ABSTRACT
RNA viruses co-opt a large number of cellular proteins that affect virus replication and, in some cases, viral genetic recombination. RNA recombination helps viruses in an evolutionary arms race with the host's antiviral responses and adaptation of viruses to new hosts. Tombusviruses and a yeast model host are used to identify cellular factors affecting RNA virus replication and RNA recombination. In this study, we have examined the role of the conserved Rpn11p metalloprotease subunit of the proteasome, which couples deubiquitination and degradation of proteasome substrates, in tombusvirus replication and recombination in Saccharomyces cerevisiae and plants. Depletion or mutations of Rpn11p lead to the rapid formation of viral RNA recombinants in combination with reduced levels of viral RNA replication in yeast or in vitro based on cell extracts. Rpn11p interacts with the viral replication proteins and is recruited to the viral replicase complex (VRC). Analysis of the multifunctional Rpn11p has revealed that the primary role of Rpn11p is to act as a “matchmaker” that brings the viral p92pol replication protein and the DDX3-like Ded1p/RH20 DEAD box helicases into VRCs. Overexpression of Ded1p can complement the defect observed in rpn11 mutant yeast by reducing TBSV recombination. This suggests that Rpn11p can suppress tombusvirus recombination via facilitating the recruitment of the cellular Ded1p helicase, which is a strong suppressor of viral recombination, into VRCs. Overall, this work demonstrates that the co-opted Rpn11p, which is involved in the assembly of the functional proteasome, also functions in the proper assembly of the tombusvirus VRCs.
IMPORTANCE RNA viruses evolve rapidly due to genetic changes based on mutations and RNA recombination. Viral genetic recombination helps viruses in an evolutionary arms race with the host's antiviral responses and facilitates adaptation of viruses to new hosts. Cellular factors affect viral RNA recombination, although the role of the host in virus evolution is still understudied. In this study, we used a plant RNA virus, tombusvirus, to examine the role of a cellular proteasomal protein, called Rpn11, in tombusvirus recombination in a yeast model host, in plants, and in vitro. We found that the cellular Rpn11 is subverted for tombusvirus replication and Rpn11 has a proteasome-independent function in facilitating viral replication. When the Rpn11 level is knocked down or a mutated Rpn11 is expressed, then tombusvirus RNA goes through rapid viral recombination and evolution. Taken together, the results show that the co-opted cellular Rpn11 is a critical host factor for tombusviruses by regulating viral replication and genetic recombination.
INTRODUCTION
Viruses and their hosts go through continuous evolution, including a genetic arms race between viral and host factors that decides if the given virus could accomplish successful infection in that host. RNA virus adaptation to the host is facilitated by the ability of RNA viruses to evolve rapidly due to high-frequency mutations and genetic RNA recombination (1–3). Viral RNA recombination is a process that could alter viral genomes by joining two or more noncontiguous segments of the same RNA or two separate RNAs together (4, 5). The consequence of viral recombination could be mutations, sequence insertions, duplications, deletions, rearrangements, or creation of new sequences. Interestingly, RNA recombination is also used by viruses to repair truncated or damaged viral RNA molecules, thus increasing the infectivity of RNA viruses (6–9). In summary, viral RNA recombination is known to change virus population dynamics and greatly contribute to virus variability (3, 4, 10).
Tomato bushy stunt virus (TBSV), a tombusvirus, is especially useful in studying viral RNA recombination based on the development of various unique approaches, including the use of a yeast (Saccharomyces cerevisiae) model host (11–18). Systematic genome-wide screens in yeast with TBSV have identified more than 30 host genes that altered various features of viral RNA recombination (14, 15, 19–21). Subsequent detailed analysis of the role of several host factors in TBSV recombination has already unveiled three different pathways. The first pathway is based on the critical roles of cellular endo- and exoribonucleases that “attack” the viral RNA, creating cleaved and partially degraded viral RNA molecules (degRNAs) (Fig. 1A). degRNAs are outstanding templates for template switching by the viral replicase, which results in viral RNA recombination (15, 22). For example, the cytosolic Xrn1p 5′-to-3′ exoribonuclease (Xrn4 in plants) suppresses TBSV recombination via rapidly degrading TBSV RNAs (degRNAs) (Fig. 1A) cleaved by cellular endoribonucleases (22–25). Thus, the degRNAs have only limited time to participate in RNA recombination in wild-type (wt) cells. The second pathway is based on the Pmr1 Ca2+/Mn2+ pump that controls Ca2+/Mn2+ levels in the cytosol and greatly affects the template-switching activity of the viral replicase. In the absence of Pmr1, the high level of cytosolic Mn2+ induces high-frequency RNA recombination in yeast or plant cells, which can also be reconstituted in a cell-free TBSV replication assay (21). The third TBSV recombination pathway is based on cellular DEAD box helicases, which are components of the viral replicase complex (VRC) and affect RNA synthesis via locally unwinding viral RNA structures and affecting the ability of the viral RNA-dependent RNA polymerase (RdRp) to dissociate from viral templates (C. Chuang, K. R. Prasanth, and P. D. Nagy, unpublished data). One of the helicases is DDX3-like Ded1p (AtRH20 in plants), which binds to the viral RNA and suppresses the replication of degRNAs and formation of RNAs formed through recombination from the degRNA template (recRNAs).
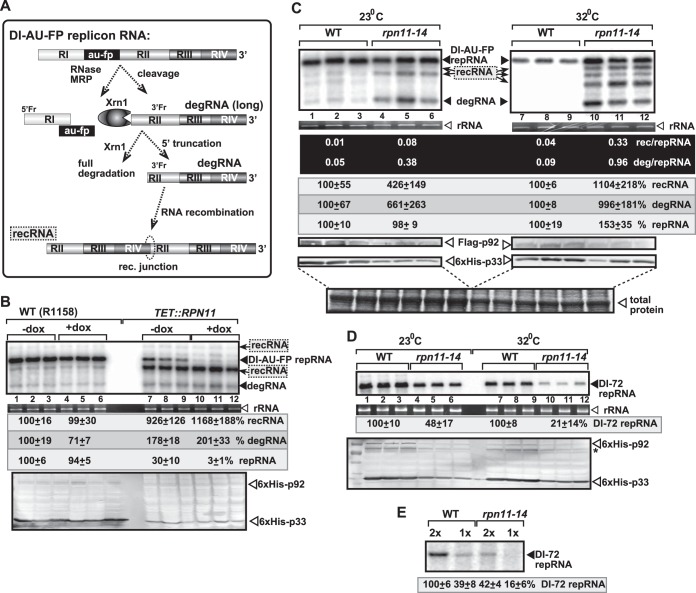
FIG 1.
The proteasomal Rpn11 metalloprotease is a suppressor of TBSV RNA recombination in yeast. (A) Scheme of the TBSV RNA recombination pathway. The replication-competent highly recombinogenic DI-AU-FP repRNA is cleaved by cellular RNase MRP endoribonuclease, followed by either limited 5′ truncations or complete degradation by the cellular Xrn1p 5′-3′ exoribonuclease. This results in the generation of a pool of replication-competent degRNAs (both the long and short forms as shown) that serve as templates in template-switching recombination events driven by the viral replicase. The sequence elements in a typical recRNA are shown schematically. (B) Depletion of Rpn11p in yeast gives rise to the rapid emergence of TBSV recRNAs and degRNAs. Addition of doxycycline (+dox samples) leads to depletion of Rpn11p expressed from the regulatable TET promoter (lanes 10 to 12). Note that expression of Rpn11p from the TET promoter in the absence of doxycycline (lanes 7 to 9) in TET::RPN11 yeast could be lower than expression from the native promoter in wt yeast. This could be the reason for the higher level of recRNA accumulation in the TET::RPN11 yeast not treated with doxycycline. Replication of the TBSV DI-AU-FP repRNA in wt and TET::RPN11 yeasts coexpressing the tombusvirus p33 and p92 replication proteins was measured by Northern blotting 24 h after launching TBSV replication. Note the emergence of different species of recRNAs and degRNA (see panel A) in samples with depleted Rpn11p. The accumulation level of repRNA was normalized based on the rRNA (middle portion). For the bottom portion, the accumulation levels of His6-p92 and His6-p33 were tested by Western blotting. The samples were obtained from independent yeast colonies. The experiments were repeated two or three times. (C) Rapid accumulation of recRNAs in yeast expressing the rpn11-14ts mutant at 23°C (permissive temperature for yeast growth) or 32°C (semipermissive temperature). The top portion shows Northern blot analysis of the accumulation of TBSV DI-AU-FP repRNA, recRNAs, and degRNA at the 24-h time point. For the middle portion, the accumulation level of repRNA was normalized based on the rRNA. The bottom portion shows Western blotting-based measurement of the accumulation levels of His6-p92 and His6-p33. Each experiment was repeated. (D) Accumulation levels of the highly efficient DI-72 repRNA in rpn11-14ts yeast were measured by Northern blotting. Western blotting was used to measure the accumulation levels of His6-p92 and His6-p33 (bottom portion). The asterisk marks the SDS-resistant p33 homodimer. Each experiment was repeated. See further details in the description for panel B. (E) CFE-based replication assay supports a role for Rpn11p DUB in TBSV replication. Purified recombinant MBP-p33 and MBP-p92pol replication proteins and in vitro-transcribed TBSV DI-72 plus-strand repRNA were added to the CFEs prepared from wt or rpn11-14ts yeast strains. Denaturing PAGE analysis of the 32P-labeled TBSV repRNA products obtained in the CFE assay is shown. Note that a comparable amount of CFEs (indicated by 1x) or double the amount of CFEs (2x) was used for the replication assay. Each experiment was repeated three times.
Tombusviruses code for two replication proteins, termed p33 and p92pol, which are translated directly from the genomic RNA (gRNA). The abundant p33 RNA chaperone functions in recruitment of viral RNA template for replication and in the assembly of the membrane-bound VRCs (26–31). p92pol is the RdRp (29, 32, 33), and it is produced through translational readthrough of the p33 stop codon (34–36). Both replication proteins are essential components of the tombusvirus VRCs (33, 37).
A recent systematic screen with TBSV based on a temperature-sensitive library of yeast mutants (K. R. Prasanth and P. D. Nagy, unpublished data) has identified the yeast Rpn11p (regulatory particle non-ATPase) metalloprotease subunit of the 19S regulatory particle, which is part of the 26S proteasome lid (38). Rpn11p is a critical component of the proteasome due to its function in coupling deubiquitination and degradation of proteasome substrates. Accordingly, polyubiquitinated proteins accumulate in yeast expressing mutated Rpn11p (39). Rpn11p is required for the stability of the proteasomes, which could get rapidly degraded in the absence of Rpn11p. In addition, Rpn11p affects the cellular localization of the proteasome via inducing the formation of proteasome storage granules under certain cellular conditions, such as the quiescent stage (40).
Rpn11p is also involved in several pathways that are independent of its catalytic activity. For example, Rpn11p regulates the Fis1-dependent fission machinery of mitochondria and peroxisomes (39, 41). Accordingly, peroxisome proliferation is halted and peroxisome distribution is abnormal in rpn11-m1 yeast (41). Another activity of Rpn11p is to regulate cell cycle progression (41, 42). Rpn11p is highly conserved protein in eukaryotes, including mammals (Rpn11p is called POH1 or PSMD14 in humans) (38). The N-terminal part contains the deubiquitinase (DUB, the catalytically active JAMM/MPN+) domain, while the C-terminal segment affects the stability of the proteasomal lid, cell cycle progression, and mitochondrial and peroxisomal fission (41, 42). Thus, mutations in the multifunctional Rpn11p could have pleiotropic effects on the cell, making it more challenging to dissect its function in viral replication and recombination.
In this work, we show evidence that Rpn11p is co-opted for tombusvirus replication. Depletion of Rpn11p or expression of Rpn11p mutants causes reduced tombusvirus replication but increased accumulation of viral recombinant RNAs in yeast cells. An in vitro recombination assay based on yeast cell extract confirmed that Rpn11p is a strong suppressor of TBSV recombination in yeast. In addition, Rpn11p contributes to the stability of the viral replication proteins. Detailed analysis of the role of Rpn11p revealed that it is less likely that Rpn11p suppresses tombusvirus recombination via inhibition of Xrn1p activity (previously shown to possess strong suppressor activity on RNA recombination) or via its role in cell cycle control or in regulating the free-ubiquitin pool via recycling of ubiquitin from proteasomal substrates. Data obtained with a fis1Δ/rpn11-m1 yeast strain suggest that the function of Rpn11p in peroxisome division could be a mechanism controlling tombusvirus replication and recombination to some extent. Also, we show that free-ubiquitin pool is important in regulation of TBSV replication and recombination, albeit independently from Rpn11p deubiquitinase function. We propose a model for the role of Rpn11p that is based on the key role of Rpn11p in controlling the recruitment of a cellular DDX3-like Ded1p DEAD box helicase, a known suppressor of tombusvirus recombination, into the membrane-bound VRCs. Overall, subversion of Rpn11p by TBSV is critical for recruitment of additional cellular factors and for the correct VRC assembly, which greatly affects both viral replication and RNA recombination.
MATERIALS AND METHODS
Yeast strains and expression plasmids.
S. cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and R1158 (wt), TET::RPN11 yeast strains (yTHC library, MATa his3-1 leu2-0 met15-0 URA3::CMV-tTA), were obtained from Open Biosystems (Huntsville, AL). Yeast strain NMY51 was obtained from Dualsystems.
Temperature-sensitive rpn11 mutants (rpn11-14 and rpn11-8) were kindly provided by Charles Boone (University of Toronto) (43). The 31-amino acid (aa) C-terminal truncation mutant of rpn11 (rpn11-m1), the double mutant strains (rpn11-RevA2, rpn11-RevA5, Δ fis1/rpn11-m1, Δmdv1/rpn11-m1), and the plasmid RPN11S119A were kindly provided by Agnès Delahodde (University of Paris-Sud) (41).
The yeast expression plasmids LpGAD-CUP1-HisFlag-p92 (LEU2 selection) and HpGBK-CUP1-His-p33/GAL1-DI-AU-FP (HIS3 selection) were constructed earlier (14). Plasmid pYES-Ubiquitin was constructed earlier (44). To produce pYC-RPN11, PCR was performed with primers 5234 (5′-CGCGGATCCATGGAACGACTACAGAGATT-3′) and 5448 (5′-CCGCTCGAGTTATTTAATTGCCACTGAATT-3′). The obtained PCR products were digested with BamHI and XhoI and cloned into pYC-NT vector. The plasmid pYC-DED1 was made by PCR amplification of yeast DED1 open reading frame (ORF) with primers 3957 (CCAGGAATTCATGGCTGAACTGAGCGAACAAG) and 4309 (CCAGCTCGAGTCACCACCAAGAAGAGTTG). The obtained PCR products were digested with EcoRI and XhoI and cloned into pMal-DED1 (45) to generate pYC-DED1.
The plant overexpression plasmids were constructed as described previously (24). Briefly, pGD-RPN11 was obtained by digesting the PCR products with BamHI and MluI and was inserted between the BamHI and MluI sites of the pGD plasmid. Plasmids pGD-p19 and pGD-CNV have been described previously (24).
Tombusvirus RNA analysis.
TBSV RNA replication and recombination were analyzed using total RNA from yeast or plants, and Northern blot analyses were performed as described previously (24). In brief, BY4741, W303, and the rpn11-14ts, rpn11-m1, RevA2, RevA5, Δfis1, Δmdv1, Δfis1/rpn11-m1, and Δmdv1/rpn11-m1 yeast strains were cotransformed with pGBK-Hisp33-CUP1/GAL1-DI-AU-FP and pGAD-His92-CUP1. The transformed yeast strains were grown at 23°C in SC-HL− (synthetic complete medium without histidine and leucine) medium with 2% glucose for 12 h at 23°C. Then, yeast cultures were resuspended in SC-HL− medium with 2% galactose containing 50 μM CuSO4. Yeasts were grown at either 23°C or 32°C for the desired times before being collected for total RNA extraction.
The yTHC strains (R1158 and RPN11) were transformed with pGBK-His33-CUP1/GAL1-DI-AU-FP, and pGAD-His92-CUP1, then inoculated into SC-ULH− medium containing 2% glucose and supplemented with 10 mg/liter of doxycycline, and cultured for 8 h at 29°C until an optical density at 600 nm of 0.8 to 1.0 was reached. The yeast cultures were subsequently centrifuged, washed, and resuspended in SC-ULH− medium supplemented with 2% galactose, 10 mg/liter of doxycycline, and 50 μM CuSO4 to induce viral replication and incubated for 24 h at 29°C.
For the complementation studies, BY4741 and the rpn11-14 strain were cotransformed with plasmids pGBK-His33-CUP1/GAL1-DI-AU-FP or DI-RIIΔ70, pGAD-His92-CUP1, and pYC-RPN11 (full-length)/pYC-RPN11S119A (null mutation in the deubiquitinase catalytic domain) (41) or pYC-Empty derivatives. For the ubiquitin complementation assay, BY4741 and the rpn11-14ts strain were cotransformed with plasmids pGBK-His33-CUP1/GAL1-DI-AU-FP, pGAD-His92-CUP1, and pYES-Ubiquitin or pYES-Empty derivatives. For the complementation study with Ded1p protein, strain RPN11-14 was cotransformed with plasmids pGBK-His33-CUP1/GAL1-DI-AU-FP, pGAD-His92-CUP1, and pYC-DED1 or pYC-Empty derivatives. The transformed yeasts were grown in SC-ULH− minimal medium supplemented with 2% glucose for 12 h at 23°C. Then, the cultures were changed to SC-ULH− minimal medium supplemented with 2% galactose and with 50 μM CuSO4 to induce virus replication and incubated for an additional 24 h at either 23°C or 32°C.
Protein analysis.
Yeast cells were grown in appropriate SC media as described above for Northern analysis. Total protein lysates were prepared by the NaOH method as described previously (46). The total protein samples were analyzed by SDS-PAGE and Western blotting with anti-His antibody (Amersham), followed by alkaline phosphatase-conjugated anti-mouse secondary antibody (Sigma) as described previously (19).
Tombusvirus replication assay based on yeast cell extracts.
The preparation of yeast cell extracts capable of supporting TBSV RNA replication and recombination in vitro was described previously (31, 47). The replication assay mixture consisted of 2 μl of yeast cell-free extract (CFE), 500 ng of DI-72 plus-strand replicon RNA (repRNA) or DI-RIIΔ70 plus-strand degRNA, 50 ng of purified maltose-binding protein (MBP)-p33, and 50 ng of purified MBP-p92 protein in a 20-μl reaction mixture. For testing the effect of recombinant Rpn11p, increasing amounts (0.5, 1.0, and 1.5 μg) of purified recombinant MBP-Rpn11 protein or MBP alone were added to the reaction mixture. The mixtures were incubated at 25°C for 3 h to support TBSV RNA replication and recombination in vitro, and the amount of newly synthesized 32P-radiolabeled repRNA was analyzed in a polyacrylamide-urea gel as described previously (33).
Viral RNA stability assay.
For the viral RNA stability studies, BY4741 and the rpn11-14ts strain were transformed with pYC2-DI-AU-FP or pYC2-RIIΔ70. The transformed yeast strains were grown at 23°C in SC-U− with 2% galactose for 24 h. Yeast cells were harvested by centrifugation and resuspended in SC-U− supplemented with 2% glucose. Samples for total RNA extraction were collected at the desired times (see below).
Analysis of protein-protein interactions using the split-ubiquitin assay.
The split-ubiquitin assay performed was based on Dualmembrane kit 3 (Dualsystems) and was performed as previously described (48). The bait constructs pGAD-BT2-N-His33 and pGAD-BT2-N-His92, expressing tombusvirus replication proteins p33 and p92, were previously described (49). Yeast strain NMY51 was cotransformed with pGAD-BT2-N-His33 or pGAD-BT2-N-His92 and pPR-N-RE (NubG) or with one of the prey constructs carrying RPN11 sequence and plated onto Trp− Leu− minimal medium plates. Transformed colonies were diluted in water and streaked onto Trp− Leu− His− Ade− plates to test for p33-RPN11 and p92-RPN11 interactions, respectively. A plasmid carrying the SSA1 ORF was used as a positive control in this assay.
Analysis of TBSV RNA recombination profile.
To observe the RNA recombination profile with DI-ΔRI RNA, BY4741 and the xrn1Δ, met22Δ, and rpn11-14ts yeast strains were cotransformed with HpGBK-His33, LpGAD-His92, and UpYC2-DI-ΔRI. The transformed cultures were grown at 23°C for 12 h in SC-ULH− medium with glucose. Yeast cultures were collected by centrifugation and dissolved in SC-ULH− medium with galactose supplemented with 50 μM CuSO4, followed by additional culturing at 23°C for 2 days before sample collection for RNA analysis.
Protein copurification from yeast.
To study if Rpn11p affects the amount of Ded1p protein in the tombusvirus replicase, BY4741 and the rpn11-14ts strain were transformed with plasmids pGBK-FLAGp33-CUP1/Gal1-DI-72 and pGAD-FLAGp92-CUP1 (or pGBK-His33-CUP1/Gal1-DI-72 and pGAD-Hisp92-CUP1 as a control) plus the pYC plasmids expressing His6-tagged Ded1 protein from the GAL1 promoter (pYC-Gal1-DED1). For copurification of Rpn11 protein with p33 and p92 replication proteins, BY4741 and the rpn11-14ts strain were cotransformed with plasmids pESC-FLAGp33-CUP1/Gal1-DI-72 and pGAD-FLAGp92-CUP1 and (or pESC-His33and pGAD-Hisp92 as a control) plus the pGBK plasmids expressing His6-tagged Rpn11 or His6-Rpn11S119A protein from the GAL1 promoter (pGBK-Gal1-RPN11/pGBK-Gal1-RPN11S119A).
After selection of transformed yeast on SC-ULH− plates, yeast cultures were pregrown in selective SC-ULH− medium with 2% glucose for 12 h at 23°C and then transferred to selective medium with 2% galactose for 24 h at either 23 or 32°C to induce His6-Ded1, His6-Rpn11, and His6-Rpn11S119A protein expression from the GAL1 promoter. Then the cultures were supplemented with 50 μM CuSO4 to induce expression of FLAG-p33 and FLAG-p92 or His6-p33 and His6-p92 from the CUP1 promoter. The yeast cultures were collected by centrifugation after 4 h, washed once with phosphate-buffered saline (PBS), and then incubated in PBS buffer containing 1% formaldehyde for 1 h on ice to cross-link proteins. Then formaldehyde was quenched by adding 2 ml of 2.5 M glycine. Finally, yeast cultures were washed in PBS and proteins were FLAG affinity purified as described previously (19, 45).
Expression of AtRpn11 in Nicotiana benthamiana.
For transient expression of Arabidopsis thaliana Rpn11 (AtRpn11) in N. benthamiana leaves, we PCR amplified the RPN11 sequence using the following pair of primers: 5984 (CGGGATCCATGGAGAGACTACAGAGAA) and 5986 (CCGCTCGAGCTAGAAGACAACAGTGTCGA). The obtained PCR product was digested with BamHI and XhoI and ligated into pGD empty vector. The resulting plasmid was transformed into Agrobacterium tumefaciens C58C1. Plants were infiltrated with A. tumefaciens carrying pGD-RPN11 or pGD (empty plasmid) along with pGD-CNV plus pGD-ΔRI or, alternatively, with pGD-p33 plus pGD-p92 plus pGD-ΔRI. Two days later, leaf samples were collected from the agroinfiltrated leaves and total RNA was extracted. DI-ΔRI RNA accumulation was analyzed by Northern blotting (23).
VIGS of RPN11 in N. benthamiana plants.
The virus-induced gene silencing (VIGS) in N. benthamiana was done as described previously (23). To generate the VIGS vector (pTRV2-RPN11), a 257-bp cDNA fragment of A. thaliana RPN11 was reverse transcription (RT)-PCR amplified from a total RNA extract of A. thaliana using primers 5696 (CGGGATCCACATCTCTTCTCTCGC) and 5697 (CGACGCGTTCCAGGATGTGAATGA) and inserted into the corresponding (BamHI and MluI) restriction sites of plasmid pTRV2. Seven days after the VIGS treatment (pTRV2-RPN11 VIGS vector together with pTRV1), the level of N. benthamiana RPN11 (NbRPN11) mRNA was determined by RT-PCR with primers 5698 (CGGGATCCGAACAAACCATCGATC) and 5699 (CGACGCGTAGATGCTTCTTTGCAT). We used tubulin mRNA as a control by RT-PCR using primers 2859 (TAATACGACTCACTATAGGAACCAAATCATTCATGTTGCTCTC) and 2860 (TAGTGTATGTGATATCCCACCAA). Subsequently, the silenced leaves were either sap inoculated with TBSV inocula or coagroinfiltrated with pGD-CNV and pGD-DI-AU-FP to launch viral replication and recombination. Samples from the agroinfiltrated leaves were collected three and 4 days after agroinfiltration, followed by total RNA extraction and Northern blot analysis as described previously (23).
RESULTS
Rpn11 mutants support increased level of tombusvirus RNA recombination in yeast.
To test the effect of Rpn11p on TBSV RNA recombination, first we depleted Rpn11p by blocking its transcription from a regulatable promoter in TET::RPN11 yeast. We coexpressed the highly recombinogenic DI-AU-FP replicon RNA (repRNA) (Fig. 1A) together with the tombusvirus p33 and p92pol replication proteins to initiate TBSV repRNA replication and recombination in these yeast strains. Depletion of Rpn11p resulted in an ∼12-fold increase in TBSV recombinant RNA (recRNA) accumulation compared with the wt yeast (Fig. 1B, lanes 10 to 12 versus 1 to 6). These RNA recombinants in the TBSV system are generated via the template-switching mechanism by the viral replicase using viral repRNA templates that are cleaved by cellular endo- and exoribonucleases (schematically shown in Fig. 1A) (14, 15, 21, 22, 24, 50, 51). In addition, yeast expressing the rpn11-14ts mutant as the only copy of Rpn11p led to an ∼11-fold increase in TBSV recRNA levels at the semipermissive temperature (Fig. 1C, lanes 10 to 12 versus 7 to 9) and ∼4-fold-higher recRNA accumulation at a lower (permissive) temperature (Fig. 1C, lanes 4 to 6). Altogether, these yeast genetic approaches have conclusively demonstrated that the wt Rpn11p is a strong suppressor of TBSV RNA recombination in yeast cells.
Interestingly, the amount of 5′-truncated viral repRNAs, called degRNAs (shown schematically in Fig. 1A), is also increased ∼6- to 10-fold in yeast with depleted Rpn11p or expressing the rpn11-14ts mutant (Fig. 1B and C). These partially degraded viral degRNA products are generated by cellular nucleases, and they serve as templates for viral RNA recombination (Fig. 1A) (14, 15). Thus, Rpn11p seems to play a role in inhibition of generation and/or accumulation of degRNAs as well.
Expression of the rpn11-14ts mutant reveals a role in viral RNA replication in yeast.
To test if Rpn11p plays a role in tombusvirus replication, we launched the highly efficient TBSV DI-72 repRNA replication in yeast expressing the rpn11-14ts mutant. Interestingly, the TBSV repRNA accumulated only to ∼20% at semipermissive temperature and to ∼50% at permissive temperature in comparison with yeast expressing the wt Rpn11p (Fig. 1D), suggesting that Rpn11p is critical for TBSV replication. Western blot analysis revealed the reduced accumulation of p33 and p92pol replication proteins in rpn11-14ts yeast (Fig. 1C and D). Therefore, it is likely that Rpn11p contributes to the stability of the viral replication proteins. Altogether, it seems that wt Rpn11p supports the replication of the full-length repRNA (such as DI-72), while it helps to suppress the accumulation of recRNAs and degRNAs in yeast.
To study the proviral function of Rpn11p in TBSV replication, we used an in vitro assay based on a cell-free extract (CFE) from yeast expressing the wt or the rpn11-14ts mutant (41). Purified recombinant p33 and p92pol and DI-72 plus-strand RNA were used to assemble the tombusvirus replicase in the CFE in vitro (45). The CFE supports a complete replication cycle resulting in both minus- and plus-strand viral RNA products (31). Denaturing PAGE analysis of the in vitro replicase products revealed that TBSV replication was decreased over 2-fold when the CFE was obtained from rpn11-14ts yeast in comparison with the wt yeast (Fig. 1E). Thus, the in vitro replication assay confirmed that Rpn11p is a critical host factor for TBSV replication.
Effect of Rpn11 on viral RNA recombination in vitro.
To further dissect the inhibitory function of Rpn11p in recRNA formation and degRNA replication, we also used an in vitro assay based on CFE from yeast expressing either wt or a C-terminally truncated Rpn11p (the rpn11-m1 mutant, missing the last 31 amino acids from the C terminus) (41). This mutant shows deficiency in maintaining a correct cell cycle and abnormal mitochondrial morphology and function (39).
Purified recombinant p33 and p92pol and the highly recombinogenic plus-strand degRNA (DI-RIIΔ70) (Fig. 2A) were used to assemble the tombusvirus replicase in the CFE in vitro (45). Denaturing PAGE analysis of the in vitro replicase products revealed that DI-RIIΔ70 degRNA replicated at an ∼2.5-fold-higher level in CFE prepared from rpn11-m1 yeast (Fig. 2B, lane 2) than from wt yeast (lane 1). In addition, the formation and accumulation of recRNAs (formed via recombination from the degRNA template) were increased ∼2.5-fold in CFE prepared from rpn11-m1 yeast (Fig. 2B, lane 2). Thus, similar to the situation in yeast, the CFE with the rpn11-m1 mutant facilitated the replication of degRNA and the production of recRNA that lacked the authentic 5′-end viral sequences (Fig. 1A).
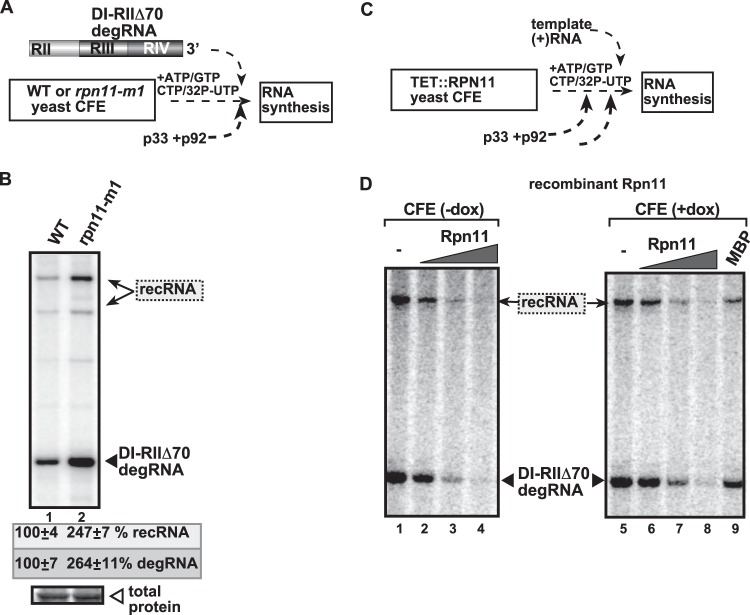
FIG 2.
High level of TBSV recRNA accumulation in an in vitro replication assay based on CFE obtained from rpn11-m1 yeast. (A) Scheme of the CFE-based in vitro replication assay. The CFE was prepared from either wt or rpn11-m1 yeasts. The purified recombinant tombusvirus p33 and p92 replication proteins from Escherichia coli were added in combination with the template RNA to program the in vitro tombusvirus replication assay. (B) The TBSV template RNA was DI-RIIΔ70 degRNA, which represents a frequently isolated degRNA species lacking RI and part of RII (see panel A). The denaturing PAGE analysis shows the emergence of recRNAs as indicated. (C) Scheme of the use of recombinant Rpn11p in a yeast CFE replication assay for TBSV replication. Note that the assembly of the TBSV replicase takes place in the CFE using recombinant viral components as shown. We used TET::RPN11 yeast with expressed (−dox) or depleted (+dox) Rpn11p to prepare the CFEs. The TBSV p33 and p92 replication proteins and Rpn11p were purified from E. coli. (D) Reduced replication of TBSV DI-RIIΔ70 degRNA in TET::RPN11 yeast CFEs with high or depleted levels of Rpn11p (see Fig. 1). Shown is denaturing PAGE analysis of the effect of purified Rpn11p on recombination and replication of DI-RIIΔ70 degRNA in the CFE assay.
In a second CFE-based assay, the addition of the purified recombinant wt Rpn11p to the CFE programmed with DI-RIIΔ70 degRNA, which lacks the authentic 5′-end sequence, led to inhibition of degRNA replication and formation of recRNAs in vitro (Fig. 2D, lanes 2 to 4 versus lane 1 and lanes 6 to 9 versus lane 5). Based on these data from CFEs, we conclude that the wt Rpn11p inhibits the replication of degRNAs and the accumulation of recRNAs missing the authentic 5′-end viral sequences.
Rpn11p is copurified with the tombusvirus replicase.
To test if Rpn11p could interact with the tombusvirus replication proteins, we used the split-ubiquitin assay that measures interaction between membrane-bound proteins in yeast (52, 53). We observed only weak interaction between the yeast Rpn11p and TBSV p33 replication protein compared with Ssa1p Hsp70, which shows strong interaction with p33 (Fig. 3A) (31, 54). However, the interaction of Rpn11p with the p92pol replication protein was more pronounced in the split-ubiquitin assay (Fig. 3B), suggesting that Rpn11p likely favors p92pol over p33 as a protein binding partner.
FIG 3.
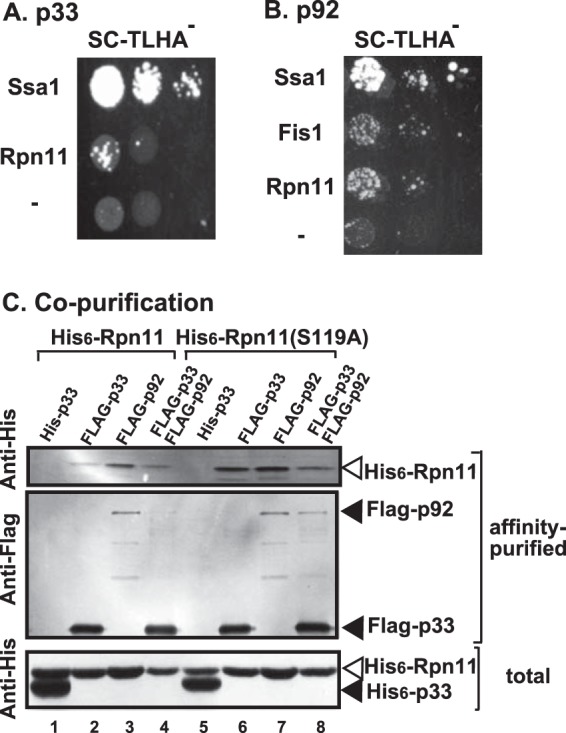
Rpn11p is recruited to the tombusvirus replicase in yeast. (A and B) Interaction between Rpn11p and the TBSV p92 replication protein. A split-ubiquitin assay was used to test binding between either p33 (A) or p92 (B) and the shown full-length yeast proteins. The bait p33 or p92 was coexpressed with the prey proteins in yeast. Ssa1p (HSP70 chaperone) and the empty prey vector (NubG, shown by a dash) were used as positive and negative controls, respectively. The image shows results for 10-fold serial dilutions of yeast cultures. (C) Copurification of Rpn11p with the p33 and p92 replication proteins from yeast. The FLAG-tagged p33 or FLAG-p92 was purified from a solubilized membranous fraction of yeast extracts using a FLAG affinity column (lanes 2 to 4 and 6 to 8). The 6×His-tagged p33 purified using a FLAG affinity column (lanes 1 and 5) was used as a negative control. (Top) Western blot analysis of 6×His-tagged Rpn11p with anti-His antibody in the affinity-purified preparations. (Middle) Western blot analysis of the same samples as in the top portion but using anti-FLAG antibody. (Bottom) Western blot analysis of 6×His-tagged p33 and 6×His-Rpn11 with anti-His antibody in the total protein extract from yeast expressing the shown proteins. Each experiment was repeated three times.
The interaction of Rpn11p with p92pol also indicates that Rpn11p might be part of the tombusvirus replicase complex (VRC). To examine if Rpn11p is present within the tombusvirus VRCs, we FLAG affinity purified the tombusvirus replicase from membrane fractions of yeast cells actively replicating TBSV repRNA (33, 48). This yeast also expressed His6-tagged Rpn11p from plasmid. We found that the purified tombusvirus replicase preparation, which is highly active on added templates in vitro (data not shown), contained Rpn11p (Fig. 3C, lane 4), while Rpn11p was undetectable in the control yeast sample obtained using the same affinity purification (Fig. 3C, lane 1). The control yeast also expressed the His6-tagged p33, which lacked a FLAG tag, and the His6-tagged Rpn11p.
Copurification of His6-tagged Rpn11p from yeast expressing either FLAG-p33 or FLAG-p92 via a FLAG affinity approach revealed a higher association with p92pol than with p33 (Fig. 3C, lanes 2 and 3). Thus, the copurification approach also indicates that the binding between Rpn11p and p92pol is more efficient than between Rpn11p and p33, although p33 is present in an ∼10-fold excess over p92pol in the VRCs (Fig. 3C). We were also able to copurify a catalytically inactive DUB mutant of Rpn11S119A with the tombusvirus VRCs (Fig. 3C, lane 8) and with either p33 or p92pol (Fig. 3C, lanes 6 and 7), suggesting that Rpn11p does not need its DUB function to bind to the tombusvirus replication proteins. Altogether, these data support the model that Rpn11p interacts with the replication proteins, and it is part of the tombusvirus VRCs.
Rpn11 suppresses recombination and replication of the 5′-truncated degRNAs independent of Xrn1p 5′-to-3′ exoribonuclease in yeast.
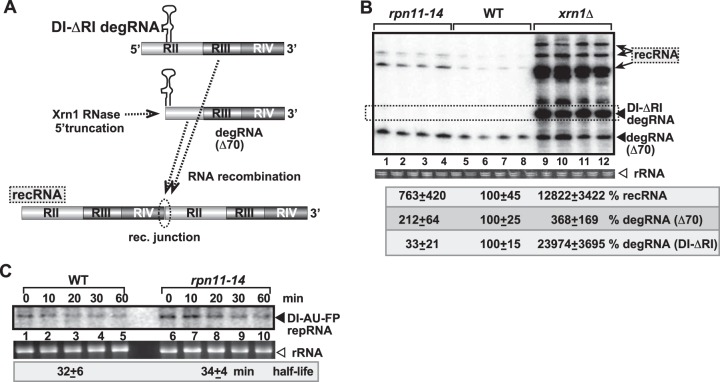
Rpn11p is a multifunctional protein (40–42), which makes it a challenging task to identify what function(s) of Rpn11p affects TBSV replication and RNA recombination. Interestingly, Rpn11p physically interacts with Xrn1p (55) 5′-to-3′ exoribonuclease, which is a key enzyme in TBSV RNA stability and for suppression of TBSV RNA recombination in yeast (22–24, 50, 56). Therefore, it is possible that Rpn11p affects TBSV recombination via controlling Xrn1p activity. To test this possibility, we expressed a 5′-truncated TBSV degRNA (DI-ΔRI) (Fig. 4A), which goes through further 5′ truncations [up to ∼70 nucleotides (nt), where the RII(+)-SL hairpin structure stops the nuclease activity of Xrn1p] in the presence of Xrn1p in wt yeast (Fig. 4A). In contrast, the 5′-truncation process with DI-ΔRI degRNA is weak in xrn1Δ yeast (Fig. 4B, lanes 9 to 12) (56). We found that due to rapid degradation, DI-ΔRI degRNA did not accumulate in rpn11-14ts yeast at a detectable extent (Fig. 4B, lanes 1 to 4), similar to the findings with wt yeast (lanes 5 to 8). On the other hand, DI-ΔRI degRNA accumulated to a high level in xrn1Δ yeast (Fig. 4B, lanes 9 to 12). In addition, the profile of recRNAs accumulating in rpn11-14ts yeast was similar to that in wt yeast and different from that in xrn1Δ yeast (Fig. 4B). Based on these results, we suggest that Rpn11p mutation does not affect TBSV RNA recombination and degRNA accumulation via inhibition of Xrn1p activity. This conclusion was further supported by RNA stability experiments that showed a half-life for DI-AU-FP plus-strand repRNA in rpn11-14ts yeasts comparable to that in the wt yeast (Fig. 4C).
FIG 4.
Suppression of viral RNA recombination by Rpn11p is independent of the Xrn1p exoribonuclease pathway. (A) Schematic representation of the TBSV RNA recombination pathway based on Xrn1p in yeast. Plasmid-driven expression of the DI-ΔRI (derived from the full-length DI-72 repRNA by deletion of the 5′ RI domain) in the presence of p33 and p92 replication proteins results in partial 5′ truncations by the cellular Xrn1p 5′-to-3′ exoribonuclease, generating DI-RIIΔ70-like degRNAs as shown. DI-Δ70RII-like degRNAs then participate in RNA recombination as indicated. (B) Recombination profile of DI-ΔRI RNA in rpn11-14ts, wt, or xrn1Δ yeasts. The original DI-ΔRI degRNA, DI-RIIΔ70-like degRNAs, and recRNAs are labeled with arrowheads and arrows. Note that the recRNA profile is dramatically different in rpn11-14ts yeast from that seen in xrn1Δ yeast. (C) Half-life of DI-AU-FP repRNA in wt and rpn11-14ts yeasts. Note that yeast did not express p33 and p92 replication proteins.
Defect in peroxisome division affects TBSV RNA recombination in yeast.
To further test if the known functions of Rpn11p affect TBSV recombination, we used cell cycle revertant mutants of Rpn11p in our recombination assay with DI-AU-FP recombinogenic repRNA. Two cell cycle revertant mutants of Rpn11p (called RevA2 and RevA5) (39, 42) supported TBSV recombination as efficiently as the cell cycle defect mutant rpn11-m1 yeast (Fig. 5, lanes 5 to 8 versus lanes 3 and 4). This finding indicates that the cell cycle defect in the rpn11-m1 mutant is unlikely to be responsible for the enhanced level of TBSV recombination.
FIG 5.
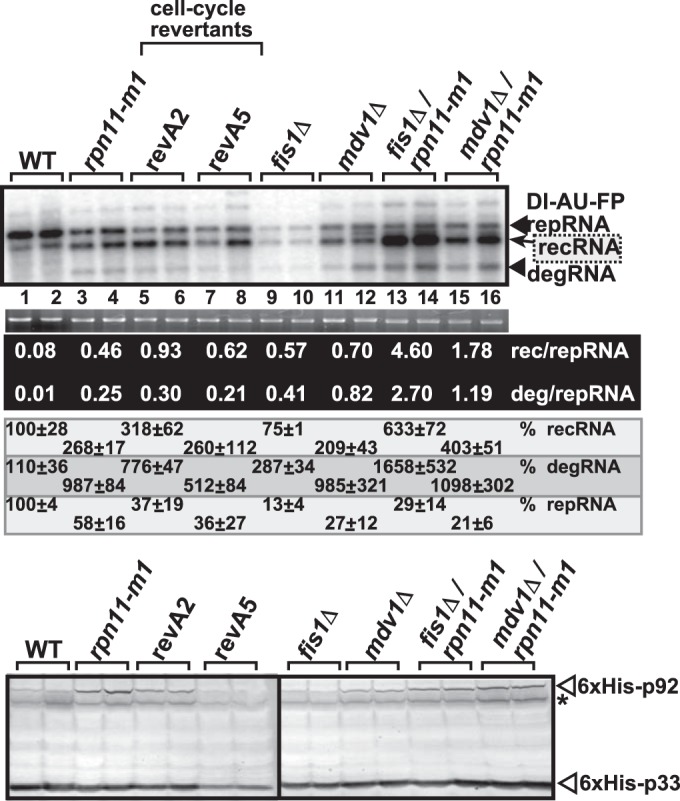
Effects of cell cycle and peroxisomal division mutant yeasts on TBSV RNA recombination. (Top) Replication of the TBSV DI-AU-FP repRNA in the shown yeast strains coexpressing the tombusvirus p33 and p92 replication proteins was measured by Northern blotting 24 h after the launch of TBSV replication. The ratio of recRNAs and degRNAs was calculated as for Fig. 1. (Middle) The accumulation level of repRNA was normalized based on the rRNA. (Bottom) The accumulation levels of His6-p92 and His6-p33 were tested by Western blotting. The samples were obtained from independent yeast colonies. The experiments were repeated three times. The asterisk indicates detergent-resistant p33 homodimers.
One of the known functions of Rpn11 is to regulate mitochondrial and peroxisomal fission and membrane proliferation (41). Since TBSV replicates on peroxisomal membrane surfaces (27, 57), it is possible that rpn11-m1 might affect TBSV recombination via this process. To test if peroxisomal division could play a role in TBSV recombination, we used fis1Δ and mdv1Δ yeasts. Both fis1Δ and mdv1Δ yeasts are defective in peroxisomal and mitochondrial fission functions (41). Interestingly, recombination increased ∼7-fold in both fis1Δ and mdv1Δ yeasts (Fig. 5, lanes 9 to 12, based on recRNA/repRNA ratios) in comparison with the wt yeast (lanes 1 and 2). Recombination was even further enhanced in the fis1Δ/rpn11-m1 double mutant and in mdv1Δ/rpn11-m1 yeasts, ∼60-fold and ∼20-fold, respectively (Fig. 5, lanes 13 to 16). Altogether, these data support, albeit indirectly, that peroxisomal division or proliferation might be a factor in TBSV recombination, which replicates on the cytosolic surface of peroxisomes. However, the additive nature of the enhanced TBSV recombination in the fis1Δ/rpn11-m1 strain suggests that rpn11-m1 does not affect TBSV recombination via its function in peroxisome division, but Rpn11p likely affects TBSV recombination via another mechanism(s). Nevertheless, these results support the role of the peroxisome and mitochondrial fission machinery, such as Fis1p and Mdv1p, in suppression of TBSV recombination in yeast. But the molecular bases of these functions of Fis1p and Mdv1p in TBSV recombination need further study.
The free-ubiquitin pool affects TBSV RNA recombination in yeast.
One of the major functions of Rpn11p is to remove the polyubiquitin chain from the proteins destined for degradation by the 26S proteasome (38). This process helps the recycling of ubiquitin before proteasomal degradation, and ultimately, this is important to maintain the free-ubiquitin pool inside the cells (38, 58). Therefore, we have overexpressed ubiquitin in rpn11-14 yeast to increase the free-ubiquitin pool in these cells, followed by testing for TBSV recombination. Interestingly, overexpression of ubiquitin greatly reduced TBSV recRNA accumulation in rpn11-14 yeast (8-fold [Fig. 6A]) at the semipermissive temperature. However, ubiquitin overexpression also resulted in reduced accumulation of the TBSV repRNA (∼3-fold [Fig. 6A]), indicating that the free-ubiquitin pool is important in regulation of TBSV replication.
FIG 6.
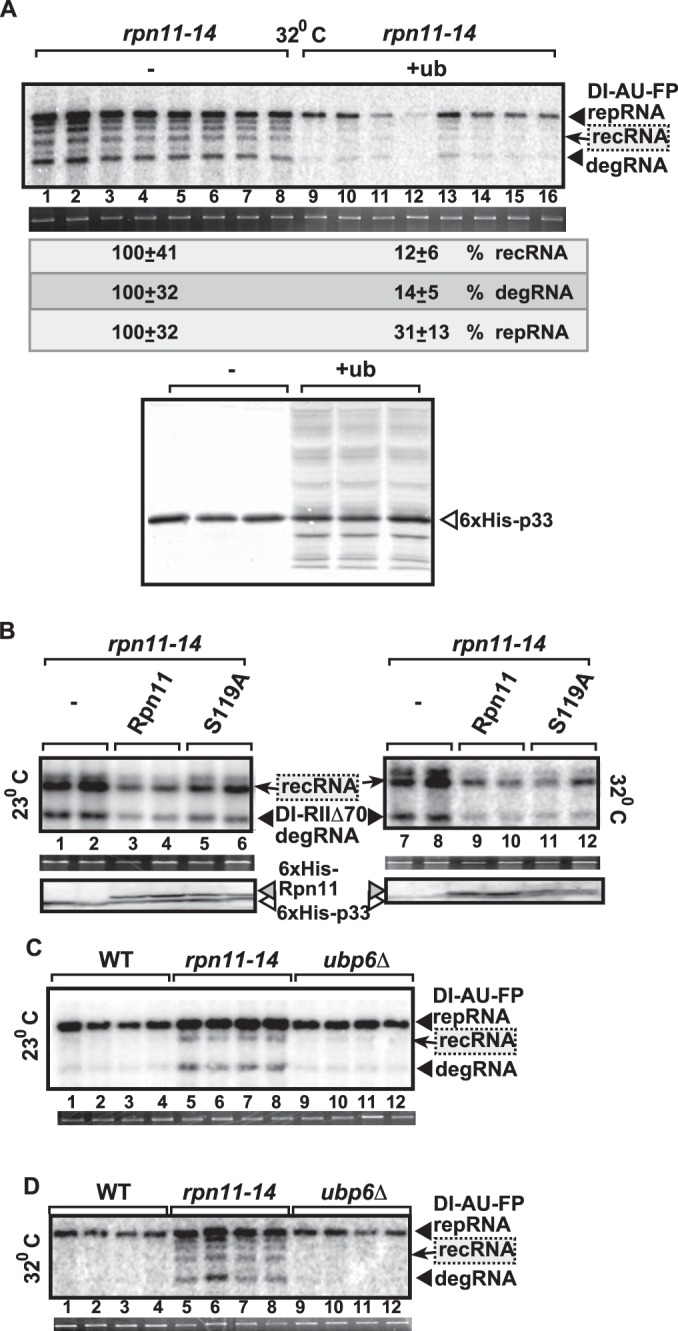
DUB function of Rpn11p does not seem to affect TBSV recombination in yeast. (A) Effect of overexpression of 6×His-tagged ubiquitin (+ub) on TBSV RNA replication and recombination in WT and rpn11-14ts yeasts at semipermissive temperature. The RNA template was the recombinogenic DI-AU-FP RNA. See further details on Northern and Western blotting in Fig. 1. The dash indicates the absence of ubiquitin overexpression. (B) The catalytically inactive Rpn11p (S119A mutation, debilitating DUB function) can complement Rpn11p function in TBSV recombination when expressed in rpn11-14ts yeast at two temperatures as shown. Dashes indicate the absence of Rpn11p expression (empty vector control). (C and D) Deletion of the second proteasomal DUB, UBP6, does not affect TBSV recombination in yeast. See further details in the description of panel A.
To test directly if the DUB function of Rpn11p plays a role in tombusvirus recombination, we complemented the defect in rpn11-14 yeast via plasmid-borne expression of Rpn11p and Rpn11S119A, which is a mutant defective in DUB function (58). Both the wt and the S119A mutant Rpn11p were able to partially suppress the accumulation of degRNA (DI-RIIΔ70) and the newly formed recRNAs at both temperatures (Fig. 6B). Therefore, these data do not support a direct role for DUB function of Rpn11p in tombusvirus recombination.
Because Rpn11p is not the only DUB in the proteasome, we also tested the effect of the second DUB, termed Ubp6p (59), which also plays a role in ubquitin recycling within the proteasome, on TBSV recombination. Replication of the highly recombinogenic DI-AU-FP repRNA and the low accumulation of recRNAs and degRNAs in ubp6Δ yeast (Fig. 6C and D), comparable to those in wt yeast, suggest that the reduction of free-ubiquitin pool via the poor recycling of ubiquitin in ubp6Δ yeast is not a major factor in promoting TBSV recombination.
Rpn11p facilitates the recruitment of the host DDX3-like Ded1p helicase for TBSV replication in yeast.
Rpn11p is a critical factor in the assembly of the proteasome, and it controls the stability of the proteasome (38, 40). Rpn11p is among the yeast proteins with the largest number of physical interactors, including ∼800 cellular partners (55, 60). Based on these features, we assumed that Rpn11p, which strongly interacts with p92pol (Fig. 3), might help the recruitment of cellular factors for viral replication. Among the previously characterized subverted host factors for tombusvirus replication and recombination, there is a DDX3-like Ded1p DEAD box helicase, which interacts with Rpn11p (45, 55, 60). To test if Rpn11p facilitates the recruitment of the cellular Ded1p helicase into the tombusvirus VRCs, we affinity purified the tombusvirus VRCs (after solubilization of the membrane-bound p33:p92pol complexes), followed by Western blotting to identify copurified cellular proteins. These experiments revealed that Ded1p helicase (Fig. 7) was inefficiently copurified with the tombusvirus replication proteins from rpn11-14 yeast in comparison with the wt yeast. These data strongly suggest that Rpn11p is involved in the recruitment of cellular Ded1p helicase into the tombusvirus VRCs. This feature of Rpn11p could be important since Ded1p is known to suppress tombusvirus recombination and promote TBSV RNA replication (45).
FIG 7.
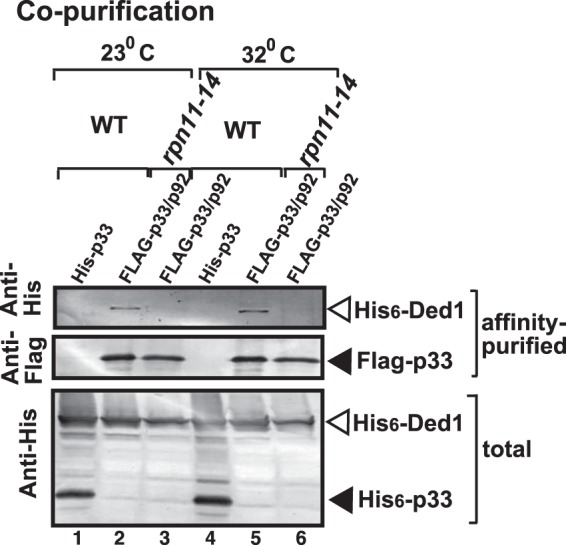
Rpn11p mutation affects the recruitment of some other host factors into the tombusvirus replicase in yeast. The Ded1p helicase was copurified with the p33 replication protein from wt or rpn11-14ts yeast. The FLAG-tagged p33 and FLAG-p92 were copurified from solubilized membranous fraction of yeast extracts using a FLAG affinity column (lanes 2 and 3 and lanes 5 and 6). The 6×His-tagged p33 and His6-p92 copurified using a FLAG-affinity column (lanes 1 and 4) were used as a negative control. (Top) Western blot analysis of 6×His-tagged Ded1p with anti-His antibody in the affinity-purified preparations. (Middle) Western blot analysis of the same samples as in the top portion but using anti-FLAG antibody. (Bottom) Western blot analysis of 6×His-tagged p33 and 6×His-Ded1 with anti-His antibody in the total protein extract from yeast expressing the shown proteins. Each experiment was repeated three times.
Overexpression of Ded1p helicase decreases TBSV recombination in rpn11-14 yeast.
Since the recruitment of yeast Ded1p helicase into the VRCs was poor in rpn11-14 yeast, we tested if we could suppress TBSV recombination by overexpressing Ded1p. These experiments revealed strong reduction in recRNA and degRNA accumulation in rpn11-14 yeast overexpressing Ded1p helicase (Fig. 8). Thus, these data support the model that overexpression of Ded1p can complement the defect observed in rpn11-14 yeast by reducing TBSV recombination.
FIG 8.
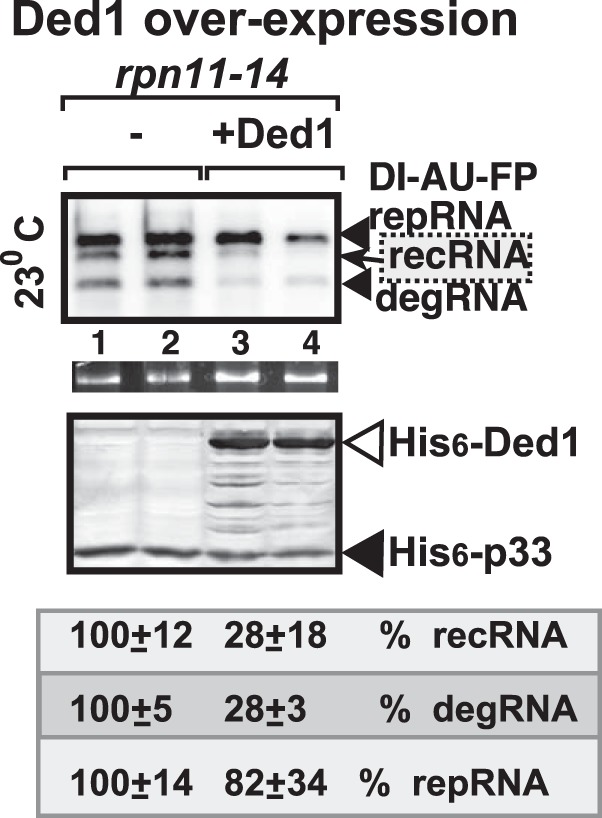
Complementation of rpn11-14ts with overexpression of Ded1p helicase in TBSV recombination in yeast. The effect of overexpression of the yeast 6×His-tagged Ded1p helicase on TBSV RNA recombination in rpn11-14ts yeasts at permissive temperature is shown. The RNA template was the recombinogenic DI-AU-FP RNA. See further details on Northern and Western blotting in the legend to Fig. 1.
Overexpression of Rpn11p decreases the accumulation of recRNAs and degRNAs in N. benthamiana plants.
To test if the orthologous plant Rpn11p (Table 1) has similar functions during tombusvirus replication and recombination, we overexpressed A. thaliana Rpn11 (AtRpn11) in N. benthamiana plants, followed by expressing the highly recombinogenic DI-ΔRI degRNA in the presence of Cucumber necrosis virus (CNV), a very closely related tombusvirus that provided p33 and p92 in trans for DI-ΔRI degRNA replication. We observed ∼5-fold reduction in recRNA accumulation and ∼5- to 8-fold reduction in degRNA accumulation in leaves overexpressing AtRpn11 (Fig. 9A). The plants did not show an obvious phenotype due to AtRpn11 expression when the samples were taken (Fig. 9A).
TABLE 1.
Sequence alignment for Rpn11 using CLUSTAL 2.1
| Protein | Sequencea |
|---|---|
| AtRPN11 | MERLQRIFGAGGGLGHASPDSPTLDTSEQVYISSLALLKMLKHGRAGVPMEVMGLMLGEF 60 |
| ScRPN11 | ------MMNSKVGSADTGRD----DTKETVYISSIALLKMLKHGRAGVPMEVMGLMLGEF 50 |
| ::.: * ..:. * **.* *****:************************* | |
| AtRPN11 | VDEYTVRVVDVFAMPQSGTGVSVEAVDHVFQTNMLDMLKQTGRPEMVVGWYHSHPGFGCW 120 |
| ScRPN11 | VDDYTVNVVDVFAMPQSGTGVSVEAVDDVFQAKMMDMLKQTGRDQMVVGWYHSHPGFGCW 110 |
| **:***.********************.***::*:************************* | |
| AtRPN11 | LSGVDINTQQSFEALNQRAVAVVVDPIQSVKGKVVIDAFRSINPQTIMLGQEPRQTTSNL 180 |
| ScRPN11 | LSSVDVNTQKSFEQLNSRAVAVVVDPIQSVKGKVVIDAFRLIDTGALINNLEPRQTTSNT 170 |
| **.**:***:*** **.*********************** *:.::: . ******** | |
| AtRPN11 | GHLNKPSIQALIHGLNRHYYSIAINYRKNELEEKMLLNLHKKKWTDGLTLRRFDTHSKTN 240 |
| ScRPN11 | GLLNKANIQALIHGLNRHYYSLNIDYHKTAKETKMLMNLHKEQWQSGLKMYDYEEKEESN 230 |
| * ***..**************: *:*:*. * ***:****::* .**.::::.::* | |
| AtRPN11 | EQTVQEMLSLAAKYNKAVQEEDELSPEKLAIVNVGRQDAKKHLEEHVSNLMSSNIVQTLG 300 |
| ScRPN11 | LAATKSMVKIAEQYSKRIEEEKELTEEELKTRYVGRQDPKKHLSETADETLENNIVSVLT 290 |
| :.:.*:.:*:*.*::**.**: *:* *****.****.* ..::..***..* | |
| AtRPN11 | TMLDTVVF- 308 |
| ScRPN11 | AGVNSVAIK 299 |
| ::::*.: |
Asterisk (*), conserved amino acid; colon (:), conservative amino acid; period (.), semiconservative amino acid.
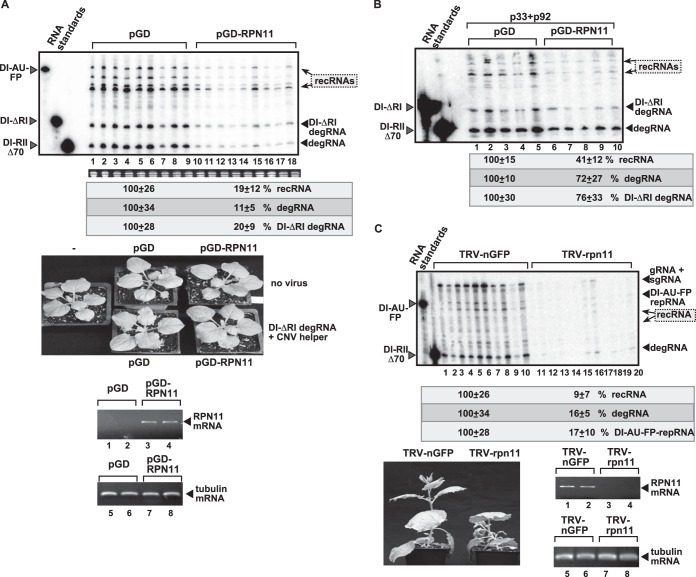
FIG 9.
Rpn11 overexpression inhibits TBSV recombination in plants. (A) Overexpression of AtRpn11 was done in N. benthamiana leaves by agroinfiltration. The same leaves were agroinfiltrated to coexpress CNV helper virus and the highly recombinogenic DI-ΔRI degRNA from the 35S promoter. The control samples were obtained from leaves not expressing AtRpn11 (lanes 1 to 9). Total RNA was extracted from leaves 2 days after agroinfiltration. The accumulation of DI-ΔRI degRNA, recRNAs, and degRNAs (5′ truncated RIIΔ70-like; see Fig. 1A) in N. benthamiana leaves was measured by Northern blotting (top). The rRNA was used as a loading control and is shown in an agarose gel stained with ethidium bromide (middle). The lack of phenotypes in N. benthamiana due to Rpn11 overexpression (bottom) is shown. The pictures were taken right before sampling. Semiquantitative RT-PCR was used to detect AtRpn11 mRNA levels in the AtRpn11 overexpression leaves. (B) Co-overexpression of AtRpn11 and TBSV p33 and p92 replication proteins and the recombinogenic DI-ΔRI degRNA from the 35S promoter was done in N. benthamiana leaves by coagroinfiltration. See further details in the description for panel A. (C) Silencing of RPN11 expression inhibits tombusvirus replication in N. benthamiana. VIGS-based silencing was induced via the TRV vector carrying RPN11 sequence or the N-terminal half of green fluorescent protein (GFP) (negative control). The upper silenced leaves were agroinfiltrated to coexpress CNV helper virus and the DI-AU-FP repRNA from the 35S promoter. Total RNA was extracted from leaves 4 days after agroinfiltration. The accumulation of repRNA, recRNAs, and degRNA in N. benthamiana leaves was measured by Northern blotting (top); see the legend to Fig. 1 for details. The rRNA was used as a loading control and is shown in an agarose gel stained with ethidium bromide (middle). Note that the CNV gRNA and the subgenomic RNAs (sgRNA) are not separated well on PAGE gels. (Bottom) The stunting phenotype in N. benthamiana due to Rpn11 silencing is shown. The pictures were taken right before agroinfiltration. Semiquantitative RT-PCR was used to detect NbRpn11 mRNA levels in the silenced versus those not silenced.
In another approach, we coexpressed the TBSV p33 and p92 replication proteins with recombinogenic DI-ΔRI degRNA, followed by testing the accumulation of recRNAs in the agroinfiltrated leaves of N. benthamiana. We found ∼2-fold-decreased recRNA accumulation in leaves overexpressing AtRpn11 in comparison with the control plants (Fig. 9B). Thus, similar to the case with yeast, we observed that Rpn11 could suppress recRNA and degRNA accumulation in plants leaves, too.
To test the importance of the plant Rpn11 in TBSV replication, we silenced N. benthamiana RPN11 in leaves via VIGS, followed by inoculation with DI-AU-FP repRNA in the presence of CNV helper virus. We observed ∼6- to 10-fold reductions in accumulation of all tombusvirus RNAs, including the repRNA, degRNAs, recRNAs, and the helper virus, in plants silenced for RPN11 (Fig. 9C). The large reduction in degRNA and recRNA levels in rpn11-silenced plants is likely due to the small amount of replication proteins supplied by the poorly replicating helper virus (see below), which is also dependent on RPN11 host factor. The RPN11-silenced plants were stunted, suggesting that RPN11 is also required for plant growth. Nevertheless, it seems that RPN11 is also an important factor in tombusvirus RNA accumulation and recombination in plants.
Rpn11p plays an important role in tombusvirus replication in N. benthamiana plants.
To further test if Rpn11p plays a role in tombusvirus replication in N. benthamiana, we tested TBSV genomic RNA and subgenomic RNA accumulation in VIGS-treated plants. Interestingly, all three TBSV-specific RNAs showed an ∼10-fold-reduced level of accumulation in RPN11-silenced plants (Fig. 10A). Similarly, the accumulation of the closely related CNV RNAs was decreased over 10-fold in RPN11-silenced plants (Fig. 10B), further supporting a key role for the plant Rpn11p in tombusvirus replication.
FIG 10.
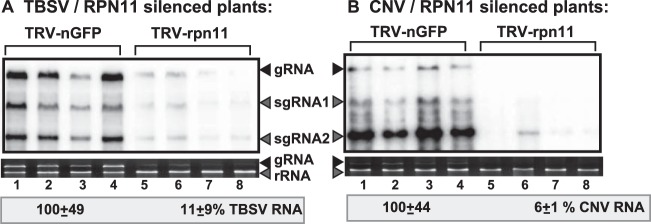
Knockdown of Rpn11 inhibits TBSV accumulation in plants. (A) Silencing of RPN11 expression was induced based on the TRV vector carrying RPN11 sequence or the N-terminal half of GFP (negative control). The upper silenced leaves of N. benthamiana were sap inoculated with infectious TBSV 7 days after agroinfiltration, followed by analysis of the accumulation of TBSV RNAs by Northern blotting (top) 3 days after inoculation. The rRNA was used as a loading control and is shown in an agarose gel stained with ethidium bromide (bottom). (B) The upper silenced leaves (7 days after the original agroinfiltration) were agroinfiltrated to express CNV from the 35S promoter. Total RNA was extracted from leaves 3 days after agroinfiltration with the CNV plasmid, followed by Northern blotting. See further details in the description for panel A.
DISCUSSION
Early works on viral RNA recombination have demonstrated the widespread occurrence of recombinants for many viruses. The roles of viral proteins, including the RdRp and other viral auxiliary factors, have been shown, suggesting template-switching-type recombination as the major mechanism (4, 61–68). Recent works on tombusviruses based on systematic genome-wide screens performed with TBSV in a yeast surrogate host (14, 15), however, have led to the identification of cellular factors with key roles in viral RNA recombination (21–24, 50, 51, 56). Tombusvirus research firmly established various cellular pathways and factors as major drivers of viral recombination.
The proteasomal Rpn11p is a major suppressor of tombusvirus RNA recombination in yeast.
This work has demonstrated an unexpected role for an essential proteasomal component, Rpn11p metalloprotease, in tombusvirus recombination. Rpn11p was originally identified in a screen using temperature-sensitive mutants of yeast as an inhibitor of TBSV recombination (Prasanth and Nagy, unpublished). Rpn11p seems to be a strong suppressor of tombusvirus recombination based on the high level of recRNA accumulation in yeast with depleted Rpn11p or expressing the rpn11-14ts mutant (Fig. 1). The direct involvement of Rpn11p can also be seen in in vitro assays, with CFE capable of assembling the tombusvirus replicase (Fig. 2). In spite of the outstanding role of Rpn11p in recombination, it seems that Rpn11p is mainly a replication factor, while its recombination suppressor activity is the consequence of the role of Rpn11p during replication (see below).
Rpn11p is a critical replicase assembly factor during viral RNA replication in yeast.
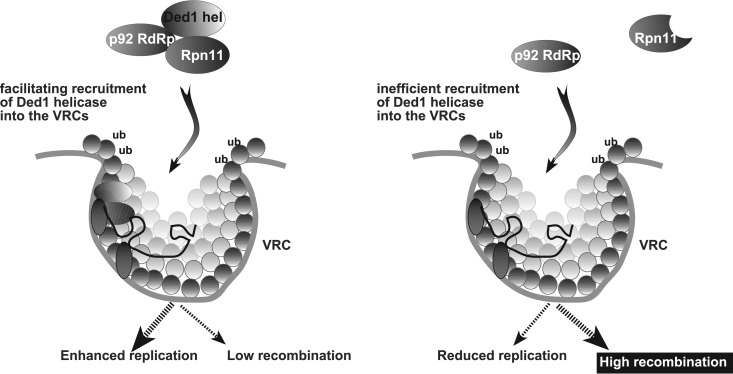
Based on the results presented in this paper, we propose that Rpn11p serves as a subverted key VRC assembly factor. The primary role of Rpn11p during VRC assembly is likely as a matchmaker that interacts with both viral p92pol and the cellular DDX3-like Ded1p (RH20 in plants) helicase to promote the subversion of Ded1p into the VRCs. Then, this complex likely interacts with additional components, such as p33 replication protein and the viral plus-strand RNA.
This VRC assembly function of Rpn11p might be analogous to its cellular role during the assembly of the proteasomal “lid” and the whole proteasome (38). Because Rpn11p is partially localized to the peroxisome (the site of TBSV replication in cells) to regulate peroxisome division (41), it might also affect the subcellular localization of tombusvirus VRCs, although this suggestion requires further studies.
Model of the viral recombination suppression activity of Rpn11p.
Although Rpn11p is a multifunctional enzyme with both proteasomal and nonproteasomal functions, the data presented here support a new activity for Rpn11p. We suggest that the recombination suppressor activity of Rpn11p is based on facilitating the recruitment of Ded1p helicase into the VRCs (Fig. 11). Then Ded1p suppresses tombusvirus recombination through inhibition of replication of degRNAs and formation of recRNAs (Chuang et al., unpublished). Accordingly, overexpression of Ded1p in rpn11-14 yeast to compensate for poor recruitment of Ded1p into VRCs has suppressed RNA recombination.
FIG 11.
Models showing the proposed function of subverted cellular Rpn11p metalloprotease in TBSV replication and viral RNA recombination. (Left) Rpn11p is proposed to facilitate the efficient recruitment of the cellular DDX3-like Ded1/AtRH20 helicases together with the viral p92pol replication protein into the VRCs. This activity of Rpn11p makes the properly assembled VRCs efficient in viral RNA replication and also results in suppression of RNA recombination. Note that AtRH20 is a functional ortholog of the yeast Ded1 in TBSV replication. (Right) When Rpn11p is depleted or mutated, then recruitment of Ded1/AtRH20 helicases into the VRCs is inefficient. This alters the VRC components, leading to low replication but highly efficient template-switching-type viral RNA recombination. Together, these models propose key recruitment and VRC assembly functions for the co-opted Rpn11p during TBSV replication and recombination.
Indirect function of Rpn11p in TBSV replication and recombination.
By systemically testing the known cellular functions of the multifunctional Rpn11p, we also observed that Rpn11p likely facilitates viral replication by maintaining the proper free-ubiquitin pool, whose excess inhibits both replication and recombination of TBSV (Fig. 6) via an unknown pathway. It has previously been shown that ubiquitination of p33 replication protein facilitates TBSV replication due to binding to Vps23p ESCRT protein (44, 54). This, in turn, promotes the recruitment of additional ESCRT proteins required for the proper formation of VRCs present within membranous invaginations called spherules (69, 70). Fusing p33 with ubiquitin destabilizes p33 (44), indicating that excess ubiquitination of replication proteins could be detrimental for TBSV replication.
Surprisingly, we also observed that the peroxisomal fission pathway, including Fis1p and Mdv1p, affects tombusvirus recombination (Fig. 5). Therefore, the role of Rpn11p in the peroxisomal fission pathway (41) could be one of the mechanisms by which Rpn11p affects TBSV recombination. However, the fis1Δ/rpn11-m1 and mdv1Δ/rpn11-m1 double mutant yeasts have shown vastly increased TBSV recombination, suggesting that Rpn11p affects TBSV recombination not only through its role in the peroxisomal fission pathway. Thus, these data support that the major suppressor activity of Rpn11p is likely the facilitation of the Ded1p helicase recruitment to the VRCs, which has an additive effect with that caused by the role of Rpn11p in peroxisomal fission.
Rpn11p has been shown to interact with a great number of cellular factors (55, 60), which might also affect TBSV recombination via unknown mechanisms. Accordingly, we have tested the possible involvement of Rpn11p-Xrn1p interaction in TBSV recombination. However, it does not seem that Rpn11p affects TBSV recombination via the previously characterized Xrn1p 5′-to-3′ exoribonuclease with TBSV recombination suppressor activity, which acts by efficiently removing degRNAs and recRNAs generated during TBSV replication (22–24, 56). This is because the profiles of degRNAs and recRNAs were vastly different in Rpn11 mutant yeast from those in xrn1Δ yeast (Fig. 4).
In summary, the example with Rpn11p clearly shows why yeast with amazing sets of available tools is so important for viral replication and recombination studies, especially in the case of multifunctional cellular factors involved in multiple pathways, such as Rpn11p. The data from our plant work (Fig. 9 and 10), however, indicate that the plant ortholog of yeast Rpn11p could have a function comparable to that of the yeast Rpn11p in TBSV replication and recombination. Overall, this work has unearthed a novel function for a co-opted cellular protein in viral replication and recombination. Rpn11p facilitates VRC assembly, which, in turn, determines the efficiency of the viral replicase during RNA replication and the ability of the replicase to produce recombinant viral RNAs.
ACKNOWLEDGMENTS
We thank Judit Pogany, C. Chuang, and Nick Kovalev for their comments on the manuscript. We also thank Charles Boone (University of Toronto) for yeast temperature-sensitive mutants and Agnès Delahodde (University of Paris-Sud) for Rpn11 mutants.
This work was supported by NSF (MCB 1122039).
REFERENCES
- 1.Aaziz R, Tepfer M. 1999. Recombination in RNA viruses and in virus-resistant transgenic plants. J Gen Virol 80(Part 6):1339–1346. [DOI] [PubMed] [Google Scholar]
- 2.Worobey M, Holmes EC. 1999. Evolutionary aspects of recombination in RNA viruses. J Gen Virol 80(Part 10):2535–2543. [DOI] [PubMed] [Google Scholar]
- 3.Sztuba-Solinska J, Urbanowicz A, Figlerowicz M, Bujarski JJ. 2011. RNA-RNA recombination in plant virus replication and evolution. Annu Rev Phytopathol 49:415–443. doi: 10.1146/annurev-phyto-072910-095351. [DOI] [PubMed] [Google Scholar]
- 4.Nagy PD, Simon AE. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 5.Bujarski JJ. 2013. Genetic recombination in plant-infecting messenger-sense RNA viruses: overview and research perspectives. Front Plant Sci 4:68. doi: 10.3389/fpls.2013.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao AL, Hall TC. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J Virol 67:969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hema M, Gopinath K, Kao C. 2005. Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus. J Virol 79:1417–1427. doi: 10.1128/JVI.79.3.1417-1427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan H, Simon AE. 2000. Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: an activity involved in 3′ end repair of viral RNAs. Proc Natl Acad Sci U S A 97:12451–12456. doi: 10.1073/pnas.97.23.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy PD, Carpenter CD, Simon AE. 1997. A novel 3′-end repair mechanism in an RNA virus. Proc Natl Acad Sci U S A 94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roossinck MJ. 2003. Plant RNA virus evolution. Curr Opin Microbiol 6:406–409. doi: 10.1016/S1369-5274(03)00087-0. [DOI] [PubMed] [Google Scholar]
- 11.Nagy PD, Pogany J. 2006. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344:211–220. doi: 10.1016/j.virol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Panavas T, Serviene E, Brasher J, Nagy PD. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A 102:7326–7331. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajendran KS, Nagy PD. 2006. Kinetics and functional studies on interaction between the replicase proteins of tomato bushy stunt virus: requirement of p33:p92 interaction for replicase assembly. Virology 345:270–279. doi: 10.1016/j.virol.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 14.Serviene E, Jiang Y, Cheng CP, Baker J, Nagy PD. 2006. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J Virol 80:1231–1241. doi: 10.1128/JVI.80.3.1231-1241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. 2005. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A 102:10545–10550. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy PD, Pogany J. 2012. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy PD, Barajas D, Pogany J. 2012. Host factors with regulatory roles in tombusvirus replication. Curr Opin Virol 2:685–692. doi: 10.1016/j.coviro.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Nagy PD, Pogany J. 2010. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against tomato bushy stunt virus. Adv Virus Res 76:123–177. doi: 10.1016/S0065-3527(10)76004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J Virol 82:6911–6926. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaag HM, Stork J, Nagy PD. 2007. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology 368:388–404. doi: 10.1016/j.virol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Jaag HM, Pogany J, Nagy PD. 2010. A host Ca2+/Mn2+ ion pump is a factor in the emergence of viral RNA recombinants. Cell Host Microbe 7:74–81. doi: 10.1016/j.chom.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Cheng CP, Serviene E, Nagy PD. 2006. Suppression of viral RNA recombination by a host exoribonuclease. J Virol 80:2631–2640. doi: 10.1128/JVI.80.6.2631-2640.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaag HM, Nagy PD. 2009. Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology 386:344–352. doi: 10.1016/j.virol.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Cheng CP, Jaag HM, Jonczyk M, Serviene E, Nagy PD. 2007. Expression of the Arabidopsis Xrn4p 5′-3′ exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology 368:238–248. doi: 10.1016/j.virol.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Cheng CP, Serviene E, Shapka N, Nagy PD. 2010. Repair of lost 5′ terminal sequences in tombusviruses: rapid recovery of promoter- and enhancer-like sequences in recombinant RNAs. Virology 404:96–105. doi: 10.1016/j.virol.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Monkewich S, Lin HX, Fabian MR, Xu W, Na H, Ray D, Chernysheva OA, Nagy PD, White KA. 2005. The p92 polymerase coding region contains an internal RNA element required at an early step in tombusvirus genome replication. J Virol 79:4848–4858. doi: 10.1128/JVI.79.8.4848-4858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panavas T, Hawkins CM, Panaviene Z, Nagy PD. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of cucumber necrosis tombusvirus. Virology 338:81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 28.Pogany J, White KA, Nagy PD. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J Virol 79:4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pogany J, Nagy PD. 2012. p33-independent activation of a truncated p92 RNA-dependent RNA polymerase of tomato bushy stunt virus in yeast cell-free extract. J Virol 86:12025–12038. doi: 10.1128/JVI.01303-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stork J, Kovalev N, Sasvari Z, Nagy PD. 2011. RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology 409:338–347. doi: 10.1016/j.virol.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pogany J, Stork J, Li Z, Nagy PD. 2008. In vitro assembly of the tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc Natl Acad Sci U S A 105:19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panaviene Z, Panavas T, Nagy PD. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J Virol 79:10608–10618. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panaviene Z, Panavas T, Serva S, Nagy PD. 2004. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J Virol 78:8254–8263. doi: 10.1128/JVI.78.15.8254-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panaviene Z, Baker JM, Nagy PD. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308:191–205. doi: 10.1016/S0042-6822(02)00132-0. [DOI] [PubMed] [Google Scholar]
- 35.Oster SK, Wu B, White KA. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J Virol 72:5845–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholthof KB, Scholthof HB, Jackson AO. 1995. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- 37.Serva S, Nagy PD. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol 80:2162–2169. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wauer T, Komander D. 2014. The JAMM in the proteasome. Nat Struct Mol Biol 21:346–348. doi: 10.1038/nsmb.2800. [DOI] [PubMed] [Google Scholar]
- 39.Rinaldi T, Hofmann L, Gambadoro A, Cossard R, Livnat-Levanon N, Glickman MH, Frontali L, Delahodde A. 2008. Dissection of the carboxyl-terminal domain of the proteasomal subunit Rpn11 in maintenance of mitochondrial structure and function. Mol Biol Cell 19:1022–1031. doi: 10.1091/mbc.E07-07-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunier R, Esposito M, Dassa EP, Delahodde A. 2013. Integrity of the Saccharomyces cerevisiae Rpn11 protein is critical for formation of proteasome storage granules (PSG) and survival in stationary phase. PLoS One 8:e70357. doi: 10.1371/journal.pone.0070357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann L, Saunier R, Cossard R, Esposito M, Rinaldi T, Delahodde A. 2009. A nonproteolytic proteasome activity controls organelle fission in yeast. J Cell Sci 122:3673–3683. doi: 10.1242/jcs.050229. [DOI] [PubMed] [Google Scholar]
- 42.Esposito M, Piatti S, Hofmann L, Frontali L, Delahodde A, Rinaldi T. 2011. Analysis of the rpn11-m1 proteasomal mutant reveals connection between cell cycle and mitochondrial biogenesis. FEMS Yeast Res 11:60–71. doi: 10.1111/j.1567-1364.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, Min R, Vandersluis B, Bellay J, Devit M, Fleming JA, Stephens A, Haase J, Lin ZY, Baryshnikova A, Lu H, Yan Z, Jin K, Barker S, Datti A, Giaever G, Nislow C, Bulawa C, Myers CL, Costanzo M, Gingras AC, Zhang Z, Blomberg A, Bloom K, Andrews B, Boone C. 2011. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol 29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barajas D, Nagy PD. 2010. Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology 397:358–368. doi: 10.1016/j.virol.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovalev N, Pogany J, Nagy PD. 2012. A co-opted DEAD-box RNA helicase enhances tombusvirus plus-strand synthesis. PLoS Pathog 8:e1002537. doi: 10.1371/journal.ppat.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panavas T, Nagy PD. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of tomato bushy stunt virus. Virology 314:315–325. doi: 10.1016/S0042-6822(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 47.Pogany J, Nagy PD. 2008. Authentic replication and recombination of tomato bushy stunt virus RNA in a cell-free extract from yeast. J Virol 82:5967–5980. doi: 10.1128/JVI.02737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. 2009. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385:245–260. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendu V, Chiu M, Barajas D, Li Z, Nagy PD. 2010. Cpr1 cyclophilin and Ess1 parvulin prolyl isomerases interact with the tombusvirus replication protein and inhibit viral replication in yeast model host. Virology 406:342–351. doi: 10.1016/j.virol.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 50.Nagy PD. 2011. The roles of host factors in tombusvirus RNA recombination. Adv Virus Res 81:63–84. doi: 10.1016/B978-0-12-385885-6.00008-0. [DOI] [PubMed] [Google Scholar]
- 51.Jaag HM, Lu Q, Schmitt ME, Nagy PD. 2011. Role of RNase MRP in viral RNA degradation and RNA recombination. J Virol 85:243–253. doi: 10.1128/JVI.01749-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iyer K, Burkle L, Auerbach D, Thaminy S, Dinkel M, Engels K, Stagljar I. 2005. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci STKE 2005:pl3. doi: 10.1126/stke.2752005pl3. [DOI] [PubMed] [Google Scholar]
- 53.Kittanakom S, Chuk M, Wong V, Snyder J, Edmonds D, Lydakis A, Zhang Z, Auerbach D, Stagljar I. 2009. Analysis of membrane protein complexes using the split-ubiquitin membrane yeast two-hybrid (MYTH) system. Methods Mol Biol 548:247–271. doi: 10.1007/978-1-59745-540-4_14. [DOI] [PubMed] [Google Scholar]
- 54.Barajas D, Li Z, Nagy PD. 2009. The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J Virol 83:11751–11764. doi: 10.1128/JVI.00789-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaake RM, Milenkovic T, Przulj N, Kaiser P, Huang L. 2010. Characterization of cell cycle specific protein interaction networks of the yeast 26S proteasome complex by the QTAX strategy. J Proteome Res 9:2016–2029. doi: 10.1021/pr1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaag HM, Nagy PD. 2010. The combined effect of environmental and host factors on the emergence of viral RNA recombinants. PLoS Pathog 6:e1001156. doi: 10.1371/journal.ppat.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rinaldi T, Pick E, Gambadoro A, Zilli S, Maytal-Kivity V, Frontali L, Glickman MH. 2004. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem J 381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guterman A, Glickman MH. 2004. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem 279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- 60.Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. 2008. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci U S A 105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng CP, Panavas T, Luo G, Nagy PD. 2005. Heterologous RNA replication enhancer stimulates in vitro RNA synthesis and template-switching by the carmovirus, but not by the tombusvirus, RNA-dependent RNA polymerase: implication for modular evolution of RNA viruses. Virology 341:107–121. doi: 10.1016/j.virol.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 62.Shapka N, Nagy PD. 2004. The AU-rich RNA recombination hot spot sequence of brome mosaic virus is functional in tombusviruses: implications for the mechanism of RNA recombination. J Virol 78:2288–2300. doi: 10.1128/JVI.78.5.2288-2300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panaviene Z, Nagy PD. 2003. Mutations in the RNA-binding domains of tombusvirus replicase proteins affect RNA recombination in vivo. Virology 317:359–372. doi: 10.1016/j.virol.2003.08.039. [DOI] [PubMed] [Google Scholar]
- 64.Cheng CP, Nagy PD. 2003. Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro. J Virol 77:12033–12047. doi: 10.1128/JVI.77.22.12033-12047.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng CP, Pogany J, Nagy PD. 2002. Mechanism of DI RNA formation in tombusviruses: dissecting the requirement for primer extension by the tombusvirus RNA dependent RNA polymerase in vitro. Virology 304:460–473. doi: 10.1006/viro.2002.1713. [DOI] [PubMed] [Google Scholar]
- 66.Figlerowicz M, Nagy PD, Tang N, Kao CC, Bujarski JJ. 1998. Mutations in the N terminus of the brome mosaic virus polymerase affect genetic RNA-RNA recombination. J Virol 72:9192–9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Figlerowicz M, Nagy PD, Bujarski JJ. 1997. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc Natl Acad Sci U S A 94:2073–2078. doi: 10.1073/pnas.94.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagy PD, Dzianott A, Ahlquist P, Bujarski JJ. 1995. Mutations in the helicase-like domain of protein 1a alter the sites of RNA-RNA recombination in brome mosaic virus. J Virol 69:2547–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barajas D, Martin IF, Pogany J, Risco C, Nagy PD. 2014. Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of tomato bushy stunt virus replicase. PLoS Pathog 10:e1004087. doi: 10.1371/journal.ppat.1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barajas D, Jiang Y, Nagy PD. 2009. A unique role for the host ESCRT proteins in replication of tomato bushy stunt virus. PLoS Pathog 5:e1000705. doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]