Abstract
Acanthamoeba is a “Trojan horse” of the microbial world. The aim of this study was to identify the presence of Pseudomonas as an amoeba-resistant microorganism in 12 isolates of Acanthamoeba. All isolates showed the genus Pseudomonas spp. as amoeba-resistant microorganisms. Thus, one can see that the Acanthamoeba isolates studied are hosts of Pseudomonas.
Keywords: Acanthamoeba, Pseudomonas, Amoeba-resistant microorganism
Abstract
Acanthamoeba é um “Cavalo de Tróia” do mundo microbiano. Este estudo teve como objetivo identificar a presença de Pseudomonas como microrganismo resistente a ameba em 12 isolados de Acanthamoeba. Todos os isolados apresentaram o gênero Pseudomonas spp. como um microrganismo resistente a ameba. Assim, podemos ver que os isolados de Acanthamoeba estudados são hospedeiros de Pseudomonas.
Acanthamoeba is an opportunistic human pathogen that is ubiquitously distributed in the environment13. It is a causative agent of cutaneous lesions, sinus infections, vision threatening keratitis and rare but fatal encephalitis, known as granulomatous amoebic encephalitis. In addition, it has the ability to act as a host/reservoir for microbial pathogens10,16.
Free-living amoebae feed by phagocytosis mainly on bacteria, fungi, and algae, and digestion occurs within phagolysosomes. Some microorganisms have evolved and have become resistant to predation by protists, since they are not internalized or are able to survive, grow, and exit free-living amoebae after internalization. Acanthamoeba is shown to be host/reservoir for numerous bacteria, including the genus Pseudomonas spp., among other bacterial pathogens13.
Pseudomonas spp. are highly adaptable bacteria that can colonize various environmental niches, including soil and marine habitats, plants and animals. Pseudomonas spp. are also opportunistic human pathogens, causing infection of the eyes, ears, skin, urethra and respiratory tract in cystic fibrosis (CF) in burned patients, as well as other immunocompromised individuals15.
In nature, free-living amoebae of the genus Acanthamoeba feed by Pseudomonas spp., which are widely distributed in the environment. Their encounter may be facilitated through better adherence of Pseudomonas spp. (than E. coli) to Acanthamoeba 2. However, some Pseudomonas spp. have evolved to become resistant to predation by amoebae, as demonstrated by the isolation of Acanthamoeba naturally infected with P. aeruginosa 6,13. Hence, free-living amoebae might also play a role as a reservoir for some amoeba-resistant strains of Pseudomonas, similar to what was shown for Legionella spp.6. This is important, given the role of Pseudomonas aeruginosa as a causative agent of pneumonia5. Acanthamoeba has been isolated from contact lens care systems contaminated with Gram-negative bacteria, including Pseudomonas aeruginosa 6.
Many studies have evaluated the interaction between Acanthamoeba spp. and Pseudomonas spp., as well as investigated the presence of these bacterial genera as amoeba-resistant bacteria3,8,11.
In this study, the conventional technique of Polymerase Chain Reaction (PCR) was used, in order to identify the presence of the genus Pseudomonas spp. as amoeba-resistant microorganisms in isolates of Acanthamoeba.
A total of 12 environmental samples existing in the laboratory were used in this study: seven isolates from air-conditioning units identified as Acanthamoeba A2, A3, A4, A5, A7, A8 and A10, and five isolates from contact lens cases, Acanthamoeba A1, A6, A9, A11 and A12. The isolates were cultured in PYG media at 30 °C (2% protease peptone, 0.2% yeast extract, and 1.5% glucose) supplemented with penicillin and streptomycin (Life Technologies). The total DNA in the sample was extracted, as described by ALJANABI & MARTINEZ1. The fresh culture containing 106 trophozoites was homogenized in 400 µL of sterile salt homogenizing buffer (0.4 M NaCl 10 mM Tris–HCl pH 8.0 and 2 mM EDTA pH 8.0), then, 40 µL of 20% SDS (2% final concentration) and 8 µL of 20 mg/mL protenase K (400 µg/mL final concentration) were added and mixed well. The samples were incubated at 65 °C for, at least, one h, after which 300 µL of 6 M NaCl (NaCl saturated H2O) was added to each sample. Samples were vortexed for 30s at maximum speed, and tubes spun down for 30 min at 10,000 × g. The supernatant was transferred to fresh tubes. An equal volume of isopropanol was added to each sample and samples were incubated at -20 °C for one h. Samples were then centrifuged for 20 min, at 4 °C and at 10,000 × g. The pellet was washed with 70% ethanol, dried and finally resuspended in 100 µL sterile dH2O.
After extraction, the isolates were screened for the presence of bacterial endosymbiont - Bacteria domain - through the 16S rDNA gene amplified by PCR, using primers fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and rP2 (5′-ACGGCTACCTTGTTACGACTT-3′) that amplify 1500 bp in size, described by WEISBURG et al. 17, under the following conditions: five min at 94 °C, followed by 35 cycles of one min at 94 °C, one min at 55 °C and one min at 72 °C.
The identification of the presence of Pseudomonas genus DNA occurred using the primers described by SPILKER et al. 14 PA-GS-F (5′- GACGGGTGAGTAATGCCTA-3′) and PA-GS-R (5′-CACTGGTGTTCCTTCCTATA-3′) that amplifies 618 pb in size. Amplification was performed in a total volume of 25 µL containing 30 ng DNA, 10 pmol each primer, 5 pmol dNTP, reaction buffer (50 mM KCl2, 10 mM Tris–HCl), 1.5 mM MgCl2, and 1 U of Platinum Taq DNA Polymerase (InvitrogenTM). The amplification reaction was carried out in a PTC-150 Minicycler MJ Research thermocycler, under the following conditions: five min at 94 °C, followed by 35 cycles of one min at 94 °C, one min at 58 °C and one min at 72 °C.
The amplification product was separated in 1% agarose gel, stained with 0.5 µM/mL ethidium bromide and observed under a UV-light transilluminator. PCR products were purified using a QIAquick purification kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions, and resolved with a MegaBace 1000 automated sequencer. Analysis of the DNA sequences was performed with the Chromas Lite program and compared to those present in GenBank (http://blast.ncbi.nlm.nih.gov/).
In the present study, all isolates of Acanthamoeba showed internalized bacteria when primers are used to amplify the Bacteria domain and all isolates showed the genus Pseudomonas spp. as amoeba-resistant microorganisms (Fig. 1). A total of six PCR products (Ap1 to Ap6) were sent for sequencing (Table 1) and all were confirmed as Pseudomonas spp.
Fig. 1. Samples of Acanthamoeba A1, A6, A9, A11 and A12 isolated from contact lens cases, and A2, A3, A4, A5, A7, A8, and A10 isolated from air conditioning units. Positive control (PC) strain of Pseudomonas aeruginosa ATCC 278532.
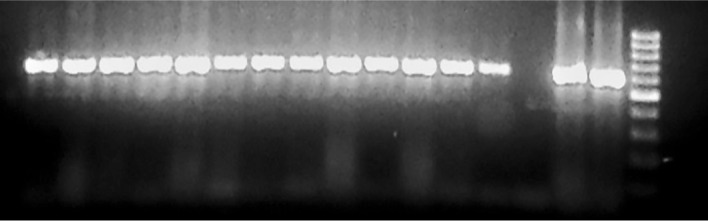
Table 1. Percentage of similarity and access number compared to GenBank sequences of identified bacteria in this study.
| Acanthamoeba | Fragment from the gel (GenBank accession) | Similarity BLAST | Access GenBank (number for access) |
|---|---|---|---|
| A1 | Ap1 (KF160336) | 98% | Pseudomonas sp. c145(2012) 16S ribosomal RNA gene, partial sequence (JQ781629.1) |
| A3 | Ap2 (KF160337) | 96% | Uncultured Pseudomonas sp. clone 3F10 16S ribosomal RNA gene, partial sequence (HM438578.1) |
| A4 | Ap3 (KF160338) | 99% | Pseudomonas sp. CJ-S-R2A3 16S ribosomal RNA gene, partial sequence (HM584286.1) |
| A6 | Ap4 (KF160339) | 99% | Pseudomonas sp. c145(2012) 16S ribosomal RNA gene, partial sequence (JQ781629.1) |
| A10 | Ap5 (KF160340) | 99% | Pseudomonas fluorescens strain C-D-TSA4 16S ribosomal RNA gene, partial sequence (HM755599.1) |
| A12 | Ap6 (KF160341) | 97% | Pseudomonas sp. c145(2012) 16S ribosomal RNA gene, partial sequence (JQ781629.1) |
CALVO et al. 3 analyzed Acanthamoeba spp. originated from natural and anthropogenic environments and recorded the presence of Pseudomonas spp. as amoeba-resistant microorganisms in 26.1% of the isolates. GARCIA et al. (4) evaluated isolates from water coming from reservoirs and obtained 32.6% positive for Pseudomonas spp. In a study on clinical isolates of Acanthamoeba spp., IOVIENO et al. 8 observed that Pseudomonas spp. were present as amoeba-resistant microorganisms in 59% of the isolates studied.
Pseudomonas spp. have also been reported to be involved in keratitis and fatal pneumonia7, among other diseases. Their presence may have a great impact on immune-suppressed individuals, since around 96% of the Pseudomonas spp. isolated from hot tubs and indoor swimming pools in a surveillance study display antimicrobial resistance9. Therefore, their prevalence in the environment, not only in recreational water but as part of biofilms in systems of distribution of drinking water, as well as their relevance in human pathogenicity led researchers to seek for its occurrence in amoeba hosts3.
The possible role of Acanthamoeba as an evolutionary precursor of pathogenicity in microbial pathogens has been suggested12. Bacteria or other microbial endosymbiont may also enhance the pathogenicity of Acanthamoeba 12. However, the results have been inconclusive. There are a few reports suggesting that amoeba-resistant microorganisms enhance the virulence of Acanthamoeba 6.
In addition to the bacteria identified in this work, the presence of other pathogenic amoeba-resistant microorganisms in the water samples tested cannot be discarded. Acanthamoeba spp. are also potential reservoirs of Mycobacterium spp.3 and Legionella spp., among others microorganisms3.
REFERENCES
- 1.Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997;25:4692–3. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottone EJ, Perez AA, Gordon RE, Qureshi MN. Differential binding capacity and internalisation of bacterial substrates as factors in growth rate of Acanthamoeba spp. J Med Microbiol. 1994;l40:148–54. doi: 10.1099/00222615-40-2-148. [DOI] [PubMed] [Google Scholar]
- 3.Calvo L, Gregorio I, García A, Fernández MT, Goñi P, Clavel A, et al. A new pentaplex-nested PCR to detect five pathogenic bacteria in free living amoebae. Water Res. 2013;47:493–502. doi: 10.1016/j.watres.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 4.Garcia A, Goñi P, Cieloszyk J, Fernandez MT, Calvo-Beguería L, Rubio E, et al. Identification of free-living amoebae and amoeba-associated bacteria from reservoirs and water treatment plants by molecular techniques. Environ Sci Technol. 2013;47:3132–40. doi: 10.1021/es400160k. [DOI] [PubMed] [Google Scholar]
- 5.Garau J, Gomez L. Pseudomonas aeruginosa pneumonia. Curr Opin Infect Dis. 2003;16:135–43. doi: 10.1097/00001432-200304000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–33. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huhulescu S, Simon M, Lubnow M, Kaase M, Wewalka G, Pietzka AT, et al. Fatal Pseudomonas aeruginosa pneumonia in a previously healthy woman was most likely associated with a contaminated hot tub. Infection. 2011;39:265–9. doi: 10.1007/s15010-011-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iovieno A, Ledee DR, Miller D, Alfonso EC. Detection of bacterial endosymbionts in clinical Acanthamoeba isolates. Ophthalmology. 2010;17:445–52. doi: 10.1016/j.ophtha.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutz JK, Lee J. Prevalence and antimicrobial-resistance of Pseudomonas aeruginosa in swimming pools and hot tubs. Int J Environ Res Public Health. 2011;8:554–64. doi: 10.3390/ijerph8020554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marciano-Cabral F, Cabral G. Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev. 2003;16:273–307. doi: 10.1128/CMR.16.2.273-307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagnier I, Raoult D, La Scola B. Isolation and identification of amoeba- resisting bacteria from water in human environment by using an Acanthamoeba polyphaga co-culture procedure. Environ Microbiol. 2008;10:1135–44. doi: 10.1111/j.1462-2920.2007.01530.x. [DOI] [PubMed] [Google Scholar]
- 12.Paterson GN, Rittig M, Siddiqui R, Khan NA. Is Acanthamoeba pathogenicity associated with intracellular bacteria? Exp Parasitol. 2011;129:207–10. doi: 10.1016/j.exppara.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba . Parasit Vectors. 2012;5:6. doi: 10.1186/1756-3305-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spilker T, Coenye T, Vandamme P, Li Puma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42:2074–9. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tashiro Y, Uchiyama H, Nomura N. Multifunctional membrane vesicles in Pseudomonas aeruginosa . Environ Microbiol. 2012;4:1349–62. doi: 10.1111/j.1462-2920.2011.02632.x. [DOI] [PubMed] [Google Scholar]
- 16.Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea . FEMS Immunol Med Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 17.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


