Abstract
The interferon (IFN)-γ response to peptides can be a useful diagnostic marker of Mycobacterium tuberculosis (MTB) latent infection. We identified promiscuous and potentially protective CD4+ T-cell epitopes from the most conserved regions of MTB antigenic proteins by scanning the MTB antigenic proteins GroEL2, phosphate-binding protein 1 precursor and 19 kDa antigen with the TEPITOPE algorithm. Seven peptide sequences predicted to bind to multiple human leukocyte antigen (HLA)-DR molecules were synthesised and tested with IFN-γ enzyme-linked immunospot (ELISPOT) assays using peripheral blood mononuclear cells (PBMCs) from 16 Mantoux tuberculin skin test (TST)-positive and 16 TST-negative healthy donors. Eighty-eight percent of TST-positive donors responded to at least one of the peptides, compared to 25% of TST-negative donors. Each individual peptide induced IFN-γ production by PBMCs from at least 31% of the TST-positive donors. The magnitude of the response against all peptides was 182 ± 230 x 106 IFN-γ spot forming cells (SFC) among TST-positive donors and 36 ± 62 x 106 SFC among TST-negative donors (p = 0.007). The response to GroEL2 (463-477) was only observed in the TST-positive group. This combination of novel MTB CD4 T-cell epitopes should be tested in a larger cohort of individuals with latent tuberculosis (TB) to evaluate its potential to diagnose latent TB and it may be included in ELISPOT-based IFN-γ assays to identify individuals with this condition.
Keywords: Mycobacterium tuberculosis, epitopes, IFN-γ ELISPOT, latent tuberculosis, Mantoux tuberculin skin test
Infection by Mycobacterium tuberculosis (MTB) remains a major public health problem in the world. In this respect, tuberculosis (TB) is the second cause of death after human immunodeficiency virus (HIV) infection and is the most common cause of death of people infected with HIV (Corbett et al. 2003, Blanc & Martinez 2007).
Most MTB-infected individuals remain asymptomatic, with a small proportion of them eventually progressing to active TB disease. Therefore, they may become a reservoir of Mycobacterium and this may constitute an obstacle to its eradication. The most common manifestation of TB is a chronic pulmonary disease and immunocompromised individuals are the most prone to developing symptomatic disease (Girardi et al. 2000, Stewart et al. 2003).
The diagnosis of latent MTB infection remains a challenge. The tuberculin skin test (TST), also known as the purified protein derivative (PPD) skin test, is still largely employed as a diagnostic tool, but it has shown low specificity, as it often displays antigenic cross-reactivity with unrelated microorganisms (Dinnes et al. 2007). The tests that measure interferon (IFN)-γ production, IFN-γ release assays (IGRA), were developed to detect latent TB and are based upon the detection of IFN-γ produced specifically from peptides identified during infection with MTB; they are considered to be more specific than TST for the diagnosis of Mycobacterium infection. One of these tests, the enzyme-linked immunospot (ELISPOT) assay, was shown to have a better diagnostic performance when compared with TST (Ewer et al. 2003, Fietta et al. 2003, Brock et al. 2004). The ELISPOT assay is closely correlated with the exposure of the individual to Mycobacterium (Ewer et al. 2003) and shows less indeterminate results than TST tests (Dogra et al. 2007).
A peptide-based IGRA test could offer several advantages, including the precise knowledge of the antigen specificity. However, such peptides should be widely recognised by the population to be tested. Until recently, the available tools for searching immunodominant epitopes was the direct testing of substantial numbers of overlapping peptides or peptide libraries. The identification of major histocompatibility complex-binding motifs allowed the prediction of potential T-cell epitopes (Rammensee 1995) and such motifs were found to cluster in certain protein regions (Meister et al. 1995). The TEPITOPE algorithm predicts binding to 25 distinct human leukocyte antigen (HLA)-DR molecules based on quantitative matrices established from HLA-DR binding assays (Sturniolo et al. 1999, Bian et al. 2003, Iwai et al. 2003, Fonseca et al. 2004, 2006, Damico et al. 2005). This advance lead to the selection of sequences enriched for high affinity-binding peptides, those with the highest chance of eliciting effective T-cell responses against immunogens (Schroers et al. 2002). Additionally, TEPITOPE also allows for the detection of sequences predicted to bind to several HLA-DR molecules simultaneously, opening the possibility of selecting promiscuous T-cell epitopes. Another advantage of the peptide-based IGRA test is the ability to test individual peptides, allowing for the analysis of peptide-specific T cell responses as well as peptide pools.
Therefore, to identify the frequently recognised immunodominant epitopes of three immunogenic proteins of MTB, we used the TEPITOPE algorithm to screen GroEL-2 (Shinnick 1987, Fleischmann et al. 2002), phosphate-binding protein 1 precursor (PBP-1) (Andersen & Hansen 1989) and 19 kDa antigen (Ashbridge et al. 1989), which are proteins that were previously found to elicit potentially strong immunological responses (Tanghe et al. 1999, Lewthwaite et al. 2001, Mendelson et al. 2005, Wilkinson et al. 2009), for multiple HLA-DR binding sequences, in a group of healthy individuals. Synthetic peptides encoding such potential CD4+ T cell epitopes were used in IFN-γ ELISPOT assays with peripheral blood mononuclear cells (PBMCs) of TST-positive and TST-negative Brazilian healthy subjects.
SUBJECTS, MATERIALS AND METHODS
Subjects - The sample comprised 32 Brazilian healthy volunteers (donors) selected from students and workers from the Advanced Laboratory of Public Health, Oswaldo Cruz Foundation (Fiocruz), Salvador, state of Bahia, Brazil, and the Immunology Laboratory, Heart Institute, University of São Paulo (USP), São Paulo, state of São Paulo, Brazil. All donors received Bacille Calmette-Guérin (BCG) vaccination during childhood. The mean age of the 32 donors was 32 ± 9 years. They were divided into two groups according to the TST response: latent MTB infection (16 subjects, TST-positive) and uninfected controls (16 subjects, TST-negative), with a mean age of 35 ± 11 years (TST-positive) or 30 ± 7 years (TST-negative). The TST was performed according to the Mantoux method with PPD, following the manufacturer’s instructions (Statens Serum Institute, Denmark). TST was considered positive when the induration was ≥ 10 mm after 48 h of PPD application. All of the individuals gave their written informed consent to participate in this study, which was approved by the Institutional Review Boards of the School of Medicine, USP (protocol 250/04) and Fiocruz (protocol 51/2004).
Peptide selection - Sequences of three important antigens expressed by MTB were used: GroEL2 protein, accession P9WPE6.1 (Fleischmann et al. 2002), PBP-1, accession P9WGU0.1 (Andersen & Hansen 1989) and 19 kDa antigen, accession S02753 (Ashbridge et al. 1989). These sequences were screened by TEPITOPE and we predicted seven epitopes that were used in the ELISPOT assay. The seven peptides were predicted to bind to at least 12 out of the 25 HLA-DR molecules in the TEPITOPE matrix, as previously described (Bian et al. 2003). They correspond to an inner nonamer core selected as the HLA-binding motif with flanking amino acids added when possible (e.g., conserved flanking sequences) at either or both the N and C-terminal ends, to increase the efficiency of in vitro peptide presentation to CD4+ T cells (Bian et al. 2003, Fonseca et al. 2006).
Peptide synthesis - Peptides of the MTB proteins were synthesised by solid phase technology using 9-fluorenylmethoxycarbonyl strategy, with the C-terminal carboxyl group in amide form. Peptide purity and quality were assessed by reverse-phase high performance liquid chromatography and mass spectrometry, as described elsewhere (Atherton & Sheppard 1989). The peptides were dissolved in dimethyl sulfoxide (DMSO) for stock solutions of 25 mg/mL.
PBMC separation - PBMC suspensions were prepared from 50 mL heparinised venous blood by density gradient centrifugation using Ficoll-Hypaque, 1.077 g/L (Pharmacia, Sweden). The cells were washed twice with phosphate-buffered saline (PBS) (Gibco, UK) and were kept frozen at -196ºC in liquid nitrogen in foetal calf serum (FCS) containing 10% DMSO (Merck, Germany) until testing. After thawing, the PBMCs were suspended in RPMI-1640 (Gibco), supplemented with 200 mM glutamine, 100 U/mL penicillin, 100 U/mL streptomycin and 10% FCS, 100 mM non-essential amino acids and 100 mM pyruvate.
IFN-γ ELISPOT assays - The IFN-γ ELISPOT assay was performed as described previously (Sun et al. 2003, Fonseca et al. 2005, Zhang et al. 2009, Xie et al. 2010). Briefly, cryopreserved PBMCs (105 cells/well) were incubated with 5 μM of each peptide on a 96-well MultiScreen ELISPOT plate (Multiscreen MAIPS4510; Millipore, USA) previously coated with 50 μL/well of capture anti-human IFN-γ monoclonal antibody (mAb) diluted 1/1,000 (IgG1/B-B1; Diaclone, France) in PBS (1 μL/mL of mAb) at 4ºC overnight. The plates were washed three times with PBS. The remaining peptide binding sites were blocked with 100 μL RPMI/10% FCS for 2 h at 37ºC. The medium was removed just before the addition of cell suspensions (100 μL) and stimulating agents (100 μL). The MTB peptides (5 μM) were added to each well of the plates. The plates were incubated for 20 h at 37ºC in 5% CO2. PBMCs incubated with 0.5 μg/mL phytohemagglutinin (Sigma, France) or RPMI/10% FCS served as the positive and negative controls, respectively. At the end of the incubation, the cells were removed from the ELISPOT plates by washing nine times: three times with PBS, three times with PBS containing 0.05% Tween-20 (Sigma) and three times again with PBS. Spots were detected after the addition of the second biotinylated anti-human IFN-γ mAb (B-G1; Diaclone) (100 μL/well, 4 h, 37ºC in 5% CO2), diluted 1/500 in PBS-bovine serum albumin (PBS-BSA) (Gibco) (1 μL/500 μL of biotinylated antibody). The plates were washed three times with PBS, followed by streptavidin-alkaline phosphatase conjugate (Amersham, France) (100 μL/well, 1 h, 37ºC in 5% CO2) diluted 1/1000 in PBS-BSA (1 μL/mL of streptavidin-alkaline phosphatase conjugate), washed three times with PBS and incubated with 50 μL/well of chromogen substrates 5-bromo-4-chloro-3-indolyl-phosphate toluidine and 4-nitroblue tetrazolium (Sigma), which were incubated at room temperature (25ºC) until the appearance of violet spots (~ 30 min). The plates were washed with tap water and dried overnight. Antigen-specific T cells, expressed as spot forming cells (SFC)/106 PBMCs frequencies, were measured using an automated stereomicroscope (KS ELISPOT; Carls Zeiss, Germany). The SFC number was calculated after subtracting the negative control values (wells with cells in the absence of peptide) from the same subject. The positivity cut-off was ≥ 30 IFN-γ SFC/106 PBMCs, which was above the mean ± 3 standard deviation of the highest response found among all uninfected control subjects to any peptide (data not shown). The magnitude of response to each given peptide or the sum of the SFC of the total peptides was calculated as the SFC/106 PBMCs of peptides with responses above the positivity cut-off.
Statistical analysis - Statistical analysis was performed using the GraphPad Prism package v.3.0. Comparisons of the number of IFN-γ SFC/106 PBMCs between the clinical groups were performed with a non-parametric one-way ANOVA, Kruskal-Wallis and Dunn’s Test. The Mann-Whitney U test was used to calculate the differences between the groups. Values of p < 0.05 were considered significant.
RESULTS
Three MTB antigenic proteins, GroEL2, PBP-1 and 19 kDa, were scanned with the TEPITOPE algorithm at a 3% threshold. Seven peptide sequences predicted to bind to multiple HLA-DR molecules were synthesised (Table I).
TABLE I. Sequences of peptides derived from Mycobacterium tuberculosis (MTB) sequences selected by TEPITOPE algorithm.
| Peptide | HLA-DR molecules predicted to bind to peptidea (%) | Sequence |
|---|---|---|
| GroEL2 (215-229) | 76 | DPYILLVSSKVSTVK |
| GroEL2 (463-477) | 60 | AEKVRNLPAGHGLNA |
| PBP-1 (59-75) | 84 | TLLYPLFNLWGPAFHER |
| PBP-1 (118-133) | 88 | KGLMNIALAISAQQVN |
| PBP-1 (351-365) | 64 | QVHFQPLPPAVVKLS |
| PBP-1 (352-371) | 84 | VHFQPLPPAVVKLSDALIAT |
| 19 kDa (127-141) | 48 | GSHYKITGTATGVDM |
a: 3% threshold; HLA: human leukocyte antigen.
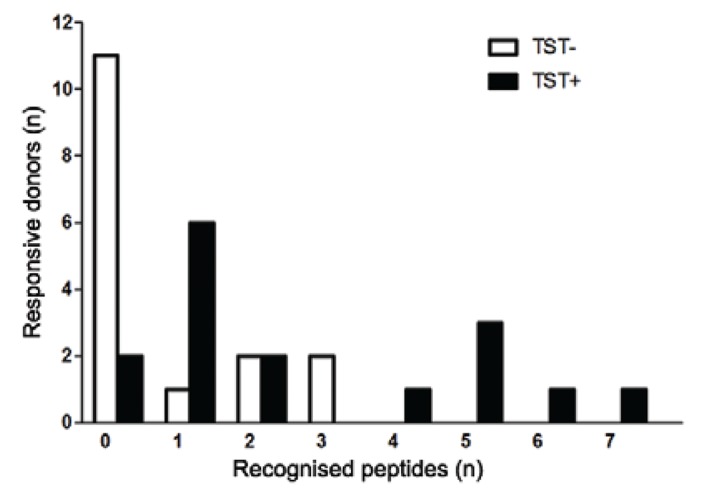
Next, we tested if the IFN-γ response to the seven peptides could discriminate between TST-positive and TST-negative healthy donors. The mean size (± standard deviation) of PPD induration detected for TST-negative individuals was 0.71 ± 2.02 mm and for TST-positive individuals was 11.14 ± 4.07 mm. PBMCs from the 16 TST-positive and 16 TST-negative individuals were tested in the presence of each peptide for an IFN-γ ELISPOT assay. The T cell responses are shown in Table II. The frequency of peptide recognition by the TST-positive group was significantly higher than the TST-negative group. Eighty-eight percent (14 out of 16 donors) of TST-positive donors recognised at least one peptide and 31% (5 out of 16 donors) had responses to three or more peptides, while in the TST-negative group, 31% (5 out of 16 donors) responded to any peptides and only 12% (2 out of 16 donors) responded to three or more peptides (p = 0.002) (Fig. 1).
TABLE II. Recognition of human leukocyte antigen (HLA)-DR promiscuous epitopes from Mycobacterium tuberculosis antigens by peripheral blood mononuclear cells (PBMC) from tuberculin skin test (TST)-positive and TST-negative subjects using the interferon (IFN)-γ ELISPOT assay.
| Peptides | IFN-γ SFC/106
PBMC (n) |
|||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TST-positive | TST-negative | |||||||||||||||||||||||||||||||
|
| ||||||||||||||||||||||||||||||||
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 | P14 | P15 | P16 | N1 | N2 | N3 | N4 | N5 | N6 | N7 | N8 | N9 | N10 | N11 | N12 | N13 | N14 | N15 | N16 | |
| PBP-1 (59-75) | 0 | 40 | 55 | 60 | 0 | 55 | 25 | 0 | 0 | 7 | 23 | 20 | 0 | 65 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 3 | 10 | 15 | 3 | 30 | 27 | 0 | 7 | 20 |
| PBP-1 (118-133) | 30 | 10 | 155 | 0 | 0 | 0 | 0 | 30 | 0 | 13 | 10 | 17 | 67 | 25 | 0 | 60 | 0 | 0 | 0 | 0 | 5 | 0 | 10 | 3 | 0 | 10 | 3 | 7 | 87 | 43 | 27 | 10 |
| PBP-1 (351-365) | 60 | 30 | 140 | 230 | 0 | 0 | 25 | 0 | 0 | 23 | 0 | 7 | 127 | 5 | 0 | 40 | 15 | 30 | 20 | 0 | 0 | 0 | 5 | 7 | 3 | 5 | 20 | 0 | 83 | 23 | 50 | 27 |
| PBP-1 (352-371) | 20 | 10 | 135 | 60 | 20 | 0 | 0 | 0 | 0 | 37 | 7 | 30 | 183 | 5 | 20 | 3 | 0 | 95 | 5 | 10 | 0 | 5 | 10 | 7 | 0 | 10 | 7 | 20 | 20 | 40 | 50 | 7 |
| GroEL2 (215-229) | 40 | 30 | 35 | 80 | 5 | 0 | 0 | 0 | 0 | 30 | 50 | 33 | 30 | 0 | 40 | 0 | 5 | 30 | 5 | 0 | 0 | 0 | 5 | 17 | 0 | 5 | 27 | 0 | 3 | 23 | 0 | 7 |
| GroEL2 (463-477) | 40 | 60 | 110 | 130 | 25 | 0 | 0 | 5 | 0 | 13 | 23 | 17 | 37 | 0 | 25 | 30 | 0 | 0 | 5 | 0 | 0 | 5 | 5 | 7 | 20 | 10 | 10 | 0 | 0 | 0 | 17 | 0 |
| 19 kDa (127-141) | 55 | 10 | 95 | 10 | 20 | 10 | 40 | 0 | 5 | 10 | 23 | 13 | 123 | 0 | 5 | 37 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 13 | 17 | 15 | 13 | 0 | 17 | 23 | 37 | 0 |
| Peptides recognised | 5 | 4 | 7 | 5 | 0 | 1 | 1 | 1 | 0 | 2 | 1 | 2 | 6 | 1 | 1 | 5 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 2 | 0 |
| Sum of spots | 225 | 160 | 725 | 560 | 0 | 55 | 40 | 30 | 0 | 67 | 50 | 63 | 567 | 65 | 40 | 267 | 0 | 155 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 30 | 170 | 83 | 137 | 0 |
SFC/106 PBMC were calculated according to the following equation: (average spots in duplicate peptide wells - average spots in duplicate control wells) x 5. Positive responses (≥ 30 SFC/106 PBMC) are highlighted by a gray background.
Fig. 1. : number of Mycobacterium tuberculosis (MTB) peptides recognised by tuberculin skin test (TST)-positive (TST+) and TST-negative (TST-) donors. Peripheral blood mononuclear cells (PBMCs) from 16 TST-positive and 16 TST-negative healthy donors were assayed with seven human leukocyte antigen-DR promiscuous peptides derived from MTB protein sequences. Each peptide was tested individually at concentration of 5 µM and the number of spot forming cells (SFC) interferon-γ/106 PBMC were calculated according to the following equation: (average spots in duplicate peptide wells - average spots in duplicate control wells) x 5. Positive responses were ≥ 30 SFC/106 PBMC. The columns represent the number of TST-positive and TST-negative donors tested that were negative or able to show positive responses in the ELISPOT assay in response to different numbers of peptides (p = 0.01, Mann-Whitney U test).
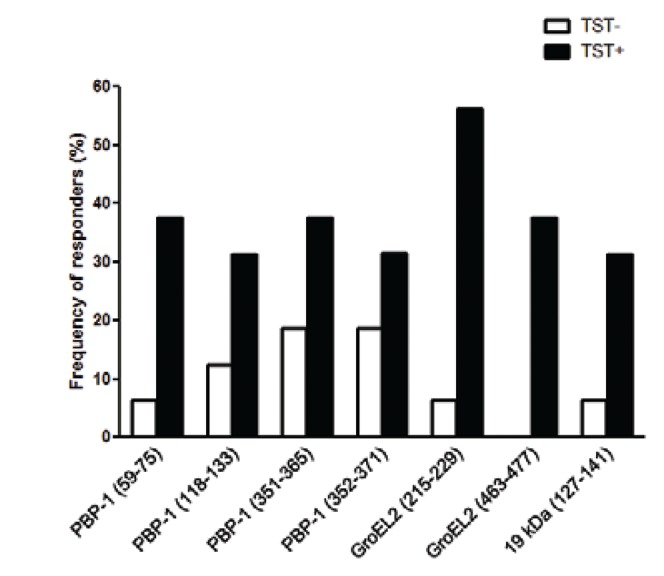
Each of the selected peptides induced IFN-γ production by PBMCs from at least 31% of the TST-positive donors. The frequency of responders to individual peptides ranged from 31% to PBP-1 (118-133), PBP-1 (352-371) and 19 kDa (127-141) to 56% to GroEL2 (215-229) (Fig. 2). In contrast, among TST-negative donors, there was no response to GroEL2 (463-477) peptide and the most frequently recognised peptides were PBP-1 (351-365) and PBP-1 (352-371), with 19% of responders.
Fig. 2. : proportion of tuberculin skin test (TST)-positive (TST+) and TST-negative (TST-) donors that recognised selected Mycobacterium tuberculosis (MTB) peptides. Peripheral blood mononuclear cells (PBMCs) from 16 TST-positive and 16 TST-negative healthy donors were tested with seven MTB derived peptides and the spot forming cells interferon-γ/106 PBMC were calculated. The columns represent the proportion of TST-positive and TST-negative individuals that recognised at list one peptide (p = 0.001, Mann-Whitney U test).
The magnitude of responses to the peptides (sum of IFN-γ SFC of all peptides with positive responses) was higher in the TST-positive group than in the TST-negative group (Fig. 3). The total number of IFN-γ SFC in response to MTB peptides was 182 ± 230 SFC/106 PBMCs among the TST-positive donors, compared to 36 ± 62 SFC/106 PBMCs in the TST-negative donors (p = 0.007). The magnitude of the responses per individual ranged from 0-725 SFC/106 PBMCs among the TST-positive individuals and 0-170 SFCs/106 PBMCs among the TST-negative individuals (Table II).
Fig. 3. : magnitude of responses to Mycobacterium tuberculosis peptides. The bars represent the magnitude of response of each peptide (overall sum of spots of all individuals with positive responses). Positive responses > 30 spot forming cells (SFC) interferon (IFN)-γ/106 peripheral blood mononuclear cells (PBMCs) (p = 0.007, Mann-Whitney U test). TST+: tuberculin skin test-positive; TST-: TST-negative.
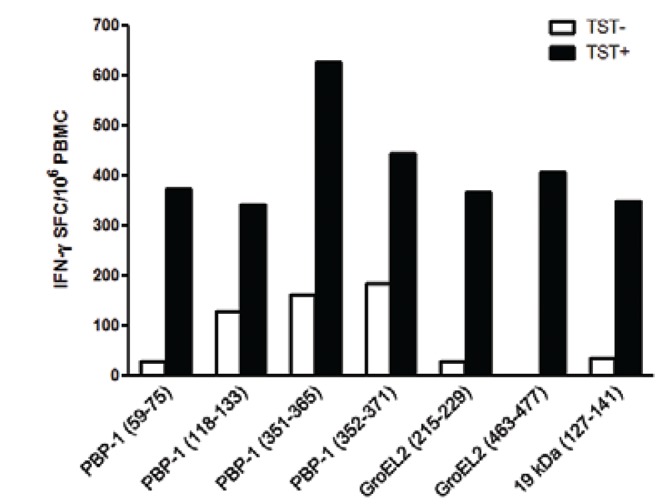
The PBP-1 (351-365) MTB peptide induced the highest magnitude of response, 627 SFC/106 PBMCs, followed by PBP-1 (352-371), with 446 SFC/106 PBMCs (Fig. 3). In the TST-negative group, the highest magnitude of response was to PBP-1 (352-371), with 185 SFC/106 PBMCs. In addition, a response to GroEL2 (463-477) was observed only in the TST-positive group (p = 0.02).
Taking into consideration that all 32 healthy donors were subjected to BCG vaccination, the combination of TST and positivity at the IFN-γ ELISPOT test raised the overall positivity value from 50% (16/32) to 59% (19/32) for recognition of MTB antigens.
DISCUSSION
Our results suggest that the peptides predicted by TEPITOPE are able to identify a significant number of individuals with latent MTB infection. Moreover, in our study, the use of these peptides allowed us to identify individuals with latent MTB infection not tracked by the TST test.
The TEPITOPE is an algorithm that uses a matrix that predicts the binding of 25 HLA-DR molecules. This allows the identification of epitopes with high binding affinity and the ability to generate a strong T cell response. The use of this algorithm can lead to the identification of a greater number of epitopes, which would display a higher sensitivity and specificity for detecting MTB, as reported elsewhere (Zhang et al. 2010).
The two methods used in this study to assess latent MTB infection, the TST and the IFN-γ ELISPOT, showed similar results in 88% of the evaluated individuals who were TST-positive. These findings are similar to the published data, in which there is a higher correlation of IGRA tests and TB when compared to TST and TB (Bartalesi et al. 2009, Girardi et al. 2009, Redelman-Sidi & Sepkowitz 2013). Furthermore, it was possible to identify TST-negative individuals (31%) who responded to peptides predicted by TEPITOPE. In this respect, Ewer et al. (2003) reported that ELISPOT offers a more accurate approach than TST for the identification of individuals who have latent TB infection and could improve TB control by more precise targeting of preventive treatment. However, possible antigenic cross-reactions or non-specific responses cannot be excluded and should be addressed. The advantage of using the IGRA with seven promiscuous peptides is that would ensure larger number of responder individuals independent of HLA class II and also a greater range or a greater number of recognised peptides.
To date, no fully effective methodology is available that can unequivocally differentiate individuals who actually have latent MTB infection from those who were BCG vaccinated or even those that have active MTB infection. The most widely and currently used test for the screening of latent MTB infection remains the TST because it is considered a low cost point-of-care-test (Ewer et al. 2003, Diel et al. 2007, Soborg et al. 2009). Therefore, it remains the method of choice for screening for latent MTB infection, primarily due to a better cost/benefit ratio when compared to tests based on IGRA (Lewinsohn et al. 2010).
Richeldi et al. (2009) report that immunocompromised persons infected with MTB have a high occurrence of false-negative results in the TST and, in such cases, they recommend to tailor the application of IGRA assays for latent MTB infection in different high-risk groups and advise caution in their use in immunosuppressed patients.
The results of the TST may show false positive results in individuals previously immunised with BCG or because the population of the study has high rates of exposure to MTB (Soborg et al. 2009). Another relevant aspect is that 70-80% of individuals with pulmonary TB have a diameter of the induration greater than 10 mm by TST and this value is reduced in individuals co-infected with HIV-1 (Idh et al. 2010). This combination of novel MTB CD4+ T-cell epitopes should be tested in a larger cohort of individuals with latent TB to confirm its potential to discriminate latent MTB infection from no TB infection and may be included in ELISPOT-based IFN-γ response assays to identify individuals with such conditions.
Footnotes
Financial support: FAPESP
REFERENCES
- Andersen AB, Hansen EB. Structure and mapping of antigenic domains of protein antigen b, a 38,000-molecular-weight protein of Mycobacterium tuberculosis. Infect Immun. 1989;57:2481–2488. doi: 10.1128/iai.57.8.2481-2488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashbridge KR, Booth RJ, Watson JD, Lathigra RB. Nucleotide sequence of the 19 kDa antigen gene from Mycobacterium tuberculosis. 1249Nucl Acids Res. 1989;17 doi: 10.1093/nar/17.3.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton E, Sheppard RC. Solid phase peptide synthesis: a practical approach. Oxford University Press; Cary: 1989. 216 [Google Scholar]
- Bartalesi F, Vicidomini S, Goletti D, Fiorelli C, Fiori G, Melchiorre D, Tortoli E, Mantella A, Benucci M, Girardi E, Cerinic MM, Bartoloni A. QuantiFERON-TB Gold and the TST are both useful for latent tuberculosis infection screening in autoimmune diseases. Eur Respir J. 2009;33:586–593. doi: 10.1183/09031936.00107608. [DOI] [PubMed] [Google Scholar]
- Bian H, Reidhaar-Olson JF, Hammer J. The use of bioinformatics for identifying class II-restricted T-cell epitopes. Methods. 2003;29:299–309. doi: 10.1016/s1046-2023(02)00352-3. [DOI] [PubMed] [Google Scholar]
- Blanc L, Martinez L. The international TB control targets. 326Bull World Health Organ. 2007;85 doi: 10.2471/07.041996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock I, Weldingh K, Lillebaek T, Follmann F, Andersen P. Comparison of tuberculin skin test and new specific blood test in tuberculosis contacts. Am J Respir Crit Care Med. 2004;170:65–69. doi: 10.1164/rccm.200402-232OC. [DOI] [PubMed] [Google Scholar]
- Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, Dye C. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- Damico FM, Cunha E, Neto, Goldberg AC, Iwai LK, Marin ML, Hammer J, Kalil J, Yamamoto JH. T-cell recognition and cytokine profile induced by melanocyte epitopes in patients with HLA-DRB1*0405-positive and negative Vogt-Koyanagi-Harada uveitis. Invest Ophtalmol Vis Sci. 2005;46:2465–2471. doi: 10.1167/iovs.04-1273. [DOI] [PubMed] [Google Scholar]
- Diel R, Nienhaus A, Loddenkemper R. Cost-effectiveness of interferon-gamma release assay screening for latent tuberculosis infection treatment in Germany. Chest. 2007;131:1424–1434. doi: 10.1378/chest.06-2728. [DOI] [PubMed] [Google Scholar]
- Dinnes J, Deeks J, Kunst H, Gibson A, Cummins E, Waugh N, Drobniewski F, Lalvani A. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol Assess. 2007;11:1–196. doi: 10.3310/hta11030. [DOI] [PubMed] [Google Scholar]
- Dogra S, Narang P, Mendiratta DK, Chaturvedi P, Reingold AL, Colford JM, Jr, Riley LW, Pai M. Comparison of a whole blood interferon-gamma assay with tuberculin skin testing for the detection of tuberculosis infection in hospitalized children in rural India. J Infect. 2007;54:267–276. doi: 10.1016/j.jinf.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Ewer K, Deeks J, Alvarez L, Bryant G, Waller S, Andersen P, Monk P, Lalvani A. Comparison of T-cell-based assay with tuberculin skin test for diagnosis of Mycobacterium tuberculosis infection in a school tuberculosis outbreak. Lancet. 2003;361:1168–1173. doi: 10.1016/S0140-6736(03)12950-9. [DOI] [PubMed] [Google Scholar]
- Fietta A, Meloni F, Cascina A, Morosini M, Marena C, Troupioti P, Mangiarotti P, Casali L. Comparison of a whole-blood interferon-gamma assay and tuberculin skin testing in patients with active tuberculosis and individuals at high or low risk of Mycobacterium tuberculosis infection. Am J Infect Control. 2003;31:347–353. doi: 10.1016/s0196-6553(02)48240-5. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, Hickey E, Kolonay JF, Nelson WC, Umayam LA, Ermolaeva M, Salzberg SL, Delcher A, Utterback T, Weidman J, Khouri H, Gill J, Mikula A, Bishai W, Jacobs WR, Jr, Venter JC, Fraser CM. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca CT, Cunha E, Neto, Kalil J, de Jesus AR, Correa-Oliveira R, Carvalho EM, Oliveira SC. Identification of immunodominant epitopes of Schistosoma mansoni vaccine candidate antigens using human T cells. Mem Inst Oswaldo Cruz. 2004;99(Suppl. I):63–66. doi: 10.1590/s0074-02762004000900011. [DOI] [PubMed] [Google Scholar]
- Fonseca SG, Coutinho-Silva A, Fonseca LA, Segurado AC, Moraes SL, Rodrigues H, Hammer J, Kallás EG, Sidney J, Sette A, Kalil J, Cunha E., Neto Identification of novel consensus CD4 T-cell epitopes from clade B HIV-1 whole genome that are frequently recognized by HIV-1 infected patients. AIDS. 2006;20:2263–2273. doi: 10.1097/01.aids.0000253353.48331.5f. [DOI] [PubMed] [Google Scholar]
- Fonseca SG, Moins-Teisserenc H, Clave E, Ianni B, Nunes VL, Mady C, Iwai LK, Sette A, Sidney J, Marin ML, Goldberg AC, Guilherme L, Charron D, Toubert A, Kalil J, Cunha E., Neto Identification of multiple HLA-A*0201-restricted cruzipain and FL-160 CD8+ epitopes recognized by T cells from chronically Trypanosoma cruzi-infected patients. Microbes Infect. 2005;7:688–697. doi: 10.1016/j.micinf.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Girardi E, Angeletti C, Puro V, Sorrentino R, Magnavita N, Vincenti D, Carrara S, Butera O, Ciufoli AM, Squarcione S, Ippolito G, Goletti D. Estimating diagnostic accuracy of tests for latent tuberculosis infection without a gold standard among healthcare workers. 19373Euro Surveill. 2009;14 [PubMed] [Google Scholar]
- Girardi E, Raviglione MC, Antonucci G, Godfrey-Faussett P, Ippolito G. Impact of the HIV epidemic on the spread of other diseases: the case of tuberculosis. AIDS. 2000;14(Suppl. 3):S47–S56. [PubMed] [Google Scholar]
- Idh J, Abate E, Westman A, Elias D, Janols H, Gelaw A, Getachew A, Alemu S, Aseffa A, Britton S, Stendahl O, Schön T. Kinetics of the QuantiFERON-TB Gold In-Tube test during treatment of patients with sputum smear-positive tuberculosis in relation to initial TST result and severity of disease. Scand J Infect Dis. 2010;42:650–657. doi: 10.3109/00365548.2010.482942. [DOI] [PubMed] [Google Scholar]
- Iwai LK, Yoshida M, Sidney J, Shikanai-Yasuda MA, Goldberg AC, Juliano MA, Hammer J, Juliano L, Sette A, Kalil J, Travassos LR, Cunha E., Neto In silico prediction of peptides binding to multiple HLA-DR molecules accurately identifies immunodominant epitopes from gp43 of Paracoccidioides brasiliensis frequently recognized in primary peripheral blood mononuclear cell responses from sensitized individuals. Mol Med. 2003;9:209–219. [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn DA, Lobato MN, Jereb JA. Interferon-gamma release assays: new diagnostic tests for Mycobacterium tuberculosis infection and their use in children. Curr Opin Pediatr. 2010;22:71–76. doi: 10.1097/MOP.0b013e3283350301. [DOI] [PubMed] [Google Scholar]
- Lewthwaite JC, Coates AR, Tormay P, Singh M, Mascagni P, Poole S, Roberts M, Sharp L, Henderson B. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60.2 (Hsp 65) and contains a CD14-binding domain. Infect Immun. 2001;69:7349–7355. doi: 10.1128/IAI.69.12.7349-7355.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister GE, Roberts CG, Berzofsky JA, de Groot AS. Two novel T cell epitope prediction algorithms based on MHC-binding motifs; comparison of predicted and published epitopes from Mycobacterium tuberculosis and HIV protein sequences. Vaccine. 1995;13:581–591. doi: 10.1016/0264-410x(94)00014-e. [DOI] [PubMed] [Google Scholar]
- Mendelson M, Walters S, Smith I, Kaplan G. Strain-specific mycobacterial lipids and the stimulation of protective immunity to tuberculosis. Tuberculosis. 2005;85:407–413. doi: 10.1016/j.tube.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Rammensee HG. Chemistry of peptides associated with MHC class I and class II molecules. Curr Opin Immunol. 1995;7:85–96. doi: 10.1016/0952-7915(95)80033-6. [DOI] [PubMed] [Google Scholar]
- Redelman-Sidi G, Sepkowitz KA. IFN-γ release assays in the diagnosis of latent tuberculosis infection among immunocompromised adults. Am J Respir Crit Care Med. 2013;188:422–431. doi: 10.1164/rccm.201209-1621CI. [DOI] [PubMed] [Google Scholar]
- Richeldi L, Losi M, D’Amico R, Luppi M, Ferrari A, Mussini C, Codeluppi M, Cocchi S, Prati F, Paci V, Meacci M, Meccugni B, Rumpianesi F, Roversi P, Cerri S, Luppi F, Ferrara G, Latorre I, Gerunda GE, Torelli G, Esposito R, Fabbri LM. Performance of tests for latent tuberculosis in different groups of immunocompromised patients. Chest. 2009;136:198–204. doi: 10.1378/chest.08-2575. [DOI] [PubMed] [Google Scholar]
- Schroers R, Huang XF, Hammer J, Zhang J, Chen SY. Identification of HLA DR7-restricted epitopes from human telomerase reverse transcriptase recognized by CD4+ T-helper cells. Cancer Res. 2002;62:2600–2605. [PubMed] [Google Scholar]
- Shinnick TM. The 65-kilodalton antigen of Mycobacterium tuberculosis. J Bacteriol. 1987;169:1080–1088. doi: 10.1128/jb.169.3.1080-1088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soborg B, Ruhwald M, Hetland ML, Jacobsen S, Andersen AB, Milman N, Thomsen VO, Jensen DV, Koch A, Wohlfahrt J, Ravn P. Comparison of screening procedures for Mycobacterium tuberculosis infection among patients with inflammatory diseases. J Rheumatol. 2009;36:1876–1884. doi: 10.3899/jrheum.081292. [DOI] [PubMed] [Google Scholar]
- Stewart GR, Robertson BD, Young DB. Tuberculosis: a problem with persistence. Nat Rev Microbiol. 2003;1:97–105. doi: 10.1038/nrmicro749. [DOI] [PubMed] [Google Scholar]
- Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F, Hammer J. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- Sun Y, Iglesias E, Samri A, Kamkamidze G, Decoville T, Carcelain G, Autran B. A systematic comparison of methods to measure HIV-1 specific CD8 T cells. J Immunol Methods. 2003;272:23–34. doi: 10.1016/s0022-1759(02)00328-9. [DOI] [PubMed] [Google Scholar]
- Tanghe A, Lefèvre P, Denis O, D’Souza S, Braibant M, Lozes E, Singh M, Montgomery D, Content J, Huygen K. Immunogenicity and protective efficacy of tuberculosis DNA vaccines encoding putative phosphate transport receptors. J Immunol. 1999;162:1113–1119. [PubMed] [Google Scholar]
- Wilkinson KA, Newton SM, Stewart GR, Martineau AR, Patel J, Sullivan SM, Herrmann JL, Neyrolles O, Young DB, Wilkinson RJ. Genetic determination of the effect of post-translational modification on the innate immune response to the 19 kDa lipoprotein of Mycobacterium tuberculosis. 93BMC Microbiol. 2009;9 doi: 10.1186/1471-2180-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Lu W, Samri A, Costagliola D, Schnuriger A, Silva BC, Blanc C, Larsen M, Theodorou I, Rouzioux C, Autran B, ALT-ANRS-CO15 study group Distinct differentiation profiles of HIV-Gag and Nef-specific central memory CD8+ T cells associated with HLA-B57/5801 and virus control. AIDS. 2010;24:2323–2329. doi: 10.1097/QAD.0b013e32833e5009. [DOI] [PubMed] [Google Scholar]
- Zhang H, Qiu Z, Jiao Y, Wang A, Li T. HIV-specific CD8+ T cell responses to HXB2 Gag and Nef peptide pools in Chinese HIV/AIDS patients. Sci China C Life Sci. 2009;52:841–846. doi: 10.1007/s11427-009-0117-1. [DOI] [PubMed] [Google Scholar]
- Zhang S, Shao L, Mo L, Chen J, Wang F, Meng C, Zhong M, Qiu L, Wu M, Weng X, Zhang W. Evaluation of gamma interferon release assays using Mycobacterium tuberculosis antigens for diagnosis of latent and active tuberculosis in Mycobacterium bovis BCG-vaccinated populations. Clin Vaccine Immunol. 2010;17:1985–1990. doi: 10.1128/CVI.00294-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


