Abstract
UL21 is a conserved protein in the tegument of alphaherpesviruses and has multiple important albeit poorly understood functions in viral replication and pathogenesis. To provide a roadmap for exploration of the multiple roles of UL21, we determined the crystal structure of its conserved N-terminal domain from herpes simplex virus 1 to 2.0-Å resolution, which revealed a novel sail-like protein fold. Evolutionarily conserved surface patches highlight residues of potential importance for future targeting by mutagenesis.
TEXT
Herpesviruses are double-stranded DNA (dsDNA), enveloped viruses that cause lifelong latent infections and ailments ranging in severity from skin lesions to blindness, encephalitis, and cancer (1, 2). A unique feature of all herpesviruses is a multiprotein tegument layer between the capsid and the envelope. Besides being necessary for viral assembly, tegument proteins also play critical roles at early stages of the viral replication cycle, in which some are released to regulate expression of viral (3) or cellular genes (4) while others remain bound to the capsid and mediate its trafficking (5–7). The involvement of tegument proteins at multiple stages during viral replication makes them attractive antiviral targets, yet few have been characterized in detail.
Herpes simplex virus 1 (HSV-1) UL21 is a 535-amino-acid tegument protein that is conserved within the Alphaherpesvirus subfamily (8) and may have analogs among other herpesviruses. UL21 is important for replication in culture because UL21-null mutants of HSV-1 and pseudorabies virus (PRV) show reductions in titer and small plaques (9, 10), whereas HSV-2 cannot replicate without UL21 (8). A lack of UL21 also leads to defects in pathogenesis of PRV in mice and pigs (11–14). UL21 is involved in secondary envelopment (15) and cell-cell spread of mature virions (16) through binding of the tegument proteins UL16 (15, 17) and UL11 (16, 17). The UL21-UL16-UL11 heterotrimer binds the cytoplasmic domain of glycoprotein E (gE), a viral glycoprotein required for viral cell spread (18, 19) and cell-cell fusion of infected cells (20), and regulates these functions (16). UL21 may also have a role in cytosolic capsid transport through association with microtubules (21). In the absence of UL21, expression of viral genes is delayed, possibly due to lower mRNA levels (8, 10). Finally, capsids of UL21-null HSV-2 are unable to undergo nuclear egress (8), suggesting a potential nuclear function for UL21, which localizes not only to the cytoplasm but also to the nucleus (17, 21), specifically the nuclear rim (8, 9, 16).
Although UL21 clearly plays multiple roles in the viral replication cycle, little is known about it due to the lack of sequence homology to other proteins and structural information. Parsing out these functions requires a roadmap in the form of a high-resolution three-dimensional structure. Toward this goal, we determined the crystal structure of the conserved N-terminal domain of UL21 (residues 1 to 216), which we termed UL21N. The crystal structure revealed a novel sail-like fold and evolutionarily conserved features and provides an important framework for elucidating the multiple roles of UL21 in the viral replication cycle and pathogenesis.
Full-length HSV-1 strain 17 UL21 expressed in Escherichia coli underwent spontaneous proteolysis, generating a stable N-terminal fragment (UL21N, residues 1 to 216). The corresponding fragment was amplified from the full-length gene and subcloned into the pET24a vector preceded by an N-terminal StrepII tag and a Gly-Ser linker. UL21N was expressed in Rosetta E. coli (Novagen) using overnight induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C. Cell pellets from 1 liter of culture were resuspended in buffer A [50 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), 100 mM NaCl, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP)] with cOmplete protease inhibitor cocktail (Roche) and egg white avidin (Sigma) and lysed using a French press. UL21N was purified from the clarified lysate with StrepTactin Sepharose resin (GE Healthcare) and size exclusion chromatography (Superdex 200, buffer A) and stored with 1× Halt protease inhibitor cocktail (Pierce).
Crystals were grown by vapor diffusion in hanging drops {2 μl protein at ∼5 mg/ml and 2 μl crystallization solution [0.8 to 1.2 M ammonium sulfate, 100 mM HEPES (pH 7.5), 7 to 10% 2-methyl-2,4-pentanediol (MPD)]} at room temperature and flash frozen in crystallization solution with 15% MPD for data collection. Hg derivative crystals were prepared from native crystals by soaking them in crystallization solution with 1.7 mM thimerosal. X-ray diffraction data were collected at 100 K on beamline X25 at the National Synchrotron Light Source and processed with the HKL2000 software package (22) (Table 1). The experimental phases were obtained using single anomalous dispersion (SAD) data collected for the thimerosal derivative crystals as implemented in the autoSHARP program (23). The model was built manually into the experimental SAD electron density using Coot (24) and improved over several cycles of phase combination and density modification using SHARP (23). The model was refined against native data in the phenix.refine structure refinement program (25) (Table 1), with 5% of the reflections set aside as a reference, and rebuilt in Coot (24). The final Rwork is 0.166, and Rfree is 22.1% (Rwork and Rfree are defined as ∑||Fobs| − |Fcalc||/∑||Fobs| for the reflections in the working or the test set, respectively). There is one UL21N molecule in the asymmetric unit, and the model contains residues 1 to 198 of UL21, the N-terminal StrepII affinity tag, and the Gly-Ser linker. Residues 38 to 46, 76 to 87, and 199 to 216 were not resolved despite being present in the crystals. S0 is the first residue of the β1 strand, and residues QEF of the StrepII tag form a 310 helix.
TABLE 1.
Data collection and refinement statistics
| Parameter | Value for: |
|
|---|---|---|
| Native crystal | Thimerosal-soaked crystal | |
| Data collectiona | ||
| Space group | P6322 | P6322 |
| Cell dimensions | ||
| a, b, c (Å) | 108.35, 108.35, 65.24 | 109.48, 109.48, 65.29 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 41.68-2.05 (2.09-2.05) | 47.41-2.80 (2.90-2.80) |
| Rsym or Rmerge | 0.061 (0.475) | 0.111 (0.651) |
| I/σI | 23.26 (2.52) | 23.96 (5.78) |
| Completeness (%) | 99.78 (99.58) | 100.00 (100.00) |
| Redundancy | 5.3 (5.5) | 18.4 (19.1) |
| Refinement | ||
| Resolution (Å) | 30.81-2.05 | |
| No. of reflections (free) | 14,225 (713) | |
| Rwork/Rfreeb | 0.166/0.221 | |
| No. of atoms | ||
| Protein | 1,643 | |
| Water | 66 | |
| B-factors | ||
| Protein | 51.80 | |
| Water | 49.40 | |
| RMSc deviations | ||
| Bond lengths (Å) | 0.007 | |
| Bond angles (°) | 1.08 | |
| Ramachandran plotd | ||
| Favored (%) | 98.49 | |
| Allowed (%) | 1.51 | |
| Outliers (%) | 0.0 | |
Values in parentheses are for the highest-resolution shell.
Rwork and Rfree are defined as ∑||Fobs| − |Fcalc||/∑||Fobs| for the reflections in the working or the test set, respectively.
RMS, root mean square.
As determined using Molprobity (molprobity.biochem.duke.edu) (34).
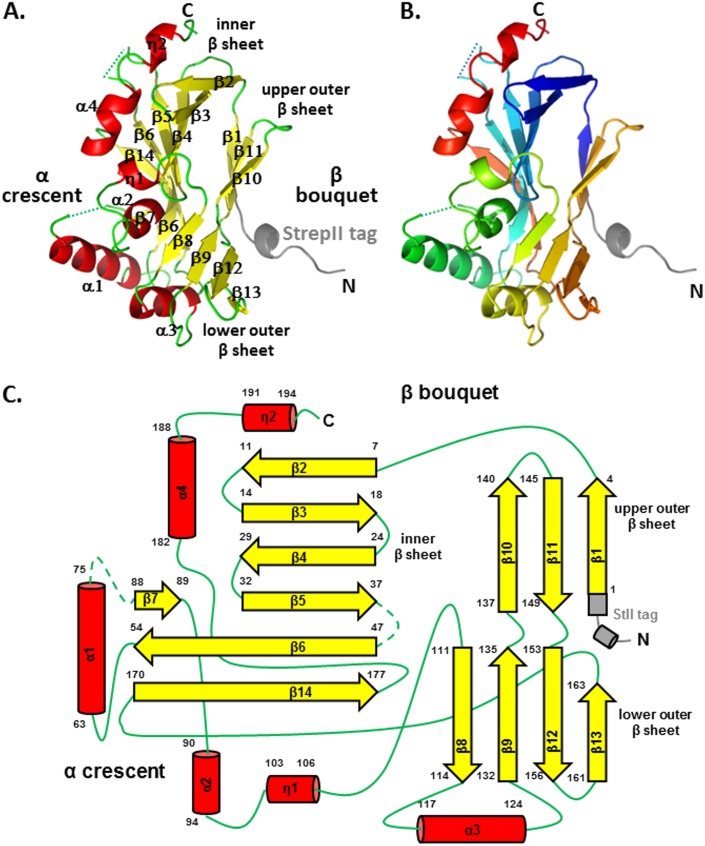
UL21N adopts a single-domain structure of an unusual α/β fold that resembles a wind-filled sail with dimensions of 50 by 30 by 30Å. A DALI search (26) revealed no appreciable overall structural similarity to other proteins. UL21N is composed of two clearly defined “halves,” segregated by the following secondary structure: the oblong β bouquet, which is composed of three antiparallel β sheets (β1 to β14), and the α crescent, which is composed of four α-helices (α1 to α4) and two 310 helices (η1 and η2) that are arranged along one face of the molecule (Fig. 1). Short 310 helix η2 and α-helix α4 look like one long helix broken by completely conserved G190 (Fig. 2). The β bouquet consists of three antiparallel β sheets oriented at an angle to the longest axis of UL21N, creating the appearance of a bouquet arranged with the following topology: an inner seven-stranded β sheet [β2-β3-β4-β5-(β7)-β6-β14], a three-stranded upper outer β sheet (β1-β11-β10), and a four-stranded lower outer β sheet (β8-β9-β12-β13) (Fig. 1). Strand β5 is shorter than the neighboring strand β6 and is effectively extended by the strand β7 such that both β5 and β7 form hydrogen bonds along the same margin of strand β6 (Fig. 1A and C). The secondary structure of UL21N matches predictions (27), except that neither strands β1 and β8 nor 310 helices η1 and η2 were predicted, long strand β14 was predicted to be a helix, and helix α2 was predicted to be a strand.
FIG 1.
UL21N structure. The crystal structure of UL21N is shown in color based on secondary structure (A) and in rainbow coloring (B), from blue (N terminus) to red (C terminus). The StrepII tag is shown in gray. Residues unresolved in the structure are shown as dashed lines. (C) Topology diagram, colored as described for panel A. Secondary structure elements are numbered sequentially, and their amino acid boundaries are given. Helices are show as cylinders, strands as arrows, loops as solid lines, and unresolved loops as dashed lines. All structures were made using Pymol (http://www.pymol.org).
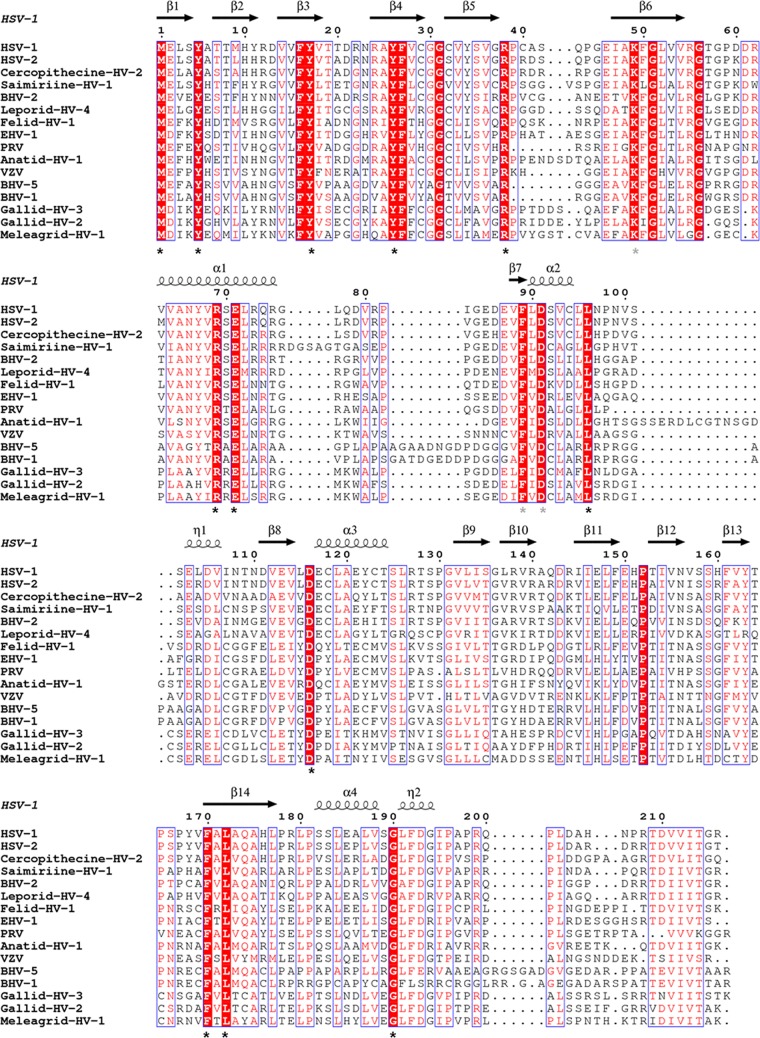
FIG 2.
Multiple-sequence alignment of UL21 homologs from 16 alphaherpesviruses. Sequence alignment was generated and analyzed using ClustalW (32) and ESPRIPT (33) using sequences from GenBank accession numbers ACM62243, AEV91359, AAU88086, ADO13807, AAK69349, AFR32463, ACT88337, AAT67298, AAA47475, ABO26208, AAT07796, AAR86141, CAA88112, AEI00223, AAF66756, and AAG45758. Only the alignment of residues corresponding to residues 1 to 216 of HSV UL21N is shown. The secondary structure of HSV-1 UL21 is shown above the aligned sequences. Similar residues are shown in red text. Identical residues are boxed in red with white text, and those exposed on the surface of UL21N are marked by asterisks. Gray asterisks identify conserved residues that are surface exposed in the model but are likely obscured in the protein by the unresolved loop containing residues 76 to 87.
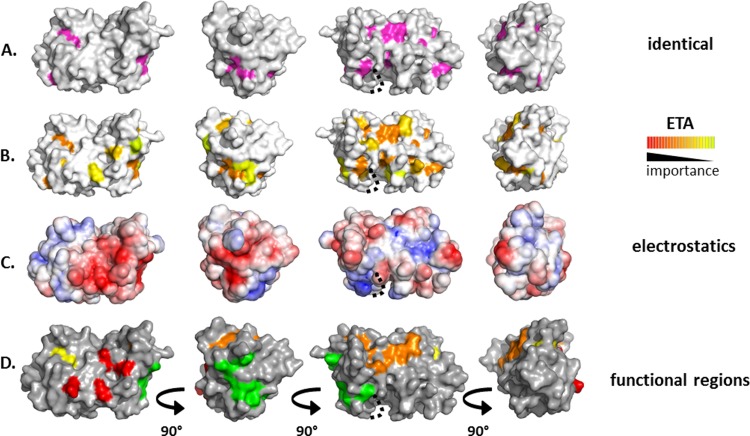
UL21 is 5.2% identical among 16 alphaherpesviruses, and 21 of 28 identical residues are within UL21N (Fig. 2). Twelve of these are surface exposed (Fig. 2 and 3A) but do not form obvious clusters, which prevented clear assignment of potentially important sites on the surface of UL21N based on sequence identity alone. Evolutionary trace analysis (ETA) (28) was performed on the same sequence alignment. ETA generates a phylogenetic tree from a sequence alignment of homologous proteins. At each partition, or branch point, closely related sequences are grouped into classes, trace residues are defined as conserved within specific classes, and these residues are assigned importance scores based on the partition at which they appear. Clustering of important trace residues on the protein surface may indicate regions of functional importance (28). ETA has been used to detect functional sites in a number of proteins (20, 29), including PRV UL37 (31). ETA of UL21N yielded several surface clusters of trace residues of increasing importance (Fig. 3B).
FIG 3.
Analysis of conservation and charge on the surface of UL21N. A dotted line marks a cavity likely obscured by unresolved residues 76 to 87. (A) Completely conserved residues on the surface of UL21N are shown in magenta. Four orientations based on 90° rotations around the vertical axis are shown. (B) Class-specific residues identified by universal evolutionary trace analysis (http://mammoth.bcm.tmc.edu/uet/) are highlighted on the surface of UL21N. The 25% of residues with the highest importance scores are shown. (C) An electrostatic surface potential map of UL21N was generated using the PBEQ Solver function in the Charmm program (http://www.charmm-gui.org/?doc=input/pbeqsolver). (D) Potential functional regions assigned on the surface of UL21N. Potential functional regions are composed of the following residues: region 1 (red), D13, D105, D111, and E113; region 2 (orange), M1, E2, R55, N156, Y163, P165, F170, and L172; region 3 (yellow), Y5, Y17; region 4 (green), Y67, R69, S70, E71, D116, E117, and E121.
Analysis of the electrostatic potential on the surface of UL21N revealed two large negatively charged patches, consistent with its calculated isoelectric point of 5.2, one of which coincides with a shallow depression and the other of which wraps around the nearby side (Fig. 3C). A number of the residues within these charged patches were also identified as important trace residues in ETA. In combination, conservation and charge patterns pinpoint several surface regions of potential functional importance (Fig. 3D), which may participate in binding the known partner UL16, in interacting with UL21C, or in binding to as yet unknown ligands. Region 1 (D13, D105, D111, E113) sits on one flat face of UL21N and is comprised of four aspartate and glutamate residues, two of which are also identified in ETA. Region 2 (M1, E2, R55, N156, Y163, P165, F170, L172) extends to three sides of the protein and contains identical and important ETA trace residues. Region 3 (Y5, Y17) comprises two conserved, surface-accessible tyrosine residues, and the large region 4 (Y67, R69, S70, E71, D116, E117, E121) wraps around one short side and contains residues identified in electrostatic and ETA analyses. These regions provide a more educated starting point for mutational analysis in the context of protein biochemistry and viral infection.
Although UL21 plays multiple roles in the viral replication cycle, including nuclear egress, cytoplasmic capsid trafficking, budding, and cell-cell spread, the molecular mechanisms by which UL21 enables these and other processes remain poorly understood. The novel sail-shaped structure of UL21N provides a three-dimensional template for targeted mutational analysis and enables structure-guided functional exploration of the multiple roles of UL21 in the replication and pathogenesis of alphaherpesviruses.
Protein structure accession number.
Atomic coordinates and structure factors for the UL21N structure have been deposited in the RCSB Protein Data Bank under accession number 4U4H.
ACKNOWLEDGMENTS
We thank John Wills for helpful discussions related to this project. We also thank A. Héroux (National Synchrotron Light Source) for collecting and processing X-ray diffraction data.
This work was funded by the Burroughs Wellcome Fund Award for Investigators in Pathogenesis (E.E.H.). P.C. was supported by NIH grant AI071286 to John W. Wills. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract no. DE-AC02-98CH10886.
C.M.M. performed all of the experiments described in this work. P.C. performed early work important for construct design and generated the full-length StrepII-UL21 construct. C.M.M. and E.E.H. designed research, analyzed data, and wrote the article.
REFERENCES
- 1.Fatahzadeh M, Schwartz RA. 2007. Human herpes simplex virus infections: epidemiology, pathogenesis, symptomatology, diagnosis, and management. J Am Acad Dermatol 57:737–763; quiz 764–766. doi: 10.1016/j.jaad.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 2.Morales-Sanchez A, Fuentes-Panana EM. 2014. Human viruses and cancer. Viruses 6:4047–4079. doi: 10.3390/v6104047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell MEM, Palfreyman JW, Preston CM. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol 180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 4.Read GS, Frenkel N. 1983. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha(immediate-early) viral polypeptides. J Virol 46:498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krautwald M, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Translocation of incoming pseudorabies virus capsids to the cell nucleus is delayed in the absence of tegument protein pUL37. J Virol 83:3389–3396. doi: 10.1128/JVI.02090-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaichick SV, Bohannon KP, Hughes A, Sollars PJ, Pickard GE, Smith GA. 2013. The herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe 13:193–203. doi: 10.1016/j.chom.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schipke J, Pohlmann A, Diestel R, Binz A, Rudolph K, Nagel CH, Bauerfeind R, Sodeik B. 2012. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol 86:3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Sage V, Jung M, Alter JD, Wills EG, Johnston SM, Kawaguchi Y, Baines JD, Banfield BW. 2013. The herpes simplex virus 2 UL21 protein is essential for virus propagation. J Virol 87:5904–5915. doi: 10.1128/JVI.03489-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baines JD, Koyama AH, Huang T, Roizman B. 1994. The UL21 gene products of herpes simplex virus 1 are dispensable for growth in cultured cells. J Virol 68:2929–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mbong EF, Woodley L, Frost E, Baines JD, Duffy C. 2012. Deletion of UL21 causes a delay in the early stages of the herpes simplex virus 1 replication cycle. J Virol 86:7003–7007. doi: 10.1128/JVI.00411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klopfleisch R, Klupp BG, Fuchs W, Kopp M, Teifke JP, Mettenleiter TC. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J Virol 80:5571–5576. doi: 10.1128/JVI.02589-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klupp BG, Lomniczi B, Visser N, Fuchs W, Mettenleiter TC. 1995. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology 212:466–473. doi: 10.1006/viro.1995.1504. [DOI] [PubMed] [Google Scholar]
- 13.de Wind N, Wagenaar F, Pol J, Kimman T, Berns A. 1992. The pseudorabies virus homology of the herpes simplex virus UL21 gene product is a capsid protein which is involved in capsid maturation. J Virol 66:7096–7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFerran JB, Dow C. 1975. Studies on immunisation of pigs with the Bartha strain of Aujeszky's disease virus. Res Vet Sci 19:17–22. [PubMed] [Google Scholar]
- 15.Klupp BG, Bottcher S, Granzow H, Kopp M, Mettenleiter TC. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J Virol 79:1510–1522. doi: 10.1128/JVI.79.3.1510-1522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci U S A 109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harper AL, Meckes DG Jr, Marsh JA, Ward MD, Yeh PC, Baird NL, Wilson CB, Semmes OJ, Wills JW. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J Virol 84:2963–2971. doi: 10.1128/JVI.02015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson DC, Webb M, Wisner TW, Brunetti C. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J Virol 75:821–833. doi: 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson DC, Huber MT. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J Virol 76:1–8. doi: 10.1128/JVI.76.1.1-8.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis-Poynter N, Bell S, Minson T, Browne H. 1994. Analysis of the contribution of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J Virol 68:7586–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takakuwa H, Goshima F, Koshizuka T, Murata T, Daikoku T, Nishiyama Y. 2001. Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in over-expressing cells. Genes Cells 6:955–966. doi: 10.1046/j.1365-2443.2001.00475.x. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- 23.de la Fortelle E, Bricogne G. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol 276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36:W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lichtarge O, Bourne HR, Cohen FE. 1996. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 29.Sowa ME, He W, Slep KC, Kercher MA, Lichtarge O, Wensel TG. 2001. Prediction and confirmation of a site critical for effector regulation of RGS domain activity. Nat Struct Biol 8:234–237. doi: 10.1038/84974. [DOI] [PubMed] [Google Scholar]
- 30.Chakravarty S, Hutson AM, Estes MK, Prasad BV. 2005. Evolutionary trace residues in noroviruses: importance in receptor binding, antigenicity, virion assembly, and strain diversity. J Virol 79:554–568. doi: 10.1128/JVI.79.1.554-568.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitts JD, Klabis J, Richards AL, Smith GA, Heldwein EE. 2014. Crystal structure of the herpesvirus inner tegument protein UL37 supports its essential role in control of viral trafficking. J Virol 88:5462–5473. doi: 10.1128/JVI.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 33.Gouet P, Courcelle E, Stuart DI, Metoz F. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 34.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and Nucleic acids. Nucleic Acids Res 35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]