Abstract
Motivation: PIWI-interacting RNAs (piRNAs), 23–36 nt small silencing RNAs, repress transposon expression in the metazoan germ line, thereby protecting the genome. Although high-throughput sequencing has made it possible to examine the genome and transcriptome at unprecedented resolution, extracting useful information from gigabytes of sequencing data still requires substantial computational skills. Additionally, researchers may analyze and interpret the same data differently, generating results that are difficult to reconcile. To address these issues, we developed a coordinated set of pipelines, ‘piPipes’, to analyze piRNA and transposon-derived RNAs from a variety of high-throughput sequencing libraries, including small RNA, RNA, degradome or 7-methyl guanosine cap analysis of gene expression (CAGE), chromatin immunoprecipitation (ChIP) and genomic DNA-seq. piPipes can also produce figures and tables suitable for publication. By facilitating data analysis, piPipes provides an opportunity to standardize computational methods in the piRNA field.
Supplementary information: Supplementary information, including flowcharts and example figures for each pipeline, are available at Bioinformatics online.
Availability and implementation: piPipes is implemented in Bash, C++, Python, Perl and R. piPipes is free, open-source software distributed under the GPLv3 license and is available at http://bowhan.github.io/piPipes/.
Contact: Phillip.Zamore@umassmed.edu or Zhiping.Weng@umassmed.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
piRNAs, a class of 23–36 nt long small silencing RNAs, suppress transposon expression in the metazoan germ line and, in some animals, the adjacent gonadal somatic cells (Luteijn and Ketting, 2013). By preventing transposition, the piRNA pathway ensures that genetic information passes faithfully to the next generation. Disruption of the piRNA pathway typically leads to transposon mobilization, double-stranded DNA breaks and sterility. High-throughput sequencing technologies have been widely deployed in the study of piRNAs. Small RNA-seq reveals the identity and abundance of piRNAs (Brennecke et al., 2007); RNA-seq detects and quantifies mRNA and transposon transcripts (Reuter et al., 2011); degradome-seq (also termed RACE-seq) detects the cleavage products of PIWI-proteins guided by piRNAs (Reuter et al., 2011); chromatin immunoprecipitation (ChIP)-seq detects chromatin modifications directed by piRNAs or transcription factor-binding events that regulate piRNA precursor or target transcription (Sienski et al., 2012); and genomic DNA sequencing detects new transposition events caused by transposons that escape piRNA repression or background differences between experimental strains and the assembled genome (Khurana et al., 2011; Sienski et al., 2012). Correctly extracting biological knowledge from such voluminous data requires significant computational expertise and effort. The repetitive nature of transposon sequences lays another layer of complexity. Moreover, different laboratories use diverse methods to analyze and interpret data (e.g. the way of treating reads that map to multiple locations in a reference genome). To provide a standardized set of tools to analyze these diverse data types, we developed piPipes, a collection of integrated pipelines for small RNA-seq, RNA-seq, degradome- and cap analysis of gene expression-seq (CAGE-seq), ChIP-seq and genome-seq analyses.
2 METHODS
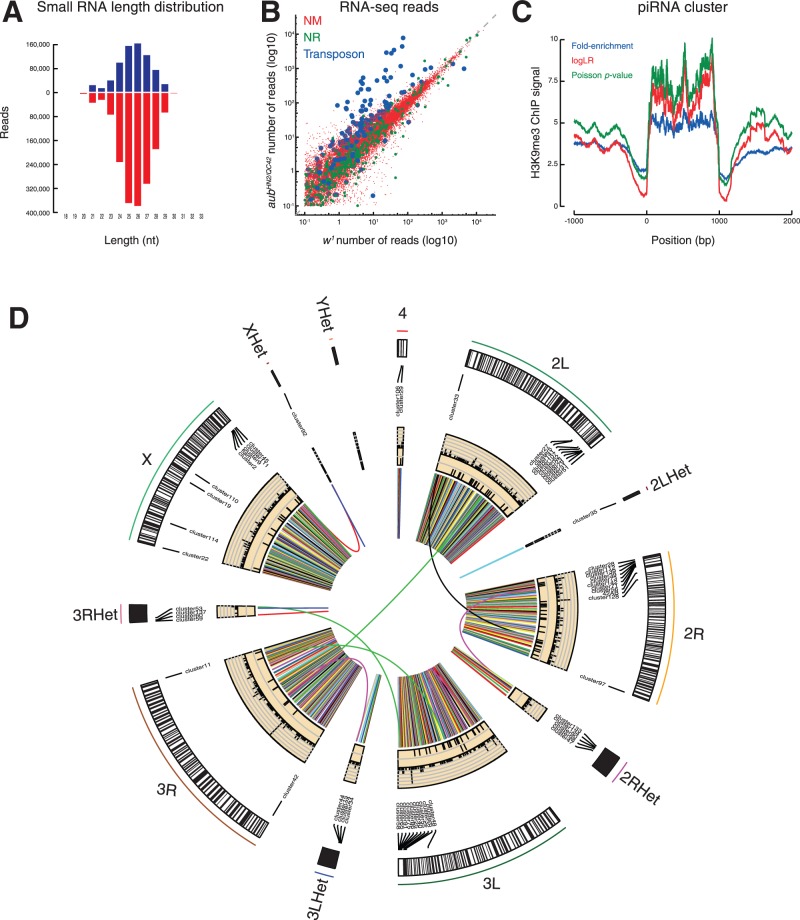
piPipes comprises five pipelines designed to analyze small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq or genome-seq data. The small RNA-seq pipeline reports the abundance, length distribution, nucleotide composition and 5′-to-5′ distance (‘Ping-Pong’ signature) of piRNAs assigned to genomic annotations, including individual transposon families and piRNA clusters, the initial sources of piRNA precursor transcripts. The RNA-seq pipeline reports the normalized abundance of transcripts from both genes and transposons. The degradome-seq pipeline offers methods to identify piRNA-directed cleavage products. This pipeline can also be used to analyze any long RNA sequencing method designed to define RNA 5′ ends, e.g. CAGE-seq. The ChIP-seq pipeline uses the widely used peak-calling algorithm MACS2 (Zhang et al., 2008), focusing on piRNA clusters and transposons. The genome-seq pipeline detects novel transposition events as well as structural variation. Supplementary Figure S1 illustrates the general piPipes workflow, using the small RNA-seq pipeline as an example. First, all reads aligned to ribosomal RNA (rRNA) sequences are removed. The remaining reads are then mapped to microRNA (miRNA) hairpin sequences to quantify the abundance and 5′- and 3′-end heterogeneity of mature miRNAs. Reads that do not match rRNAs or miRNAs are then mapped to the reference genome. piPipes next assigns reads to different genomic features (e.g., transposons, piRNA clusters and genes) by their coordinates. To achieve maximal speed, piPipes parallelizes this step on multiple threads using ParaFly software from the Trinity package (Grabherr et al., 2011). For the reads assigned to each genomic feature, piPipes draws publication-quality graphs of length distribution and nucleotide composition, as well as the distance between the 5′ ends of two small RNAs from opposite strands of the same locus, a standard method for detecting piRNA ‘Ping-Pong’ amplification or siRNA phasing (Fig. 1A and Supplementary Fig. S1B). Furthermore, piPipes generates a table that summarizes the number of unique and multiple mappers counted as species (distinct sequences) or reads. The RNA-seq pipeline also starts with rRNA removal. The remaining reads are then mapped to the genome using STAR (Dobin et al., 2013). piPipes quantifies transcript abundance from genomic alignment by both Cufflinks (Trapnell et al., 2010) and HTSeq-count (Anders et al., 2014). In addition, direct mapping of the reads to the transcriptome is performed using Bowtie2 followed by eXpress quantification (Roberts and Pachter, 2013). Degradome-seq and CAGE-seq share the same pipeline because both methods aim to characterize the 5′ ends of RNAs. This pipeline discards reads that can only be mapped to the genome via soft clipping of their 5′ ends (i.e. the prefixes of these reads do not map to the genome). The alignment procedure is otherwise similar to that used for RNA-seq data. The nucleotide composition for each genomic feature is calculated as in the small RNA pipeline (Supplementary Fig. S3B). The ChIP-seq pipeline aligns the ChIP and input libraries to the genome using Bowtie2. piPipes calls peaks using MACS2 (Zhang et al., 2008), which supports both narrow (such as transcription factors) and broad (such as histone 3 trimethyl lysine 9, H3K9me3) peaks. Transcription start site, transcription end site and metagene analyses of different genomic features are implemented by bwtool (Pohl and Beato, 2014). The genome-seq pipeline applies different algorithms, including BreakDancer (Chen et al., 2009), RetroSeq (Hormozdiari et al., 2010; Keane et al., 2013) and TEMP (Zhuang et al., 2014), to discover transposon insertion, deletion and other structural variation events (Supplementary Fig. S5). piPipes uses a Circos plot (Zhang et al., 2013) to represent the variant loci discovered by each algorithm across different chromosomes (Fig. 1D).
Fig. 1.
Gallery of piPipes Figures (A) Barplot representing length distribution of Drosophila w1 ovary small RNAs assigned to sense (blue) and antisense (red) strands of transposons. (B) Scatterplot comparing w1 to aubHN2/QC42 Drosophila ovary RNA-seq reads assigned to mRNA (NM; red), non-coding RNA (NR; green) and transposons (blue). (C) Metagene plot of H3K9me3 ChIP-seq of piRNA clusters from flies in which piwi mRNA was depleted by double-stranded RNA-triggered RNA driven by a triple Gal4 driver (SRX215630). (D) Circos plot representing the locations of, from the periphery to the center, cytological position, piRNA clusters, SV discovered by TEMP (tiles), retroSeq (tiles) and VariationHunter (links) using genomic sequencing of 2–4-day-old ovaries
The small RNA-seq, RNA-seq and ChIP-seq pipelines can each be run in two modes, allowing analysis of a single sample or a pair of samples. The dual-sample mode uses the output from the single-sample mode and performs pair-wise comparison as illustrated by balloonplots and scatterplots (Supplementary Fig. S1C and D). The comparison can be performed on miRNA, piRNA or mRNA. Figure 1B illustrates a scatterplot showing the mRNA abundance in an RNA-seq dataset analyzed by the RNA-seq pipeline in the dual-sample mode. The dual-sample mode of the RNA-seq pipeline also uses Cuffdiff (Trapnell et al., 2013) to perform differential analysis on genic transcripts. In the dual-sample mode, the ChIP-seq pipeline uses MACS2 to identify differentially enriched loci (Supplementary Fig. S4).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the Zamore and Weng laboratories for helpful discussions, and Jia Xu, Jui-Hung Hung and Soo Lee for their initial work.
Funding: This work was funded by National Institutes of Health grants [GM62862 and GM65236] to P.D.Z. and [U41HG007000] to Z.W.
Conflicts of interest: none declared.
REFERENCES
- Anders S, et al. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;6:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;9:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;1:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;7:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, et al. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. Bioinformatics. 2010;12:i350–i357. doi: 10.1093/bioinformatics/btq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, et al. RetroSeq: transposable element discovery from next-generation sequencing data. Bioinformatics. 2013;3:389–390. doi: 10.1093/bioinformatics/bts697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, et al. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;7:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat. Rev. Genet. 2013;8:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- Pohl A, Beato M. bwtool: a tool for bigWig files. Bioinformatics. 2014;11:1618–1619. doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;7376:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;1:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, et al. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;5:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;5:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;1:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14:244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, et al. TEMP: a computational method for analyzing transposable element polymorphism in populations. Nucleic Acids Res. 2014;11:6826–6838. doi: 10.1093/nar/gku323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.