ABSTRACT
In late 2011, an A(H3N8) influenza virus infection resulted in the deaths of 162 New England harbor seals. Virus sequence analysis and virus receptor binding studies highlighted potential markers responsible for mammalian adaptation and a mixed receptor binding preference (S. J. Anthony, J. A. St Leger, K. Pugliares, H. S. Ip, J. M. Chan, Z. W. Carpenter, I. Navarrete-Macias, M. Sanchez-Leon, J. T. Saliki, J. Pedersen, W. Karesh, P. Daszak, R. Rabadan, T. Rowles, W. I. Lipkin, MBio 3:e00166-00112, 2012, http://dx.doi.org/10.1128/mBio.00166-12). Here, we present a detailed structural and biochemical analysis of the surface antigens of the virus. Results obtained with recombinant proteins for both the hemagglutinin and neuraminidase indicate a true avian receptor binding preference. Although the detection of this virus in new species highlights an increased potential for cross-species transmission, our results indicate that the A(H3N8) virus currently poses a low risk to humans.
IMPORTANCE Cross-species transmission of zoonotic influenza viruses increases public health concerns. Here, we report a molecular and structural study of the major surface proteins from an A(H3N8) influenza virus isolated from New England harbor seals. The results improve our understanding of these viruses as they evolve and provide important information to aid ongoing risk assessment analyses as these zoonotic influenza viruses continue to circulate and adapt to new hosts.
INTRODUCTION
Influenza A viruses (IAVs) are classified into subtypes according to the serological reactivity of their surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) (1, 2). While 16 HA (H1 to H16) and 9 NA (N1 to N9) subtypes have been identified in wild aquatic birds in the last 100 years, only 3 have adapted to the human population, resulting in four pandemics: H1N1 in 1918 and 2009, H2N2 in 1957, and H3N2 in 1968 (3–6). Although aquatic birds are believed to be the natural reservoir for influenza viruses (7), two novel subtypes of influenza A viruses, H17N10 and H18N11, were recently described in New World bats, which thus may be an alternate reservoir of influenza viruses in nature (8, 9).
Since the first isolation of H1N1 IAV in swine (10) and the subsequent observation that ferrets were susceptible to IAV (11), it became evident that multiple IAV subtypes of either avian or human descent can infect a number of mammalian species (1). Indeed, over the last decade, the perception and impact of influenza virus infections in animals have changed dramatically as a result of a number of recent outbreaks in poultry involving viruses from the H5, H7, and H9 subtypes that resulted in >1,000 human infections, as well as the swine origin of the recent A(H1N1)pdm09 pandemic virus (4, 12–14). This has prompted some governments to invest heavily in global surveillance, research, and capacity building. Public health officials now have a keen interest in all IAVs, but particularly in whether animals, such as pigs, that are closely associated with humans can represent a possible pathway for increased interspecies virus adaptation and transmission.
Similar to the infection of pigs, IAV (i.e., H7N7, H4N5, H3N3, H13N2, H13N9, and H4N6 subtypes) and influenza B virus infections have been detected in marine mammals, including seals, whales, and sea otters (15–22). In November 2011, a marine mammal “unusual mortality event” was declared for Maine, New Hampshire, and northern Massachusetts in response to a large number of deaths in the New England harbor seal population. Sequence analysis revealed that an avian-like H3N8 influenza A virus, A/harbor seal/Massachusetts/1/2011 (seal11), was the cause of the outbreak (23). Although A(H3N8), usually found in avian, canine, equine, and swine hosts (24–27), had been isolated from a seal prior to this event (28), the high mortality observed increased interest from public health officials.
Here, we focus our studies on the two surface antigens, HA and NA, critical to host cell entry and release/spread. To better understand HA and NA at the molecular level, we expressed recombinant HA (recHA) and NA (recNA) antigens to analyze HA receptor binding interactions using glycan microarray and biolayer interferometry analyses, NA cleavage activity, and binding to current antivirals. In addition, three-dimensional atomic structures were determined, including an HA complex with an avian receptor analog (3′-sialyl-N-acetyllactosamine [3-SLN]) and NA complexes with the NA inhibitors oseltamivir carboxylate (Ose) and zanamivir (Zan). Although the results suggest that the A(H3N8) virus currently poses a low risk to humans, it is important to maintain continued surveillance to assess these viruses as they evolve.
MATERIALS AND METHODS
Recombinant HA and NA cloning and expression.
The cDNAs for the ectodomains of the H3 HA (residues 1 to 503 in mature-protein numbering) and N8 NA (residues 80 to 470) from A/harbor seal/Massachusetts/1/2011 (GenBank accession numbers JQ433879 and JQ433881) were synthesized by GenScript USA Inc. (Piscataway, NJ) as a codon-optimized gene for insect cell expression and subcloned into the baculovirus transfer vector pAcGP67-B (BD Biosciences). To improve expression and purification, the N terminus of the HA sequence (QNPSENNNNTATL) was modified at two positions (underlined) (QKPSENDNNTATL), and a C-terminal thrombin site followed by a trimerizing sequence (foldon) from the bacteriophage T4 fibritin and a His tag were added (29). The recombinant seal11 NA protein contained an N-terminal His tag, a tetramerization domain from the human vasodilator-stimulated phosphoprotein (30), and a thrombin cleavage site (31). Residue 84 of seal11 NA was mutated (Asn84Gln) to remove the potential glycosylation site (NNT to QNT). Secreted proteins were recovered from the culture supernatant and purified by metal affinity chromatography and size exclusion chromatography (SEC) as described previously (29, 32–35). For structural analyses, the proteins were further subjected to trypsin cleavage and SEC. The trypsin-treated proteins were buffer exchanged into 10 mM Tris-HCl, 50 mM NaCl, pH 8.0, and the HAs and NAs were concentrated to 16 mg/ml and 3 mg/ml, respectively, for crystallization trials.
Crystallization and data collection.
Initial crystallization trials were set up using a Topaz Free Interface Diffusion (FID) Crystallizer system (Fluidigm Corporation, San Francisco, CA). Crystals were observed under conditions containing various polyethylene glycol (PEG) polymers of different molecular weights. Following optimization, diffraction quality crystals for HA were obtained at 20°C using a modified method for microbath under oil (36) by mixing the protein with reservoir solution containing 0.2 M ammonium iodide, 20% PEG 3350. For receptor analog complexes, crystals were soaked for 1 h in the crystallization buffer containing 10 mM 3-SLN or 6′-sialyl-N-acetyllactosamine (6-SLN) (V-Labs Inc., Covington, LA). However, the soaking of 6-SLN dramatically decreased the crystal diffraction, and no diffraction quality data could be collected. Crystals were flash-cooled at 100 K using 20% glycerol as a cryoprotectant.
While diffraction quality crystals for NA were obtained at 20°C using the same method, the crystallization conditions differed (0.01 M NiCl2, 0.1 M Tris-HCl, pH 8.5, 20% PEG 2000 MME). Crystals of the NA-Ose and NA-Zan complexes were obtained by soaking in 20 mM inhibitor for 3 h. The crystals were flash-cooled at 100 K. Data were collected at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7-1 and Advanced Photon Source (APS) beamline 22-ID at 100 K and processed with the Denzo-Scalepack suite (37). The statistics for data collection are presented in Table 1.
TABLE 1.
Data collection and refinement
| Parameter | Value(s)a for: |
||||
|---|---|---|---|---|---|
| HA | HA–3-SLN | NA | NA-Ose | NA-Zan | |
| Data collection statistics | |||||
| Space group | P1 | P1 | I4 | I4 | I4 |
| Cell dimensions (Å) | 80.06, 103.16, 110.86 | 79.54, 102.33, 109.67 | 90.82, 90.82, 110.45 | 90.67, 90.67, 108.354 | 90.45, 90.45, 108.98 |
| Cell angle (°) | 89.95, 90, 90.28 | 90.02, 90.02, 89.51 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–1.9 (1.95–1.9) | 50–2.5 (2.59–2.5) | 50–1.8 (1.85–1.8) | 50–1.95 (2–1.95) | 50–1.95 (2–1.95) |
| Rsym or Rmerge | 0.111 (0.646) | 0.101 (0.597) | 0.088 (0.291) | 0.115 (0.421) | 0.075 (0.272) |
| I/σ | 12.4 (2.6) | 15.4 (2.3) | 20.5 (3.2) | 22.7 (4.2) | 25.9 (5.1) |
| Completeness (%) | 97.1 (93.1) | 96.5 (92.2) | 94.9 (84.5) | 99.1 (92.7) | 97.5 (99.3) |
| Redundancy | 3.8 (3.6) | 3.7 (3.5) | 2.7 (2.1) | 4.6 (3.9) | 2.8 (2.6) |
| Refinement | |||||
| Resolution (Å) | 50–1.9 (1.95–1.9) | 50–2.5 (2.59–2.5) | 50–1.8 (1.85–1.8) | 50–1.95 (2–1.95) | 50–1.95 (2–1.95) |
| No. of reflections (total) | 1,032,389 | 423,475 | 103,969 | 146,772 | 85,252 |
| No. of reflections (test) | 272,212 | 115,418 | 39,209 | 32,134 | 31,115 |
| Rwork/Rfree | 16.9/19.5 | 18.5/23.6 | 14.6/17.1 | 17.3/20.1 | 20.4/24.2 |
| No. of atoms | 26,664 | 24,811 | 3,344 | 3,323 | 3,033 |
| RMSD, bond length (Å) | 0.007 | 0.015 | 0.021 | 0.008 | 0.019 |
| RMSD, bond angle (°) | 1.12 | 1.74 | 2.04 | 1.15 | 2.18 |
| MolProbity scoresb | |||||
| Favored (%) | 97 | 96 | 94 | 96 | 91 |
| % Outliersc | 0 | 0.24 (7/2,922) | 0 | 0 | 0.26 (1/386) |
| PDB code | 4WA1 | 4WA2 | 4WA3 | 4WA4 | 4WA5 |
The numbers in parentheses refer to the highest-resolution shell unless otherwise specified.
Data from the Molprobity server (39).
For the outlier values, the numbers in parentheses are the number of outlier residues and the total number of residues.
Structure determination and refinement.
The seal HA and NA structures were determined by molecular replacement with Phaser (38). For the HA structure, A/Finland/486/2004 (H3N2), Protein Data Bank identifier (PDB ID) 2YP2 (HA1, 80% identity; HA2, 93% identity), was used as a search model. Six hemagglutinin monomers forming one noncrystallographic trimer and three monomers that form one-third and two-thirds of two crystallographic trimers occupy the asymmetric unit with an estimated solvent content of 49.1% based on a Matthews' coefficient (Vm) of 2.42 Å3/Da. The model was then “mutated” to the correct sequence and rebuilt with Coot (39), and then the protein structures were refined with REFMAC (40) using TLS refinement (41) and Phenix refine (42). The HA–3-SLN complex structure was refined using the same strategy. Of the six monomers in the asymmetric unit, due to the low occupancy of the binding sites, only two 3-SLN molecules could be built into the final structure.
The seal NA structure was solved using the NA structure from A/duck/Ukraine/1/63 (H3N8), PDB ID 2HT5 (sequence identity is 90%), as a search model. One neuraminidase monomer occupied the asymmetric unit with an estimated solvent content of 53.4% based on a Vm of 2.64 Å3/Da. The apo form NA and two complexed NA structures were refined using the same strategy as for the seal HA structure. All final models were assessed using MolProbity (43), and the statistics for data processing and refinement are presented in Table 1.
Glycan binding analyses.
A/harbor seal/New Hampshire/179629/2011 virus was propagated in 10-day-old embryonated chicken eggs. The virus was inactivated by treatment with β-propiolactone (1 part β-propiolactone to 1,000 parts infected allantoic fluid) at 4°C for 20 h, with virus inactivation confirmed by two rounds of passage in eggs with no detectable red blood cell hemagglutination titers in the allantoic fluid. The inactivated virus was diluted in phosphate-buffered saline containing 2% (wt/vol) bovine serum albumin (PBS-BSA) to an HA titer of 256 and incubated on the microarray slide on ice for 1 h with gentle agitation. Unbound virus was washed off by sequential washes with PBS containing 0.05% Tween 20 (PBST) and PBS. The slide was then subjected to successive 1-h incubations with anti-A/harbor seal/New Hampshire/179629/2011 ferret immune serum (1:1,000 dilution in PBS-BSA), biotinylated anti-ferret IgG (1:200 dilution; Rockland Immunochemicals), and streptavidin-Alexa Fluor 488 (1:1,000; Life Technologies) with PBST/PBS washes between incubations.
For analysis of recHA, HA-antibody precomplexes were prepared by mixing HA (10 μl; 1 mg/ml), mouse anti-penta-His–Alexa Fluor 488 (17.5 μl; 0.2 mg/ml; Qiagen Inc.), and anti-mouse IgG-Alexa Fluor 488 (1.2 μl; 2 mg/ml; Life Technologies) in a molar ratio of 4:2:1, respectively. The mixtures were incubated for 60 min on ice and then diluted with 500 μl PBS-BSA containing streptavidin-Alexa Fluor 488 (1:1,000; Life Technologies) and incubated on the microarray slide on ice for 90 min.
After the final incubation step, the slides were washed by successive rinses in PBST, PBS, and deionized water and then dried. Fluorescence intensities were detected using a ProScanArray HT microarray scanner (Perkin-Elmer). Image analyses were carried out using ImaGene 8 image analysis software (BioDiscovery Inc.). Table 2 shows the specific glycans on the array and lists the glycans used in the experiments, as well as a tabulated qualitative assessment of binding for each protein analyzed.
Table 2.
Glycan microarray for A/Harbor Seal/New Hampshire/179629/2011 H3 HA
| Glycan no.a | Glycan structureb | Bindingc |
|
|---|---|---|---|
| Seal H3 recHA | Seal H3 virus | ||
| 1 | Neu5Acα | NB | NB |
| 2 | Neu5Acα | NB | NB |
| 3 | Neu5Acβ | NB | NB |
| 4 | Neu5Acα2-3(6OSO3)Galβ1-4GlcNAcβ | NB | + |
| 5 | Neu5Acα2-3Galβ1-3[6OSO3]GalNAcα | +++ | ++ |
| 6 | Neu5Acα2-3Galβ1-4[6OSO3]GlcNAcβ | +++ | ++ |
| 7 | Neu5Acα2-3Galβ1-4(Fucα1-3)[6OSO3]GlcNAcβ-propyl-NH2 | +++ | ++ |
| 8 | Neu5Acα2-3Galβ1-3[6OSO3]GlcNAc-propyl-NH2 | +++ | ++ |
| 9 | Neu5Acα2-3Galβ1-3(Neu5Acα2-3Galβ1-4)GlcNAcβ | +++ | +++ |
| 10 | Neu5Acα2-3Galβ1-3(Neu5Acα2-3Galβ1-4GlcNAcβ1-6)GalNAcα | +++ | +++ |
| 11 | Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | +++ | +++ |
| 12 | Neu5Acα(2-3)-Galβ(1-4)-GlcNAcβ(1-3)-Galβ(1-4)-GlcNAcβ(1-2)-Manα(1-3)-[Neu5Acα(2-3)-Galβ(1-4)-GlcNAcβ(1-3)-Galβ(1-4)-GlcNAcβ(1-2)-Manα(1-6)]-Manβ(1-4)-GlcNAcβ(1-4)-GlcNAcβ | +++ | ++ |
| 13 | Neu5Acα2-3Galβ | +++ | ++ |
| 14 | Neu5Acα2-3Galβ1-3GalNAcα | +++ | +++ |
| 15 | Neu5Acα2-3Galβ1-3GlcNAcβ | +++ | ++ |
| 16 | Neu5Acα2-3Galβ1-3GlcNAcβ | +++ | ++ |
| 17 | Neu5Acα2-3Galβ1-4Glcβ | +++ | + |
| 18 | Neu5Acα2-3Galβ1-4Glcβ | +++ | + |
| 19 | Neu5Acα2-3Galβ1-4GlcNAcβ | +++ | ++ |
| 20 | Neu5Acα2-3Galβ1-4GlcNAcβ | +++ | ++ |
| 21 | Neu5Acα2-3GalNAcβ1-4GlcNAcβ | NB | +++ |
| 22 | Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | +++ | ++ |
| 23 | Neu5Aca2-3Galβ1-3GlcNAcβ1-3Galβ1-4GlcNAcβ | +++ | ++ |
| 24 | Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | +++ | + |
| 25 | Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-3GlcNAcβ | +++ | ++ |
| 26 | Neu5Acα2-3Galβ1-3GalNAcα | +++ | ++ |
| 27 | Galβ1-3(Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-6)GalNAcα | NB | NB |
| 28 | Neu5Acα2-3Galβ1-3(Fucα1-4)GlcNAcβ | +++ | +++ |
| 29 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ | +++ | +++ |
| 30 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ | +++ | +++ |
| 31 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ | +++ | +++ |
| 32 | Neu5Acα2-3Galβ1-3[Fucα1-4]GlcNAcβ1-3Galβ1-4[Fucα1-3]GlcNAcβ | +++ | +++ |
| 33 | Neu5Acα2-3Galβ1-3[Fucα1-3]GlcNAcβ1-3Galβ1-4[Fucα1-3]GlcNAcβ | +++ | +++ |
| 34 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ | +++ | +++ |
| 35 | Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcNAcβ | NB | NB |
| 36 | Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcNAcβ | NB | + |
| 37 | Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glcβ | NB | NB |
| 38 | Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ | NB | NB |
| 39 | Fucα1-2Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ | NB | NB |
| 40 | Fucα1-2Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ | NB | NB |
| 41 | Neu5Acα2-6Galβ1-4[6OSO3]GlcNAcβ | NB | NB |
| 42 | Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB |
| 43 | Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB |
| 44 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-2Manα1-3[Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-2Manα1-6]Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB |
| 45 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-2Manα1-3[Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-2Manα1-6]-Manβ1-4GlcNAcβ1-4GlcNAcβ | + | + |
| 46 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3[Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-6]GalNAca | NB | NB |
| 47 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3[Neu5Acα2-6Galβ1-4GlcNAcβ1-6]GalNAca | NB | NB |
| 48 | Neu5Acα2-6GalNAcα | NB | NB |
| 49 | Neu5Acα2-6Galβ | NB | NB |
| 50 | Neu5Acα2-6Galβ1-4Glcβ | NB | NB |
| 51 | Neu5Acα2-6Galβ1-4Glcβ | NB | NB |
| 52 | Neu5Acα2-6Galβ1-4GlcNAcβ | NB | NB |
| 53 | Neu5Acα2-6Galβ1-4GlcNAcβ | NB | NB |
| 54 | Neu5Acα2-6GalNAcβ1-4GlcNAcβ | NB | NB |
| 55 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3GalNAcα | NB | NB |
| 56 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | NB | NB |
| 57 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3GalNAcα | NB | NB |
| 58 | Neu5Aca2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | NB | NB |
| 59 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ | NB | NB |
| 60 | Galβ1-3(Neu5Acα2-6)GlcNAcβ1-4Galβ1-4Glcβ-Sp10 | NB | NB |
| 61 | Neu5Acα2-6[Galβ1-3]GalNAca | NB | NB |
| 62 | Neu5Acα2-6Galβ1-4GlcNAcβ1-6[Galβ1-3]GalNAca | NB | + |
| 63 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-6[Galβ1-3]GalNAca | NB | NB |
| 64 | Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB |
| 65 | Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | +++ | ++ |
| 66 | Neu5Acα2-3Galβ1-3(Neu5Acα2-6)GalNAcα | +++ | +++ |
| 67 | Neu5Acα2-3(Neu5Acα2-6)GalNAcα | NB | NB |
| 68 | Neu5Gcα | NB | NB |
| 69 | Neu5Gcα2-3Galβ1-3(Fucα1-4)GlcNAcβ | NB | NB |
| 70 | Neu5Gcα2-3Galβ1-3GlcNAcβ | +++ | + |
| 71 | Neu5Gcα2-3Galβ1-4(Fucα1-3)GlcNAcβ | +++ | +++ |
| 72 | Neu5Gcα2-3Galβ1-4GlcNAcβ | NB | NB |
| 73 | Neu5Gcα2-6GalNAcα | NB | NB |
| 74 | Neu5Gcα2-6Galβ1-4GlcNAcβ | NB | NB |
| 75 | Neu5Acα2-8Neu5Acα | NB | NB |
| 76 | Neu5Acα2-8Neu5Acα2-8Neu5Acα | NB | NB |
| 77 | Neu5Acα2-8Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glcβ | NB | NB |
| 78 | Neu5Acα2-8Neu5Acα2-3Galβ1-4Glcβ | NB | NB |
| 79 | Neu5Acα2-8Neu5Acα2-8Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glcβ | NB | NB |
| 80 | Neu5Acα2-8Neu5Acα2-8Neu5Acα2-3Galβ1-4Glcβ | NB | NB |
| 81 | Neu5Acα2-8Neu5Acβ-Sp17 | NB | NB |
| 82 | Neu5Acα2-8Neu5Acα2-8Neu5Acβ | NB | NB |
| 83 | Neu5Acβ2-6GalNAcα | NB | NB |
| 84 | Neu5Acβ2-6Galβ1-4GlcNAcβ | NB | NB |
| 85 | Neu5Gcβ2-6Galβ1-4GlcNAc | NB | NB |
| 86 | Galβ1-3(Neu5Acβ2-6)GalNAcα | NB | NB |
| 87 | [9NAc]Neu5Acα | NB | + |
| 88 | [9NAc]Neu5Acα2-6Galβ1-4GlcNAcβ | NB | NB |
| 89 | Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | NB | NB |
| 90 | Galβ1-3GlcNAcβ1-3Galβ1-3GlcNAcβ | NB | NB |
| 91 | Galβ1-4GlcNAcβ1-2Manα1-3[Galβ1-4GlcNAcβ1-2Manα1-6]Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB |
| 92 | GalNAcα1-3(Fucα1-2)Galβ1-3GlcNAcβ | NB | NB |
| 93 | GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAcβ | NB | NB |
| 94 | Galα1-3(Fucα1-2)Galβ1-3GlcNAcβ | NB | NB |
| 95 | Galα1-3(Fucα1-2)Galβ1-4(Fucα1-3)GlcNAcβ | NB | NB |
| 96 | Galβ1-3GalNAcα | NB | NB |
Different categories of glycans on the array are grouped together as follows: glycans 1 to 3, sialic acid; glycans 4 to 40, α2-3 sialosides; glycans 41 to 63, α2-6 sialosides; glycans 64 to 67, mixed α2-3/α2-6 biantennaries; glycans 68 to 74, N-glycolylneuraminic acid-containing glycans; glycans 75 to 82, α2-8-linked sialosides; glycans 83 to 88, β2-6-linked and 9-O-acetylated sialic acids; glycans 89 to 96, asialo glycans.
Neu5Ac, sialic acid; Neu5Gc, N-glycolylneuraminic acid; OSO3, sulfate; Gal, galactose; Fuc, fucose; Glc, d-glucose; GlcNAc, N-acetyl-d-glucosamine; GalNAc, N-acetyl-d-galactosamine; Man, d-mannose; 9NAc, 9-O-acetyl.
Significant binding of samples to glycans was qualitatively estimated based on the relative strength of the signal for the data shown. Fluorescence intensity, >10,000, +++; 5,000 to 9,999, ++; 1,250 to 4,999, +; <1,250, NB.
For kinetic studies, biotinylated receptor analogs, Neu5Ac(α2-3)Gal(β1-4)GlcNAcb-biotin (3-SLN-b), Neu5Ac(α2-3)Gal(β1-4)GlcNAc(β1-3)Gal(β1-4)GlcNAcb-biotin (3-SLNLN-b), Neu5Ac(α2-6)Gal(β1-4)GlcNAcb-biotin (6-SLN-b), and Neu5Ac(α2-6)Gal(β1-4)GlcNAc(β1-3)Gal(β1-4)GlcNAcb-biotin (6-SLNLN-b), were obtained from the Consortium for Functional Glycomics (http://www.functionalglycomics.org) through the resource request program. The glycans were precoupled to streptavidin-coated biosensors (Fortebio Inc.), and the binding of recombinant HA, diluted to 5.4 μM trimer in kinetics buffer (PBS containing 0.02% Tween 20 and 100 μg/ml bovine serum albumin), was analyzed by biolayer interferometry (BLI) using an Octet Red instrument (Fortebio, Inc.) according to the manufacturer's instructions. Data were analyzed using the system software and fitted to a 1:1 binding model.
NA activity and drug susceptibility assays.
RecNA activities were assessed using the the fluorescent 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA) (Sigma-Aldrich Inc.) as the substrate (44). Briefly, recNA protein was mixed with MUNANA in a reaction buffer containing 32.5 mM 2-(N-morpholino)ethanesulfonic acid (MES), 4 mM CaCl2, and 50 μg/ml of bovine serum albumin. The reaction mixture was incubated at 37°C for 30 min and terminated by the addition of stop solution containing 25% ethanol and 0.1 M glycine. The fluorescence of the enzyme-cleaved product was measured using a Synergy H1 hybrid multimode microplate reader (BioTek) with excitation and emission wavelengths of 360 nm and 460 nm, respectively. To determine the enzyme kinetics of recNA measured as a function of the substrate amount, a series of 2-fold-diluted recNAs were mixed with 200 μM MUNANA, and a sigmoidal curve of NA activity was generated. The NA concentration corresponding to the midpoint of the linear section of the curve was chosen for the kinetics assay. Different concentrations of MUNANA in a series of 2-fold dilutions were mixed with recNA of seal N8 NA or A/Perth/16/2009 N2 NA at the fixed concentrations of 3.13 ng/ml and 31.3 ng/ml, respectively. Reaction parameters (Km, Vmax, and kcat) were calculated by fitting the data to Michaelis-Menten equations using GraphPad Prism software (GraphPad Software Inc.).
RecNA and the egg-propagated A/harbor seal/New Hampshire/179629/2011 A(H3N8) virus were tested in the fluorescent neuraminidase inhibition assay with an NA-Fluor kit (Life Technologies), as described previously (45). The NA inhibitors zanamivir (GlaxoSmithKline) and oseltamivir carboxylate (Roche Diagnostics GmbH) were kindly provided by the respective manufacturers. A drug concentration (nM) required to inhibit enzyme activity by 50% (IC50) was determined for each drug using JASPR v1.2 curve-fitting software (45). Mean IC50 and standard deviation (SD) values were calculated based on the results of two independent tests, each conducted in duplicate. An oseltamivir-sensitive A/California/12/2012 A(H1N1)pdm09 virus and an oseltamivir-resistant A/Texas/23/2012 A(H1N1)pdm09 virus carrying the H275Y NA substitution were used as reference viruses.
Protein structure accession numbers.
The atomic coordinates and structure factors of seal HA (apo and in complex with 3-SLN) and NA (apo and in complex with oseltamivir and zanamivir) are available from the RCSB PDB under accession codes 4WA1, 4WA2, 4WA3, 4WA4, and 4WA5.
RESULTS AND DISCUSSION
Structure of seal11 HA.
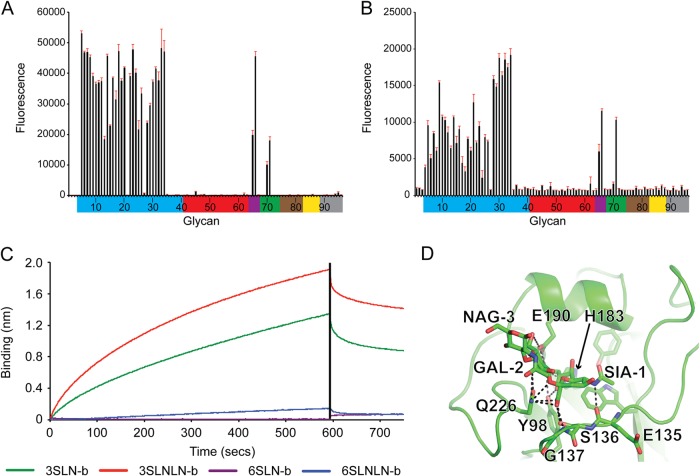
The three-dimensional HA structure of the trimeric ectodomain from seal11 HA was determined by X-ray crystallography at 1.9-Å resolution (Table 1). As expected, the overall structure of the HA monomer is composed of a globular head containing the receptor binding site (RBS) and a membrane-proximal domain that includes a central helical stalk and the HA1/HA2 cleavage site (32, 33, 35, 46–55). Although seven asparagine-linked glycosylation sites (NXS/T) are predicted in the seal11 HA monomer, one at residue 2 (NPS) is not believed to be utilized, due to the presence of the proline at the X position in the sequon (56). Of the remaining 6 sites (residues 8, 22, 38, 165, 285, and 483), interpretable carbohydrate electron density, containing only one or two N-acetylglucosamines, was observed at only three sites, Asn38, Asn165, and Asn285, in HA1 (Fig. 1A).
FIG 1.
Structure of seal HA. (A) Overall structure, with one monomer HA highlighted in green (HA1 domain) and cyan (HA2 domain). Potential glycosylation sites are labeled, and those sites/glycans that could be visualized in the structure are shown as sticks (magenta). (B) Expanded view of the seal HA RBS with its three structural elements comprising the binding site—the 130 loop, the helix, and the 220 loop—colored yellow, red, and purple. Conserved residues are shown as sticks. (C) Comparison of the seal HA RBS (green) with overlapping equivalent structures from avian (cyan) and human (magenta) H3 HAs. Residue differences are shown as sticks. Seal HA is shown in cartoon form, while 3-SLN and interacting HA residues are shown as sticks. All structural figures were generated with MacPyMol (83).
The HA protein is synthesized as a single-chain precursor (HA0) during viral replication and then cleaved by a specific host protease into the infectious HA1 and HA2 forms. In the baculovirus expression system used for these studies, seal11 HA was produced in the HA0 form. For structural studies, seal11 HA was digested with trypsin to remove the trimerization tag, which also cleaves the HA into the active HA1/HA2 form. Comparison of the seal11 HA monomer to an avian H3 HA (PDB ID 1MQL) (49) and a recent human H3 HA (PDB ID 2YP2) (57) revealed a highly similar structure, with the C-α atoms superimposed to give root mean square deviations (RMSD) of 0.86 Å and 0.76 Å, respectively.
Functional and structural analyses of the seal11 HA receptor binding site.
The seal11 HA RBS is at the membrane-distal end of each HA monomer (Fig. 1A), and its specificity for sialic acid and the nature of its linkage to the vicinal galactose residue in host cell surface complex carbohydrates contribute to host range restriction (58). The consensus RBS in all influenza A virus HAs is composed of three structural elements: a 190 helix (residues 188 to 194), a 220 loop (residues 221 to 228), and a 130 loop (residues 134 to 138). In addition, the highly conserved residues Tyr98, Trp153, His183, and Tyr195 form the base of the pocket (Fig. 1B). Comparison of the seal11 HA RBS with those from avian and human H3 HAs (PDB IDs 1MQL and 2YP2) revealed an almost identical RBS (Fig. 1C). Interestingly, while the 220 loop of the human RBS is positioned ∼1.5 Å away from the avian loop, resulting in a more open RBS, the 220 loop of the seal11 HA aligns with the avian RBS (Fig. 1C), suggesting a closer structural relationship to avian than to human HAs. Sequence alignment of only the seal11 residues that comprise the RBS (Fig. 2) with A(H3N8) and A(H3N2) HAs from other species also suggests that the RBS is more avian-like. While it shows 91% identity to the consensus RBS for avian viruses, it shows only 82% and 48% identity to the human A(H3N2) HA RBS from 1968 and 2013, respectively (Table 3). This is in agreement with previous phylogenetic analysis of the seal11 HA that showed it to be closest to the HA from the A/blue-winged teal/Ohio/926/2002 H3N8 virus (23).
FIG 2.
Sequence alignment of the amino acid sequence of seal11 HA with those of the avian A(H3N8) virus A/Duck/Ukraine/1/1963 (dkUkr63; PDB ID 1MQL) and two human HA sequences from A/Aichi/2/1968 (Aichi68; PDB ID 3HMG) and the seasonal H3 vaccine strain from A/Hong Kong/4443/2005 (HK05; PDB ID 2YP7; Gisaid no. EPI397688) (68–71). The sequence alignment is also annotated with additional lines to indicate residues comprising the receptor binding site (Receptor) (labeled r), the solvent-accessible surface residues (Srf res) (labeled s; determined by Areaimol, part of the CCP4 program suite [84, 85]), and the antigenic sites (labeled a, b, c, d, and e) (68–71).
TABLE 3.
Comparison of seal11 RBS to consensus H3 sequences from other species
| RBS | % identity to seal RBS |
|---|---|
| Avian H3N8 (n = 518)a | 91 |
| Equine H3N8 (n = 713) | 82 |
| Canine H3N8 (n = 181) | 86 |
| 1968 Human H3N2 (n = 96) | 82 |
| 2013 Human H3N2 (n = 433) | 48 |
Number of sequences in the database used to generate a consensus sequence.
Previously, a double mutation (Gln226Leu and Gly228Ser) in the HA receptor binding domains of the H2N2 and H3N2 subtypes was shown to be responsible for adaptation of these viruses from birds to humans (59, 60), while recent gain-of-function experiments highlighted their use as a possible route for adaptation if A(H5N1) evolved to infect and transmit efficiently in humans (61, 62). In the seal HA, the 226/constellation is avian-like (Gln226/Gly228). The vicinal Leu222 in the seal11 HA, however, is a Trp in avian and equine A(H3N8) viruses, as well as early human A(H3N2) virus. Interestingly, Leu222 is present in canine A(H3N8) viruses and has been implicated in the adaptation of the equine viruses to a canine host (63, 64), and thus, it may be similarly important in A(H3N8) virus adaptation to seals.
A previous study suggested that the virus bound less efficiently to avian α2-3 receptors than avian A(H3N8) viruses and, based on hemagglutination assays, also concluded it bound to human-type α2-6 receptors (23). More recent data using solid-phase glycan binding assays also suggested greater binding affinity for α2-6 receptors (65). To assess this observation and to gain further insight into the interactions of this A(H3N8) virus with host receptors, glycan microarray analyses of both seal11 recHA and a virus from the same outbreak, A/harbor seal/New Hampshire/179629/2011, were performed. The results for both seal11 recHA and the virus revealed a strong binding preference for the α2-3-linked sialosides, as well as mixed α2-3/α2-6 branched sialosides (glycans 65 to 66). However, little binding to human α2-6-linked sialosides was detected in the assay (Fig. 3A and B and Table 2). Glycan binding to seal11 recHA was further analyzed by BLI using an Octet Red system (ForteBio Inc.). The results of the binding of seal11 HA to biotinylated glycans (3-SLN-b, 3-SLNLN-b, 6-SLN-b, and 6-SLNLN-b) preloaded onto streptavidin-coated biosensors confirmed the glycan array data in that the recHA bound only to the α2-3-linked analogs, with apparent KD (equilibrium dissociation constant) values of approximately 139 and 8 μM for 3-SLN-b and 3-SLNLN-b glycan analogs, respectively (Fig. 3C and Table 4).
FIG 3.
Glycan binding to seal recHA. (A and B) Glycan array analysis of seal11 recHA (A) and A/harbor seal/New Hampshire/179629/2011 (B). Glycans on the microarray are grouped according to SA linkage: α2-3 SA (blue), α2-6 SA (red), α2-6–α2-3 mixed SA (purple), N-glycolyl SA (green), α2-8 SA (brown), β2-6 and 9-O-acetyl SA (yellow), and asialo glycans (gray). The error bars reflect the standard error in the signal for six independent replicates on the array. The structures of the numbered glycans are found in Table 2. Specific glycan structures that were used in biosensor assays are represented on the array as glycans 19/20, 22, 52/53, and 56. (C) The binding kinetics of seal11 recHA protein to specific biotinylated glycans, 3-SLN-b, 3-SLNLN-b, 6-SLN-b, and 6-SLNLN-b, immobilized on streptavidin-coated biosensors were analyzed by BLI. The black vertical line indicates the switch in the experiment from collecting association to collecting dissociation data. (D) 3-SLN glycan interactions with seal11 HA RBS. Seal HA is shown in cartoon form, while 3-SLN and interacting HA residues are shown as sticks. Hydrogen bonds between the glycan and HA are shown as dashed lines. The structural figure was generated with MacPyMol (83).
TABLE 4.
Kinetic results for glycan binding to seal recHAa
| Glycan | Apparent KD (μM) | kon (ms−1) | kobs ± SD (10−3 s−1) | koff ± SD (10−2 s−1) |
|---|---|---|---|---|
| 3-SLN-b | 14.19 | 131 | 2.59 ± 0.014 | 1.86 ± 0.010 |
| 3-SLNLN-b | 18.25 | 166 | 3.95 ± 0.017 | 3.03 ± 0.012 |
| 6-SLN-b | NB | NB | NB | NB |
| 6-SLNLN-b | NB | NB | NB | NB |
KD, equilibrium dissociation constant; kon, kobs, and koff, association, observed, and dissociation rates, respectively; NB, no detectable binding.
To understand from a structural perspective how seal11 HA interacts with host receptors, we determined the structure of seal11 HA in complex with an avian receptor analog, 3-SLN, to 2.5-Å resolution (Table 1). All attempts to obtain diffraction data for seal11 HA in complex with the human receptor analog, 6-SLN, were unsuccessful. For the avian receptor analog, 3-SLN, the electron density maps revealed well-ordered features for SIA-1, GAL-2, and NAG-3 in the seal11 HA complex structure. Most of the hydrogen bonds were formed between SIA-1 and residues (Tyr98, Thr135, Thr136, Ser137, His183, Glu190, and Gln226) within the pocket (Fig. 3D). Tyr98 is highly conserved in all influenza A viruses, and its interaction with sialic acid is critical for receptor interaction and virus function (66). Residue Gln226, one of the key residues in receptor specificity and host adaptation, also binds to GAL-2 in 3-SLN. Structural comparison of seal11 HA–3-SLN to avian H3 HA complexes (PDB ID 1MQM) revealed high similarity. The trans conformation of the α2-3 linkage in the avian receptor analog points out of the RBS, and the terminal SIA-1 and GAL-2 are positioned almost identically in the two structures. Like all previously reported complexes with α2-3-linked analogs, the seal11 H3 HA–3-SLN structure revealed no hydrogen bonding between the HA and NAG-3, which suggests that for α2-3 linkage glycans, only the first two saccharides are required for host receptor binding to avian HAs (33, 35, 48, 50, 67).
Seal HA antigenic sites.
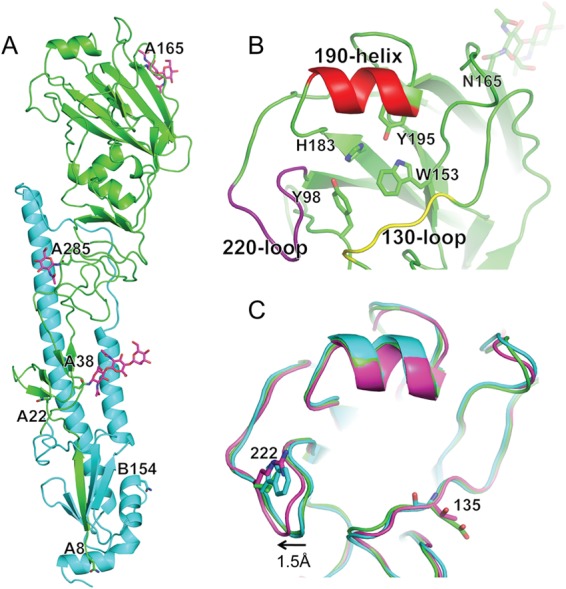
Human seasonal A(H3N2) HA molecules have five distinct antigenic sites (AS): AS-A, AS-B, AS-C, AS-D, and AS-E (68–71) (Fig. 2 and 4A). Although these sites have been under antibody-mediated immune pressure since 1968, the sites nearest the RBS, AS-A, AS-B, and AS-D, are reported to be immunodominant (70). To investigate the structures of these antigenic sites on the seal HA, we compared each site to equivalent positions on the HAs from an A/duck/Ukraine/1/1963 (dkUkr63) H3N8 avian virus and the A/Aichi/2/1968 pandemic virus (Aichi68), as well as a recent human virus, A/Hong Kong/4443/2005 (HK05), H3N2 HA source (Fig. 4B). While seal11 HA differed significantly in shape and charge at all 5 antigenic sites compared to the more recent HK05 HA, it does appear to be antigenically related to both the avian and the early human 1968 pandemic virus HAs (Fig. 4B). Comparison of seal11 HA to consensus sequences for avian H3N8 and H3N2 HAs and human HAs from 1968 and 2013 showed agreement with our structural analysis. While the seal11 HA maintained 73% identity to that of 2013 AS-D, the major antigenic sites, AS-A and AS-B, were at only 52% and 32% identity, respectively (Table 5). In contrast, seal11 HA identity with the avian AS H3N2 and H3N8 HAs ranged from 89 to 96% and 88 to 95%, respectively, across the 5 AS. Thus, current vaccines against the contemporary seasonal H3N2 viruses circulating appear to be a poor match for the seal11 virus should it ever successfully adapt to humans. The recent publication by Karlsson et al. showed that recent seasonal vaccination of humans failed to elicit cross-reactive antibody against the virus, suggesting a lack of population immunity (65).
FIG 4.
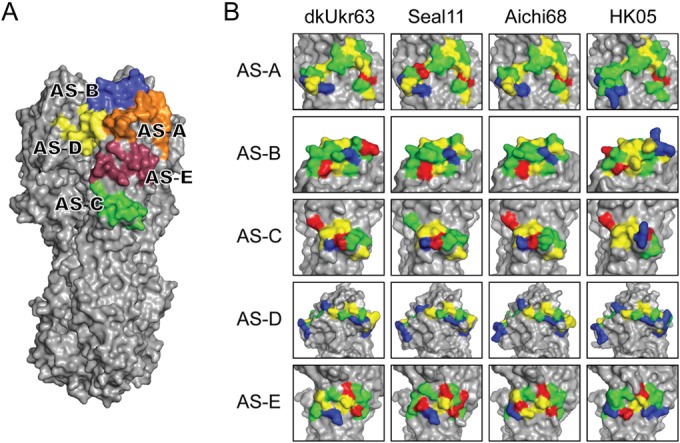
Comparison of seal11 HA antigenic sites with other HA structures. (A) Recognized AS mapped onto the seal11 HA structure, shown as a surface representation and colored individually. (B) Structural comparison of the antigenic sites on the HA molecules between A(H3N8) (avian dkUkr63 and seal11) and seasonal H3N2 (Aichi68 and HK05). Three-dimensional models of the H3 HA molecules of each HA were used and are shown as surface representations. Expanded views of the antigenic sites (A through E) are shown. Amino acids are colored as follows: positive (Arg and Lys), blue; negative (Asp and Glu), red; hydrophobic (Ala, Phe, Gly, Ile, Leu, Met, Val, and Trp), yellow; polar (His, Asn, Gln, Ser, Thr, and Tyr), green.
TABLE 5.
Comparison of seal11 antigenic sites with those of avian A(H3N2) and A(H3N8), early human pandemic A(H3N2) 1968, and recent human seasonal A(H3N2) virus HAs
| AS | No. of amino acids in AS | % identity of consensus sequence to seal11 sequencea |
|||
|---|---|---|---|---|---|
| Avian |
Human pandemic A(H3N2) |
||||
| A(H3N2) | A(H3N8) | 1968 | 2013 | ||
| A | 25 | 96 | 88 | 84 | 52 |
| B | 19 | 89 | 95 | 89 | 32 |
| C | 12 | 92 | 92 | 92 | 42 |
| D | 22 | 91 | 91 | 86 | 73 |
| E | 13 | 92 | 92 | 77 | 46 |
Sequences were downloaded from the GISAID database and aligned/analyzed using CLUSTALX (81).
Structure of seal11 N8 NA.
The recombinant seal11 N8 NA was expressed in a baculovirus expression system. The purified protein was crystallized in the I4 space group, and its structure was determined to 1.8-Å resolution (Table 1) with good electron density for all residues. The seal11 NA structure is a typical “box-shaped” tetrameric association of identical monomers containing six four-stranded, antiparallel β-sheets that form a propeller-like arrangement (Fig. 5A), as previously described for influenza A virus subtypes N1, N2, N4, N5, N8, and N9 and influenza B virus NA (72–76). A comparison of seal11 NA with an avian N8 NA (PDB ID 2HT5) revealed a highly similar structure, and all C-α atoms were superimposed with an RMSD of only 0.36 Å (74). In addition, one calcium ion binding site, which is conserved in all known influenza A and influenza B virus NAs, was observed in seal11 NA (Fig. 5A). Ca2+ is bound through interactions with two water molecules and four backbone carbonyl oxygen atoms from Asn292, Gly296, Asp322, and Tyr344 (residues 293, 297, 324, and 347 in N2 numbering, based on the structural sequence alignment). Calcium ions have been shown to be critical for the thermostability and activity of influenza virus NAs (77, 78), and this conserved metal site was proposed to be important in stabilizing a reactive conformation of the active site by otherwise flexible loops (79). Although seal11 NA has four potential N-linked glycosylation sites at Asn84, Asn144, Asn293, and Asn398 (residues 86, 146, 294, and 402 in N2 numbering), our expression construct included an Asn84Gln substitution that removed one of these sites. For the remaining three sites, the final model had interpretable glycan density at only one site, Asn144 (residue 146 in N2 numbering), which is situated on the membrane-distal surface close to the active site and is the only glycosylation site conserved among all other influenza A and B NAs (72, 74, 80).
FIG 5.
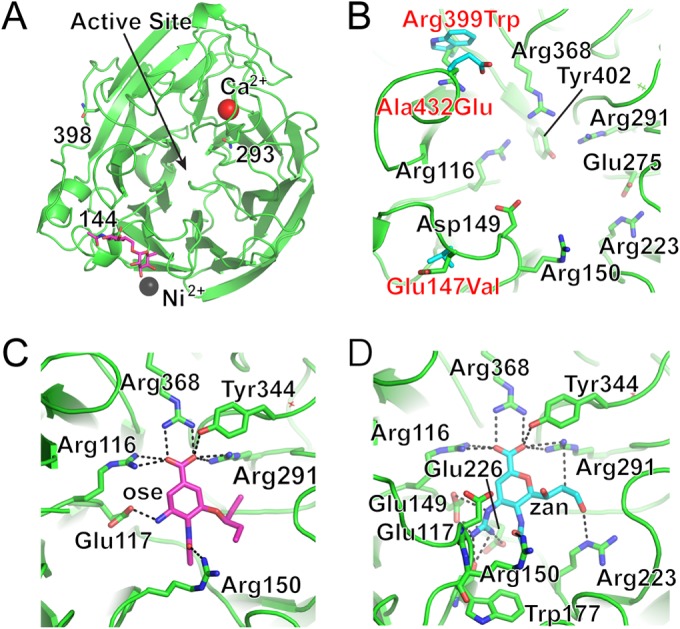
Structure of seal NA. (A) Overall structure of one NA monomer, looking down on the monomer, with the position of the enzyme active site indicated. While only one glycosylation site was occupied in the final structure (pink sticks), two others that are visible in this view are labeled and shown as sticks. The fourth position, Asn84, was substituted in the protein but would be at the back of this view. (B) NA active site, with highly conserved residues shown as sticks (labeled in black text). The amino acid substitutions between seal11 NA (green) and the avian N8 NA (cyan) are labeled and shown as sticks. The seal residue is indicated first in the labels and the avian residue second. (C and D) Exploded views of the enzyme active site with NA antivirals Ose (C) and Zan (D) bound. The NA residues that interact with each drug (dashed lines) are shown as sticks.
Active site of seal11 NA and antiviral drug susceptibility with oseltamivir and zanamivir.
Among all of the NA subtypes, the enzyme active site includes eight highly conserved residues, Arg116, Asp149, Arg150, Arg223, Glu275, Arg291, Arg368, and Tyr402 (residues 118, 151, 152, 224, 276, 292, 371, and 406 in N2 numbering) (Fig. 5B). They are all charged/polar residues that directly interact with the substrate in the catalytic site. The geometry of the catalytic site is structurally stabilized through a network of hydrogen bonds and salt bridges by a constellation of largely conserved framework residues, Glu117, Arg154, Trp177, Ser178, Asp197, Ile221, Glu226, His273, Glu276, Asn293, and Glu425 (residues 119, 156, 178, 179, 198, 222, 227, 274, 277, 294, and 425 in N2 numbering) (76). The active site of seal11 NA has a characteristic open conformation with the 150 cavity and has high similarity to the previously published avian N8 NA and other group 1 neuraminidases (74). Comparing seal11 NA with the avian N8 NA, there are three residue differences positioned near the active site: Glu/Val147, Arg/Trp399, and Ala/Glu432 (the residue differences are listed as seal/avian). While their locations on the 150 loop and 430 loop are not close enough to interact directly with the substrate and thus may not affect NA activity, these substitutions may contribute to the local stability of the active site (Fig. 5B).
The NA activity of seal N8 NA was assessed using the fluorescent substrate MUNANA (44). Compared to a seasonal N2 recNA from A/Perth/16/2009, the seal11 NA appeared more active, with a 10-fold-increased kcat (Table 6). In the NA inhibition assay, the activity of recNA was effectively inhibited by the current antivirals, Ose and Zan, which indicates a drug-susceptible phenotype of seal11 virus (Table 6). The IC50s for seal11 recNA were comparable to those of the A/harbor seal/New Hampshire/179629/2011 virus that was isolated from the same outbreak (0.66 nM for Ose and 1.29 nM for Zan). Furthermore, we solved the crystal structures of seal11 NA in complex with the two widely used antiviral drugs. The interactions between seal11 NA and both inhibitors are similar (Fig. 5C and D), although more hydrogen bonds are formed between NA and Zan than between NA and Ose. The carboxyl group of both Ose and Zan hydrogen bonds to Arg116, Arg291, Tyr344, and Arg368 (Arg118, Arg292, Tyr346, and Arg371 in N2 numbering), while the carbonyl oxygen of the N-acetyl group hydrogen bonds with Arg150 (Arg152 in N2 numbering). The bulky 4-guanidino group of Zan is buried underneath the 150 loop and hydrogen bonds to Glu117, Glu149, Trp177, and Glu226 (Glu119, Glu151, Trp178, and Glu227 in N2 numbering). There are two additional hydrogen bond interactions between the Zan 8-hydroxyl group and Arg291 (Arg292 in N2 numbering) and 9-O with Arg223 (Arg224 in N2 numbering) in the NA-Zan complex structure.
TABLE 6.
Seal11 NA kinetic analysis and drug susceptibility assessment
| Virus name | Subtype | Parameter value |
||||
|---|---|---|---|---|---|---|
| kcat (s−1) ± SD | Km (μM) ± SD | kcat/Km (M−1 s−1) | IC50 (nM) ± SDa |
|||
| Oseltamivir carboxylate | Zanamivir | |||||
| RecNA | ||||||
| A/harbor seal/Massachusetts/1/2011 | N8 | 328,281 ± 5,769 | 3.227 ± 0.205 | 1.02 × 1011 | 1.42 ± 0.12 | 2.14 ± 0.70 |
| A/Perth/16/2009 | N2 | 32,039 ± 501 | 7.985 ± 0.365 | 4.01 × 1010 | 0.30 ± 0.07 | 0.44 ± 0.10 |
| Virus | ||||||
| A/harbor seal/New Hampshire/179629/2011 | A(H3N8) | NDb | ND | 0.66 ± 0.14 | 1.29 ± 0.39 | |
| Reference virus | ||||||
| A/California/12/2012 | A(H1N1)pdm09 | ND | ND | 0.28 ± 0.02 | 0.13 ± 0.02 | |
| A/Texas/23/2012c | A(H1N1)pdm09 | ND | ND | 160.82 ± 18.59 | 0.16 ± 0.02 | |
Average value from at least three measurements.
ND, not determined.
Oseltamivir resistant with His275Tyr substitution in the NA.
Conclusions.
Here, we report a detailed molecular characterization of seal11 HA and NA. The results for the seal11 HA suggest that while it possesses a specific avian receptor binding preference in our assays, thus reducing its potential to easily infect humans, results from recent transmission studies highlight additional HA mutations that may need to be introduced to increase its ability for respiratory-droplet transmission in mammalian species (65). The analysis of the known human antigenic sites on this HA suggests that immunity conferred by infection or vaccination with currently circulating human A(H3N2) viruses may be insufficient to protect humans, should human adaptation of the A(H3N8) virus occur. However, the seal11 NA is sensitive to currently available antivirals, and their use may be beneficial while a vaccine is developed and manufactured. With increasing evidence of infections by avian and human influenza viruses in marine mammals (21, 81, 82), it is important to continue the study and surveillance of these animals to assess whether these hosts are a possible route by which influenza viruses can adapt to humans, leading to new viruses with pandemic potential.
ACKNOWLEDGMENTS
This work was funded by the Centers for Disease Control and Prevention. The U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357, supports use of the Advanced Photon Source at Argonne National Laboratory. The Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory, is operated for the U.S. Department of Energy Office of Science by Stanford University and is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). Glycan microarray slides were produced under contract for the Centers for Disease Control and Prevention using a glycan library generously provided by the Consortium for Functional Glycomics (CFG) (http://www.functionalglycomics.org), funded by National Institute of General Medical Sciences grant GM62116.
We thank Hon Ip of the National Wildlife Health Center, United States Geological Survey, Madison, WI, USA, for providing the A/harbor seal/New Hampshire/179629/2011 virus. We also thank the staff of SER-CAT sector 22 at the APS and the SSRL beamline 7-1 for their help in data collection. We also thank the CFG for supplying the biotinylated glycans used here through their resource request program.
The findings and conclusions in this report are ours and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
REFERENCES
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. 1980. A revision of the system of nomenclature for influenza viruses: a WHO memorandum. Bull World Health Organ 58:585–591. [PMC free article] [PubMed] [Google Scholar]
- 3.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawaoka Y, Bean WJ, Webster RG. 1989. Evolution of the hemagglutinin of equine H3 influenza viruses. Virology 169:283–292. doi: 10.1016/0042-6822(89)90153-0. [DOI] [PubMed] [Google Scholar]
- 6.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. 1978. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 7.Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 8.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shope RE. 1931. Swine influenza: III. Filtration experiments and etiology. J Exp Med 54:373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith W, Andrewes CH, Laidlaw PP. 1933. A virus obtained from influenza patients. Lancet ii:66–68. [Google Scholar]
- 12.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 13.de Jong JC, Rimmelzwaan GF, Bartelds AI, Wilbrink B, Fouchier RA, Osterhaus AD. 2003. The 2002/2003 influenza season in the Netherlands and the vaccine composition for the 2003/2004 season. Ned Tijdschr Geneeskd 147:1971–1975. (In Dutch.) [PubMed] [Google Scholar]
- 14.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. 1999. Human infection with influenza H9N2. Lancet 354:916–917. doi: 10.1016/S0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 15.Webster RG, Hinshaw VS, Bean WJ, Van Wyke KL, Geraci JR, St Aubin DJ, Petursson G. 1981. Characterization of an influenza A virus from seals. Virology 113:712–724. doi: 10.1016/0042-6822(81)90200-2. [DOI] [PubMed] [Google Scholar]
- 16.Webster RG, Geraci J, Petursson G, Skirnisson K. 1981. Conjunctivitis in human beings caused by influenza A virus of seals. N Engl J Med 304:911. [DOI] [PubMed] [Google Scholar]
- 17.Geraci JR, St Aubin DJ, Barker IK, Webster RG, Hinshaw VS, Bean WJ, Ruhnke HL, Prescott JH, Early G, Baker AS, Madoff S, Schooley RT. 1982. Mass mortality of harbor seals: pneumonia associated with influenza A virus. Science 215:1129–1131. doi: 10.1126/science.7063847. [DOI] [PubMed] [Google Scholar]
- 18.Hinshaw VS, Bean WJ, Webster RG, Rehg JE, Fiorelli P, Early G, Geraci JR, St Aubin DJ. 1984. Are seals frequently infected with avian influenza viruses? J Virol 51:863–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinshaw VS, Bean WJ, Geraci J, Fiorelli P, Early G, Webster RG. 1986. Characterization of two influenza A viruses from a pilot whale. J Virol 58:655–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callan RJ, Early G, Kida H, Hinshaw VS. 1995. The appearance of H3 influenza viruses in seals. J Gen Virol 76:199–203. doi: 10.1099/0022-1317-76-1-199. [DOI] [PubMed] [Google Scholar]
- 21.White CL, Schuler KL, Thomas NJ, Webb JL, Saliki JT, Ip HS, Dubey JP, Frame ER. 2013. Pathogen exposure and blood chemistry in the Washington, U S A population of northern sea otters (Enhydra lutris kenyoni). J Wildl Dis 49:887–899. doi: 10.7589/2013-03-053. [DOI] [PubMed] [Google Scholar]
- 22.Bodewes R, Morick D, de Mutsert G, Osinga N, Bestebroer T, van der Vliet S, Smits SL, Kuiken T, Rimmelzwaan GF, Fouchier RA, Osterhaus AD. 2013. Recurring influenza B virus infections in seals. Emerg Infect Dis 19:511–512. doi: 10.3201/eid1903.120965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anthony SJ, St Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, Navarrete-Macias I, Sanchez-Leon M, Saliki JT, Pedersen J, Karesh W, Daszak P, Rabadan R, Rowles T, Lipkin WI. 2012. Emergence of fatal avian influenza in New England harbor seals. mBio 3:e00166-00112. doi: 10.1128/mBio.00166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford PC, Dubovi EJ, Castleman WL, Stephenson I, Gibbs EP, Chen L, Smith C, Hill RC, Ferro P, Pompey J, Bright RA, Medina MJ, Johnson CM, Olsen CW, Cox NJ, Klimov AI, Katz JM, Donis RO. 2005. Transmission of equine influenza virus to dogs. Science 310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 25.Guo Y, Wang M, Kawaoka Y, Gorman O, Ito T, Saito T, Webster RG. 1992. Characterization of a new avian-like influenza A virus from horses in China. Virology 188:245–255. doi: 10.1016/0042-6822(92)90754-D. [DOI] [PubMed] [Google Scholar]
- 26.Wilcox BR, Knutsen GA, Berdeen J, Goekjian V, Poulson R, Goyal S, Sreevatsan S, Cardona C, Berghaus RD, Swayne DE, Yabsley MJ, Stallknecht DE. 2011. Influenza-A viruses in ducks in northwestern Minnesota: fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS One 6:e24010. doi: 10.1371/journal.pone.0024010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu J, Zhou H, Jiang T, Li C, Zhang A, Guo X, Zou W, Chen H, Jin M. 2009. Isolation and molecular characterization of equine H3N8 influenza viruses from pigs in China. Arch Virol 154:887–890. doi: 10.1007/s00705-009-0381-1. [DOI] [PubMed] [Google Scholar]
- 28.Bogomolni AL, Gast RJ, Ellis JC, Dennett M, Pugliares KR, Lentell BJ, Moore MJ. 2008. Victims or vectors: a survey of marine vertebrate zoonoses from coastal waters of the Northwest Atlantic. Dis Aquat Organ 81:13–38. doi: 10.3354/dao01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. 2013. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol 22:12433–12446. doi: 10.1128/JVI.01854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhnel K, Jarchau T, Wolf E, Schlichting I, Walter U, Wittinghofer A, Strelkov SV. 2004. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc Natl Acad Sci U S A 101:17027–17032. doi: 10.1073/pnas.0403069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X, Zhu X, Dwek R, Stevens J, Wilson I. 2008. Structural characterization of the 1918 influenza H1N1 neuraminidase. J Virol 82:10493–10501. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Carney P, Stevens J. 2010. Structure and receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr 2:RRN1152. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, Carney PJ, Donis RO, Stevens J. 2012. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J Virol 86:8645–8652. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H, Chang JC, Guo Z, Carney PJ, Shore DA, Donis RO, Cox NJ, Villanueva JM, Klimov AI, Stevens J. 2014. Structural stability of influenza A(H1N1)pdm09 virus hemagglutinins. J Virol 88:4828–4838. doi: 10.1128/JVI.02278-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. 2010. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog 6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chayen NE, Shaw-Steward PD, Blow DM. 1992. Microbatch crystallization under oil—a new technique allowing many small volume crystallization experiments. J Cryst Growth 122:176–180. doi: 10.1016/0022-0248(92)90241-A. [DOI] [Google Scholar]
- 37.Otwinowski Z, Minor W. 1997. Scalepack manual. HKL Research Inc., Charlottesville, VA. [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Winn MD, Isupov MN, Murshudov GN. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57:122–133. doi: 10.1107/S0907444900014736. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal Biochem 94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 45.Okomo-Adhiambo M, Sleeman K, Lysen C, Nguyen HT, Xu X, Li Y, Klimov AI, Gubareva LV. 2013. Neuraminidase inhibitor susceptibility surveillance of influenza viruses circulating worldwide during the 2011 Southern Hemisphere season. Influenza Other Respir viruses 7:645–658. doi: 10.1111/irv.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417. doi: 10.1016/S0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 47.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. 1988. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 48.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci U S A 98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2003. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology 309:209–218. doi: 10.1016/S0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 50.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. 2009. Structures of receptor complexes formed by hemagglutinins from the Asian influenza pandemic of 1957. Proc Natl Acad Sci U S A 106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell RJ, Gamblin SJ, Haire LF, Stevens DJ, Xiao B, Ha Y, Skehel JJ. 2004. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325:287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 52.Russell RJ, Kerry PS, Stevens DJ, Steinhauer DA, Martin SR, Gamblin SJ, Skehel JJ. 2008. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A 105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 54.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 55.Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 56.Shakin-Eshleman SH, Spitalnik SL, Kasturi L. 1996. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J Biol Chem 271:6363–6366. doi: 10.1074/jbc.271.11.6363. [DOI] [PubMed] [Google Scholar]
- 57.Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu J, Skehel JJ, Gamblin SJ, Hay AJ, Daniels RS, McCauley JW. 2012. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A 109:21474–21479. doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74:8502–8512. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 60.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 61.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collins PJ, Vachieri SG, Haire LF, Ogrodowicz RW, Martin SR, Walker PA, Xiong X, Gamblin SJ, Skehel JJ. 2014. Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci U S A 111:11175–11180. doi: 10.1073/pnas.1406606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang G, Li S, Blackmon S, Ye J, Bradley KC, Cooley J, Smith D, Hanson L, Cardona C, Steinhauer DA, Webby R, Liao M, Wan XF. 2013. Mutation tryptophan to leucine at position 222 of haemagglutinin could facilitate H3N2 influenza A virus infection in dogs. J Gen Virol 94:2599–2608. doi: 10.1099/vir.0.054692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karlsson EA, Ip HS, Hall JS, Yoon SW, Johnson J, Beck MA, Webby RJ, Schultz-Cherry S. 2014. Respiratory transmission of an avian H3N8 influenza virus isolated from a harbour seal. Nat Commun 5:4791. doi: 10.1038/ncomms5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bradley KC, Galloway SE, Lasanajak Y, Song X, Heimburg-Molinaro J, Yu H, Chen X, Talekar GR, Smith DF, Cummings RD, Steinhauer DA. 2011. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J Virol 85:12387–12398. doi: 10.1128/JVI.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 68.Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 69.Daniels RS, Douglas AR, Skehel JJ, Wiley DC. 1983. Analyses of the antigenicity of influenza haemagglutinin at the pH optimum for virus-mediated membrane fusion. J Gen Virol 64:1657–1662. doi: 10.1099/0022-1317-64-8-1657. [DOI] [PubMed] [Google Scholar]
- 70.Popova L, Smith K, West AH, Wilson PC, James JA, Thompson LF, Air GM. 2012. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS One 7:e41895. doi: 10.1371/journal.pone.0041895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stray SJ, Pittman LB. 2012. Subtype- and antigenic site-specific differences in biophysical influences on evolution of influenza virus hemagglutinin. Virol J 9:91. doi: 10.1186/1743-422X-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varghese JN, Laver WG, Colman PM. 1983. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature 303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 73.Baker AT, Varghese JN, Laver WG, Air GM, Colman PM. 1987. Three-dimensional structure of neuraminidase of subtype N9 from an avian influenza virus. Proteins 2:111–117. doi: 10.1002/prot.340020205. [DOI] [PubMed] [Google Scholar]
- 74.Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 75.Wang M, Qi J, Liu Y, Vavricka CJ, Wu Y, Li Q, Gao GF. 2011. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol 85:8431–8435. doi: 10.1128/JVI.00638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burmeister WP, Ruigrok RW, Cusack S. 1992. The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J 11:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chong AK, Pegg MS, Taylor NR, von Itzstein M. 1992. Evidence for a sialosyl cation transition-state complex in the reaction of sialidase from influenza virus. Eur J Biochem 207:335–343. doi: 10.1111/j.1432-1033.1992.tb17055.x. [DOI] [PubMed] [Google Scholar]
- 78.Burmeister WP, Cusack S, Ruigrok RW. 1994. Calcium is needed for the thermostability of influenza B virus neuraminidase. J Gen Virol 75:381–388. doi: 10.1099/0022-1317-75-2-381. [DOI] [PubMed] [Google Scholar]
- 79.Smith BJ, Huyton T, Joosten RP, McKimm-Breschkin JL, Zhang JG, Luo CS, Lou MZ, Labrou NE, Garrett TP. 2006. Structure of a calcium-deficient form of influenza virus neuraminidase: implications for substrate binding. Acta Crystallogr D Biol Crystallogr 62:947–952. doi: 10.1107/S0907444906020063. [DOI] [PubMed] [Google Scholar]
- 80.Janakiraman MN, White CL, Laver WG, Air GM, Luo M. 1994. Structure of influenza virus neuraminidase B/Lee/40 complexed with sialic acid and a dehydro analog at 1.8-A resolution: implications for the catalytic mechanism. Biochemistry 33:8172–8179. doi: 10.1021/bi00193a002. [DOI] [PubMed] [Google Scholar]
- 81.Boyce WM, Mena I, Yochem PK, Gulland FMD, Garcia-Sastre A, Moreno N, Perez DR, Gonzalez-Reiche AS, Stewart BS. 2013. Influenza A(H1N1)pdm09 virus infection in marine mammals in California. Emerg Microbes Infect 2:e40. doi: 10.1038/emi.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li ZN, Ip HS, Trost JF, White CL, Murray MJ, Carney PJ, Sun XJ, Stevens J, Levine MZ, Katz JM. 2014. Serologic evidence of influenza A(H1N1)pdm09 virus infection in Northern Sea otters. Emerg Infect Dis 20:915–917. doi: 10.3201/eid2005.131890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeLano WL. 2002. The PyMol molecular graphics systems. http://www.pymol.org.
- 84.Lee B, Richards FM. 1971. The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55:379–400. doi: 10.1016/0022-2836(71)90324-X. [DOI] [PubMed] [Google Scholar]
- 85.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]