ABSTRACT
Mason-Pfizer monkey virus (M-PMV) is a prototypical betaretrovirus responsible for simian AIDS (SAIDS) in rhesus macaques. It has been shown previously that mouse cells are resistant to infection by HIV-1 and other primate lentiviruses. However, the susceptibility of mouse cells to primate retroviruses such as M-PMV remains unexplored. In the present study, using single-round green fluorescent protein (GFP) reporter viruses, we showed that various mouse cell lines are unable to support the early stages of M-PMV replication. The block to infection occurs postentry and is independent of the viral envelope. Using quantitative real-time PCR, we showed that the block to infection occurs after reverse transcription but before formation of circular DNA or proviral DNA. Finally, we showed that the M-PMV block in mouse cells is not attributable to the previously characterized mouse restriction factor Fv1. Overall, these findings suggest that mouse cells exhibit a previously uncharacterized block to M-PMV infection.
IMPORTANCE Here we document a novel postentry restriction to M-PMV infection in mouse cells. The block occurs after reverse transcription but before the formation of circular or proviral DNA and is independent of the previous characterized mouse restriction factor Fv1.
INTRODUCTION
Retroviruses are capable of infecting diverse vertebrates, and successful infection requires cellular factors to support each phase of the retroviral life cycle. Retroviral tropism is therefore partly dictated by whether the infected cell possesses the necessary cofactors to assist viral replication. For example, human immunodeficiency virus type 1 (HIV-1) infects human T cells and macrophages efficiently but is unable to infect mouse cells. Multiple steps in the viral life cycle are blocked in mouse cells. First, the mouse homologues of the human T cell receptors and coreceptors CD4, CXCR4, and CCR5 (1, 2) cannot bind HIV env-encoded gp120, preventing HIV-1 from entering the cell. This entry block can be bypassed in engineered mouse cell lines expressing the human T cell receptors (3, 4) or by pseudotyping HIV-1 virions with foreign viral envelope proteins such as glycoprotein G of vesicular stomatitis virus (VSV-G). Upon establishment of the provirus in mouse cells, the late phase of the HIV-1 life cycle is hindered by additional cellular blocks. Transcription initiated in the long terminal repeat (LTR) is defective due to the incompatibility of the HIV-1 Tat transactivator with mouse cyclin T1 (5–8). In addition, viral assembly is defective due to the absence of necessary factors required for late stages of the retroviral life cycle, a phenotype that can be rescued by the fusion of uninfected human cells with infected mouse cells (9–11). Study of blocks to HIV-1 replication in mouse cells has led to the identification and characterization of critical cellular factors necessary for infection.
Species-specific retroviral tropism is also governed by the presence of species-specific restriction factors. For example, human and primates are known to be resistant to N-tropic murine leukemia virus (MLV) infection (12, 13). The human protein mediating this restriction was identified as TRIM5α (14). In addition to restricting N-tropic MLV, rhesus macaque TRIM5α has the remarkable ability to restrict HIV-1 infection (15, 16). This observation was later attributed to species-specific variations in the TRIM5α sequence (17–19), supporting the notion that species-specific restriction factors are a major determinant of retroviral tropism. Mechanistically, TRIM5α has been proposed to interact with the viral capsid (CA) (20), leading to premature dissociation of viral cores (21) and terminating infection prior to nuclear entry (16). In mice, the Friend virus susceptibility factor 1 (Fv1) gene functions similarly to TRIM5α. Fv1 encodes a gag-like protein with homologies to the ERV-L family of endogenous retroviruses (22). There are two major naturally occurring alleles of Fv1. NIH Swiss mice carry the Fv1n allele, which restricts the replication of B-tropic MLVs but not N-tropic MLVs. In contrast, BALB/c mice carry the Fv1b allele, which confers resistance to N-tropic MLV infection but not B-tropic MLVs (23, 24). A few laboratory mice and some wild mice carry a third Fv1 allele, Fv1nr, which restricts the replication of B-tropic MLVs and a subset of N-tropic MLVs (25, 26).
Mason-Pfizer monkey virus (M-PMV) is a prototypical betaretrovirus capable of causing simian AIDS (SAIDS) in rhesus macaques (27). Though mouse cells are known to be resistant to lentiviral infections, it was not known whether they would also be resistant to M-PMV. In the present study, we showed that M-PMV-based vectors are unable to transduce various mouse cell lines. Mouse resistance to M-PMV infection is envelope independent, with the strongest block occurring after reverse transcription but before the formation of circular or proviral DNAs. The block to M-PMV infection is not attributable to the Fv1 gene, and additional tests ruled out the possibility of transcriptional silencing of the M-PMV LTR in mouse cells. Taken together, the results demonstrate that infection by M-PMV is strongly blocked in mouse cells during the early phase of the life cycle, most likely at the time of import of the viral DNA into the nucleus.
MATERIALS AND METHODS
Cell culture.
HEK-293T, HeLa, COS-7, L929, NIH 3T3, and RAW264.7 cells, mouse embryonic fibroblasts (MEFs), and Mus dunni tail fibroblasts (MDTFs) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, 1,000 U/ml of penicillin, and 100 mg/ml of streptomycin. Ba/F3 cells were cultured in RPMI medium supplemented with 10% heat-inactivated FBS and 1.6 ng/ml of recombinant interleukin 3 (IL-3). All cells were maintained in a 37°C incubator with 5% CO2.
Plasmids.
Plasmids pCIG3-B and pCMV-intron express gag and pol from B-tropic (28) and NB-tropic (29) MLVs, respectively. pMD.G expresses the vesicular stomatitis virus (VSV) envelope glycoprotein. pNCA-GFP is a replication-defective single-round MLV vector described previously (30). pNCA-Luc is a firefly luciferase reporter virus in which the green fluorescent protein (GFP) cassette in pNCA-GFP is replaced with the firefly luciferase gene via BamHI/XhoI restriction sites. pSARM-EGFP is a replication-defective single-round M-PMV vector in which the env gene has been replaced with the gene for enhanced GFP (EGFP) as described previously (31). pSARM-Luc is a firefly luciferase reporter virus in which the GFP cassette in pSARM-GFP has been replaced with the firefly luciferase gene via XhoI/BlpI restriction sites. pCI-Fv1n and pCI-Fv1b express the n and b alleles of the mouse Fv1 gene as described previously (22). pHA-mCAT is a plasmid encoding the mouse cationic amino acid transporter (mCAT), which serves as a receptor for MLV ecotropic envelope (gift from Walther Mothes, Yale University School of Medicine). MLV-LTR-Luc and MPMV-LTR-Luc are constructs expressing luciferase under the control of the viral LTRs. The U3-R-U5 regions of the respective viral LTRs were amplified by PCR using the following primer pairs: for MLV, 5′-AGATCTGCGATCTAAGTAAGCTTAATGAAAGACCCCACCTGTAGGT-3′ and 5′-CCAACAGTACCGGAATGCCAAGCTTGGTGGTCCCTGGGCAGGG-3′, and for M-PMV, 5′-AGATCTGCGATCTAAGTAAGCTTTGTCCGGAGCCGTGCTG-3′ and 5′-CCAACAGTACCGGAATGCCAAGCTTACTGTCCCGACCCGCGG-3′. The PCR products were cloned into the HindIII site of pGL3-Basic vector (Promega) using sequence- and ligation-independent cloning (32). All constructs were verified by DNA sequencing.
Retroviral transduction assay.
M-PMV–GFP reporter viruses were produced by transfection of 293T cells with 8 μg of pSARM-EGFP and 8 μg of pMD.G per 100-mm plate using polyethylenimine (PEI). Reporter viruses were harvested 48 h later, filtered (0.45 μm), and used directly for transduction assays. Similarly, NB-tropic MLV-GFP reporter viruses were produced by 293T cell transfection with 8 μg of pNCA-GFP, 4 μg of pCMV-intron, and 4 μg of pMD.G DNAs. For transduction assays, the desired cell types were seeded in 12-well plates at a density of 1 × 105 cells per well and infected with GFP reporter viruses for 5 h at 37°C. Forty-eight hours postinfection, cells were trypsinized, diluted using flow cytometry buffer (PBS with 1% BSA), and subjected to flow cytometry using an automated cell analyzer (LSRII; BD Bioscience). For Fv1 overexpression experiments, HeLa cells were transfected with 0.5 μg of pCI-Fv1n or pCI-Fv1b together with 0.1 μg of pHA-mCAT for 24 h, followed by transduction experiments outlined above using MLV or M-PMV luciferase reporter viruses.
Quantitative real-time PCR analysis of viral replication intermediates.
At various time points following infection, cells were washed with PBS, and total DNA was isolated using a Qiagen DNeasy kit according to the manufacturer's instructions. For analysis of early and late reverse transcription (RT) intermediates, 80 ng of total DNA was combined with M-PMV-specific TaqMan primer/probe sets targeting minus-strand strong-stop DNA and GFP, respectively, as described previously (33). For the detection of 2-LTR circles, 80 ng of total DNA was mixed with SYBR green PCR master mix (Roche) containing 15 pmol of both forward (5′-TCCTCCAGGTTCCTACTGTT-3′) and reverse (5′-ACGGAGAAGAACCAGGAAATAC-3′) primers. PCRs were performed in 96-well plates using a 7900 Fast real-time PCR system (Applied Biosystems) with the following reaction conditions: 10 min at 95°C, followed by 40 cycles of 30 s at 95°C and 1 min at 60°C. For all reactions, relative expression was quantified using the threshold cycle (2−ΔΔCT) method (34) normalized to the human and mouse Tert genes.
Dual-luciferase assay.
HeLa cells (1 × 105 cells/well) and NIH 3T3 and L929 cells (2 × 105 cells/well) were seeded in a 12-well plate 1 day before transfection. The next day, each well was transfected with 1 μg of various LTR firefly luciferase constructs and 10 ng of herpes simplex virus thymidine kinase (HSV-TK) Renilla luciferase control plasmid using Lipofectamine 2000 (Invitrogen) at a 1:4 ratio. Thirty-six hours later, cells were lysed in 100 μl of 1× reporter lysis buffer (Promega) and luciferase levels were quantified using a POLARstar Omega multimode plate reader (BMG Labtech) according to the manufacturer's instructions. Relative luciferase expression was calculated by first dividing the firefly luciferase signal by the Renilla luciferase signal in a given cell, and the resulting ratio was then normalized to the ratio obtained from HeLa cells transfected with the simian virus 40 (SV40)-firefly luciferase construct and HSV-TK Renilla luciferase (set to 1).
RESULTS
M-PMV is unable to transduce mouse cells.
To determine whether mouse cells are susceptible to M-PMV infection, we utilized a single-round M-PMV reporter genome in which the viral env gene was replaced with the GFP gene (31). Virions capable of infecting a wide range of cell lines were prepared by collecting culture medium after transfection of 293T cells with the M-PMV reporter genome and a DNA encoding the VSV-G envelope protein. To assess the susceptibility of mouse cells to M-PMV infection, NIH 3T3 cells were infected with VSV-G-pseudotyped M-PMV-GFP, and the efficiency of infection was monitored by flow cytometry analysis 48 h later. HeLa cells, previously shown to be permissive to M-PMV (33), were infected in parallel. As positive controls, we also transduced both cell lines with VSV-G-pseudotyped MLV-GFP viruses. While NIH 3T3 and HeLa cells were equally and highly susceptible to MLV infection, NIH 3T3 cells showed a 100-fold reduction in the formation of M-PMV GFP+ cells compared to that by HeLa cells (Fig. 1A). Next, we extended our analysis of M-PMV infectivity to other mouse cell lines, including MEFs, MDTFs, and L929, RAW264.7, and Ba/F3 cells (Fig. 1B). These cell lines were selected on the basis of differences in their genetic backgrounds and variations in the Fv1 allele, as well as cell type (Table 1). Interestingly, while all cell lines supported MLV replication to similar levels, M-PMV transduction was potently blocked in all the mouse cell lines tested (Fig. 1B). Since our reporter viruses were pseudotyped with VSV-G, the observed effect is not likely due to defective viral entry. Moreover, the fact that the cells were susceptible to MLV indicates that they were dividing adequately for infection by viruses that require division. These results suggest that mouse cells broadly exhibit a postentry block to M-PMV infection.
FIG 1.
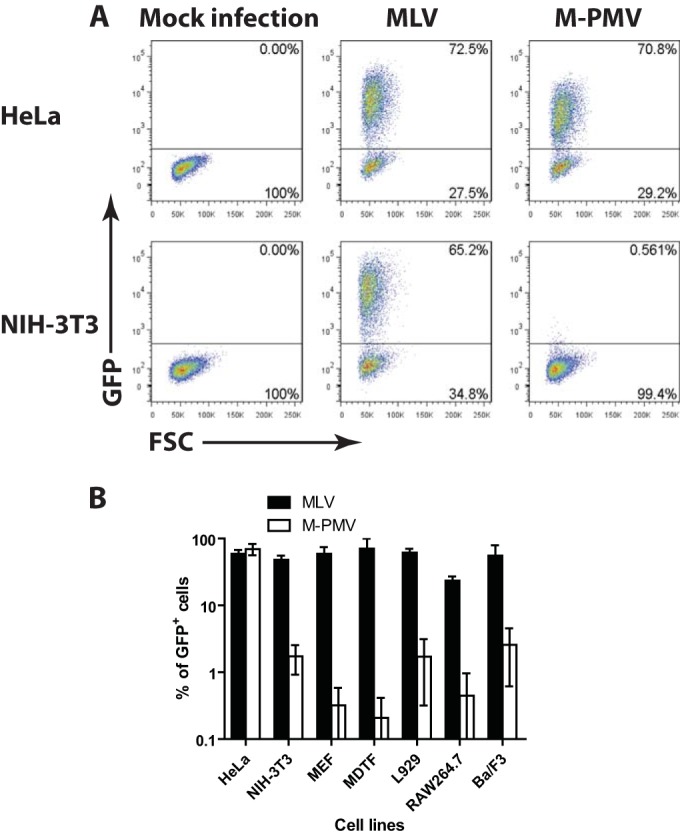
M-PMV is unable to transduce mouse cells. (A) Flow cytometry analysis of HeLa or NIH 3T3 cells infected with VSV-G-pseudotyped single-round MLV or M-PMV–GFP reporter viruses 2 days postinfection. The x axis shows forward scatter (FSC), and the y axis shows GFP intensity. One representative experiment of five independent experiments is shown. (B) Experiment similar to that in panel A examining the susceptibility of other mouse cell lines to MLV and M-PMV–GFP infection. A logarithmic graph shows the percentage of GFP+ cells 2 days postinfection. Results shown are means ± SD from two independent experiments.
TABLE 1.
Mouse cell lines used in this study
| Cell line | Mouse strain | Fv1 genotype | Cell type |
|---|---|---|---|
| NIH 3T3 | NIH Swiss | n/n | Fibroblast |
| MEF | C57BL/6 | b/b | Fibroblast |
| MDTF | Mus dunni | −/− | Fibroblast |
| L929 | C3H | n/n | Fibroblast |
| RAW267.4 | BALB/c | b/b | Macrophage |
| Ba/F3 | C3H | n/n | B cell |
M-PMV replication block in mouse cells is not envelope dependent.
Different viral envelopes mediate virus entry into the target cell via distinct mechanisms. For example, VSV-G promotes viral entry via endocytosis, while the amphotropic (ampho) MLV envelope may facilitate direct fusion with the plasma membrane (35) and has very recently been suggested to promote entry into some cells by macropinocytosis (36). In the case of the ecotropic (eco) MLV envelope, there exists evidence consistent with both endocytosis (i.e., pH dependence) and direct cell fusion (i.e., pH independence) (37, 38). To evaluate whether the M-PMV block in mouse cells is dependent on the route of entry, we repeated the experiment outlined above using M-PMV–GFP reporter viruses pseudotyped with either MLV ecotropic or amphotropic envelopes. As controls, infections with pseudotyped MLV-GFP reporter viruses were carried out in parallel. M-PMV–GFP–VSV-G and M-PMV–GFP–ampho were able to infect the permissive HeLa cells efficiently, with undiluted virus preparations resulting in 64% and 6.5% GFP+ cells, respectively (Fig. 2A). The reduced titer of the M-PMV–GFP–ampho virus was presumably due to decreased stability of the amphotropic envelope compared to that of VSV-G. The M-PMV–GFP–eco virus was unable to infect HeLa cells, consistent with the fact that the MLV ecotropic glycoprotein cannot bind the human homologue of the mouse ecotropic MLV receptor (mouse cationic amino acid transporter [mCAT]) (39) (Fig. 2A). In contrast to HeLa cells, NIH 3T3 cells were resistant to all three pseudotyped M-PMV reporter viruses (Fig. 2A). As expected, pseudotyping MLV with each of the three envelopes conferred infectivity to NIH 3T3 cells (Fig. 2B). Overall, these findings suggest that the M-PMV block in mouse cells occurs in an envelope-independent manner.
FIG 2.
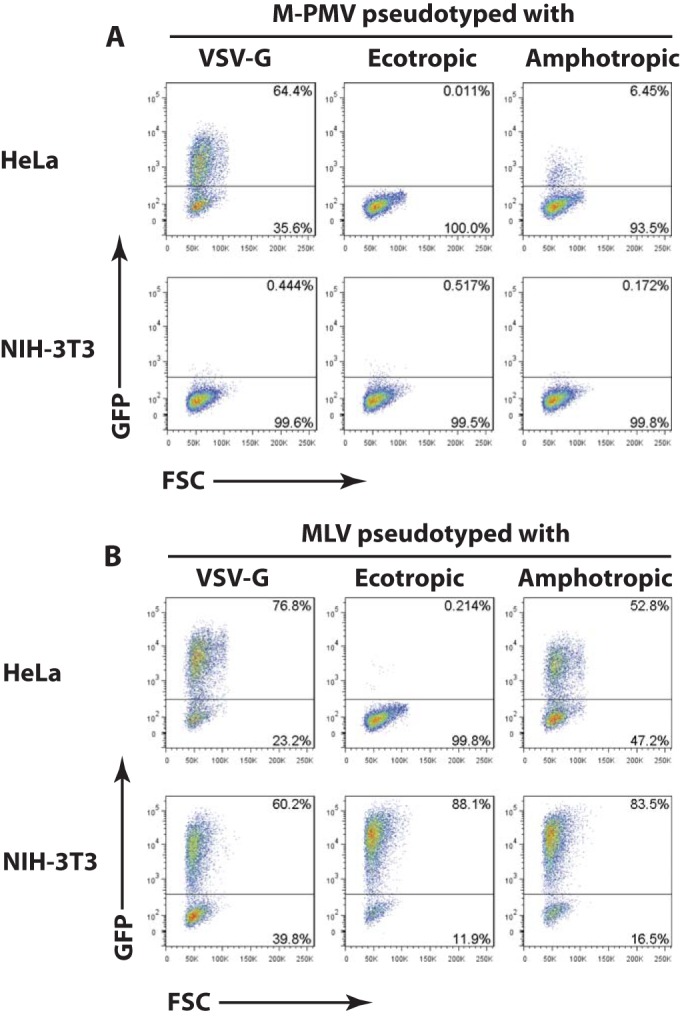
M-PMV replication block in mouse cells is not envelope dependent. (A) Flow cytometry analysis of HeLa or NIH 3T3 cells infected with M-PMV–GFP reporter virus pseudotyped with either VSV-G or the envelope of ecotropic or amphotropic MLV. Results shown are obtained 2 days postinfection (p.i). The x axis shows forward scatter (FSC), and the y axis shows GFP intensity. One representative experiment of two independent experiments is shown. (B) Experiment similar to that in panel A, infecting HeLa or NIH 3T3 cells with MLV-GFP reporter viruses pseudotyped with VSV-G, ecotropic MLV, or amphotropic MLV. Results shown are obtained 2 days postinfection. The x axis shows forward scatter (FSC), and the y axis shows GFP intensity. One representative experiment of two independent experiments is shown.
Characterizing the M-PMV block in mouse cells.
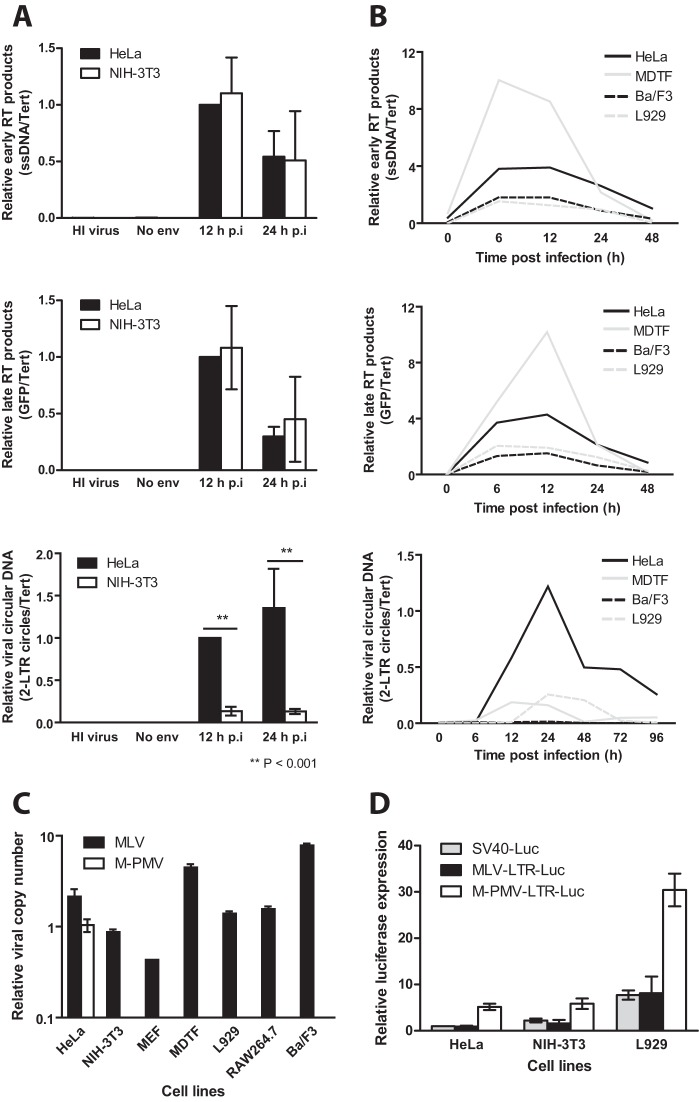
Our single-round GFP reporter viruses score for all the early events of infection, including reverse transcription, nuclear entry, integration, and reporter gene expression. To define the step of M-PMV infection blocked in mouse cells, NIH 3T3 or HeLa cells were infected with M-PMV–GFP, total DNA was harvested at 12 or 24 h postinfection, and the formation of viral replication intermediates was monitored by quantitative real-time PCR (Fig. 3A). At both 12 and 24 h postinfection, HeLa and NIH 3T3 cells showed comparable levels of early (minus-strand strong-stop DNA) and late (GFP) reverse transcription (RT) intermediates, suggesting that reverse transcription occurred normally in NIH 3T3 cells (Fig. 3A, top and middle graphs). Strikingly, at both 12 and 24 h postinfection, NIH 3T3 cells showed a 10-fold reduction in 2-LTR circle formation compared to that of HeLa cells, implying a defect in the formation of viral circular DNA (Fig. 3A, bottom graph). To extend these observations, we performed similar kinetic studies of viral DNA formation using MDTFs and Ba/F3 and L929 cells (Fig. 3B). All cell lines showed comparable and high levels of early and late viral DNAs. For example, MDTFs showed a 2-fold increase in early and late RT products at 12 h postinfection, while L929 and Ba/F3 cells showed a 2- to 3-fold reduction, compared to HeLa cells (Fig. 3B, top and middle graphs). However, as in NIH 3T3 cells, 2-LTR circle formation was profoundly reduced in all three mouse cell lines, albeit to various degrees (8- to 40-fold for MDTFs, 60- to 200-fold for Ba/F3 cells, and 5- to 50-fold for L929 cells) (Fig. 3B, bottom graph). Taken together, our data indicate that although there were subtle variations in the efficiency of M-PMV reverse transcription, a more potent block in 2-LTR circle formation was observed across all the mouse cell lines tested.
FIG 3.
Characterizing the M-PMV block in mouse cells. (A) HeLa or NIH 3T3 cells were infected with VSV-G-pseudotyped M-PMV–GFP reporter virus. To control for potential plasmid DNA carryover in the viral supernatant, heat-inactivated (HI) virus and M-PMV–GFP with no envelope (“No env”) were used in parallel. Total DNA from infected cells was isolated at indicated time points, followed by real-time quantitative PCR to quantify the levels of viral replication intermediates. The graph shows detection of early RT products (minus-strand strong-stop DNA), the middle graph shows detection of late RT products (GFP), and the bottom graph shows detection of viral circular DNA (2-LTR circles). Levels of various viral replication intermediates were first normalized using the 2−ΔΔCT method to the value for the human or mouse Tert gene; the obtained values were then normalized to the HeLa cell 12-h time point (set to 1). Results shown are means ± SEMs from three independent experiments performed in triplicate. Student's t test was used for statistical analysis. **, P < 0.001. (B) Viral kinetic study similar to that in panel A using additional mouse cell lines. (C) Various mouse cell lines were infected with either VSV-G pseudotyped MLV or M-PMV-GFP reporter viruses for 5 h at 37°C. Infected cells were then propagated for 21 days, after which genomic DNA was extracted and real-time quantitative PCR was carried out using primers specific for GFP. The extent of viral integration (relative viral copy number) was quantified using the 2−ΔΔCT method by normalizing the GFP signal to the human or mouse Tert gene. Results shown are means ± SD from two independent experiments performed in triplicate. (D) Indicated cell lines were transfected with SV40 promoter-, MLV LTR-, or M-PMV LTR-driven firefly luciferase reporter plasmid for 36 h. Relative luciferase expression was calculated by first dividing the firefly luciferase signal by the Renilla luciferase signal in a given cell, and the resulting ratio was then normalized to the ratio obtained from HeLa cells transfected with the SV40 firefly luciferase construct and HSV-TK Renilla luciferase (set to 1). Results shown are means ± SEMs from three independent experiments performed in duplicate.
We extended the above-described findings by analyzing the extent of viral integration following infection. Human and mouse cell lines were infected with MLV or M-PMV vectors, as outlined for Fig. 1B, and passaged for 21 days in culture, after which genomic DNA was extracted and viral DNA was measured by quantitative real-time PCR using GFP-specific primers. MLV-GFP proviral DNA was readily detectable and at comparable levels in all cell lines tested (Fig. 3C), consistent with the flow cytometry data showing similar levels of MLV transduction among these cells (Fig. 1B). In contrast, while integrated M-PMV–GFP provirus was readily detected in HeLa cells, no M-PMV proviral DNA was detected in any of the mouse cell lines (Fig. 3C). This is consistent with the observed defects in circular DNA formation and suggestive of a block to nuclear entry preceding integration (Fig. 3A and B, bottom graphs). Finally, to rule out the possibility that the M-PMV LTR is transcriptionally silent in mouse cells, we performed luciferase reporter assays with HeLa, NIH 3T3, and L929 cells after transfection with plasmids encoding firefly luciferase under the control of either the SV40 promoter, the MLV LTR, or the M-PMV LTR. A thymidine kinase (TK) promoter-driven Renilla luciferase plasmid was cotransfected for normalization. Transfected cells were harvested 36 h later, and lysates were subjected to a dual-luciferase assay. The M-PMV LTR directed high-level expression of firefly luciferase in NIH 3T3 and L929 cells, compared to the levels expressed from the SV40 promoter or the MLV LTR (Fig. 3D). This finding suggests that there are no blocks to M-PMV infection in mouse cells at the level of viral gene expression.
Fv1 does not contribute to M-PMV restriction in mouse cells.
The timing of the M-PMV block before nuclear entry partially overlaps the timing of the previously described Fv1 restriction. Among inbred mouse strains, two major naturally occurring Fv1 alleles exist. NIH Swiss mice carry the Fv1n allele, which blocks the replication of B-tropic MLVs and not N-tropic MLVs; BALB/c mice carry the Fv1b allele, restricting N-tropic MLVs and not B-tropic MLVs. To explore the potential antiviral activities of mouse Fv1 against M-PMV, HeLa cells were transfected with plasmids expressing either the N-tropic or B-tropic forms of the Fv1 protein and the MLV ecotropic receptor mCAT. Twenty-four hours later, cells were challenged with either B-tropic MLV-eco–luciferase or M-PMV-eco–luciferase, and they were scored by luciferase assay 48 h later to monitor the extent of restriction. This experimental design ensures that only cells expressing the Fv1 allele are infected, thereby minimizing background signals. As expected, no infection of HeLa cells lacking the mCAT-expressing plasmid was observed. Introduction of Fv1n, but not Fv1b, reduced B-tropic MLV infection 10-fold (Fig. 4). Overexpression of either Fv1n or Fv1b failed to confer resistance to M-PMV infection. This phenotype is consistent with our above-described observations that mouse cells carrying Fv1n (NIH 3T3, L929, and Ba/F3 cells) or Fv1b (MEFs and RAW264.7 cells), as well as Fv1-null MDTFs, all restricted M-PMV (Fig. 1B).
FIG 4.
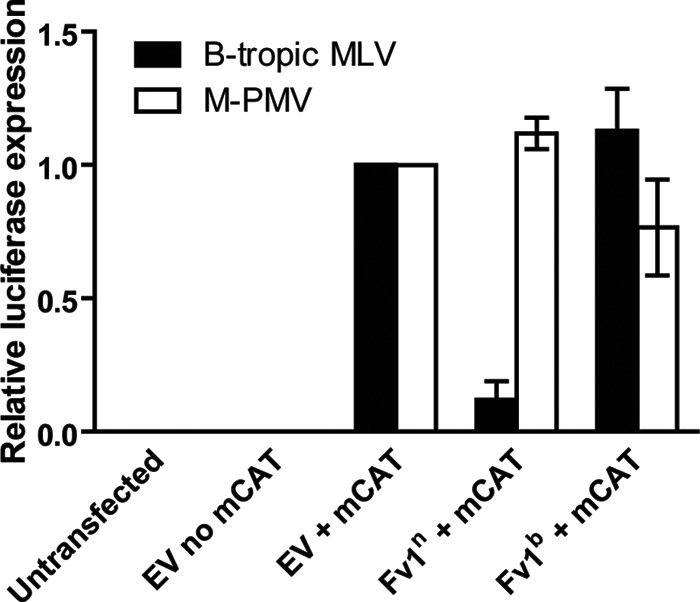
Fv1 does not contribute to M-PMV restriction in mouse cells. HeLa cells were transfected with the indicated plasmids for 24 h prior to infection with the indicated luciferase reporter viruses. Infected cells were harvested 48 h postinfection and subjected to luciferase assay to quantify the extent of infection. Luciferase units obtained from B-tropic MLV or M-PMV infections were then normalized to values obtained from infection of empty vector (EV) plus mCAT (set to 1). Results shown are means ± SEMs from three independent experiments.
DISCUSSION
Mouse cells do not support the replication of HIV-1 due to multiple blocks acting at the levels of entry (3, 4), gene expression (5–8), and viral assembly (9–11). In this study, we showed that mouse cells are resistant to infection by M-PMV, a prototypical simian betaretrovirus. Resistance to M-PMV appears to be widespread, seen among all mouse cell types tested, including hematopoietic B cells (Ba/F3 cells), macrophages (RAW247.6 cells), and fibroblasts (NIH 3T3 and L929 cells, MEFs, and MDTFs) (Fig. 1). The observation that mouse cells blocked M-PMV reporter viruses pseudotyped with various envelope proteins suggests that the block occurs postentry (Fig. 2). Moreover, the process of reverse transcription, as measured by the levels of minus-strand strong-stop DNA and the reporter GFP DNA, occurred normally in all the mouse cell lines tested (Fig. 3A and B, top and middle graphs). The levels of 2-LTR circles, however, were drastically reduced, suggesting a significant block at the time of nuclear entry (Fig. 3A and B, bottom graphs). This conclusion is further supported by diminished levels of integrated proviral DNA remaining in mouse cells after long-term passage (Fig. 3C). It is not known if infection of permissive cells by M-PMV requires cell division as is the case for MLVs, but the cells used in all our experiments were dividing, ensuring that this is not the basis for the block. Given that the defect in viral circular DNA formation partially coincides with the timing of Fv1 restriction, we hypothesized that the Fv1 alleles present in the various mouse lines might account for the block in M-PMV infection. But overexpression of the Fv1n or Fv1b alleles in human cells failed to induce any resistance to M-PMV (Fig. 4). This observation was consistent with the inability of M-PMV to transduce mouse cells harboring different Fv1 alleles (Table 1), making Fv1 an unlikely candidate for M-PMV restriction. Interestingly, it has been reported that Fv1 also failed to block primate lentiviruses (40). Of note, although our data suggest a major defect at the approximate time of nuclear entry, we cannot exclude the possibility that additional blocks at the level of integration per se may also exist in mouse cells.
In summary, the present study revealed a potent block to M-PMV infection occurring after reverse transcription in all mouse cells tested. The inability of M-PMV to replicate in mouse cells may be due either to the absence or incompatibility of critical host factors or to the presence of active viral restriction factors exemplified by Fv1. Infection of heterokaryons formed by the fusion between permissive and nonpermissive cells can be used to distinguish between the two possibilities. In preliminary tests, we found that heterokaryons formed by the fusion of HeLa and NIH 3T3 cells remained resistant to M-PMV infection (data not shown), implying that mouse cells may contain a novel dominant restriction factor with Fv1-like activity. We cannot rule out the possibility, however, that the fusion process itself causes virus resistance and that the heterokaryons are resistant for other reasons. Recently, the effects of human and mouse TRIM proteins on the early and late events of HIV and MLV life cycle identified numerous family members with antiviral activities (41). With that in mind, it is possible that a ubiquitously expressed mouse-specific TRIM protein is responsible for the M-PMV resistance. Preliminary tests of a panel of mouse TRIMs expressed in HeLa cells did not reveal any TRIM capable of blocking M-PMV. Identification of the putative dominant mouse restriction factor, if present, remains a subject for future investigations. Finally, although mouse cells are nonpermissive to M-PMV transduction, we note that they are susceptible to infection by other betaretroviruses such as mouse mammary tumor virus (MMTV). It remains to be determined whether this is due to MMTV's ability to escape host restriction or the presence of compatible cofactors necessary for infection in mouse cells.
ACKNOWLEDGMENTS
This work was supported by NCI grant R01 CA 30488 from the National Cancer Institute. S.P.G. is an investigator of the Howard Hughes Medical Institute.
We thank Eric Hunter (Emory University School of Medicine) and Walther Mothes (Yale University School of Medicine) for their generosity with reagents.
REFERENCES
- 1.Atchison RE, Gosling J, Monteclaro FS, Franci C, Digilio L, Charo IF, Goldsmith MA. 1996. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science 274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 2.Landau NR, Warton M, Littman DR. 1988. The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature 334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- 3.Browning J, Horner JW, Pettoello-Mantovani M, Raker C, Yurasov S, DePinho RA, Goldstein H. 1997. Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc Natl Acad Sci U S A 94:14637–14641. doi: 10.1073/pnas.94.26.14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton LK, Hussey RE, Steinbrich R, Ramachandran H, Husain Y, Reinherz EL. 1988. Substitution of murine for human CD4 residues identifies amino acids critical for HIV-gp120 binding. Nature 335:363–366. doi: 10.1038/335363a0. [DOI] [PubMed] [Google Scholar]
- 5.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462. doi: 10.1016/S0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 6.Fujinaga K, Taube R, Wimmer J, Cujec TP, Peterlin BM. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci U S A 96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwak YT, Ivanov D, Guo J, Nee E, Gaynor RB. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J Mol Biol 288:57–69. doi: 10.1006/jmbi.1999.2664. [DOI] [PubMed] [Google Scholar]
- 9.Bieniasz PD, Cullen BR. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J Virol 74:9868–9877. doi: 10.1128/JVI.74.21.9868-9877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mariani R, Rasala BA, Rutter G, Wiegers K, Brandt SM, Krausslich HG, Landau NR. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J Virol 75:3141–3151. doi: 10.1128/JVI.75.7.3141-3151.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariani R, Rutter G, Harris ME, Hope TJ, Krausslich HG, Landau NR. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J Virol 74:3859–3870. doi: 10.1128/JVI.74.8.3859-3870.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Besnier C, Ylinen L, Strange B, Lister A, Takeuchi Y, Goff SP, Towers GJ. 2003. Characterization of murine leukemia virus restriction in mammals. J Virol 77:13403–13406. doi: 10.1128/JVI.77.24.13403-13406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towers G, Bock M, Martin S, Takeuchi Y, Stoye JP, Danos O. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci U S A 97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci U S A 101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yap MW, Nisole S, Lynch C, Stoye JP. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci U S A 101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 17.Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A 102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stremlau M, Perron M, Welikala S, Sodroski J. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol 79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap MW, Nisole S, Stoye JP. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol 15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 20.Sebastian S, Luban J. 2005. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A 103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 23.Goff SP. 1996. Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell 86:691–693. doi: 10.1016/S0092-8674(00)80141-5. [DOI] [PubMed] [Google Scholar]
- 24.Stoye JP. 1998. Fv1, the mouse retrovirus resistance gene. Rev Sci Tech 17:269–277. [DOI] [PubMed] [Google Scholar]
- 25.Pincus T, Hartley JW, Rowe WP. 1971. A major genetic locus affecting resistance to infection with murine leukemia viruses. I Tissue culture studies of naturally occurring viruses. J Exp Med 133:1219–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak CA. 1985. Analysis of wild-derived mice for Fv-1 and Fv-2 murine leukemia-virus restriction loci—a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J Virol 55:281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marx PA, Maul DH, Osborn KG, Lerche NW, Moody P, Lowenstine LJ, Henrickson RV, Arthur LO, Gilden RV, Gravell M, London WT, Sever JL, Levy JA, Munn RJ, Gardner MB. 1984. Simian AIDS: isolation of a type D retrovirus and transmission of the disease. Science 223:1083–1086. doi: 10.1126/science.6695196. [DOI] [PubMed] [Google Scholar]
- 28.Bock M, Bishop KN, Towers G, Stoye JP. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J Virol 74:7422–7430. doi: 10.1128/JVI.74.16.7422-7430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res 23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, Bestor TH. 2010. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin 3:17. doi: 10.1186/1756-8935-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman RM, Hall L, Connole M, Chen GL, Sato S, Yuste E, Diehl W, Hunter E, Kaur A, Miller GM, Johnson WE. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci U S A 103:19134–19139. doi: 10.1073/pnas.0605838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 33.Diehl WE, Stansell E, Kaiser SM, Emerman M, Hunter E. 2008. Identification of postentry restrictions to Mason-Pfizer monkey virus infection in New World monkey cells. J Virol 82:11140–11151. doi: 10.1128/JVI.00269-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.McClure MO, Sommerfelt MA, Marsh M, Weiss RA. 1990. The pH independence of mammalian retrovirus infection. J Gen Virol 71(Part 4):767–773. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen I, Vilhardt F. 26November2014. Macropinocytosis is the entry mechanism of amphotropic murine leukemia virus (A-MLV). J Virol doi: 10.1128/JVI.02343-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mothes W, Boerger AL, Narayan S, Cunningham JM, Young JA. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679–689. doi: 10.1016/S0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 38.Nussbaum O, Roop A, Anderson WF. 1993. Sequences determining the pH dependence of viral entry are distinct from the host range-determining region of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol 67:7402–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimoto T, Yoshimoto E, Meruelo D. 1993. Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J Virol 67:1310–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatziioannou T, Cowan S, Bieniasz PD. 2004. Capsid-dependent and -independent postentry restriction of primate lentivirus tropism in rodent cells. J Virol 78:1006–1011. doi: 10.1128/JVI.78.2.1006-1011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. 2008. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog 4:e16. doi: 10.1371/journal.ppat.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]