, Olah I, Fukumori LMI, Smid J, Penalva de Oliveira AC, Kanzaki LIB, et al. Low DNA HTLV-2 proviral load among women in São Paulo City. Virus Res. 2008;135:22–5. doi: 10.1016/j.virusres.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 96.Montenegro RA, Stephens C. Indigenous health in Latin America and the Caribbean. Lancet. 2006;367:1859–69. doi: 10.1016/S0140-6736(06)68808-9. [DOI] [PubMed] [Google Scholar]
- 97.Murphy EL, Wang B, Sacher RA, Fridey J, Smith JW, Nass CC, et al. Respiratory and urinary tract infections, arthritis, and asthma associated with HTLV-I and HTLV-II infection. Emerg Infect Dis. 2004;10:109–16. doi: 10.3201/eid1001.020714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Murphy EL, Lee TH, Chafets D, Nass CC, Wang B, Loughlin K, et al. Higher human T lymphotropic virus (HTLV) provirus load is associated with HTLV-I versus HTLV-II, with HTLV-II subtype A versus B, and with male sex and a history of blood transfusion. J Infect Dis. 2004;190:504–10. doi: 10.1086/422398. [DOI] [PubMed] [Google Scholar]
- 99.Nakauchi CM, Linhares AC, Maruyama K, Kanzaki LI, Macedo JE, Azevedo VN, et al. Prevalence of human T cell leukemia virus-I (HTLV-I) antibody among populations living in the Amazon region of the Brazil. Mem Inst Oswaldo Cruz. 1990;85:29–33. doi: 10.1590/s0074-02761990000100004. [DOI] [PubMed] [Google Scholar]
- 100.Nakauchi CM, Maruyama K, Kanzadi LI, Linhares AC, Azevedo VN, Fukushima T, et al. Prevalence of HTLV-I antibody among two distinct ethnic groups inhabiting the Amazon region of Brazil. Rev Inst Med Trop Sao Paulo. 1992;34:323–8. doi: 10.1590/s0036-46651992000400009. [DOI] [PubMed] [Google Scholar]
- 101.Neel JV, Biggar RJ, Sukernik RI. Virologic and genetic studies relate Amerind origins to the indigenous people of the Mongolia/Manchuria/ southeastern Siberia region. Proc Natl Acad Sci USA. 1994;91:10737–41. doi: 10.1073/pnas.91.22.10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Novoa P, Granato GFH, Baruzzi RG, Hall WW. International Conference on Human retrovirology: HTLV. 8th. Rio de Janeiro: 1997. Evidence for and the rate of mother-to-child transmission of human T-cell leukaemia/lymphoma virus type II among Kaiapo Indians, Brazil. [Google Scholar]
- 103.Oger JJ, Werker. DM Foti J, Dekaban GA. HTLVI- associated myelopathy: an endemic disease of Canadian aboriginals of the northwest Pacific coast? Can J Neurol Sci. 1993;20:302–6. [PubMed] [Google Scholar]
- 104.Ohkura S, Yamashita M, Cartier L, Tanabe DG, Hayami M, Sonoda S, et al. Identification and phylogenetic characterization of a human T-cell leukaemia virus type I isolate from a native inhabitant (Rapa Nui) of Easter Island. J Gen Virol. 1999;80:1995–2001. doi: 10.1099/0022-1317-80-8-1995. [DOI] [PubMed] [Google Scholar]
- 105.Pardi D, Switzer W, Hadlock KG, Kaplan JE, Lal RB, Folks TM. Complete nucleotide sequence of an Amerindian human T-cell lymphotropic virus type II (HTLV-II) isolate: identification of a variant HTLV-II subtype b from a Guaymi Indian. J Virol. 1993;67:4659–64. doi: 10.1128/jvi.67.8.4659-4664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perez GE, Leon-Ponte M, Noya O, Botto C, Gallo D, Bianco N. First description of endemic HTLV-II infection among Venezuelan Amerindians. J Acquir Immune Defic Syndr Hum Retrovirol. 1993;6:1368–72. [PubMed] [Google Scholar]
- 107.Peters AA, Coulthart MB, Oger JJ, Waters DJ, Crandall KA, Baumgartner AA. HTLV Type I/II in British Columbia Amerindians: a seroprevalence study and sequence characterization of an HTLV type IIa isolate. AIDS Res Hum Retroviruses. 2000;16:883–92. doi: 10.1089/08892220050042828. [DOI] [PubMed] [Google Scholar]
- 108.Picard FJ, Coulthart MB, Oger J, King EE, Kim S, Arp J, et al. Human T-lymphotropic virus type 1 in coastal natives of British Columbia: phylogenetic affinities and possible origins. J. Virol. 1995;69:7248–56. doi: 10.1128/jvi.69.11.7248-7256.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Power C, Weinshenker BG, Dekaban GA, Ebers GC, Francis GS, Rice GP. HTLV-1 associated myelopathy in Canada. Can J Neurol Sci. 1989;16:330–5. doi: 10.1017/s0317167100029176. [DOI] [PubMed] [Google Scholar]
- 110.Proietti FA, Carneiro-Proietti AB, Catalan-Soares BC, Murphy EL. Global epidemiology of HTLV-I infection and associated diseases. Oncogene. 2005;24:6058–68. doi: 10.1038/sj.onc.1208968. [DOI] [PubMed] [Google Scholar]
- 111.Reeves WC, Cutler JR, Gracia F, Kaplan JT, Catillo L, Hartley TM, et al. Human T cell lymphotropic virus infection in Guaymi Indians from Panama. Am J Trop Med Hyg. 1990;43:410–8. doi: 10.4269/ajtmh.1990.43.410. [DOI] [PubMed] [Google Scholar]
- 112.Reich DE, Goldstein DB. Genetic evidence for a paleolithic human population expansion in Africa. Proc Natl Acad Sci USA. 1998;95:8119–23. doi: 10.1073/pnas.95.14.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SG. IMGT/HLA Database - a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29:210–3. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roucoux DF, Murphy EL. The epidemiology and disease outcomes of human T-lymphotropic virus type II. AIDS Rev. 2004;6:144–54. [PubMed] [Google Scholar]
- 115.Salemi M, Van Dooren S, Audenaert E, Delaporte E, Goubau P, Desmyter J, et al. Two new human T-lymphotropic virus type I phylogenetic subtypes in seroindeterminates, a Mbuti pygmy and a Gabonese, have closest relatives among African STLV-I strains. Virology. 1998;246:277–87. doi: 10.1006/viro.1998.9215. [DOI] [PubMed] [Google Scholar]
- 116.Salemi M, Vandamme AM, Desmyter J, Casoli C, Bertazzoni U. The origin and evolution of human T-cell lymphotropic virus type II (HTLV-II) and the relationship with its replication strategy. Gene. 1999;234:11–21. doi: 10.1016/s0378-1119(99)00169-9. [DOI] [PubMed] [Google Scholar]
- 117.Sanchez-Palacios C, Gotuzzo E, Vandamme AM, Maldonado Y. Seroprevalence and risk factors for human T-cell lymphotropic virus (HTLV-I) infection among ethnically and geographically diverse Peruvian women. Int J Infect Dis. 2003;7:132–7. doi: 10.1016/s1201-9712(03)90009-9. [DOI] [PubMed] [Google Scholar]
- 118.Segurado AA, Biasutti C, Zeigler R, Rodrigues C, Damas CD, Jorge ML, et al. Identification of human T-lymphotropic virus type I (HTLV-I) subtypes using restricted fragment length polymorphism in a cohort of asymptomatic carriers and patients with HTLV-I-associated myelopathy/tropical spastic paraparesis from São Paulo, Brazil. Mem Inst Oswaldo Cruz. 2002;97:329–33. doi: 10.1590/s0074-02762002000300009. [DOI] [PubMed] [Google Scholar]
- 119.Shindo N, Alcantara LCJ, Van Dooren S, Salemi M, Costa MCR, Kashima S, et al. Human retroviruses (HIV and HTLV) in Brazilian Indians: seroepidemiological study and molecular epidemiology of HTLV type 2 isolates. AIDS Res Hum Retroviruses. 2002;18:71–7. doi: 10.1089/088922202753394736. [DOI] [PubMed] [Google Scholar]
- 120.Sonoda S, Li HC, Tajima K. Ethnoepidemiology of HTLV-1 related diseases: ethnic determinants of HTLV-1 susceptibility and its worldwide dispersal. Cancer Sci. 2011;102:295–301. doi: 10.1111/j.1349-7006.2010.01820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Switzer WM, Pieniazek D, Swanson P, Samdal HH, Soriano V, Khabbaz RF, et al. Phylogenetic relationship and geographic distribution of multiple human T-cell lymphotropic virus type II subtypes. J Virol. 1995;69:621–32. doi: 10.1128/jvi.69.2.621-632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Talarmin A, Vion B, Ureta-Vidal A, Du Fou G, Marty C, Kazanji M. First seroepidemiological study and phylogenetic characterization of human T-cell lymphotropic virus type I and II infection among Amerindians in French Guiana. J Gen Virol. 1999;80:3083–8. doi: 10.1099/0022-1317-80-12-3083. [DOI] [PubMed] [Google Scholar]
- 123.The HUGO Pan-Asian SNP Consortium. Mapping human genetic diversity in Asia. Science. 2009;326:1541–5. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 124.Toro C, Rodes B, Bassani S, Jimenez V, Tuset C, Brugal MT, et al. Molecular epidemiology of HTLV-2 infection among intravenous drug users in Spain. J Clin Virol. 2005;33:65–70. doi: 10.1016/j.jcv.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 125.Vallinoto ACR, Azevedo VN, Santos DEM, Caniceiro S, Mesquita FCL, Hall WW, et al. Serological evidence of HTLV-I and HTLV-II coinfections in HIV-1 positive patients in Belém, State of Pará, Brazil. Mem Inst Oswaldo Cruz. 1998;93:407–9. doi: 10.1590/s0074-02761998000300026. [DOI] [PubMed] [Google Scholar]
- 126.Vallinoto ACR, Ishak MOG, Azevedo VN, Vicente ACP, Otsuki K, Hall WW, et al. Molecular epidemiology of human T-Lymphotropic virus type II infection in Ameridian and urban populations of the Amazon region of Brazil. Hum Biol. 2002;74:633–44. doi: 10.1353/hub.2002.0059. [DOI] [PubMed] [Google Scholar]
- 127.Vallinoto AC, Muto NA, Pontes GS, Machado LF, Azevedo VN, dos Santos SE, et al. Serological and molecular evidence of HTLV-I infection among Japanese immigrants living in the Amazon region of Brazil. Jpn J Infect Dis. 2004;57:156–9. [PubMed] [Google Scholar]
- 128.Van Dooren S, Gotuzzo E, Salemi M, Watts D, Audenaert E, Duwe S, et al. Evidence for a post-Columbian introduction of human T-cell lymphotropic virus in Latin America. J Gen Virol. 1998;79:2695–708. doi: 10.1099/0022-1317-79-11-2695. [DOI] [PubMed] [Google Scholar]
- 129.Van Dooren S, Pybus OG, Salemi M, Liu HF, Goubau P, Remondegui C, et al. The low evolutionary rate of human T-cell lymphotropic virus type-1 confirmed by analysis of vertical transmission chains. Mol Biol Evol. 2004;21:603–11. doi: 10.1093/molbev/msh053. [DOI] [PubMed] [Google Scholar]
- 130.Vandamme AM, Liu HF, Goubau P, Desmyter J. Primate T-lymphotropic virus type I LTR sequence variation and its phylogenetic analysis: compatibility with an African origin of PTLV-1. Virology. 1994;202:212–23. doi: 10.1006/viro.1994.1337. [DOI] [PubMed] [Google Scholar]
- 131.Visoná K, Yamaguchi K, Bonilla J. In: International Conference on Human Retrovirology: HTLV. 8th. Rio de Janeiro: 1997. Human T-cell leukaemia/lymphoma virus type I and type II infections in Costa Rica. [Google Scholar]
- 132.Vitek CR, Gracia FI, Giusti R, Fukuda K, Green DB, Castillo LC, et al. Evidence for sexual and mother-to-child transmission of human T lymphotropic virus type II among Guaymi Indians, Panama. J Infect Dis. 1995;171:1022–6. doi: 10.1093/infdis/171.4.1022. [DOI] [PubMed] [Google Scholar]
- 133.Wolfe ND, Heineine W, Carr JK, Garcia AD, Shanmugam V, Tamoufe U, et al. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc Natl Acad Sci USA. 2005;102:7994–9. doi: 10.1073/pnas.0501734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yamashita M, Picchio G, Veronesi R, Ohkura S, Bare P, Hayami M. HTLV-Is in Argentina are phylogenetically similar to those of other South American countries, but different from HTLV-Is in Africa. J Med Virol. 1998;55:152–60. doi: 10.1002/(sici)1096-9071(199806)55:2<152::aid-jmv12>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 135.Yanagihara R, Jenkins CL, Ajdukiewicz AB, Lai RB. Serological discrimination of HTLV I and II infection in Melanesia. Lancet. 1991;337:617–8. doi: 10.1016/0140-6736(91)91684-m. [DOI] [PubMed] [Google Scholar]
- 136.Zamora T, Zaninovic V, Kajiwara M, Komoda H, Hayami M, Tajima K. Antibody to HTLV-I in indigenous inhabitants of the Andes and Amazon regions in Colombia. Jpn J Cancer Res. 1990;81:715–9. doi: 10.1111/j.1349-7006.1990.tb02633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zaninovic V, Sanzon F, Lopez F, Velandia G, Blank A, Blank M, et al. Geographic independence of HTLV-I and HTLV-II foci in the Andes highland, the Atlantic coast, and the Orinoco of Colombia. AIDS Res Hum Retroviruses. 1994;10:97–101. doi: 10.1089/aid.1994.10.97. [DOI] [PubMed] [Google Scholar]
- 138.Zella D, Mori L, Sala M, Ferrante P, Casoli C, Magnani G, et al. HTLV-II infection in Italian drug abusers. Lancet. 1990;336:575–6. doi: 10.1016/0140-6736(90)92140-d. [DOI] [PubMed] [Google Scholar]
- 139.Zihlmann KF, de Alvarenga AT, Casseb J. Living invisible: HTLV-1-infected persons and the lack of care in public health. PLOS Negl Trop Dis. 2012;6: doi: 10.1371/journal.pntd.0001705. doi:10.1371/journal.pntd.0001705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zihlmann KF, Alvarenga AT, Casseb J. Reproductive decisions among people living with human T-cell lymphotropic virus type 1 (HTLV-1) J Infect Dis Ther. 2013;1:108. Available from: http://dx.doi.org/10.4172/2332-0877.1000108. [Google Scholar]