ABSTRACT
Perturbation of protein-protein interactions relies mostly on genetic approaches or on chemical inhibition. Small RNA viruses, such as influenza A virus, do not easily lend themselves to the former approach, while chemical inhibition requires that the target protein be druggable. A lack of tools thus constrains the functional analysis of influenza virus-encoded proteins. We generated a panel of camelid-derived single-domain antibody fragments (VHHs) against influenza virus nucleoprotein (NP), a viral protein essential for nuclear trafficking and packaging of the influenza virus genome. We show that these VHHs can target NP in living cells and perturb NP's function during infection. Cytosolic expression of NP-specific VHHs (αNP-VHHs) disrupts virus replication at an early stage of the life cycle. Based on their specificity, these VHHs fall into two distinct groups. Both prevent nuclear import of the viral ribonucleoprotein (vRNP) complex without disrupting nuclear import of NP alone. Different stages of the virus life cycle thus rely on distinct nuclear localization motifs of NP. Their molecular characterization may afford new means of intervention in the virus life cycle.
IMPORTANCE Many proteins encoded by RNA viruses are refractory to manipulation due to their essential role in replication. Thus, studying their function and determining how to disrupt said function through pharmaceutical intervention are difficult. We present a novel method based on single-domain-antibody technology that permits specific targeting and disruption of an essential influenza virus protein in the absence of genetic manipulation of influenza virus itself. Characterization of such interactions may help identify new targets for pharmaceutical intervention. This approach can be extended to study proteins encoded by other viral pathogens.
INTRODUCTION
The replication cycle of influenza A virus (IAV) is complex. The virus attaches to susceptible host cells via its hemagglutinin (HA), a homotrimeric type I membrane glycoprotein that recognizes sialoconjugates (1–3). The virus then enters the endocytic pathway, and upon arrival in acidified late endosomes, the HA trimer undergoes a conformational transition that renders it fusogenic. The M2 ion channel is responsible for acidification of the virus lumen, which results in dissociation of the eight viral ribonucleoproteins (vRNPs) (comprised of PB1, PB2, PA, NP, and genomic RNA) from the M1 protein and release of the vRNPs into the host cytosol (4–6). These vRNPs translocate into the nucleus via one of at least two nuclear localization sequences, NLS1 and NLS2, in NP (7–11). mRNA generated from vRNP-dependent synthesis of viral genomic RNA (vRNA) is exported from the nucleus and translated in the cytoplasm. Newly synthesized PB1, PB2, PA, and NP translocate into the nucleus as monomers (NP and PB2) or dimers (PB1-PA), where they assemble with newly synthesized vRNA to yield the vRNP complex (12, 13). These vRNP complexes are exported from the nucleus for incorporation into budding virus particles (14).
In the course of a single replication cycle, influenza virus NP interacts with viral RNA and with viral proteins, including PB1, PB2, and M1 (15, 16). Several host proteins also interact with NP, including importin-α, BAT1, F-actin, and CRM1 (17–20). Mapping such interactions and assessing their relevance for virus replication remains a challenge because of their often-essential role in the replication cycle. With rare exceptions, the influenza virus genome has resisted genetic manipulation, because many such changes cause a complete loss of a particular function (21–23) and compromise viral fitness.
The variable domains of heavy-chain-only antibodies (VHHs) isolated from camelids are small, ∼15 kDa, and their ability to bind their cognate ligand is largely independent of modifications such as disulfide bonds and glycosylation (24, 25). These properties allow the VHHs to be expressed in the cytosol of eukaryotic cells with retention of the antigen binding capabilities. This in turn permits the specific targeting of host or viral proteins recognized by VHHs, thus enabling possible perturbation of target protein function (26–32; for a review, see reference 33). VHHs are therefore unique tools for analysis of essential proteins encoded by RNA viruses in living cells.
We generated a VHH library against influenza virus and isolated VHHs specific for NP (αNP-VHHs). Interaction of αNP-VHHs with NP occurred when both proteins were coexpressed in the cytosol of mammalian cells. Expression of αNP-VHHs during infection disrupted the replication cycle at an early stage and prevented nuclear import of vRNPs. This αNP-VHH-dependent inhibition of import was specific for vRNPs, as nuclear import of NP alone was unperturbed, as was infection with an unrelated pathogen, vesicular stomatitis virus (VSV). We conclude that influenza virus utilizes separate features of structure for import of NP and vRNPs.
MATERIALS AND METHODS
Antibodies and plasmids.
GAPDH-HRP (horseradish peroxidase)-conjugated antibody was purchased from Abcam (ab105428). Anti-mCherry antibody was purchased from Abcam (ab167453). Anti-HA antibody was purchased from Sigma (HA-7). Protein G-agarose was purchased from Roche Diagnostics. Nickel-nitrilotriacetic acid (NTA) beads were purchased from Qiagen. FluB1 antibody was purified from FluBI hybridoma cells using published methods (34). Anti-WSN serum was obtained from mice immunized with the A/WSN/33 strain of influenza virus. Expression of NP-HA, NP-GFP (green fluorescent protein), VHH-Ch, and mCherry constructs were achieved by cloning into pCAGGs. Expression vectors encoding PB1, PB2, PA, and NP were a kind gift from Adolfo Garcia-Sastre. Anti-NP antibody was a kind gift from Ari Helenius.
Virus.
VSV-GFP was propagated and titrated over Vero cells. A/WSN/33 strain of influenza virus was propagated and titrated over MDCK cells. All infections, unless otherwise noted, were performed as follows: cells were washed in phosphate-buffered saline (PBS) and then incubated with virus resuspended in PBS (0.2% bovine serum albumin [BSA]). After 1 h at room temperature, the inoculum was replaced with Dulbecco's modified Eagle medium (DMEM) (0.2% BSA), and cells were shifted to 37°C with 5% CO2.
Cells.
Stable integration of mCherry or VHH-Ch constructs in MDCK cells was achieved by cotransfection of pCAGGS expressing vectors and a plasmid expressing the zeocin resistance cassette with subsequent selection for zeocin resistance. Thereafter, cells were maintained in DMEM (10% fetal bovine serum [FBS]). A549 cells were cultivated in DMEM with 10% FBS. A549 cell lines inducibly expressing HA-tagged VHHs were generated using lentivirus produced with derivatives of pInducer20 (35) and selected in the presence of 500 μg/ml G418.
VHH library generation.
To obtain VHHs specific for influenza virus, two alpacas underwent five rounds of immunization with alcohol-fixed influenza virus PR8. Peripheral blood cells were collected from both alpacas, and total RNA was isolated from each sample using a Qiagen RNeasy Plus minikit (Qiagen). Purified RNA was then used for cDNA synthesis using random hexamers, oligo(dT), and primers specific for the constant region of the alpaca heavy chain gene. cDNA was then used as a template with primers specific to the VHH region (36). PCR products from both alpacas were pooled, digested with NotI-HF and AscI (NEB), gel purified, ligated into a M13 phagemid vector (pJSC), and transformed via electroporation into TG1 bacteria for phage display-based panning. Three separate methods of panning were utilized to isolate influenza virus-specific VHHs. The first method used intact biotinylated HAsrt virus (37) immobilized on streptavidin beads. The second method utilized recombinant influenza virus protein with a C-terminal sortase motif expressed in 293T cells, biotinylated, and immobilized on streptavidin beads. A third method utilized a Staphylococcus aureus display platform, based on a strategy developed for Staphylococcus carnosus (38), which will be described in detail elsewhere.
Recombinant protein expression and purification.
NP from influenza virus A/WSN/33 was cloned with a C-terminal His tag into pET30b+ expression vector. Transformed Escherichia coli BL21(DE3) cells were cultured in Terrific Broth at 37°C before the temperature was reduced to 25°C at an optical density at 600 nm (OD600) of 0.5. Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an OD600 of 0.6, and cells were grown for an additional 3 h. The resulting pellet was resuspended in 25 mM Tris (pH 7.5), 1 M NaCl, 0.2% NP-40, 10 units/ml Benzonase, and 0.1 mg/ml lysozyme. Cells were lysed by sonication, and NP was purified on Ni-NTA agarose, cation exchange, and Superdex 200 gel filtration columns.
Recombinant VHHs were cloned with a C-terminal sortase recognition site (LPETG) followed by a 6×His tag into the expression vector pHEN6 for periplasmic expression and transformed into E. coli. strain WK6. Protein expression was induced by addition of 1 mM IPTG at an OD600 of 0.6, and cells were grown overnight at 30°C. The periplasm fraction was harvested by osmotic shock, and VHHs were purified by Ni-NTA affinity and size exclusion chromatography using a Superdex 75 column.
Sortase modification of recombinant proteins.
A sortase A pentamutant of S. aureus (5 μM final concentration) and probe (GGG-biotin; 250 mM final concentration) were added to the VHH (30 μM final concentration) in sortase buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 10 mM CaCl2). The mixture was incubated at 4°C overnight. The His-tagged sortase and unreacted VHH were removed by being passed over a Ni-NTA column, and the biotinylated VHH was purified on a Superdex 75 size exclusion column and analyzed by gel electrophoresis.
Competition assay.
Immunoprecipitations for competition assays were performed with 2 μg of biotinylated VHH bound to streptavidin magnetic beads (MyOne Dynabeads; Invitrogen) and 7.5 μg of recombinant NP. Before addition to the beads, NP was blocked with 50 μg of the individual His-tagged VHHs from the panel of NP-specific (VHH1 to -6) and control (VHH7 and F8) VHHs. Bound NP was eluted in 0.2 M glycine, pH 2.2, and analyzed by SDS-PAGE and immunoblotting with anti-His-HRP.
Metabolic labeling and immunoprecipitation.
Pulse-chase experiments were performed as described in references 39 and 40. Briefly, MDCK cells (∼1 × 107/sample) were trypsinized and incubated at 37°C in starvation medium (methionine/cysteine-free DMEM supplemented with 10% dialyzed fetal calf serum) for 30 min. Metabolic labeling was performed at 37°C using starvation medium plus 10 mCi/ml [35S]methionine/cysteine (PerkinElmer). Cell pellets were lysed in NP-40 containing lysis buffer and precleared with protein G-agarose beads for 1 h at 4°C. Immunoprecipitations were performed for 1 h at 4°C with gentle agitation. Samples were eluted by boiling in reducing sample buffer, run on SDS-PAGE, and visualized by autoradiography.
Flow cytometry-based infection assays.
To quantify infection by flow cytometry, MDCK or A549 cells were seeded in 24-well plates 16 or 40 h before infection (3 × 104 cells/well). A549 cells were treated with 1 μg/ml doxycycline 24 h before infection to induce VHH expression or left untreated. Cells were infected with appropriate amounts of IAV (in 0.2% BSA–DMEM) or VSV-GFP (in DMEM) to infect ca. 20% of wild-type cells. At 30 min postinfection, inocula were removed, and cells were cultivated for 5.5 h in DMEM (10% serum). Cells were trypsinized, fixed in 4% formaldehyde–PBS, and stained in the presence of 0.05% saponin with 1 μg/ml Alexa Fluor (AF) 647-coupled anti-NP VHH1 or -2 (IAV-infected cells) and/or mouse anti-HA and goat anti-mouse AF 488/AF 647 (A549-based cell lines), all under permeabilizing conditions. Levels of infection were normalized to the wild-type (WT) sample (for MDCK cells) or the noninduced sample (for A549 samples). Fluorescence was quantified using a BD Biosciences LSRFortessa flow cytometer and the FlowJo software package.
RESULTS
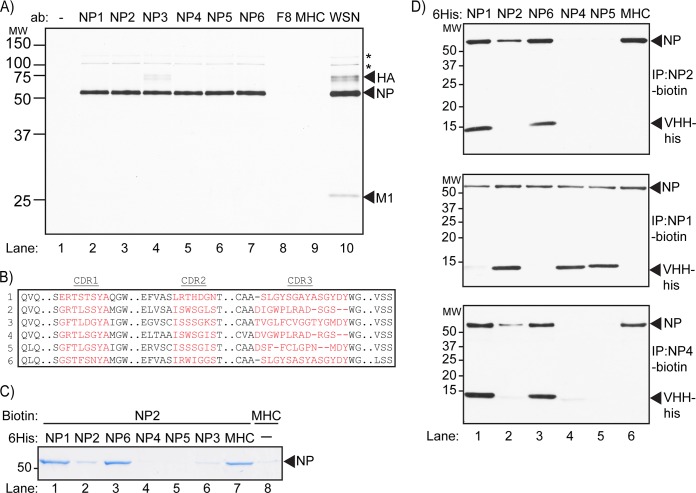
To generate VHHs specific for influenza virus, we immunized alpacas with an inactivated preparation of influenza A virus PR8, a mouse-adapted H1N1 strain of influenza virus. After five rounds of immunization, peripheral mononuclear cells were collected. VHH sequences were amplified from purified RNA and cloned into an M13 phagemid vector. We used phage display to identify VHHs that bound virus immobilized on streptavidin beads via biotinylated HA (37) or recombinant HA-tagged influenza virus protein immobilized on anti-HA Sepharose beads. After two consecutive rounds of phage display and panning, we obtained four unique VHHs specific for influenza virus. Three additional VHHs specific for influenza virus were then obtained by selection using staphylococcal surface expression (38). All seven VHHs were equipped with a C-terminal sortase motif (41, 42) followed by a 6×His tag, expressed in bacteria, and purified on a Ni-NTA affinity matrix followed by size exclusion chromatography. To determine the identity of their corresponding antigen(s), we immobilized the VHHs on Ni-NTA beads and immunoprecipitated the target antigen from lysates of influenza virus-infected MDCK cells metabolically labeled with [35S]cysteine/methionine for 2 h. The presence of HA, NP, M1, PB1, PB2, and PA proteins in the lysate was confirmed by immunoprecipitation using serum from influenza virus-infected mice (Fig. 1A, lane 10). As a negative control, we used anti-major histocompatibility complex class II (MHCII) VHH7, a single-domain antibody that recognizes murine class II MHC products (43) (Fig. 1A, lane 9), which are absent from the MDCK lysate. Immunoprecipitation with six of the seven VHHs isolated from the influenza virus library yielded the influenza virus NP protein (Fig. 1A, lanes 2 to 7). These VHHs are referred to here as αNP-VHH1, -2, -3, -4, -5, and -6. Interestingly, small amounts of HA coprecipitated with NP in the αNP-VHH3 sample, the significance of which is not currently known. VHH F8 did not recover NP by immunoprecipitation (Fig. 1A, lane 8) and was used as an additional control in subsequent experiments.
FIG 1.
VHHs isolated from the influenza virus VHH library are specific for NP. (A) Lysate from 35S-labeled MDCK cells infected with influenza virus were subjected to immunoprecipitation with purified VHH containing a C-terminal 6×His tag immobilized on Ni-NTA beads. Samples were analyzed using SDS-PAGE and autoradiography. Asterisks denote expected mobility for PB2 (upper asterisk) and PB2/PA (lower asterisk). (B) ClustalW-based alignment of αNP-VHH CDR1, CDR2, and CDR3 amino acid sequences (in red). (C) His-tagged NP was incubated briefly with His-tagged versions of the given VHHs. Biotinylated αNP-VHH2 immobilized on streptavidin beads was then used to precipitate available NP-His. Precipitated NP-His was run on SDS-PAGE and visualized by Coomassie staining. (D) His-tagged NP was incubated briefly with His-tagged versions of the given VHHs. Biotinylated αNP-VHH2 (top), VHH4 (middle), and VHH1 (bottom) immobilized on streptavidin beads were then used to pull down NP-His. Precipitated protein was run on SDS-PAGE and detected via immunoblotting with αHis-HRP antibody.
To determine whether these αNP-VHHs bind distinct epitopes on NP, we performed cross-competition assays, as follows. We tested the ability of biotinylated αNP-VHH2 to retrieve recombinant NP equipped with a C-terminal 6×His tag (NP-His) preincubated with an excess of each of the individual VHHs. Recovery of VHH2 on streptavidin agarose would occur only if preincubation of NP with any of the other VHHs failed to occlude the epitope recognized by VHH2. Material recovered on streptavidin-agarose was analyzed by SDS-PAGE and Coomassie staining. As expected, recovery of NP was poor in samples preincubated with αNP-VHH2 compared to that in an irrelevant control (Fig. 1C, lanes 2 and 6). We also observed competition upon preincubation of NP with αNP-VHH3, -4, and -5 (Fig. 1C, lanes 4 to 6), suggesting that these VHHs occlude the epitope recognized by αNP-VHH2. Preincubation with αNP-VHH1 and -6 did not affect recovery of NP by αNP-VHH2 (Fig. 1C, lanes 1 and 3), indicating that these two VHHs recognized an epitope(s) distinct from that recognized by αNP-VHH2.
We next tested biotinylated αNP-VHH1 and -4 in a similar assay. Recovery of NP-His was detected by SDS-PAGE and immunoblotting using an anti-His antibody. αNP-VHH2-biotin precipitated NP-His after preincubation with αNP-VHH1 and -6 but not αNP-VHH2, -4, or -5 (Fig. 1D, top). Coprecipitation of αNP-VHH1 and αNP-VHH6 with αNP-VHH2-biotin further confirmed their recognition of distinct epitopes. A similar profile was observed with αNP-VHH4-biotin (Fig. 1D, bottom). In contrast, retrieval of αNP-VHH1-biotin-bound material showed recovery of NP-His from samples preincubated with αNP-VHH2, -4, -5, and -6. Neither αNP-VHH1 nor αNP-VHH6 recognized NP in the presence of αNP-VHH1-biotin, indicating that αNP-VHH1 and -6 compete for the same epitope or similar epitopes. From these results, we conclude that the αNP-VHHs in our panel comprise two groups, αNP-VHH1 and -6 and αNP-VHH2, -3, -4, and -5, each of which binds distinct nonoverlapping epitopes on NP. None of the αNP-VHHs reacted with recombinant NP in immunoblots, suggesting that they recognize conformation-sensitive epitopes.
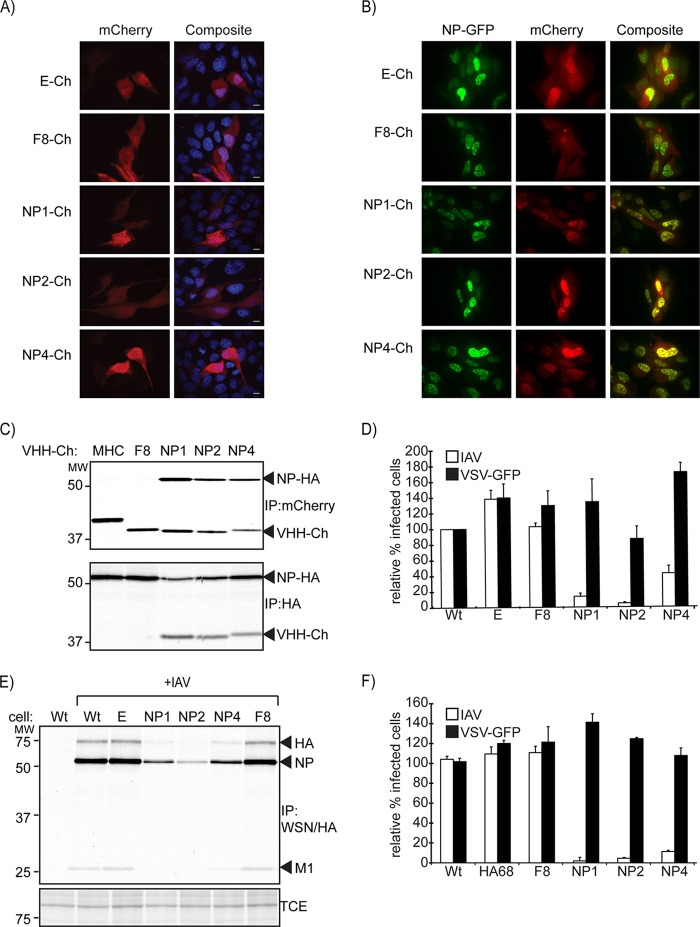
We next examined the anti-NP VHH panel for intracellular expression and manipulation of virus replication, in order to identify regions of NP essential for virus replication. As a first step, we confirmed that αNP-VHHs interact with NP when expressed in the cytosol of mammalian cells in the absence of infection. We cotransfected MDCK cells with expression vectors that encode enhanced green fluorescent protein (eGFP) fusions of NP (NP-GFP) and mCherry fusions of αNP-VHH1, -2, or -4 (αNP-VHH-Ch). Since αNP-VHH1, -2, and -4 represent both groups of αNP-VHHs, we omitted αNP-VHH3, -5, and -6 from this experiment and from the remainder of the study because of their comparatively poor expression in the cytosol (data not shown). Coexpressed fluorescent fusion proteins were visualized by live-cell imaging. In the absence of NP expression, mCherry and all VHH-mCherry fusions were diffusely expressed throughout the cell (Fig. 2A). In the absence of αNP-VHH expression, NP-GFP fusion protein localized to the nucleus, indicative of the presence of a functional nuclear localization signal (NLS) (Fig. 2B, top two left panels). None of the αNP-VHHs tested altered the nuclear localization of NP-GFP (Fig. 2B, lower three left panels). Furthermore, while VHH F8-mCherry or mCherry alone was observed throughout the cell, αNP-VHH1 and -4 colocalized in the nucleus with NP-GFP, consistent with a specific interaction of αNP-VHHs with NP inside the cell (Fig. 2B, panels in middle column).
FIG 2.
αNP-VHH-Ch and NP-GFP colocalize in the nucleus of transfected MDCK cells. (A) Plasmids expressing αNP-VHH-Ch were transfected into MDCK cells. At 24 posttransfection, cells were fixed and permeabilized with saponin. DNA was stained with Hoechst, and cells were imaged via confocal microscopy. (B) Plasmids expressing αNP-VHH-Ch and NP-GFP were cotransfected into MDCK cells. At 24 h posttransfection, cells were imaged using live confocal microscopy. (C) NP and αNP-VHH-Ch interact in the cytosol. 293T cells were transfected with expression plasmids for αNP-VHH-Ch and NP-HA. At 24 posttransfection, cells were metabolically labeled with [35S]cysteine/methionine for 20 min and subsequently lysed in NP-40-containing lysis buffer. The lysate was split into two samples and subjected to immunoprecipitation using antibodies against HA or mCherry. Samples were analyzed via SDS-PAGE and autoradiography. (D) αNP-VHH-Ch expression in the cytosol specifically inhibits influenza virus replication. MDCK cells constitutively expressing αNP-VHH-Ch were challenged with influenza virus or VSV-GFP. At 6 h postinfection, cells were fixed, and IAV-infected cells were stained with Alexa Fluor (AF) 647-coupled αNP-VHH1 under permeabilizing conditions. Levels of infection were quantified by flow cytometry and normalized to that in the WT sample. Means and standard errors of the means (SEM) from three independent experiments are shown. (E) Expression of αNP-VHH-Ch in MDCK cells results in decreased influenza virus expression upon virus challenge. MDCK cells stably expressing VHH-Ch were challenged with influenza virus (MOI, 1.0). Two hours postinfection, cells were metabolically labeled with [35S]methionine for 20 min. Cells were lysed in NP-40-containing lysis buffer, and the lysate was subsequently probed with WSN serum and FluB1 antibody. Samples were analyzed by SDS-PAGE and autoradiography. (F) αNP-VHH-mediated inhibition is independent of cell type or mCherry fusion. A549 cells inducibly expressing αNP-VHH-HA were treated with 1 μg/ml doxycycline for 24 h or left untreated. Cells were subsequently challenged with influenza virus or VSV-GFP. At 6 h postinfection, cells were fixed, and IAV-infected cells were stained with AF 647-coupled α-NP-VHH1. Levels of infection were quantified by flow cytometry, and infections obtained in induced samples were normalized to their uninduced counterpart. Means and SEM from three independent experiments are shown.
To confirm this interaction, we transiently expressed αNP-VHH1-Ch, -2-Ch, or -4-Ch with C-terminally HA-tagged NP (NP-HA) in 293T cells. As a negative control, we included αMHCII-VHH7 fused to mCherry and VHH F8-mCherry. At 24 h after transfection, we metabolically labeled the cells with [35S]cysteine/methionine for 20 min and then lysed them in buffer containing NP-40. We performed immunoprecipitations with anti-mCherry or anti-HA antibody. Anti-mCherry coimmunoprecipitated NP only in lysates expressing an αNP-VHH–mCherry fusion protein (Fig. 2C, top). Furthermore, the reciprocal immunoprecipitation using anti-HA coimmunoprecipitated only those VHHs specific for NP (Fig. 2C, bottom). Based on these results, we concluded that epitope recognition and binding of the αNP-VHHs were retained when expressed in the cytosol.
Having demonstrated the ability of αNP-VHHs to interact with NP in living cells, we wondered whether αNP-VHHs can target and disrupt influenza virus replication. We generated MDCK cells that stably express αNP-VHH-mCherry and measured virus replication using two independent methods. To quantify infection and expression of αNP-VHHs by flow cytometry, MDCK cell lines were infected for 6 h, then stained for NP protein using Alexa Fluor 647-labeled αNP-VHH1, and examined by flow cytometry. MDCK cells that express αNP-VHH1, -2, and -4 supported influenza virus replication far worse than wild-type (wt) MDCK cells or cell lines that express only mCherry or VHH F8-mCherry (Fig. 2D).
We next infected wt MDCK cells or cells expressing αNP-VHHs with influenza virus at a multiplicity (MOI) of 1.0. Two hours postinfection, cells were pulsed with [35S]cysteine/methionine for 10 min and then lysed in buffer containing NP-40. Viral antigens were immunoprecipitated from the lysates using a mixture of anti-WSN polyclonal mouse serum and a mouse monoclonal antibody (FluBI) which recognizes HA (44). Infection, as measured by the amount of newly synthesized NP, HA, and M1, was robust in wt cells and in cells that expressed mCherry or VHH F8-mCherry (Fig. 2E, lanes 2, 3, and 7). In contrast, cells that expressed αNP-VHH1, -2, and -4 produced substantially less viral antigen, indicating that infection was disrupted in these cells (Fig. 2E, lanes 4, 5, and 6).
To rule out the possibility that inhibition was cell type specific or dependent on the relatively large mCherry fusion, we used lentiviral transduction to generate A549 cell lines with doxycycline-inducible expression of αNP-VHHs. Instead of mCherry, the doxycycline-inducible αNP-VHHs carry a C-terminal HA epitope tag. As an additional negative control for these experiments, we included cells that express αHA-VHH68 (44). Infection with influenza virus was assessed by flow cytometry as described above. Infection proceeded normally in the absence of αNP-VHH expression (no doxycycline) but was essentially blocked in doxycycline-induced cells (Fig. 2F). To demonstrate the selectivity of the αNP-VHHs for influenza virus, we challenged the MDCK and A549 cell lines with a strain of vesicular stomatitis virus (VSV) that expresses eGFP (VSV-GFP) (45) and quantified infection by flow cytometry using the GFP signal as a proxy for infection. No difference in infection with VSV was observed in cell lines induced for expression of αNP-VHHs compared to uninduced cells (Fig. 2D and F). The inhibition of infection imposed by αNP-VHHs observed in influenza virus-challenged cells was therefore specific for influenza virus and not a virus-independent feature of the infected state.
In the course of infection, NP engages in a number of interactions with host proteins and viral components (46). Binding of αNP-VHHs to NP could disrupt any one of them to account for the observed inhibition of virus replication. To explore how the αNP-VHHs block virus replication, we established a mini-genome assay in 293T cells. In this assay, replication and transcription of a model vRNA segment are dependent on NP expression, nuclear import, and association with RNA and the polymerase complex. Cells were transfected with expression plasmids for NP, PBI, PB2, and PA. The model vRNA segment encoding an HA-tagged version of influenza virus NA was transcribed from a cotransfected plasmid. Nuclear localization of αNP-VHH-Ch (and by extension the NP ligand) was confirmed by confocal microscopy (data not shown). At 24 h after transfection, cells were lysed and NA protein levels were measured by SDS-PAGE and immunoblot analysis. Coexpression of neither VHH F8 nor αMHCII VHH7 fused to mCherry inhibited expression of NA in the mini-genome assay (Fig. 3, lanes 6 and 7). For the three αNP-VHHs tested, only αNP-VHH1 inhibited NA protein expression, whereas αNP-VHH2 and -4 were inactive (Fig. 3, lanes 3, 4, and 5). We thus conclude that expression of αNP-VHH1 inhibits genome replication, whereas αNP-VHH2 and -4 must block infection at a step upstream from replication.
FIG 3.

αNP-VHH2 and -4 do not influence NP activity in a mini-genome assay. 293T cells were transfected with plasmids expressing influenza virus PB1, PB2, PA, NP, and VHH-Ch and a plasmid transcribing a model genome segment encoding neuraminidase with a sortag motif inserted at the C terminus (NAsrt) (under the control of the pPol1 promoter). At 24 h posttransfection, cells were lysed in NP-40-containing lysis buffer. Samples were analyzed via Western blotting using antibody against the HA epitope, NP, mCherry, and GAPDH.
We next assayed nuclear import of vRNPs in MDCK cells that express αNP-VHHs and challenged these cells with a high dose of influenza virus. We included the translation inhibitor cycloheximide to ensure that all NP (used here as a proxy for vRNPs) was derived from the input virus and not from newly synthesized protein. After 4 h of infection, cells were fixed and stained for NP to determine the extent of vRNP nuclear import. In wt cells and in cells expressing mCherry or VHH F8, input NP protein is localized predominantly in the nucleus (Fig. 4). In contrast, in cells that express αNP-VHH1, -2, and -4, the NP signal was mostly absent from the nucleus, indicating that nuclear import of the vRNPs was restricted. Thus, αNP-VHH1 interferes with replication in the mini-genome assay and with nuclear import, whereas αNP-VHH2 and -4 affect only nuclear import. Consequently, the segments of NP recognized by αNP-VHH1 and by αNP-VHH2 and -4 not only are distinct epitopes but also participate in different aspects of its function in the course of infection.
FIG 4.
αNP-VHH-Ch inhibits nuclear import of the incoming vRNP complex. MDCK cells stably expressing VHH-Ch were infected with influenza virus (MOI, 500) in the presence of cycloheximide. Four hours after infection, cells were fixed and permeabilized with 4% paraformaldehyde and 0.5% NP-40. Staining was performed with Hoechst (blue) and anti-NP (green) before visualization by confocal microscopy.
DISCUSSION
We exploited the unique properties of single-domain antibodies that target influenza virus NP in the cytosol of influenza virus-infected cells and studied their impact on influenza virus replication. The αNP-VHHs expressed in this fashion interacted with influenza virus NP in intact cells, consistent with the fact that epitope recognition is independent of lumen-associated posttranslational modifications of the VHHs, such as glycosylation and disulfide bond formation (modifications not present in the bacterial system used for phage display). Interactions between αNP-VHHs and NP in the course of infection disrupted influenza virus replication at an early step. The primary mechanism by which this occurs is a halt in nuclear import of the incoming vRNPs. The αNP-VHH1 recognizes an epitope distinct from that recognized by αNP-VHH2 and -4 and had an additional inhibitory activity at the level of genome replication. None of the αNP-VHHs tested interfered with nuclear localization of NP when it was expressed on its own. Thus, we conclude that the inhibitory activity of αNP-VHHs results from interference with the nuclear import pathway utilized specifically by influenza virus vRNPs and not by NP alone, consistent with separate functions for the two nuclear localization signals identified for NP (7, 47).
Analysis of the NLS motifs in NP has resisted identification of separable functions for these motifs in the course of infection. The complexity of interactions of NP with host and viral proteins and the lack of tools available to analyze essential gene function have surely contributed to this state of affairs (8–10, 48). Mutational inactivation of either or both motifs failed to inhibit nuclear import of NP when it was expressed alone, leading to the suggestion that the protein contains a third NLS (48). Likewise, import of purified vRNPs in a digitonin-permeabilized cell system was partially inhibited by preincubation with conventional antibodies specific for NLS1 and NLS2 (49). In the latter case, nuclear import was assessed by immunocytochemistry with an anti-NP antibody, and vRNP-independent import of NP may have confounded these results. Our panel of αNP-VHHs allowed us to perturb nuclear import of vRNPs in living cells. This critical step in the virus life cycle could thus be analyzed under physiological conditions, i.e., during an infection.
Could an influenza virus-infected cell avoid nuclear reimport of vRNPs recently exported from the nucleus and destined for incorporation into budding virions? In a similar vein, what determines nuclear import of incoming vRNPs? Our data, along with the model proposed by Wu and Pante (47), suggest that directionality of transport for vRNPs may be accomplished by regulating access to NLS1 and NLS2 sites on NP. Thus, RNA-associated NP, as found on the incoming vRNPs, might be available for the nuclear import machinery via NLS2, while NLS1 remains masked in vRNPs. An additional modification(s) to NP would have to occur to prevent reimport of newly exported vRNPs via NLS2 in the course of infection. How this occurs is unclear but likely involves interactions with influenza virus M1 and nuclear export protein (NEP) (50). Further manipulation of this pathway using the αNP-VHHs in hand should improve our understanding of the virus life cycle. For example, cocrystallization studies of αNP-VHHs and NP should allow the molecular identification of the precise regions of NP recognized by αNP-VHHs and thus the features of structure of NP essential for vRNP import.
Nuclear import is an essential step that must occur at least twice during the life cycle of the virus: once during nuclear entry of vRNPs, so that the incoming virus can establish a foothold, and then to enable assembly of newly replicated RNA with newly synthesized NP in the course of producing progeny virions. Given the essential nature of these steps and the relatively conserved nature of NP across different strains of influenza virus, nuclear import may be a promising target for development of broad-spectrum antivirals. A recent screen for small molecules capable of inhibiting influenza virus replication identified nucleozin as a potent inhibitor of NP nuclear import (51). Nucleozin treatment does not specifically affect nuclear import of vRNPs, indicating that it must target an aspect of NP function different from that blocked by our αNP-VHHs.
A detailed examination of the function of influenza virus NP protein in replication beyond its identification as an essential protein has been challenging. Excepting some temperature-sensitive influenza virus strains, efforts to generate viable influenza virus with mutant versions of NP displaying complete loss of function in, for example, nuclear import or nuclear localization have so far proven unsuccessful (7, 52, 53). In addition, only a few small molecules to date have been identified that targets NP function (51, 54–58). The use of intracellularly expressed single-domain antibodies represents an alternative for studying the function of essential proteins encoded by RNA viruses. The specificity of VHHs and their high affinity and ease of expression in both prokaryotic and eukaryotic cells, paired with their suitability as crystallization chaperones (59), make them attractive tools to unlock aspects of the influenza virus life cycle that have been difficult to access by other means.
ACKNOWLEDGMENTS
F.I.S. and M.C. were supported by an Early Postdoc.Mobility Fellowship from the Swiss National Science Foundation (SNSF).
REFERENCES
- 1.Gottschalk A, Fezekas De St Groth S. 1960. On the relationship between the indicator profile and prosthetic group of mucoproteins inhibitory for influenza virus haemagglutinin. J Gen Microbiol 22:690–697. doi: 10.1099/00221287-22-3-690. [DOI] [PubMed] [Google Scholar]
- 2.Graham ER, Gottschalk A. 1960. Studies on mucoproteins. I. The structure of the prosthetic group of ovine submaxillary gland mucoprotein. Biochim Biophys Acta 38:513–524. [DOI] [PubMed] [Google Scholar]
- 3.Hirst GK. 1948. The nature of the virus receptors of red cells; evidence on the chemical nature of the virus receptors of red cells and of the existence of a closely analogous substance in normal serum. J Exp Med 87:301–314. doi: 10.1084/jem.87.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matlin KS, Reggio H, Helenius A, Simons K. 1981. Infectious entry pathway of influenza virus in a canine kidney cell line. J Cell Biol 91:601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doms RW, Helenius A, White J. 1985. Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change. J Biol Chem 260:2973–2981. [PubMed] [Google Scholar]
- 6.Martin K, Helenius A. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117–130. doi: 10.1016/0092-8674(91)90576-K. [DOI] [PubMed] [Google Scholar]
- 7.Ozawa M, Fujii K, Muramoto Y, Yamada S, Yamayoshi S, Takada A, Goto H, Horimoto T, Kawaoka Y. 2007. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J Virol 81:30–41. doi: 10.1128/JVI.01434-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cros JF, Garcia-Sastre A, Palese P. 2005. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 6:205–213. doi: 10.1111/j.1600-0854.2005.00263.x. [DOI] [PubMed] [Google Scholar]
- 9.Weber F, Kochs G, Gruber S, Haller O. 1998. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 250:9–18. doi: 10.1006/viro.1998.9329. [DOI] [PubMed] [Google Scholar]
- 10.Neumann G, Castrucci MR, Kawaoka Y. 1997. Nuclear import and export of influenza virus nucleoprotein. J Virol 71:9690–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Neill RE, Jaskunas R, Blobel G, Palese P, Moroianu J. 1995. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem 270:22701–22704. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 12.Huet S, Avilov SV, Ferbitz L, Daigle N, Cusack S, Ellenberg J. 2010. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J Virol 84:1254–1264. doi: 10.1128/JVI.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol 14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 14.Chou YY, Heaton NS, Gao Q, Palese P, Singer RH, Lionnet T. 2013. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog 9:e1003358. doi: 10.1371/journal.ppat.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas SK, Boutz PL, Nayak DP. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J Virol 72:5493–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol 88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elton D, Simpson-Holley M, Archer K, Medcalf L, Hallam R, McCauley J, Digard P. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J Virol 75:408–419. doi: 10.1128/JVI.75.1.408-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Digard P, Elton D, Bishop K, Medcalf E, Weeds A, Pope B. 1999. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J Virol 73:2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Neill RE, Palese P. 1995. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology 206:116–125. doi: 10.1016/S0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 20.Momose F, Basler CF, O'Neill RE, Iwamatsu A, Palese P, Nagata K. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J Virol 75:1899–1908. doi: 10.1128/JVI.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackenzie JS, Dimmock NJ. 1973. A preliminary study of physiological characteristics of temperature-sensitive mutants of influenza virus. J Gen Virol 19:51–63. doi: 10.1099/0022-1317-19-1-51. [DOI] [PubMed] [Google Scholar]
- 22.Palese P, Tobita K, Ueda M, Compans RW. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 24.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. 1993. Naturally occurring antibodies devoid of light chains. Nature 363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 25.Holliger P, Hudson PJ. 2005. Engineered antibody fragments and the rise of single domains. Nat Biotechnol 23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 26.Cardoso FM, Ibanez LI, Van den Hoecke S, De Baets S, Smet A, Roose K, Schepens B, Descamps FJ, Fiers W, Muyldermans S, Depicker A, Saelens X. 2014. Single-domain antibodies targeting neuraminidase protect against an H5N1 influenza virus challenge. J Virol 88:8278–8296. doi: 10.1128/JVI.03178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibanez LI, De Filette M, Hultberg A, Verrips T, Temperton N, Weiss RA, Vandevelde W, Schepens B, Vanlandschoot P, Saelens X. 2011. Nanobodies with in vitro neutralizing activity protect mice against H5N1 influenza virus infection. J Infect Dis 203:1063–1072. doi: 10.1093/infdis/jiq168. [DOI] [PubMed] [Google Scholar]
- 28.Acharya P, Luongo TS, Georgiev IS, Matz J, Schmidt SD, Louder MK, Kessler P, Yang Y, McKee K, O'Dell S, Chen L, Baty D, Chames P, Martin L, Mascola JR, Kwong PD. 2013. Heavy chain-only IgG2b llama antibody effects near-pan HIV-1 neutralization by recognizing a CD4-induced epitope that includes elements of coreceptor- and CD4-binding sites. J Virol 87:10173–10181. doi: 10.1128/JVI.01332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vercruysse T, Pardon E, Vanstreels E, Steyaert J, Daelemans D. 2010. An intrabody based on a llama single-domain antibody targeting the N-terminal alpha-helical multimerization domain of HIV-1 rev prevents viral production. J Biol Chem 285:21768–21780. doi: 10.1074/jbc.M110.112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, Leonhardt H. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods 3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 31.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, Leonhardt H, Hopfner KP, Rothbauer U. 2010. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 32.Caussinus E, Kanca O, Affolter M. 2012. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol 19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 33.Vanlandschoot P, Stortelers C, Beirnaert E, Ibanez LI, Schepens B, Depla E, Saelens X. 2011. Nanobodies(R): new ammunition to battle viruses. Antiviral Res 92:389–407. doi: 10.1016/j.antiviral.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Avalos AM, Bilate AM, Witte MD, Tai AK, He J, Frushicheva MP, Thill PD, Meyer-Wentrup F, Theile CS, Chakraborty AK, Zhuang X, Ploegh HL. 2014. Monovalent engagement of the BCR activates ovalbumin-specific transnuclear B cells. J Exp Med 211:365–379. doi: 10.1084/jem.20131603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, Bernardi RJ, Fu X, Bland CS, Cooper TA, Schiff R, Rosen JM, Westbrook TF, Elledge SJ. 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A 108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maass DR, Sepulveda J, Pernthaner A, Shoemaker CB. 2007. Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). J Immunol Methods 324:13–25. doi: 10.1016/j.jim.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popp MW, Karssemeijer RA, Ploegh HL. 2012. Chemoenzymatic site-specific labeling of influenza glycoproteins as a tool to observe virus budding in real time. PLoS Pathog 8:e1002604. doi: 10.1371/journal.ppat.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleetwood F, Devoogdt N, Pellis M, Wernery U, Muyldermans S, Stahl S, Lofblom J. 2013. Surface display of a single-domain antibody library on Gram-positive bacteria. Cell Mol Life Sci 70:1081–1093. doi: 10.1007/s00018-012-1179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claessen JH, Mueller B, Spooner E, Pivorunas VL, Ploegh HL. 2010. The transmembrane segment of a tail-anchored protein determines its degradative fate through dislocation from the endoplasmic reticulum. J Biol Chem 285:20732–20739. doi: 10.1074/jbc.M110.120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 41.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. 2007. Sortagging: a versatile method for protein labeling. Nat Chem Biol 3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 42.Popp MW, Antos JM, Ploegh HL. 2009. Site-specific protein labeling via sortase-mediated transpeptidation. Curr Protoc Protein Sci Chapter 15:Unit 15.13. doi: 10.1002/0471140864.ps1503s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witte MD, Cragnolini JJ, Dougan SK, Yoder NC, Popp MW, Ploegh HL. 2012. Preparation of unnatural N-to-N and C-to-C protein fusions. Proc Natl Acad Sci U S A 109:11993–11998. doi: 10.1073/pnas.1205427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dougan SK, Ashour J, Karssemeijer RA, Popp MW, Avalos AM, Barisa M, Altenburg AF, Ingram JR, Cragnolini JJ, Guo C, Alt FW, Jaenisch R, Ploegh HL. 2013. Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. Nature 503:406–409. doi: 10.1038/nature12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stojdl DF, Lichty BD, ten Oever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275. doi: 10.1016/S1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe T, Watanabe S, Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu WW, Pante N. 2009. The directionality of the nuclear transport of the influenza A genome is driven by selective exposure of nuclear localization sequences on nucleoprotein. Virol J 6:68. doi: 10.1186/1743-422X-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bullido R, Gomez-Puertas P, Albo C, Portela A. 2000. Several protein regions contribute to determine the nuclear and cytoplasmic localization of the influenza A virus nucleoprotein. J Gen Virol 81:135–142. [DOI] [PubMed] [Google Scholar]
- 49.Wu WW, Sun YH, Pante N. 2007. Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol J 4:49. doi: 10.1186/1743-422X-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu T, Takizawa N, Watanabe K, Nagata K, Kobayashi N. 2011. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP. FEBS Lett 585:41–46. doi: 10.1016/j.febslet.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 51.Kao RY, Yang D, Lau LS, Tsui WH, Hu L, Dai J, Chan MP, Chan CM, Wang P, Zheng BJ, Sun J, Huang JD, Madar J, Chen G, Chen H, Guan Y, Yuen KY. 2010. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol 28:600–605. doi: 10.1038/nbt.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noton SL, Simpson-Holley M, Medcalf E, Wise HM, Hutchinson EC, McCauley JW, Digard P. 2009. Studies of an influenza A virus temperature-sensitive mutant identify a late role for NP in the formation of infectious virions. J Virol 83:562–571. doi: 10.1128/JVI.01424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Watanabe T, Hatta M, Watanabe S, Nanbo A, Ozawa M, Kakugawa S, Shimojima M, Yamada S, Neumann G, Kawaoka Y. 2009. Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein. J Virol 83:4153–4162. doi: 10.1128/JVI.02642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagiwara K, Kondoh Y, Ueda A, Yamada K, Goto H, Watanabe T, Nakata T, Osada H, Aida Y. 2010. Discovery of novel antiviral agents directed against the influenza A virus nucleoprotein using photo-cross-linked chemical arrays. Biochem Biophys Res Commun 394:721–727. doi: 10.1016/j.bbrc.2010.03.058. [DOI] [PubMed] [Google Scholar]
- 55.Shen YF, Chen YH, Chu SY, Lin MI, Hsu HT, Wu PY, Wu CJ, Liu HW, Lin FY, Lin G, Hsu PH, Yang AS, Cheng YS, Wu YT, Wong CH, Tsai MD. 2011. E339.R416 salt bridge of nucleoprotein as a feasible target for influenza virus inhibitors. Proc Natl Acad Sci U S A 108:16515–16520. doi: 10.1073/pnas.1113107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su CY, Cheng TJ, Lin MI, Wang SY, Huang WI, Lin-Chu SY, Chen YH, Wu CY, Lai MM, Cheng WC, Wu YT, Tsai MD, Cheng YS, Wong CH. 2010. High-throughput identification of compounds targeting influenza RNA-dependent RNA polymerase activity. Proc Natl Acad Sci U S A 107:19151–19156. doi: 10.1073/pnas.1013592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerritz SW, Cianci C, Kim S, Pearce BC, Deminie C, Discotto L, McAuliffe B, Minassian BF, Shi S, Zhu S, Zhai W, Pendri A, Li G, Poss MA, Edavettal S, McDonnell PA, Lewis HA, Maskos K, Mortl M, Kiefersauer R, Steinbacher S, Baldwin ET, Metzler W, Bryson J, Healy MD, Philip T, Zoeckler M, Schartman R, Sinz M, Leyva-Grado VH, Hoffmann HH, Langley DR, Meanwell NA, Krystal M. 2011. Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers. Proc Natl Acad Sci U S A 108:15366–15371. doi: 10.1073/pnas.1107906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lejal N, Tarus B, Bouguyon E, Chenavas S, Bertho N, Delmas B, Ruigrok RW, Di Primo C, Slama-Schwok A. 2013. Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob Agents Chemother 57:2231–2242. doi: 10.1128/AAC.02335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tereshko V, Uysal S, Koide A, Margalef K, Koide S, Kossiakoff AA. 2008. Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold. Protein Sci 17:1175–1187. doi: 10.1110/ps.034892.108. [DOI] [PMC free article] [PubMed] [Google Scholar]