Abstract
Microbes have recently been recognized as dominant forces in nature, with studies benefiting from gene markers that can be quickly, informatively, and universally surveyed. Viruses, where explored, have proven to be powerful modulators of locally and globally important microbes through mortality, horizontal gene transfer, and metabolic reprogramming. However, community-wide virus studies have been challenged by the lack of a universal marker. Here, I propose that viral metagenomics has advanced to largely take over study of double-stranded DNA viruses.
INTRODUCTION
There was a time, not so long ago, when exploring diversity in wild viral communities required researchers to focus on gene markers. Because viruses do not share a single gene, such gene markers can target only specific viral groups, including the T4-like myoviruses (e.g., major capsid and portal proteins), T7-like podoviruses (e.g., DNA polymerase), and phycodnaviridae (e.g., DNA polymerase). Gene marker studies complemented fluorescence or electron microscopy-based counts of virus-like particles to help advance the field one step closer to considering the details of natural viral community diversity and interactions. By assessing genetic variability in these target groups, clear links were made between diversity and how it changes over space and time, particularly by leveraging throughput to examine high-resolution temporal dynamics in concert with parallel measurements for microbes (see, e.g., reference 1).
Here, I propose that it is time to experimentally evaluate whether amplicon-derived operational taxonomic unit abundances correlate with their actual abundances in nature. Why do we need to evaluate this if microbial ecologists routinely conduct similar gene marker studies? Unlike our microbial counterparts, for whom nondegenerate primers commonly exactly match community sequences, viral ecologists are often forced to employ highly degenerate primer sets designed from insufficient databases and low PCR annealing temperatures (details appear in reference 2). Even with fewer concerns, microbial ecologists also remain conservative in using such data only in unweighted analyses, thereby implicitly acknowledging that absolute abundances might be significantly altered by the PCR process in ways yet undocumented. Such a practice should perhaps also become mainstream in viral ecology. Fortunately, the sample-to-sequence viral metagenome (virome) pipeline, at least for the double-stranded DNA (dsDNA) viruses that pass through a 0.2-μm filter, offers a means to experimentally evaluate the quantitative performance of PCR-based studies. Further, drawing upon examples from the study of oceanic viral communities, I suggest that, with the right experimental design, viromics offers advantages over the study of gene markers for inferring biology in complex systems and testing a priori and ad hoc hypotheses (summarized in Fig. 1 and detailed below).
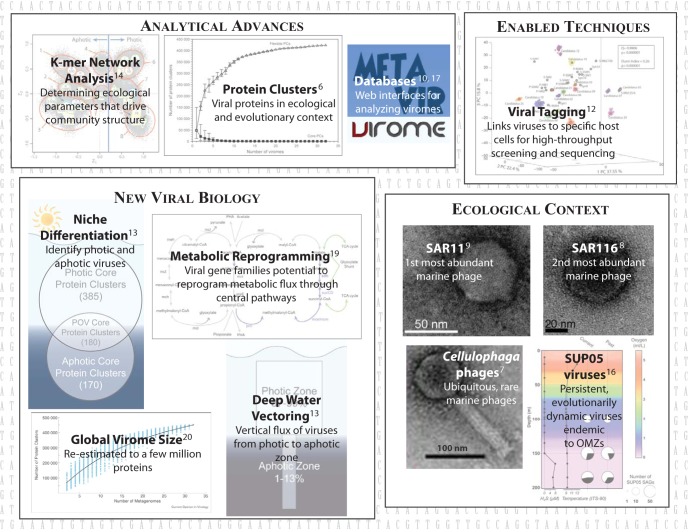
FIG 1.
Viromics is now optimized for studying dsDNA viruses in nature. Enabled by an optimized sample-to-sequence pipeline (4) and analytical advances (6, 13, 14), databases and tools (16, 17), and quantitative viromes and virome-enabled techniques (12) are now revealing significant new biological information for natural viral communities (13, 19, 20) and ecological context for newly isolated and genome-sequenced (7–9) or virome-determined viruses (10). POV, Pacific Ocean viromes; OMZs, oxygen minimum zone.
Why is viromics not yet mainstream? First, sampling for environmental viruses often yields too little material for standard sequencing libraries. However, numerous groups have now shown that sequencing libraries can be made from far less than 1 nanogram of DNA (3–5) and that linker-amplified viromes are quantitative (±1.5-fold) from so little DNA (4).
Second, unknown sequences dominate viromes, as ∼63 to 93% of the reads often lack functional or taxonomic annotations (reviewed in reference 6). However, new approaches now illuminate this “viral dark matter.” For example, coverage of viral-genome sequence space is rapidly improving through genome sequencing of isolates in culture collections that represent abundant and rare ocean viruses (see, e.g., references 7, 8, and 9), as well as through extraction of sequence data of novel viruses from single-cell genomics projects (10). Further, smaller viral genomes are being assembled from marine metagenomes (see, e.g., references 11 and 10), and strategies to simplify more-complex viral communities are enabling assembly of large genomic regions of larger viruses (12).
Today, deeper sequencing combined with extrapolation of “population-level” variability from wild T4-like phage genome variability (12) enables ocean researchers to identify and quantify populations in a virome even when a database representative is completely lacking. One study (10) mined 186 microbial and viral metagenomes to show that uncultivated SUP05 viruses are persistent and evolutionarily dynamic over 3 years but endemic to the particular study site. Further, the genomic context for these new SUP05 viruses suggested that they manipulate the central and defining metabolism of their SUP05 bacterial hosts through virus-encoded sulfur-cycling genes (10), a feature shared with a prior study of viruses assembled from microbiomes (11). Thus, viromes can simultaneously map the spatiotemporal variation of many target groups while also providing genome-enabled hypotheses about ecological drivers of any particular target group. This is, of course, limited to those populations that are abundant in the data set, but as sequencing improves, many populations already qualify.
Two other approaches have emerged to help viral ecologists make meaningful inferences from virome “dark matter.” Protein clustering organizes viral sequence space into arbitrarily defined bins that can be quantified. Such PC-based “gene ecology” advances include establishing (i) a sampling effort and community diversity (6), (ii) core gene sets as windows into the functions (known and unknown) required across viral communities (13), and (iii) flexible gene sets that define niche-differentiating functions across viral communities (13). Additionally, new system-level phenomena can be unveiled. For example, it now appears that vertical vectoring of viral particles from the surface to the depths of oceans strongly influences the genetic diversity of deep-sea viral communities (13). Yet, these powerful PC-based approaches are only part of the picture, as currently they map only ∼70% of virome reads and are dependent upon front-end assembly and gene prediction. To remedy this, kmer abundances can be derived from every virome read and, when coupled to networking algorithms, used to develop ecological models that explore drivers of viral-community niche differentiation (14). Both areas are enabling new scientific discovery, development, and application, and when combined with population analyses, they lead to a triumvirate of gene, population, and community ecology at a system-wide level (reviewed in reference 15).
Finally, experimentally linking viruses (and associated viromes) to host cells is now possible through viral tagging (12). This enables exploration of viral-genome sequence space in association with particular cultivable host cells. When this process is coupled with other recently developed virus-host linkage methods (e.g., phageFISH, microfluidic digital PCR, fosmids, and SAGs [reviewed in reference 15]), it is clear that viral ecologists have an emerging toolkit for evaluating virus-host interaction dynamics at unprecedented spatiotemporal scales and for specific virus-host pairings—all as virome-enabled alternatives to PCR-based gene marker studies.
Placing these advances aside, the reality is that two additional roadblocks keep viromics from widespread use. First, user-friendly tools and extensive databases for analyzing and interpreting viromes are underdeveloped compared to their microbial counterparts. Fortunately, new tools are emerging; they include MetaVir (16) and VIROME (17), which offer Web-based interfaces for analyzing and interpreting viromes and large and growing publicly available virome databases. Complementarily, iVirus (http://ivirus.us/) is a recent effort leveraging the iPlant cyberinfrastructure to a point where a user can access cutting-edge, high-performance computing capability without computer science knowledge. Moreover, this platform enables users to design and make their own tools available to the community using iPlant computational resources and an application programming interface. New viromic “apps” (software programs), curated data sets, and query-able metadata are now being made accessible in iPlant (http://www.iplantcollaborative.org/) through the iVirus/iMicrobe project using either a graphical user interface or command line interface, depending upon user capabilities (http://www.moore.org/grants/list/GBMF4491). These emerging resources mean that interpreting viromes is getting easier. The second bottleneck is cost. I will simply suggest that at some point, with plummeting sequencing costs, viromes will become a more cost-effective means to document viral-community structure than gene markers, at least for abundant viral populations. Arguably, if researchers consider personnel and reagent costs for primer development, optimization, and analyses for PCR-based studies and weigh the relative differences in the resulting biological inferences that can be made from the data, then that time is now. If not now, then it will be soon, likely as major community-available informatics tools and databases mature.
Undoubtedly, PCR-based gene marker studies remain critical (i) where viromics remains poorly developed (e.g., with single-stranded DNA [ssDNA] and RNA viruses and with dsDNA viruses of >0.2 μm), (ii) for targeted studies for which gene markers are mined from quantitative viromes to assess population-level spatiotemporal variability (see, e.g., reference 18), and (iii) where high-resolution data sets for a target viral population are needed to answer a particular research question (see, e.g., reference 1). However, for the smaller dsDNA viral communities, I suggest that viromes are largely ready to quantitatively evaluate and likely replace PCR-based gene marker surveys to address most of the fundamental questions needing answers in viral ecology.
ACKNOWLEDGMENTS
Grants from the Gordon and Betty Moore Foundation (2631, 3790), as well as Tucson Marine Phage Laboratory members past and present, are acknowledged for enabling method developments that led to the quantitative sample-to-sequence pipeline for <0.2-μm dsDNA viruses.
Bonnie Hurwitz, Simon Roux, and Eric Wommack are thanked for their development of viromics analytical tools, and Christine Schirmer is thanked for preparing Fig. 1.
REFERENCES
- 1.Chow CE, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. 2014. Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J 8:816–829. doi: 10.1038/ismej.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duhaime M, Sullivan MB. 2012. Ocean viruses: rigorously evaluating the metagenomic sample-to-sequence pipeline. Virology 434:181–186. doi: 10.1016/j.virol.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 3.Adey A, Morrison HG, Asan Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, Shendure J. 2010. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duhaime MB, Deng L, Poulos BT, Sullivan MB. 2012. Towards quantitative metagenomics of wild viruses and other ultra-low concentration DNA samples: a rigorous assessment and optimization of the linker amplification method. Environ Microbiol 14:2526–2537. doi: 10.1111/j.1462-2920.2012.02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkinson NJ, Maslau S, Ferneyhough B, Zhang G, Gregory L, Buck D, Ragoussis J, Ponting CP, Fischer MD. 2012. Preparation of high-quality next-generation sequencing libraries from picogram quantities of target DNA. Genome Res 22:125–133. doi: 10.1101/gr.124016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurwitz BH, Sullivan MB. 2013. The Pacific Ocean Virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One 8:e57355. doi: 10.1371/journal.pone.0057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmfeldt K, Solonenko N, Shah M, Corrier K, Riemann L, Verberkmoes NC, Sullivan MB. 2013. Twelve previously unknown phage genera are ubiquitous in global oceans. Proc Natl Acad Sci U S A 110:12798–12803. doi: 10.1073/pnas.1305956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang I, Oh HM, Kang D, Cho JC. 2013. Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc Natl Acad Sci U S A 110:12343–12348. doi: 10.1073/pnas.1219930110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, Temperton B, Thrash J C, Schwalbach MS, Vergin KL, Landry ZC, Ellisman M, Deerinck T, Sullivan MB, Giovannoni SJ. 2013. Abundant SAR11 viruses in the ocean. Nature 494:357–360. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
- 10.Roux S, Hawley AK, Beltran MT, Scofield M, Schwientek P, Stepanauskas R, Woyke T, Hallam SJ, Sullivan MB. 2014. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell and environmental genomics. Elife 3:03125. doi: 10.7554/eLife.03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anantharaman K, Duhaime MB, Breier J A, Wendt KA, Toner BM, Dick GJ. 2014. Sulfur oxidation genes in diverse deep-sea viruses. Science 344:757–760. doi: 10.1126/science.1252229. [DOI] [PubMed] [Google Scholar]
- 12.Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, Sullivan MB. 2014. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513:242–245. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
- 13.Hurwitz BH, Brum JR, Sullivan MB. 5August2014. Depth-stratified functional and taxonomic niche specialization in the ‘core’ and ‘flexible’ Pacific Ocean Virome. ISME J doi: 10.1038/ismej.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurwitz BL, Westveld AH, Brum J R, Sullivan MB. 2014. Modeling ecological drivers in marine viral communities using comparative metagenomics and network analyses. Proc Natl Acad Sci U S A 111:10714–10719. doi: 10.1073/pnas.1319778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brum JR, Sullivan MB. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat Rev Microbiol, in press. [DOI] [PubMed] [Google Scholar]
- 16.Roux S, Tournayre J, Mahul A, Debroas D, Enault F. 2014. MetaVir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinformatics 15:76. doi: 10.1186/1471-2105-15-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wommack KE, Polson SW, Bhaysar J, Srinivasiah S, Jamindar S, Dumas M. 2011. VIROME: a standard operating procedure for classification of viral metagenome sequences. Stand Genomic Sci 4:427–439. doi: 10.4056/sigs.2945050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakowski EG, Munsell EV, Hyatt M, Kress W, Williamson SJ, Nasko DJ, Polson SW, Wommack KE. 2014. Ribonucleotide reductases reveal novel viral diversity and predict biological and ecological features of unknown marine viruses. Proc Natl Acad Sci U S A 111:15786–15791. doi: 10.1073/pnas.1401322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurwitz BL, Hallam SJ, Sullivan MB. 2013. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol 14:R123. doi: 10.1186/gb-2013-14-11-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ignacio-Espinoza JC, Solonenko SA, Sullivan MB. 2013. The global virome: not as big as we thought? Curr Opin Virol 3:566–571. doi: 10.1016/j.coviro.2013.07.004. [DOI] [PubMed] [Google Scholar]