Abstract
We present here three expression plasmids for Trypanosoma cruzi adapted to the Gateway® recombination cloning system. Two of these plasmids were designed to express trypanosomal proteins fused to a double tag for tandem affinity purification (TAPtag). The TAPtag and Gateway® cassette were introduced into an episomal (pTEX) and an integrative (pTREX) plasmid. Both plasmids were assayed by introducing green fluorescent protein (GFP) by recombination and the integrity of the double-tagged protein was determined by western blotting and immunofluorescence microscopy. The third Gateway adapted vector assayed was the inducible pTcINDEX. When tested with GFP, pTcINDEX-GW showed a good response to tetracycline, being less leaky than its precursor (pTcINDEX).
Keywords: Trypanosoma cruzi, expression vectors - Gateway® system, tandem affinity purification, pTcINDEX
Trypanosoma cruzi, Trypanosoma brucei and Leishmania major (together named as TriTryp) genome-sequencing projects were completed in 2005 (El-Sayed et al. 2005). Currently, there are 16 genomes from Leishmania spp: two from T. brucei and nine from T. cruzi strains available in the TriTryp database (tritrypdb.org/tritrypdb). To fully exploit this vast amount of information, it is essential to improve and extend the range of genetic tools available for analysing the function of genes in vivo.
Among the TriTryp species, T. cruzi is the most technically limited. Unlike T. brucei for which RNAi-based procedures have been widely exploited for exploring gene function, in T. cruzi the molecular machinery necessary for RNAi has not been detected (da Rocha et al. 2004). Additionally, in contrast to T. brucei and Leishmania spp, conventional gene knockouts in T. cruzi are notably less efficient and hindered by the fact that most genes are encoded by multiple-copy gene families, making this technique laborious and time-consuming (Clayton 1999).
Since the 1980s, several groups have attempted to genetically manipulate trypanosomatids. One of the first reports of stable transfection in T. cruzi was achieved by the electroporation of epimastigotes with the pTEX expression vector. This episomal plasmid was constructed using flanking sequences derived from the glycosomal glyceraldehyde-3-phosphate dehydrogenase gene (Kelly et al. 1992). The pTEX plasmid was improved a few years later by insertion of an 800 bp ribosomal DNA fragment upstream of the multiple cloning site. The result, the pRIBOTEX integrative plasmid, had a shorter selection time and a mild increase in expression levels (Martínez-Calvillo et al. 1997). pRIBOTEX was further enhanced by the insertion of the upstream region of the TcP2beta H1 gene (HX1) next to the putative ribosomal promoter element to generate pTREX (Vazquez & Levin 1999). All of the vectors described above promote the constitutive expression of cloned genes, which is a limitation for studying gene function. To overcome this limitation, systems for expressing transgenes in a controlled and repressible manner were developed. One of these vectors is pTcINDEX, a stable tetracycline-regulated expression vector, which integrates into a ribosomal non-transcribed spacer region and has a T7 promoter upstream of the polymerase I transcription initiation site. pTcINDEX is used together with the episomal pLew plasmid, which constitutively expresses the T7 polymerase and tetR genes (Taylor & Kelly 2006).
Other plasmids adapted to T. brucei and Leishmania sp. are those used for tandem affinity purification (TAP) (Estévez et al. 2001, Panigrahi et al. 2003). The classical TAP tag contains two immunoglobulin G (IgG)-binding domains for the Staphylococcus aureus protein A (PA) and calmodulin-binding peptide (CBP) epitopes separated by a spacer region and a cleavage site for Tobacco Etch Virus (TEV) protease. In consecutive steps, TAP is achieved by the binding of a TAP-tagged protein to an IgG column, release of the protein by TEV protease cleavage, binding of the CBP-tagged protein to a calmodulin column and final elution of the bound protein by a buffer containing ethylene glycol-bis(beta-aminoethyl ether)-N,N,N’,N’-tetra acetic acid. One of the requirements for successful TAP is the expression of the tagged protein at its natural level (Rigaut et al. 1999) because over-expression can lead to artificial complex formation.
The Gateway® Technology is a universal cloning method that takes advantage of the site-specific recombination properties of bacteriophage lambda (Landy 1989) to provide a rapid and highly efficient way to move a gene of interest into multiple vector systems. Such site-specific recombination systems increase cloning efficiency and decrease time spent at the workbench. Gateway® Technology has been recently employed to construct a wide variety of vectors for T. cruzi gene expression. Xu et al. (2009) developed a Multisite Gateway strategy to rapidly produce efficient knockout constructs that generate specific gene deletions. Batista et al. (2010) constructed a flexible platform based on the Gateway® cloning system that allows rapid exchange of different elements such as promoters, fusion tags, resistant markers and intergenic regions. Finally, Westergaard et al. (2010) constructed the pTREX-A GW plasmid as a tool to analyse the nuclear localisation signal of the p14 splicing factor in T. cruzi. Here, we present three new T. cruzi vectors adapted to the Gateway® system, two TAPtag plasmids (pTEX-TAPtag-GW and pTREX-TAPtag-GW) and an inducible pTcINDEX-GW plasmid.
To construct the pTEX-TAPtag-GW and pTREX-TAPtag-GW vectors, the TAPtag cassette was first polymerase chain reaction (PCR)-amplified from the pBS1479 TAPtag plasmid (Rigaut et al. 1999) and then inserted into the HindIII-XhoI restriction sites. The pTEX-TAPtag plasmid has been successfully used in T. cruzi to express different fused proteins (Dallagiovanna et al. 2008, P Cribb, unpublished observations).
Then, the Gateway® cassette was PCR-amplified from the pDEST™17 vector (Invitrogen) and inserted into EcoRV-HindIII sites (Fig. 1A, B). To determine the functionality of pTEX-TAPtag-GW and pTREX-TAPtag-GW, the green fluorescent protein (GFP) sequence was transferred by recombination (LR) with the LR Clonase® II enzyme mix (Invitrogen) into the TAPtag-GW plasmids as described by the manufacturer from the previously constructed pENTR3C-GFP vector. The plasmids were maintained in the DB3.1 Escherichia coli strain.
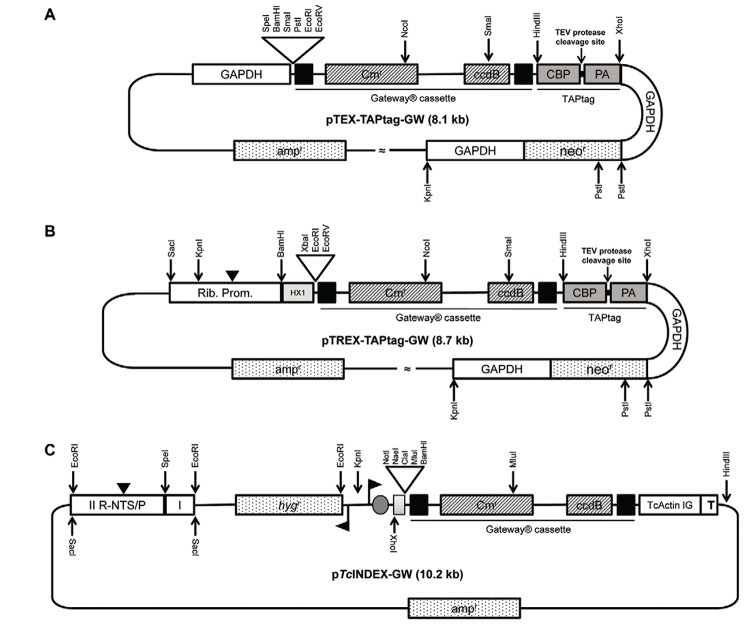
Fig. 1. : restriction maps and genetic elements of the Gateway® plasmids (A) pTEX-TAPtag-GW map. The ampicillin resistance gene (neo r), Gateway® cassette and TAPtag sequence are flanked by the 5’-upstream and 3-downstream regions of the Trypanosoma cruzi glycosomal glycosomal glyceraldehyde-3-phosphate dehydrogenase gene (gGAPDH) genes. The Gateway® cassette consists of the attR1 and R2 sites (black boxes), the chloramphenicol resistance gene (Cmr) and the ccdB gene. The TAPtag cassette consists of the calmodulin-binding peptide (CBP), protein A (PA) and a spacer region with a Tobacco Etch Virus (TEV) protease cleavage site; B: pTREX-TAPtag-GW map. This vector has the same features as pTEX with the addition of a ribosomal promoter (with a transcription starting point, black arrow head) and the HX1 fragment; C: inducible expression vector pTcINDEX-GW. The grey box indicates the HX1 fragment. The T. cruzi actin intergenic region (TcActin IG), the T7 transcriptional terminator (T) and the Gateway® cassette are shown. The black flags indicate the T7 promoter and the grey circle denotes the location of the tetracycline operator. R-NTS/P is the ribosomal non-transcribed spacer and promoter region used for targeting. The Roman numerals I and II indicate the two halves of the targeting sequence cloned in the opposite orientation of that in the genome. The black arrowhead indicates the location of the polymerase I transcription start site.
The two obtained GFP expressing plasmids were transfected into the T. cruzi Dm28c and CL Brener strains. Epimastigotes were grown at 28ºC in liver infusion tryptose (LIT) medium supplemented with 10% foetal bovine serum (FBS), 100 μg mL-1 streptomycin and 100 μg mL-1 penicillin. The parasites were maintained by subculture and maintained at cell densities ranging between 106-108 cells mL-1. T. cruzi epimastigotes were transfected by electroporation. The parasites were harvested by centrifugation at 2,000 g for 5 min at room temperature, washed once in phosphate-buffered saline (PBS) and resuspended in 0.4 mL electroporation buffer, pH 7.5 (140 mM NaCl, 25 mM HEPES, 0.74 mM Na2HPO4) to a density of 108 cells mL-1. Cells were then transferred to a 0.2 cm gap cuvette (Bio-Rad, USA) and 15-100 μg of DNA was added. The mixture was placed on ice for 15 min and then subjected to two pulses of 400 V and 500 μF using GenePulser II (Bio-Rad). After electroporation, cells were maintained on ice until being transferred into 4-10 mL of LIT medium containing 10% FBS and then incubated at 28ºC. After 24 h of incubation, a selective antibiotic was added to an initial concentration of 125 μg mL-1. Then, 72-96 h after electroporation, cultures were diluted 1:10 and the antibiotic concentration was doubled. Stable resistant cells were obtained approximately 30 days after transfection.
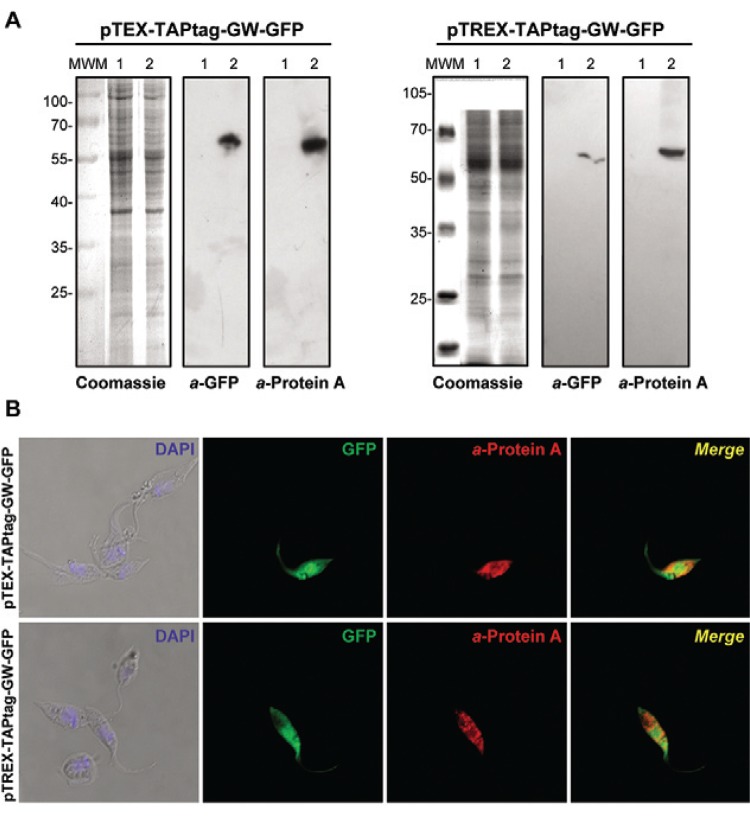
Once the lines were selected with G418, we monitored GFP expression by western blotting and fluorescence microscopy (Fig. 2). Immunoblot analysis with commercial anti-GFP (Santa Cruz Biotechnology) and anti-Protein A (Sigma) antibodies showed a single band of ~60 kDa, which corresponds to the full fused protein that is present only in transfected parasites, but not in the wild type. No partially synthesised or proteolytic products were observed (Fig. 2A). Immunofluorescence microscopy of transfected epimastigotes demonstrated expression of GFP and the TAP tag, which was detected with mouse anti-Protein A antibodies and Alexa Fluor 555 anti-mouse (Invitrogen) as a secondary antibody. We observed a positive signal for PA only in parasites that expressed GFP, which have a similar fluorescence pattern in the whole cell body, but do not completely co-localise. This difference is likely because the red signal is indirect and comes from the antibodies and the GFP signal is more intense (Fig. 2B).
Fig. 2. : pTEX and pTREX-TAPtag-GW experimental validation (A) Coomassie and western blot analysis of total lysates from CL Brener wild type cells (Lane 1) and those transfected (2) with pTEX-TAPtag-GW-GFP or pTREX-TAPtag-GW-GFP. Anti-green fluorescent protein (GFP) (a-GFP) and anti-protein A (a-protein A) antibodies were used. Bound antibodies were detected using peroxidase-labelled anti-mouse or anti-rabbit immunoglobulin G (IgG) (GE Healthcare) and ECL Prime (GE Healthcare) using standard protocols; B: immunofluorescence microscopy of CL Brener transfected with pTEX-TAPtag-GW-GFP and pTREX-TAPtag-GW-GFP. Parasites were adhered to poly-L-lysine coated slides and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), permeabilised with 0.1% Triton X-100 in PBS and then incubated with a-protein A antibodies in 1% bovine serum albumin-PBS for 3 h. Then, the slides were incubated with anti-mouse IgG antibody conjugated to Alexa Fluor 555 secondary antibodies for 1 h. The slides were mounted with VectaShield (Vector Laboratories) in the presence of 2 µg mL-1 4’-6-diamidino-2-phenylindole (DAPI) in PBS. Images were acquired with a Nikon Eclipse TE-2000-E2 confocal microscope. Adobe Photoshop CS v.8.0.1 (Adobe System Incorporated) and Nikon EZ-C1 FreeViewer v.3.70 (Nikon Corporation) software were used to analyse the images.
Both pTEX and pTREX-derived plasmids showed similar expression rates (data not shown). Finally, it is worth mentioning that the expression levels observed for the episomal pTEX and integrated pTREX plasmids in CL Brener appeared to be less homogeneous than that in Dm28c (data not shown). To address the biological functions of an unknown or hypothetical protein, it is necessary to determine its interacting partners. Co-immunoprecipitation using polyclonal antibodies directed against the protein of interest depends on the antibody specificity and it is a laborious and inefficient method. Conversely, TAP is a useful technique for studying protein-protein interactions at or close to natural expression levels with efficient recovery. In principle, the use of a generic tag allows for parallel sample preparation without the need for optimising the purification protocol for each protein complex, making it suitable for large-scale studies. The conversion of pTEX and pTREX-TAPtag to the Gateway® System results in an easier and faster approach. The pTREX-TAPtag plasmid was used by our group to identify T. cruzi Bromodomain Factor 2 interaction partners (V Villanova, unpublished observations).
To convert pTcINDEX to the Gateway® system, the plasmid was digested (5 μg of DNA) with the NruI restriction enzyme. The 5’-phosphates were removed with calf intestinal alkaline phosphatase and the plasmid was ligated with the Gateway® cassette RfA (Gateway® Vector Conversion System, Invitrogen) to generate the destination vector (Fig. 1C), which was maintained in the ccdB Survival™ 2 T1R E. coli strain. To determine the functionality of the obtained pTcINDEX-GW vector, the GFP open reading frame was transferred by recombination (LR) with LR clonase II enzyme (Invitrogen) from the entry vector pENTR3C-GFP as previously described.
T. cruzi epimastigotes (CL Brener and Dm28c strains) were transfected by electroporation with pTcINDEX-GW-GFP and pTcINDEX-Red (Taylor & Kelly 2006) as previously described. Selected CL Brener pTcINDEX-GW-GFP transformants were cloned by limiting dilution. One hundred microlitres of diluted culture (with the addition of wild type parasites) was transferred to a 48-well plate and the cells were allowed to grow for 24 h prior to addition of the selective antibiotic hygromycin.
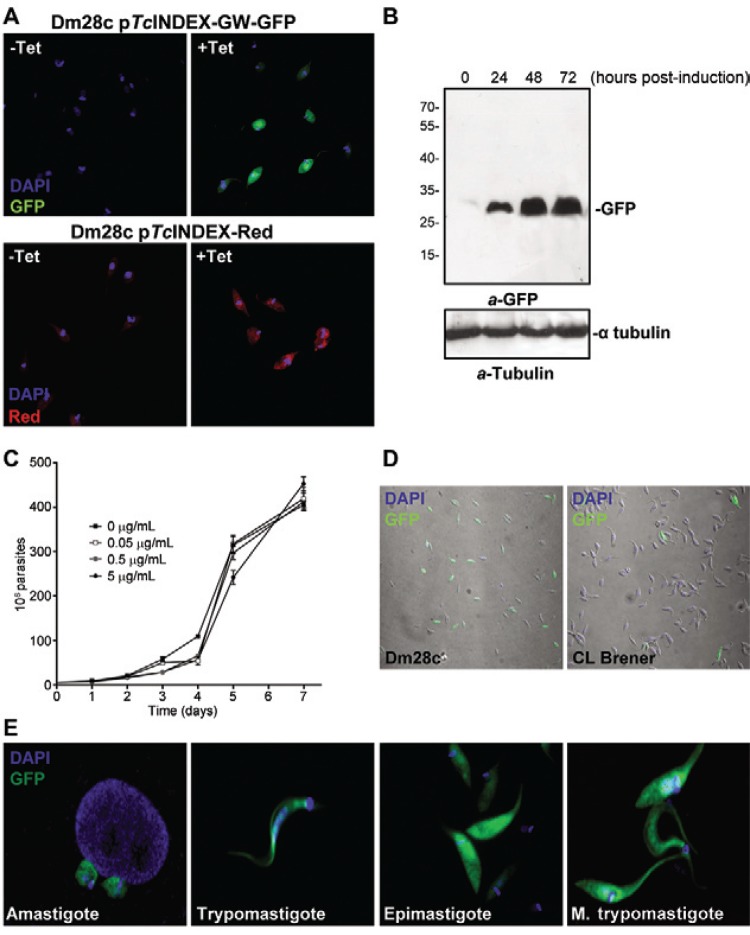
Once the lines were selected, we monitored the expression of the fluorescent proteins in induced and un-induced cultures by fluorescence microscopy (Fig. 3A). GFP was observed in the cultures induced with tetracycline, but not in those under un-induced conditions. In contrast, the pTcINDEX-Red strain (used as a transfection control) showed leaky expression because we detected red fluorescence in un-induced parasites. The expression of GFP was also tested by western blot assays (Fig. 3B). We observed that between 48-72 h post-induction, there were no significant changes in the expression level of GFP. Then, we performed growth curves in the presence of different concentrations of tetracycline (Fig. 3C) and observed that the parasites grew normally using up to 5 µg mL-1 of the antibiotic. We demonstrated that the attB1 and attB2 sequences generated by recombination did not affect the expression of exogenous proteins in T. cruzi. If anything, these sequences appear to have made the vector more efficient because there was no leaky expression in the Dm28c pTcINDEX-GW-GFP strain.
Fig. 3. : pTcINDEX-GW experimental validation. A: fluorescence microscopy of Dm28c epimastigotes transfected with pTcINDEX-GW-green fluorescent protein (GFP) and pTcINDEX-Red before and after induction with tetracycline (0.5 µg mL-1 for 96 h). Parasites were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), permeabilised with 0.1% Triton X-100 in PBS and incubated with 2 µg mL-1 4’-6-diamidino-2-phenylindole (DAPI) in PBS before mounting the slides with VectaShield (Vector Laboratories). Images were acquired with a Nikon Eclipse TE-2000-E2 confocal microscope; B: western blot analysis of Dm28c pTcINDEX-GW-GFP total lysates induced with 0.5 µg mL-1 tetracycline for 0, 24, 48 and 72 h. Anti-GFP (a-GFP) and anti-α tubulin antibodies were used. Bound antibodies were detected with peroxidase-labelled anti-mouse or anti-rabbit immunoglobulin G (GE Healthcare) and ECL Prime (GE Healthcare) using standard protocols; C: growth curve of Dm28c pTcINDEX-GW-GFP induced with different concentrations of tetracycline (0-5 µg mL-1); D: expression of GFP after induction (0.5 µg mL-1 of tetracycline for 96 h) in Dm28c and CL Brener strains transfected with pTcINDEX-GW-GFP; E: fluorescence microscopy of Dm28c pTcINDEX-GW-GFP at different stages of the life cycle. Parasites were fixed with 4% paraformaldehyde in PBS, permeabilised with 0.1% Triton X-100 in PBS and incubated with 2 µg mL-1 DAPI in PBS before mounting the slides with VectaShield (Vector Laboratories). Images were acquired with a Nikon Eclipse E300 microscope.
Again, we observed irregular expression of GFP in transfected CL Brener epimastigotes (Fig. 3D). Even after performing dilution cloning of this cell line, the expression of GFP was not homogeneous and had a wide range of fluorescence intensity among the induced parasites, including some cells in which no fluorescence was detected. More homogeneous results were observed with Dm28c, which is a cloned strain with a smaller genome with more homogeneous experimental behaviour. This strain also presents good rates of in vitro metacyclogenesis and efficiently infects common laboratory mammalian cells. In addition, the genomic sequence of Dm28c was recently obtained and incorporated into the TriTryp database (Grisard et al. 2014).
We also tested the expression of induced GFP throughout the T. cruzi life cycle. Dm28c pTcINDEX-GW-GFP epimastigotes were washed with PBS and differentiated in vitro to trypomastigotes following the procedure described by Contreras et al. (1988). Briefly, cells were washed with PBS and incubated in triatomine artificial urine (TAU) medium (190 mM NaCl, 17 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 8 mM phosphate buffer pH 6.0), reaching a density of 5 x 108 parasites mL-1 at 28ºC for 2 h. Then, the cells were diluted 1:100 in TAU3AAG Medium (TAU medium supplemented with 10 mM glucose, 2 mM L-aspartic acid, 50 mM L-glutamic acid and 10 mM L-proline) and incubated at 28ºC for 72 h. Metacyclic trypomastigotes were recovered from the supernatant, whereas non-differentiated epimastigotes remained adhered to the flask surface. Intracellular forms (amastigotes) and trypomastigotes were obtained by infecting Vero cells with the metacyclic trypomastigotes as previously described (Tonelli et al. 2004). The Vero cell line was routinely cultivated in Dulbecco’s Modified Eagle’s Medium medium (Gibco) supplemented with 10% heat-inactivated FBS, 0.15% (w/v) NaHCO3, 100 U mL-1 penicillin and 100 mg mL-1 streptomycin at 37ºC in a humidified atmosphere containing 5% CO2.
Tetracycline induction was tested in the different life cycle stages. First, epimastigotes were induced for 72 h and tetracycline was removed during the PBS washing step prior to performing the in vitro metacyclogenesis. We also assayed the addition of tetracycline in the TAU medium of un-induced epimastigotes and Vero cells (with intracellular amastigotes) infected by un-induced metacyclic trypomastigotes. Finally, we tested the induction of trypomastigotes released from infected cells. The expression of GFP in the four stages was monitored by fluoresce microscopy (Fig. 3D). We observed that each developmental stage responded to tetracycline induction. Once the expression of GFP was induced, it was maintained throughout the cycle. Moreover, the expression was maintained for at least one round of infection when induced metacyclic trypomastigotes were used. These results suggest that induction with tetracycline promotes protein expression in all stages (including the non-replicative trypomastigotes) and renders an important amount of product that, in the case of a stable protein such as GFP, remains in cells during the differentiation processes.
In conclusion, we constructed and tested three new destination vectors for T. cruzi that employ Gateway® Technology. This recombination cloning system is a rapid and highly efficient way to move DNA sequences into multiple destination vectors. All plasmids are available for the scientific community and can be requested from the authors.
Footnotes
VLA and CR contributed equally to this work.
REFERENCES
- Batista M, Marchini FK, Celedon PA, Fragoso SP, Probst CM, Preti H, Ozaki LS, Buck GA, Goldenberg S, Krieger MA. A high-throughput cloning system for reverse genetics in Trypanosoma cruzi. 259BMC Microbiol. 2010;10 doi: 10.1186/1471-2180-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton CE. Genetic manipulation of kinetoplastida. 372-378Parasitol Today. 1999;15 doi: 10.1016/s0169-4758(99)01498-2. [DOI] [PubMed] [Google Scholar]
- Contreras VT, Araujo-Jorge TC, Bonaldo MC, Thomaz N, Barbosa HS, Meirelles MNSL, Goldenberg S. Biological aspects of the Dm28c clone of Trypanosoma cruzi after metacyclogenesis in chemically defined media. 123-133Mem Inst Oswaldo Cruz. 1988;83 doi: 10.1590/s0074-02761988000100016. [DOI] [PubMed] [Google Scholar]
- da Rocha WD, Otsu K, Teixeira SM, Donelson JE. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol Biochem Parasitol. 2004;133:175–186. doi: 10.1016/j.molbiopara.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Dallagiovanna B, Correa A, Probst CM, Holetz F, Smircich P, de Aguiar AM, Mansur F, da Silva CV, Mortara RA, Garat B, Buck GA, Goldenberg S, Krieger MA. Functional genomic characterization of mRNAs associated with TcPUF6, a pumilio-like protein from Trypanosoma cruzi. 8266-8273J Biol Chem. 2008;283 doi: 10.1074/jbc.M703097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NM Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD, Andersson B. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Estévez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisard EC, Teixeira SM, de Almeida LG, Stoco PH, Gerber AL, Talavera-Lopez C, Lima OC, Andersson B, de Vasconcelos AT. Trypanosoma cruzi clone Dm28c draft genome sequence. Genome Announc. 2014;2: doi: 10.1128/genomeA.01114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JM, Ward HM, Miles MA, Kendall G. A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania. Nucleic Acids Res. 1992;20:3963–3969. doi: 10.1093/nar/20.15.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Martínez-Calvillo S, López I, Hernández R. pRIBOTEX expression vector: a pTEX derivative for a rapid selection of Trypanosoma cruzi transfectants. Gene. 1997;199:71–76. doi: 10.1016/s0378-1119(97)00348-x. [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Taylor MC, Kelly JM. pTcINDEX: a stable tetracycline-regulated expression vector for Trypanosoma cruzi. 32BMC Biotechnol. 2006;6 doi: 10.1186/1472-6750-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli RR, Silber AM, Almeida-de-Faria M, Hirata IY, Colli W, Alves MJ. L-proline is essential for the intracellular differentiation of Trypanosoma cruzi. Cell Microbiol. 2004;6:733–741. doi: 10.1111/j.1462-5822.2004.00397.x. [DOI] [PubMed] [Google Scholar]
- Vazquez MP, Levin MJ. Functional analysis of the intergenic regions of TcP2beta gene loci allowed the construction of an improved Trypanosoma cruzi expression vector. Gene. 1999;239:217–225. doi: 10.1016/s0378-1119(99)00386-8. [DOI] [PubMed] [Google Scholar]
- Westergaard GG, Bercovich N, Reinert MD, Vazquez MP. Analysis of a nuclear localization signal in the p14 splicing factor in Trypanosoma cruzi. Int J Parasitol. 2010;40:1029–1035. doi: 10.1016/j.ijpara.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Xu D, Brandan CP, Basombrio MA, Tarleton RL. Evaluation of high efficiency gene knockout strategies for Trypanosoma cruzi. 90BMC Microbiol. 2009;9 doi: 10.1186/1471-2180-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]