ABSTRACT
Microtubule transport of circovirus from the periphery of the cell to the nucleus is essential for viral replication in early infection. How the microtubule is recruited to the viral cargo remains unclear. In this study, we observed that circovirus trafficking is dependent on microtubule polymerization and that incoming circovirus particles colocalize with cytoplasmic dynein and endosomes. However, circovirus binding to dynein was independent of the presence of microtubular α-tubulin and translocation of cytoplasmic dynein into the nucleus. The circovirus capsid (Cap) subunit enhanced microtubular acetylation and directly interacted with intermediate chain 1 (IC1) of dynein. N-terminal residues 42 to 100 of the Cap viral protein were required for efficient binding to the dynein IC1 subunit and for retrograde transport. Knockdown of IC1 decreased virus transport and replication. These results demonstrate that Cap is a direct ligand of the cytoplasmic dynein IC1 subunit and an inducer of microtubule α-tubulin acetylation. Furthermore, Cap recruits the host dynein/microtubule machinery to facilitate transport toward the nucleus by an endosomal mechanism distinct from that used for physiological dynein cargo.
IMPORTANCE Incoming viral particles hijack the intracellular trafficking machinery of the host in order to migrate from the cell surface to the replication sites. Better knowledge of the interaction between viruses and virus proteins and the intracellular trafficking machinery may provide new targets for antiviral therapies. Currently, little is known about the molecular mechanisms of circovirus transport. Here, we report that circovirus particles enter early endosomes and utilize the microtubule-associated molecular motor dynein to travel along microtubules. The circovirus capsid subunit enhances microtubular acetylation, and N-terminal residues 42 to 100 directly interact with the dynein IC1 subunit during retrograde transport. These findings highlight a mechanism whereby circoviruses recruit dynein for transport to the nucleus via the dynein/microtubule machinery.
INTRODUCTION
Porcine circovirus (PCV) belongs to the genus Circovirus of the family Circoviridae. This small icosahedral nonenveloped virus is 17 nm in diameter and has circular single-stranded DNA (1). Two genotypes of PCV have been identified: PCV type 1 (PCV1), which is nonpathogenic to pigs (2), and PCV type 2 (PCV2), which is the etiological agent of PCV2-associated disease leading to swine immunosuppression (3–7). Antibodies (Ab) in humans share antigenic epitopes with PCV (8). Unexpectedly, PCV1 contamination was recently detected in live poliovirus seeds and commercial live-attenuated human rotavirus vaccines (9, 10), and infectious PCV1 was found in the human hepatocellular carcinoma Huh-7 cell line (11). Undoubtedly, PCV exposure poses a potential risk to public health.
Of the 11 potential open reading frames (ORF) within the PCV genome, four encode viral proteins (12–15). ORF1 encodes a replicase (Rep) that is responsible for the rolling-circle replication of PCV DNA (16–18). The capsid protein (Cap), encoded by ORF2, is responsible for the successive packaging of the PCV genome and is the main structural and immunogenic protein with linear and conformational epitopes (19–23). ORF3 and ORF4 proteins were identified by Liu and He, respectively, and shown to be involved in productive PCV2 infection but not in PCV2 replication (14, 15). Specifically, ORF3 is involved in viral pathogenesis via apoptotic induction (14), while ORF4 functions in antagonizing apoptosis (15). PCV2 is the smallest known virus, and the virion binds to heparan sulfate and chondroitin sulfate B glycosaminoglycans (GAGs) on the cell surface (24–26) before entering the cell via clathrin-mediated endocytosis in monocytic cells or via an actin- and small-GTPase-dependent pathway in epithelial cells (27). Recent research showed that the PCV2 Cap protein binds to α-tubulin in PCV2-infected cells (28), and the Hsp70/Cap complex activated the NF-κB pathway and reduced caspase-3 activity in PCV2-infected monocytic cells (29). However, the mechanism of intracellular transport of the PCV2 virion remains unclear.
The cytoskeleton plays several crucial roles in the life cycle of the virus, including attachment, internalization, endocytosis, nuclear trafficking, transcription, replication, assembly of progeny subviral particles, exocytosis, and cell-to-cell spreading (30). Different virus families engage different receptors and utilize endocytosis to infect host cells (31). It is becoming increasingly apparent that many viral proteins interact with actin directly or via actin-binding proteins during cell entry by endocytosis (30). Endocytosis is regulated by several Rab proteins, which constitute the largest family of small GTPases. Rab5 and Rab7 are generally regarded as markers of early and late endosomes, respectively. Endosomal trafficking of the virus involves actin- and microtubule-dependent processes (32–34), and endosomes are transported via the motor protein dynein along microtubules toward the nucleus for virus replication (35, 36). Cargo transport throughout the cell is a finely regulated process, and recent studies showed that viral proteins interact directly with the molecular motors (37, 38). In the microtubule/dynein machinery, microtubules are stabilized by polymerization and posttranslational modification of tubulin, along with many microtubule-associated proteins (39–41). Dynein and kinesin motors use microtubules as tracks to move cargo throughout the cytoplasm (42). The dynein motor is a huge protein complex with a molecular mass of ∼1.5 MDa, consisting of two heavy chains (DHCs), two intermediate chains (ICs), two light-intermediate chains (LICs), and several light chains (LCs), which are regulated by dynactin (43–46). Several dynein subunits have been observed to interact with viral proteins, such as light-chain 8 (LC8), which interacts with the p54 protein of African swine fever virus (47), and the adenovirus hexon binds similarly to both ICs and LICs (48). Although the dynein/dynactin motor complex is implicated in viral transport, molecular events underlying viral recruitment of this complex remain poorly defined. Understanding the biology of dynein-based transport is especially important for studying immunosuppressive circoviruses.
The current study explored the mechanism by which incoming porcine circovirus particles recruit and hijack dynein for intracellular transport purposes. We found that inhibition of microtubular function or dynein activity decreased the efficiency of nuclear trafficking and reduced PCV2 replication. Additionally, we demonstrated a direct interaction between the virus Cap protein and the IC1 subunit of dynein. During viral replication, IC1 occupied an abnormal nuclear location, and IC1 knockdown decreased PCV2 replication. Based on these findings, we infer that the Cap protein of PCV2 is active when bound to dynein IC1 and promotes sustained retrograde microtubule transport in cells. These results provide a detailed mechanism for a virus-motor protein interaction and a potential avenue for antiviral intervention.
MATERIALS AND METHODS
Cell cultures and virus infection.
Permanent porcine kidney epithelial cell line PK15, which was free of PCV and kindly provided by the China Institute of Veterinary Drugs Control, was cultured in minimal essential medium (MEM; Life Technologies/Gibco, Carlsbad, CA). Cells of continuous porcine monocytic cell line 3D4/31 (CRL-2844; ATCC, Rockville, MD) were cultured in RPMI 1640 (Gibco). Human embryonic kidney epithelial (HEK) 293T cells (CRL-3216, ATCC) were cultured in Dulbecco modified Eagle medium (DMEM; Gibco). All cells were maintained in a suitable medium supplemented with 10% heat-inactivated fetal bovine serum (Gibco).
PCV2 strain HZ0201 (106.45 50% tissue culture infective doses [TCID50]/0.1 ml), isolated from pig farms with naturally occurring postweaning multisystemic wasting syndrome (PMWS) (13), was propagated in PK15 cells. To analyze virus replication, cells were infected with PCV2 at a multiplicity of infection (MOI) of the TCID50 and cultured for the indicated times. To investigate virus nuclear trafficking, cells were infected with PCV2 at an MOI of 25 for the indicated times.
Treatment of cell cultures with chemicals and recombinant soluble proteins.
Chemicals used to pretreat PK15 or 293T cells included the microtubule-destabilizing drug nocodazole (NOC) (S1765; Beyotime, Haimen, China), the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) (T1952; Sigma, St. Louis, MO), the HDAC inhibitor sodium butyrate (NaBut) (B5887; Sigma), and the dynein inhibitor sodium orthovanadate (Na3VO4) (S6508; Sigma). Before treatment, cells were cultured overnight to 60% to 70% confluence and transferred to fresh medium in the absence or presence of the drug. The solvent dimethyl sulfoxide (DMSO) was used as a control.
The PCV2-soluble recombinant Cap (rCap) protein was recombinantly expressed by baculovirus using pFast-BacHTB (Invitrogen, Carlsbad, CA) and purified by ultrasonic treatment and sucrose density gradient centrifugation. The protein concentration was quantified using a bicinchoninic acid (BCA) assay kit (Thermo, Waltham, MA), and the reaction mixture was filtered through a 0.22-μm-pore-size filter (Millipore, Bedford, MA). The soluble rCap protein at a concentration of 6 μg/ml was used to treat 293T cells to study Cap-induced α-tubulin acetylation.
Plasmid construction and DNA transfection.
Using specific primers (Table 1), full-length ORF1 (Rep) and ORF2 (Cap) were cloned from the PCV2 genome. Dynein IC1, dynactin-P50, HDAC6, Rab5, TUBA1A, TUBA1B, and TUBB2A were amplified from PK15 cells and 293T cells. The construction strategy used for the nuclear localization signal (NLS; amino acids [aa] 1 to 41)-deleted ORF2 and the ORF2-truncated variants (see Fig. 6) resulted in gene fragments that were cloned into vectors pCMV-N-myc (Clontech, Palo Alto, CA), pCMV-N-flag (Clontech), pcDNA3.0 (Invitrogen), pCI-neo (Promega, Madison, WI), pEGFP-C3 (Clontech), pGEX4T-1 (GE Healthcare Biosciences, Piscataway, NJ), and pET-28a (Novagen, Madison, WI) to construct recombinant expression vectors. All constructs were verified by DNA sequencing and were designated pCI-neo-Cap, pCMV-N-myc-CapΔ1-41 (dCap), pcDNA3.0-CapΔ42-100, pCI-neo-Rep, pCMV-Nflag-IC1, pCMV-Nflag-P50, pCMV-Nflag-HDAC6, pEGFP-C3-Rab5, pGEX4T-1-dCap, pET-28a-IC1, pET-28a-TUBA1A, pET-28a-TUBA1B, pET-28a-TUBB2A, pGEX4T-1-dCapM1(GST-234-), pGEX4T-1-dCapM2(GST-23--), pGEX4T-1-dCapM3(GST--34-), and pGEX4T-1-dCapM4(GST-2---) (a hyphen[s] at the end of each designation indicates the removed domain[s]).
TABLE 1.
Primers used for cloning and quantitative real-time PCR
| Gene product | Sense primer (5′ to 3′) | Antisense primer (5′ to 3′) |
|---|---|---|
| Rep | ATGCCCAGCAAGAAGAA | TCAGTAATTTATTTCATATGG |
| Cap | ATGACGTATCCAAGGAGGC | TTAAGGGTTAAGTGGGGGG |
| IC1 | ATGTCTGACAAAAGTGACT | CTAGGCGGTTAACTCAACAGTGCCT |
| P50 | ATGGCGGACCCTAAATACGC | TCACTTTCCCAGCTTCTTCATCCGT |
| HDAC6 | ATGACCTCAACCGGCCAGGATT | TTAGTGTGGGTGGGGCATATCCTCC |
| TUBA1A | ATGCGTGAGTGCATCTCAGTCC | CTAGTATTCGTCCCCCTCTTCCTCT |
| TUBA1B | ATGCGTGAGTGCATCTCCATCCA | TTAGTATTCCTCGCCTTCTTCCT |
| TUBB2A | ATGAGGGAAATCGTGCATATCC | TTAGGCCTCCTCTTCGGCCTCCTCA |
| Rab5 | ATGGCTAATCGAGGAGCAACAA | TTAATTACTACAACACTGACTCCTG |
| dCap | ATGAATGGCATCTTCAACA | TTAAGGGTTAAGTGGGGGG |
| dCapM1 | AATGGCATCTTCAACAC | AGTGCCGAGGCCTACGTGGTCCACA |
| dCapM2 | AATGGCATCTTCAACAC | TATGGTATGGCGGGAGGAGTAGTTT |
| dCapM3 | AATGGCATCTTCAACAC | CTTTCTTATTCTGTAGTATTCAAAG |
| dCapM4 | GTTAAGGTTGAATTCTGGC | AGTGCCGAGGCCTACGTGGTCCACA |
| Cap1-41 | ATGACGTATCCAAGGAGGCG | TTTCCTTCTCCAGCGGTAACGGTGGC |
| Cap101-233 | GCCACCGTTACCGCTGGAGAAGGAAA | TTAAGGGTTAAGTGGGGGGTCTTTA |
| RT-Rep | TGATGACTTTTATGGCTGGCT | TCCTCCGTGGATTGTTCTGT |
| RT-Cap | TGTAGTATTCAAAGGGCACAGA | CGGATATACTATCAAGCGAACCAC |
FIG 6.
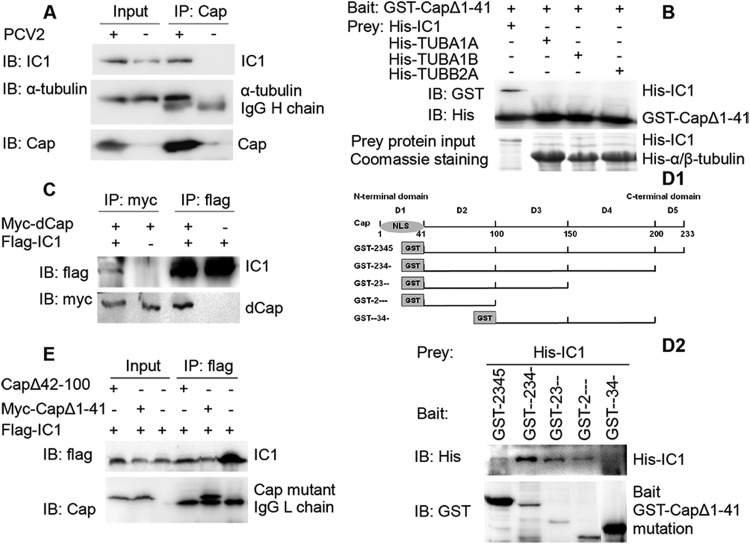
Interaction of PCV2 Cap with dynein intermediate-chain IC1. (A) The lysates of PCV2 and mock-infected PK15 cells were immunoprecipitated with mouse anti-Cap IgG. Immunoblotting was then performed to determine the presence of IC1 and α-tubulin in the Cap immunoprecipitate. (B) Recombinant GST-dCap protein was immobilized on glutathione-Sepharose beads and incubated with recombinant His-IC1, His-TUBA1A, His-TUBA1B, or His-TUBB2A. His-tagged proteins in GST-pulldown assays were examined by immunoblotting with anti-His antibody. The levels of His-tagged proteins were determined by Coomassie blue staining. (C) 293T cells were cotransfected with a myc-tagged dCap expression plasmid together with a flag-IC1 expression plasmid. Only cells expressing myc-dCap or flag-IC1 were included as controls. Cell lysates were immunoprecipitated with an anti-flag antibody or an anti-myc antibody. The resulting precipitates were examined by immunoblotting using an anti-flag or an anti-myc antibody to examine the interaction between myc-dCap and flag-IC1. (D1 and D2) Identification of the IC1 interaction domain on Cap. (D1) Schematic representation of various truncated forms of the PCV2 Cap that were tagged with GST and used to identify the IC1 interaction domain. Constructs are named for each intact domain number, with a hyphen(s) indicating the removed domain(s). (D2) Bacterially expressed His-IC1 was incubated with various truncated forms of Cap tagged with GST and immobilized on glutathione-Sepharose beads. Immunoblotting was then performed to characterize the IC1 interaction domain on Cap. (E) Cells were cotransfected with plasmids expressing dCap (CapΔ1-41) or CapΔ42-100, together with a flag-IC1 expression plasmid. Immunoprecipitation and immunoblotting were then performed to examine the interactions between various forms of Cap and flag-IC1.
One day prior to DNA transfection, PK15 and 293T cells were seeded into 35-mm-diameter glass bottom dishes or common culture dishes (Corning Incorporated, Corning, NY), and the resultant constructs were transfected using Lipofectamine 2000 reagent (Invitrogen) for transient protein expression.
Indirect IFA, confocal microscopy, and gated stimulated emission depletion microscopy (gSTED).
The inoculation procedure and immunofluorescence assay (IFA) protocol were performed as previously described (49). Briefly, PK15 cells were mixed with PCV2 virus dilutions and seeded in 96-well flat-bottom microtiter plates at 100 μl/well. After incubation at 37°C for 72 h, cells were washed twice with phosphate-buffered saline (PBS) and fixed with a methanol-acetone mixture (1:1 [vol/vol]) at −20°C for 20 min. The cells were blocked with PBS containing 5% skimmed milk at 37°C for 1 h. For IFA, cells were incubated with mouse monoclonal antibody (MAb) against Cap (23) for 1.5 h at 37°C, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Kirkegaard & Perry Laboratories Inc. [KPL], Gaithersburg, MD) for 1 h. Titers were determined by observing infected cells under a fluorescence microscope and calculating the TCID50 per 0.1 ml.
For confocal microscopy, cells grown on glass coverslips were fixed, permeabilized with 0.2% Triton X-100, and incubated at 4°C with primary antibodies overnight. Cells were then incubated with FITC-conjugated secondary antibodies (KPL) and/or Alexa Fluor 546-conjugated secondary antibodies (Invitrogen) at 37°C for 1 h. Cellular nuclei were stained with 10 μg/ml DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (10236276001; Roche, Mannheim, Germany) for 5 min, coverslipped, and viewed with a LSM780 laser scanning confocal microscope (Zeiss, Oberkochen, Germany). FITC fluorescence was detected after excitation at 488 nm with an emission long-band filter at 505 to 530 nm (green). Alexa Fluor 546 fluorescence was detected after excitation at 561 nm with an emission long-pass filter at 550 to 600 nm (red). DAPI was detected after excitation at 405 nm with an emission long-pass filter at 445 to 450 nm (blue). Images were acquired and analyzed using Zen 2012 software (Zeiss). The primary antibodies used included mouse anti-IC1 MAb (ab6304; Abcam, Cambridge, MA), rabbit anti-α-tubulin polyclonal antibody (pAb) (ab15246; Abcam), mouse anti-Cap MAb, rabbit anti-IC1 pAb, pig anti-Rep pAb (50), and pig anti-Cap pAb (23).
For gSTED superresolution microscopy, PK15 cells were transfected with the green fluorescent protein (GFP)-Rab5 plasmid for 24 h and were subsequently infected with PCV2 at an MOI of 25. Virus was incubated with cells in an initial cold binding step to synchronize the infection process. At 6 hpi, the cells were fixed, incubated with mouse anti-Cap IgG and then with anti-mouse Alexa Fluor 546, and viewed using a Leica TCS SP8 gated STED (gSTED) microscope equipped with an HCX PL apochromatic (APO) 100-by-1.40-numerical-aperture (NA) oil objective. PCV2 gSTED images (red) were acquired using wavelengths of 546 nm (excitation) and 660 nm (depletion) and collected at between 554 and 600 nm. GFP-Rab5 images (green) were obtained using wavelengths of 488 nm (excitation) and 592 nm (depletion) and collected at between 497 and 581 nm. The widths of fluorescence peaks at half-maximum values were ∼0.3 μm in the confocal mode and ∼70 to ∼80 nm for the same image captured in gSTED mode, providing a measure of the increase in resolution with gSTED imaging. Images were deconvoluted with Huygens Essential software (Scientific Volume Imaging, Hilversum, Netherlands).
Immunoblotting, glutathione S-transferase (GST) pulldown, and coimmunoprecipitation.
Common immunoblotting (IB) was performed as previously described (28). Briefly, cells were lysed in lysis buffer after infection or other treatments for the indicated times. Lysates were collected, and proteins were separated by standard SDS-PAGE gels and electrotransferred onto 0.22-μm-pore-size nitrocellulose membranes (GE Healthcare) that were subsequently blocked in PBS containing 0.05% Tween 20 (PBST) and 5% skimmed milk for 1 h. Membranes were then incubated with primary antibody overnight at 4°C, washed three times with PBST for 5 min each time, and incubated with an appropriate secondary antibody conjugated to horseradish peroxidase (HRP) (KPL) for 1 h at 37°C. Finally, membranes were washed three more times with PBST before visualization was performed using SuperSignal West Pico chemiluminescent substrate (Thermo) under the conditions recommended by the manufacturer. Images were captured on a chemiluminescent imaging system, and protein band densities were normalized against the β-actin signal and quantified using AlphaView SA software (Cell Biosciences, Santa Clara, CA). The primary antibodies used to probe membranes were mouse anti-Cap MAb, mouse anti-IC1 MAb, rabbit anti-α-tubulin pAb, mouse anti-acetylated α-tubulin MAb (ab24610; Abcam), rabbit anti-acetylated histone 3 pAb (06-599; Merck Millipore, Darmstadt, Germany), rabbit anti-histone 3 pAb (R1105-1; Huaan Biological Technology Co., Ltd., Hangzhou, China), mouse anti-β-actin (ab49846; Abcam), and rabbit anti-HDAC6 (our unpublished data).
Interactions between recombinant proteins expressed in Escherichia coli were determined using a Pierce GST protein interaction pulldown kit (21516; Thermo, Rockford, IL). For GST-pulldown experiments, GST-fusion proteins immobilized on glutathione-Sepharose beads were incubated with His-fusion proteins at 4°C for 8 h. The beads were washed extensively and boiled in SDS loading buffer, and the precipitated proteins were subjected to SDS-PAGE and detected by immunoblotting with mouse anti-GST MAb (M0807-1; Huaan Biological Technology) and mouse anti-His MAb (our unpublished data).
For coimmunoprecipitation, cell lysates were prepared using NP-40 lysis buffer (P0013F; Beyotime) in the presence of phenylmethylsulfonyl fluoride (PMSF) protease inhibitor (ST506; Beyotime). After centrifugation at 12,000 × g for 10 min, the supernatant was pretreated with protein A/G Plus-agarose (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at 4°C to eliminate nonspecific binding to the agarose gel. The supernatant was incubated with immunoprecipitation (IP) antibody at 4°C for 8 h, and immune complexes were precipitated by incubation with fresh agarose for another 8 h at 4°C. Beads were washed five times with PBS, and immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotting as previously described (28). In the coimmunoprecipitation assay, the IP antibodies used were mouse anti-Cap MAb, mouse anti-Flag M2 MAb (F1804; Sigma), and rabbit anti-Myc pAb (R1208-1; Huaan Biological Technology). Precipitated proteins on membranes were probed with the IP antibodies named above, along with mouse anti-α-tubulin MAb and mouse anti-IC1 MAb.
Knockdown by lentivirus-mediated RNA interference.
The construction of IC1-knockdown cells was performed as previously stated (51). Briefly, pLenti6.3/V5-DEST (Invitrogen)-based lentiviral particles for knockdown of IC1 (catalog no. 12MR0103A-LR-1; targeting sequence AAGCCATTCCGGTAACAGCCA) and nontarget small hairpin RNA (shRNA) control transduction particles (targeting sequence AAATGTACTGCGCGTGGAGAC) were purchased from Invitrogen. PK15 cells were plated in a 24-well plate with MEM overnight, and lentivirus transduction was carried out according to the manufacturer's instructions. Polybrene (H9268; Sigma) was added to the medium (final concentration, 8 μg/ml) to enhance transduction efficiency. Lentiviral particles (MOI = 0.5) were added, and the plates were gently mixed. After 12 h of incubation, infected cells were maintained in fresh MEM and supplemented with 5 to 10 mg/ml Blasticidin S HCl (BSD) (R210-01; Invitrogen) for a week of selection to obtain a stabilizing effect of shRNA. The shRNA-expressing cells were counted by flow cytometry (FC500 MPL; Beckman Coulter, Brea, CA). After transduction of the lentiviral vectors into the target cells, translation of IC1 was analyzed by immunoblotting. The viability of PK15 cells expressing IC1 shRNA was determined using Cell Counting Kit-8 (CCK-8) (C0037; Beyotime). To further analyze the function of IC1 in PCV2 infection, the IC1-knockdown PK15 cell line was infected with PCV2 at an MOI of 1 and cultured for 72 h. Cells were freeze-thawed three times and centrifuged at 8,000 × g for 10 min. Viral titers were determined in PK15 cells, and TCID50 values were calculated by the Reed-Muench method (52). PCV2 genomic DNA was quantified by absolute real-time PCR as previously described (53). To analyze the influence of IC1 knockdown on virus transport, PK15 cells expressing shRNA were infected with PCV2 (MOI = 25), and the subcellular location of viral particles was examined by confocal microscopy after infection for 9 h. To investigate whether the PCV2 replication cycle was affected, PK15 cells expressing shRNA were infected with PCV2 at an MOI of 1 for 8 h and then treated with or without cycloheximide (CHX) (S1560; Beyotime) at a dose of 100 μg/ml for another 8 h as described previously (51). Total cellular RNA was isolated from the resultant cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Finally, mRNA transcripts from the viral PCV2 Rep and Cap genes were quantified using a threshold cycle relative-quantification method (2−ΔΔCT) and specific primers for reverse transcription of Rep and Cap genes.
Subcellular proteome extraction.
Isolation of nuclear components was performed using a nuclear and cytoplasmic protein extraction kit (P0027; Beyotime) as previously stated (54). Briefly, PCV2-infected PK15 cells were treated with 200 μl of cytoplasmic protein extraction buffer A containing complete protease inhibitor cocktail for 5 to 10 min on ice and then added to 10 μl buffer B. After vigorous vortex mixing and centrifugation at 12,000 × g, the nucleus-debris pellet was resuspended in 50 μl nuclear protein extraction buffer for 30 min and centrifuged for 5 min. SDS-PAGE and Western blot analysis were performed on the supernatant. Expression of the histone 3 nucleus marker was assessed using rabbit anti-histone 3 pAb.
Statistical analysis.
All results are presented as means ± standard deviations. Significant differences between treated and control groups were analyzed using Student t tests. A P value of <0.05 was considered significant.
Ethics statement.
The animal study proposal was approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang University, permit number SYXK 2012-0178. All animal experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People's Republic of China.
RESULTS
Viral protein Cap-triggered α-tubulin acetylation enhances PCV2 replication.
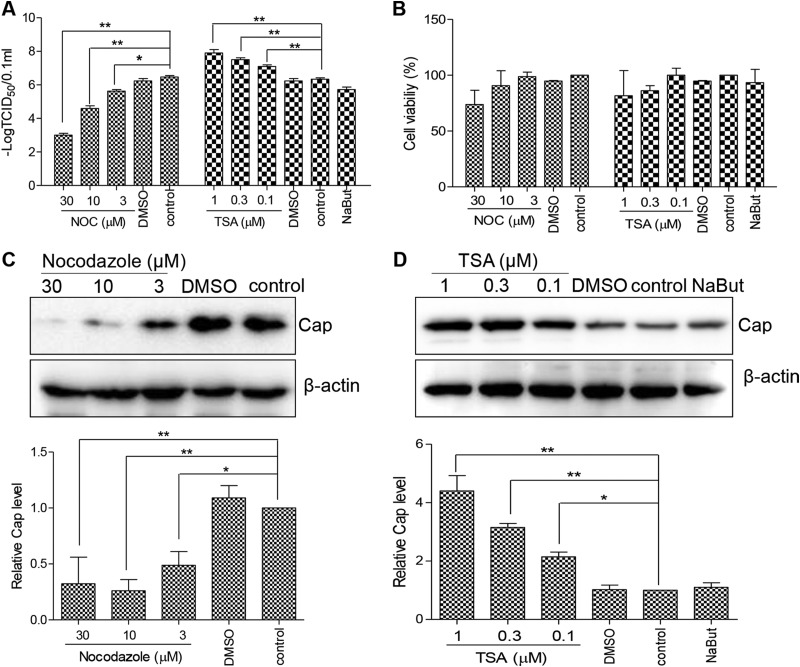
To determine whether microtubules participate in PCV2 replication, PK15 cells were pretreated for 3 h with nocodazole, a microtubule-destabilizing agent, and then infected with PCV2 at a multiplicity of infection (MOI) of 1 for 48 h postinfection (hpi). Compared to DMSO-treated controls and mock pretreated cells (Fig. 1A and C), virus titers in nocodazole pretreated cells decreased 10-fold to 1,000-fold in a dose-dependent manner (P < 0.05 or P < 0.01). Cap expression was also significantly decreased (P < 0.05). This suggested that microtubular stability is important for PCV2 production. To assess whether microtubular chemical modification was involved in PCV2 replication, PK15 cells were pretreated with TSA for 3 h or with NaBut for 6 h and then infected with PCV2. PK15 cells exposed to TSA exhibited enhanced PCV2 replication in a dose-dependent manner, as evidenced by increased viral titers (P < 0.01) (Fig. 1A) and increased Cap protein expression (P < 0.05; Fig. 1D). In contrast, PK15 cells exposed to NaBut did not exhibit increased PCV2 replication (P > 0.05) (Fig. 1A) or Cap protein expression (P > 0.05; Fig. 1D), indicating that acetylation of α-tubulin enhanced PCV2 replication. In order to exclude the possibility that the viability of drug-pretreated cells affected PCV2 replication, the cell viability of the cells treated with drug for 3 or 6 h was determined using the CCK-8 assay. This showed that the cell viability of drug-treated cells was decreased to some extent at a high drug dose, although there was no significant difference from mock-treated cell results (P > 0.05) (Fig. 1B). Furthermore, cells treated with a high dose of drug quickly recovered viability (data not shown), indicating that the drug treatment did not significantly affect cell proliferation. Collectively, these results suggest that acetylation of microtubular α-tubulin is required for PCV2 replication.
FIG 1.
Depolymerization and deacetylation of microtubules inhibits PCV2 infection. (A) PK15 cells were treated with different concentrations of nocodazole (NOC) or trichostatin A (TSA) or DMSO (control) for 3 h or with 3 mM sodium butyrate (NaBut) for 6 h and were then infected with PCV2 at an MOI of 1 for 48 h. Virus titers were determined and are represented by TCID50 values. (B) PK15 cells were treated for 3 or 6 h, and cell viability was analyzed by a CCK-8 assay. (C and D) Lysates of the cells described for panel A were probed with anti-Cap and anti-β-actin antibodies in immunoblotting experiments.
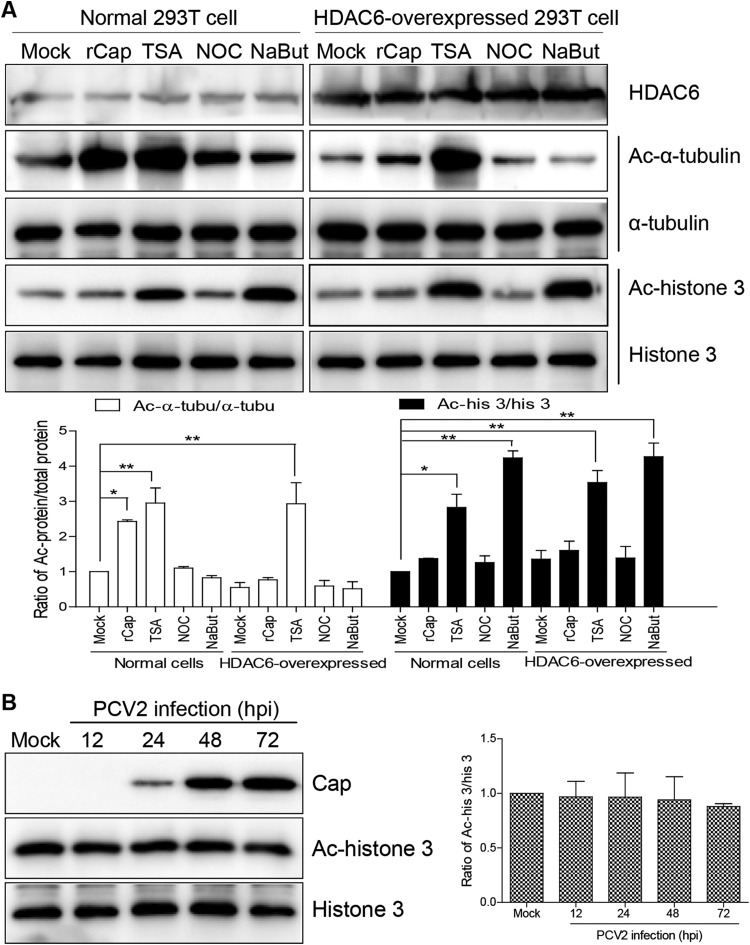
Unfortunately, during the course of the experiment, it was not possible to detect acetylated α-tubulin in PCV2-infected cells using a commercial anti-acetylated α-tubulin antibody. To establish which viral protein(s) participates in α-tubulin acetylation, acetylated α-tubulin was investigated using 293T cells as a model. Western blot analysis (Fig. 2A) revealed that acetylated α-tubulin was significantly upregulated in 293T cells incubated with recombinant rCap and TSA (P < 0.01) but not when the cells were treated with NaBut. However, rCap treatment increased levels of acetylated α-tubulin in 293T cells overexpressing HDAC6 only slightly, and TSA significantly upregulated acetylated α-tubulin in these cells, indicating that Cap may inhibit HDAC6 activity. Notably, acetylation of histone 3 was not significantly altered in PCV2-infected cells, rCap-treated cells, or HDAC6-overexpressing cells following Cap treatment (P > 0.05; Fig. 2). However, compared with mock-treated cell results, TSA and NaBut stimulation significantly upregulated acetylation of histone 3 (P < 0.01), including in HDAC6-overexpressing cells (P < 0.05; Fig. 2A). This demonstrated that HDAC6 is not relevant to histone 3 acetylation. In addition, there was no significant difference in the levels of acetylated histone 3 between NaBut-treated and TSA-treated cells (P > 0.05; Fig. 2A). These data suggest that α-tubulin acetylation was triggered by the PCV2 Cap protein and regulated by HDAC6 but not by histone 3 acetylation.
FIG 2.
Acetylation modifications of α-tubulin and histone 3. (A) 293T cells and HDAC6-overexpressing 293T cells were treated with purified rCap for 6 h, TSA for 3 h, NOC for 3 h, or NaBut for 6 h, and immunoblots of cell lysates was probed with mouse anti-acetylated α-tubulin MAb and rabbit anti-acetylated histone 3 antibody to analyze the level of acetylated α-tubulin and histone 3. Data are represented as means ± standard deviations (SD) (n = 3; one asterisk [*] represents P < 0.05, and two asterisks [**] represent P < 0.01) Ac, acetylated. (B) PK15 cells were infected with PCV2 for 12, 24, 48, and 72 h. Cell lysates were probed with anti-Cap, anti-acetylated histone 3, and anti-histone 3 antibodies in immunoblotting experiments.
Microtubules mediate nuclear trafficking of PCV2.
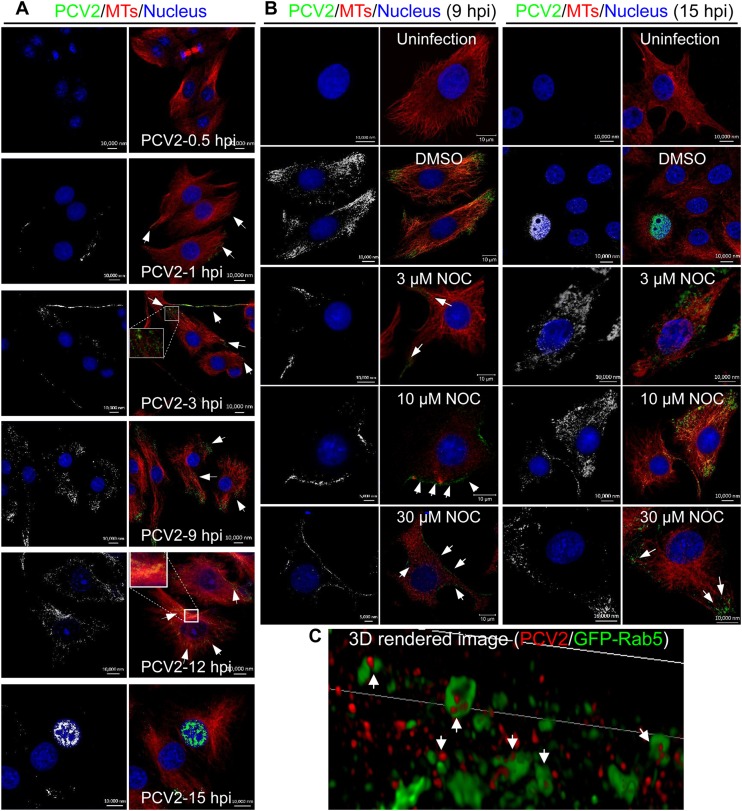
Our previous study demonstrated the interplay between the PCV2 Cap protein and α-tubulin (28). It is known that microtubules function as superhighways to mediate the transport of various cargoes. To determine whether PCV2 virions travel toward the nucleus in a microtubule-dependent manner, intracellularly invading viral particles were examined in the early stages of PCV2 infection. Dynamic analysis (Fig. 3A) showed that no visible viral particles had adhered to the cell surface at 0.5 hpi and that viral particles began to enter at 1 hpi. At 3 hpi, a number of viral particles had entered the cytoplasm and were near the cell membrane, where they began minus-end trafficking. At 9 hpi, a number of viral particles had been transported closer to the perinuclear region, and at 12 hpi, numerous viral particles were localized to the microtubule organizing centers (MTOC) near the nucleus, although no PCV2 virions were present in the nucleus itself. At 15 hpi, PCV2 Cap protein was present in the nucleus and most PCV2 virions were no longer in the cytoplasm, suggesting that the PCV2 genome was being delivered to the nucleus and was beginning to replicate. Additionally, many cytoplasmic PCV2 virions were adjacent, distributed along and within microtubules (Fig. 3A). In contrast, when cells were pretreated with different concentrations of nocodazole, the viral particles remained near the PCV2-infected cell membrane at 9 hpi and were diffusely distributed in the cytoplasm or remained near the cell membrane at 15 hpi (Fig. 3B, white arrows). This suggested that the intracellular transportation of PCV2 virions was inefficient in cells with a compromised microtubular network. To further investigate whether early endosomes were involved in microtubule-dependent nuclear trafficking of PCV2, we analyzed their colocalization at 6 hpi using gSTED. The gSTED image (Fig. 3C) showed that PCV2 particles localized predominantly in Rab5-positive endosomes in PK15 cells, indicating that PCV2 virions were taken up by the endosomes. Similar gSTED images were observed in PCV2-infected Na3VO4-pretreated cells (data not shown), indicating that Na3VO4 did not interfere with endosomal uptake of PCV2 virions. Collectively, these results suggest that nuclear trafficking of PCV2 is dependent on the motility of endosomes and that intact microtubules appear to play a railway track-like role in PCV2 transport.
FIG 3.
Superresolution microscopy of PCV2 in early endosomes. (A) PK15 cells were infected with PCV2 at an MOI of 25 and cultured for 0.5, 1, 3, 9, 12, and 15 h. Confocal microscopy was performed to examine viral particles (green) with mouse anti-Cap IgG, microtubules (MTs; red) with rabbit anti-α-tubulin antibodies, and the nucleus with DAPI (blue). The white arrows show magnification of the virus enrichment area in the infected cells. (B) PK15 cells were treated with different concentrations of NOC or DMSO for 3 h and then infected with PCV2 at an MOI of 25 for 9 h or 15 h. Mock-infected PK15 cells served as controls. Confocal microscopy was performed to examine the subcellular localization of the viral particles using rabbit anti-α-tubulin and mouse anti-Cap IgG overnight at 4°C followed by staining with anti-rabbit Alexa 546 and anti-mouse FITC secondary antibodies. White arrows represent the subcellular localization of viral particles (green). (C) PK15 cells were transfected with GFP-Rab5 plasmid for 24 h and infected with PCV2 at an MOI of 25. Virus was added to the cells in an initial cold binding step to synchronize the infection process. At 6 hpi, cells were incubated with mouse anti-Cap IgG overnight at 4°C, followed by staining with anti-mouse Alexa Fluor 546. Rendered three-dimensional (3D) images of PCV2 particles (red) within endosomes (green) are shown.
Cytoplasmic dynein is necessary for nuclear trafficking of PCV2.
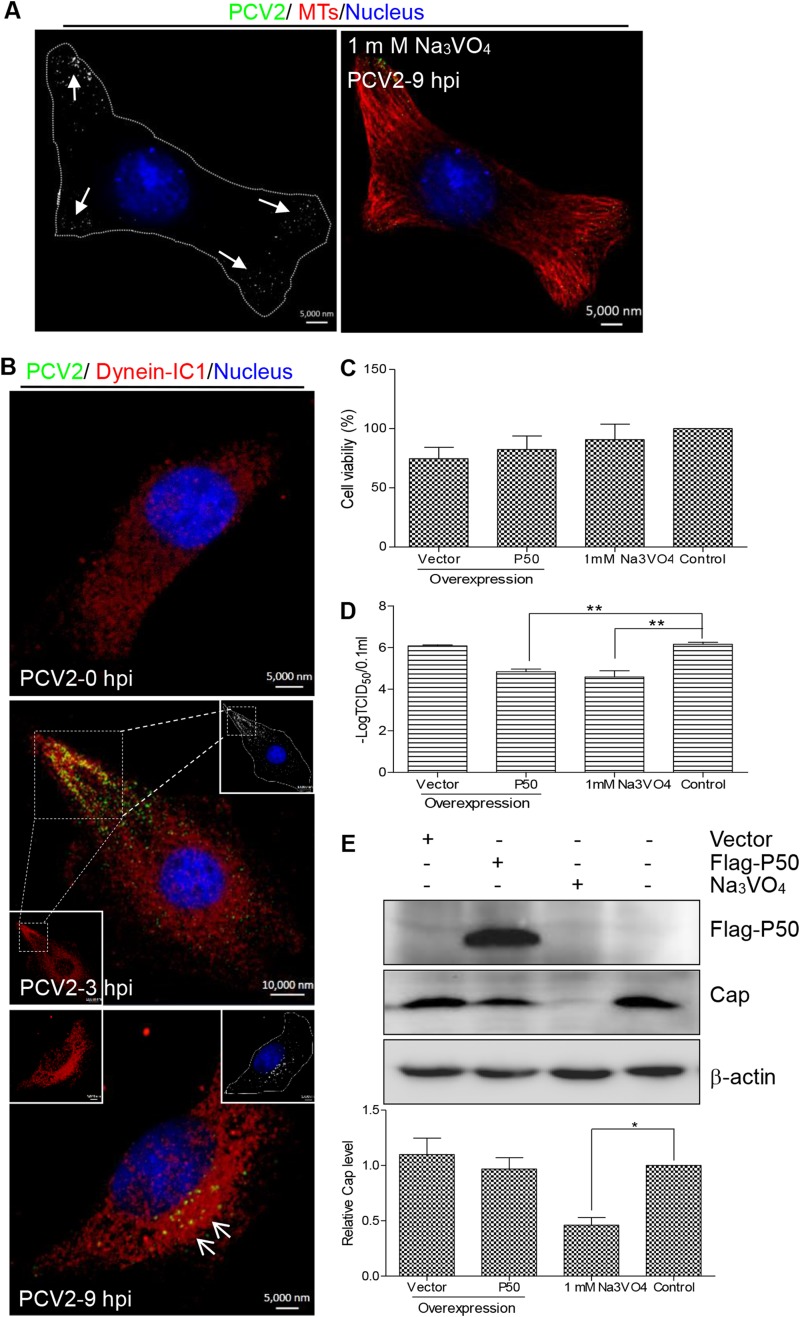
Cytoplasmic dynein is required for the movement of endosomes from the cell periphery to the perinuclear cytoplasm (55, 56). To further investigate whether cytoplasmic dynein is involved in endosomal minus-end trafficking of PCV2, we pretreated PK15 cells with Na3VO4, a well-characterized inhibitor of dynein activity, and examined the subcellular localization of invading viral particles. As shown in Fig. 4A, the viral particles remained in the peripheral region, adjacent to the cell membrane, indicating that internalization of viral particles was not affected by Na3VO4, although the efficiency of nuclear trafficking was compromised as a consequence of dynein inhibition. Likewise, we analyzed dynein subcellular localization in PCV2-infected PK15 cells. As shown in Fig. 4B, intermediate chain 1 (IC1) of the dynein complex was distributed in the cytoplasm of mock-infected PK15 cells but was present as granular-like accumulations in PCV2-infected cells which overlapped with the distribution of the PCV2 Cap protein. Moreover, we analyzed the effect of dynein activity on PCV2 production. PK15 cells were treated with Na3VO4 or transfected with a construct for overexpressing dynamitin (P50; ∼ 23.5% transfection rate), which causes P150Glued to dissociate from the dynein complex, thereby interfering with dynein function (57). These two cell types were then infected with PCV2. To investigate the possible toxicity of P50 overexpression and drug treatments, the viability of drug-treated and plasmid-transfected cells was determined using the CCK-8 assay. This showed that there were no significant differences from mock-transfected cell results (P > 0.05) (Fig. 4C), indicating that drug treatment and P50 overexpression did not significantly affect cell proliferation. Measurement of viral titers showed that the production of PCV2 infectious virions decreased significantly in both Na3VO4-treated and P50-overexpressing cells (P < 0.01) (Fig. 4D). Additionally, Western blot analysis confirmed that Na3VO4 treatment significantly decreased Cap expression (P < 0.05) (Fig. 4E). These results demonstrated that dynein clearly performs a positive role in microtubule-dependent transportation of PCV2 particles and productive infection.
FIG 4.
Incoming PCV2 capsids recruited cytoplasmic dynein. (A) PK15 cells were pretreated with 1 mM Na3VO4 for 3 h and then infected with PCV2 at an MOI of 25 for 9 h. Subcellular localization of viral particles (green) was examined by confocal microscopy with mouse anti-Cap IgG, microtubules (red) with rabbit anti-α-tubulin antibodies, and the nucleus with DAPI (blue). (B) PK15 cells were infected with PCV2 at an MOI of 25 for 3 h or 9 h and then stained for confocal microscopy using rabbit anti-IC1 antibody (red) and mouse anti-Cap IgG (green). Mock-infected cells served as controls. The white box shows magnification and colocalization of the virus enrichment area in infected cells after 3 h. (C) PK15 cells were transfected with vector pCMV-Nflag-P50 for 24 h or pretreated with Na3VO4 for 3 h, and cell viability was analyzed by a CCK-8 assay. (D and E) PK15 cells were transfected with vector pCMV-Nflag-P50 or pretreated with Na3VO4. At 24 h posttransfection or at 3 h posttreatment with Na3VO4, cells were infected with PCV2 at an MOI of 1 and cultured for 48 h. Viral titers were detected by TCID50 (D), and Cap expression in the cell lysates was analyzed by immunoblotting using mouse anti-Cap IgG (E). Data represented are means ± SD (n = 3; two asterisks [**] represent P < 0.01).
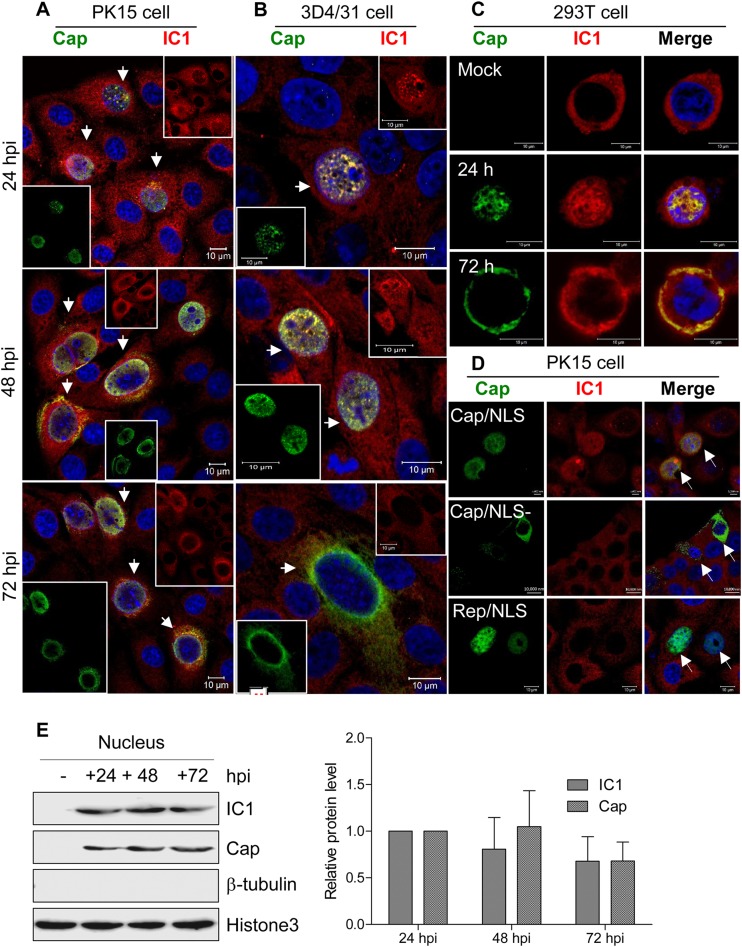
Dynein IC1 enters the nucleus via the nuclear localization signal of PCV2 Cap.
Cytoplasmic dynein can mediate cargo transport only through the cytoplasm, and its subunits are all localized in the cytoplasm (58, 59). We believe that the intracellular dynein distribution is altered in PCV2-infected cells. Previous studies have shown that during a productive PCV2 infection, viral transcripts are detected at 18 hpi and cell-free progeny viruses begin to appear at about 30 hpi (60). Therefore, we determined the subcellular localization of the dynein IC1 subunit in PCV2-infected cells. Surprisingly, confocal analysis revealed that the IC1 protein aggregates within the nucleus of PCV2-infected PK15 and 3D4/31cells at 24 hpi and at 48 hpi. At 72 hpi, IC1 protein aggregation was decreased in the nucleus; however, IC1 was expressed only in the cytoplasm of mock-infected cells (Fig. 5A and B). Likewise, viral Cap was colocalized with endogenous IC1 within the nucleus of infected cells at 24 hpi and was rarely observed in the cytoplasm of infected cells. Cap was colocalized with the IC1 mainly in the cytoplasm and nuclei of infected cells at 48 hpi, and Cap levels were markedly decreased in the nuclei of infected cells at 72 hpi (Fig. 5A and B). The subcellular distribution of the cytoplasmic IC1 subunit was therefore identical to that of the viral Cap protein throughout the PCV2 infection cycle. Furthermore, dynein IC1 was used by newly invading PCV2 particles for transport to the nuclei and by new progeny virions for transport through the cytoplasm from the nucleus at 24 hpi. This may explain why IC1 and Cap were not colocalized in the cytoplasm of infected cells at 24 hpi. Western blot analysis also revealed that IC1 was present in the fractionated nuclei of PCV2-infected cells but not in mock-infected cells (Fig. 5E), indicating that IC1 is translocated during PCV2 infection. In order to exclude the possibility of other viral proteins participating in the nuclear translocation of intracellular IC1, we transfected 293T cells with a plasmid expressing Cap. Consistent with the observation above, endogenous IC1 was colocalized with exogenously expressed Cap proteins and translocated into the nucleus (Fig. 5C). These data suggest that the Cap protein independently induces the recruitment and nuclear translocation of IC1 during PCV2 infection.
FIG 5.
Dynamic colocalization of Cap with dynein IC1 subunit. (A to C) Cells immunostained with pig anti-Cap IgG and mouse anti-IC1 antibody were stained with anti-pig FITC and anti-mouse Alexa 546, respectively. Colocalization of Cap with IC1 was determined at the indicated time points, including in PCV2-infected PK15 cells (A), PCV2-infected 3D4/31 cells (B), and 293T cells expressing exogenous Cap (C). (D) PK15 cells were transfected with the indicated plasmid expressing Cap (Cap/NLS [with NLS]), dCap (Cap/NLS− [without NLS]), or Rep (Rep/NLS [with NLS]). At 24 h posttransfection, cells were examined for the subcellular location of exogenous Cap, dCap, and Rep and of endogenous IC1 with pig anti-Cap IgG, pig anti-Rep pAb, and mouse anti-IC1 antibody, followed by staining with anti-pig FITC and anti-mouse Alexa 546. (E) The samples described for panel A were subjected to separation of subcellular components. The indicated proteins in the nuclear fraction were then tested by immunoblotting to validate intranuclear colocalization of Cap with IC1 in PCV2-infected PK15 cells.
Previous studies have demonstrated that the nuclear localization signal (NLS) of Cap is responsible for the nuclear location of the Cap protein (61). To investigate whether intranuclear translocation of endogenous IC1 of dynein is associated with the NLS in PCV2-infected cells, PK15 cells were transfected with a plasmid expressing wild-type Cap (Cap/NLS) or NLS-deleted Cap (Cap/NLS-). Confocal microscopy showed that NLS-deleted Cap disrupted IC1 entry into the nucleus. In addition, Rep did not induce IC1 localization in the nucleus (Fig. 5D), even though the Rep protein includes three identical NLS signals at the N terminus (62). These findings confirmed that intranuclear localization of IC1 is mediated by the NLS of PCV2 Cap.
Involvement of the binding domain in the direct interaction between PCV2 Cap and dynein IC1.
To investigate the colocalization mechanism of IC1 and PCV2 Cap, coimmunoprecipitation was performed on lysates from PCV2-infected cells precipitated with antibody against Cap. IC1 and α-tubulin were detected in the precipitates of PCV2-infected cells but not in uninfected cells (Fig. 6A), indicating that Cap interacts simultaneously with both IC1 and α-tubulin in infected cells. However, in pulldown assays of GST-Cap, only His-IC1 was detected and not α-tubulin type 1A (TUBA1A), α-tubulin type 1B (TUBA1B), or β-tubulin type 2A (TUBB2A) (Fig. 6B). Similarly, the interaction between Cap and IC1 was also validated in precipitates from 293T cells cotransfected with plasmids expressing myc-dcap and flag-IC1 (Fig. 6C), further confirming that Cap directly interacts with dynein IC1 but not tubulin.
To identify the IC1 interaction domain on Cap, we constructed four truncated versions of Cap tagged with GST and analyzed the interaction between the various truncated forms of the Cap protein and IC1 in a GST-pulldown assay. These results showed that deletions of Cap domain 1 (D1), D3, D4, or D5 did not destroy the interaction between IC1 and the Cap protein, whereas deletion of the D2 domain (N-terminal residues 42 to 100) significantly inhibited binding to the dynein IC1 subunit (Fig. 6D). Likewise, coimmunoprecipitation assays revealed that deletion of the N-terminal residues 1 to 41 (CapΔ1-41) enabled the interaction between Cap and IC1, while deletion of N-terminal residues 42 to 100 (CapΔ42-100) inhibited binding with IC1 (Fig. 6E), confirming that Cap residues 42 to 100 are involved in IC1 binding.
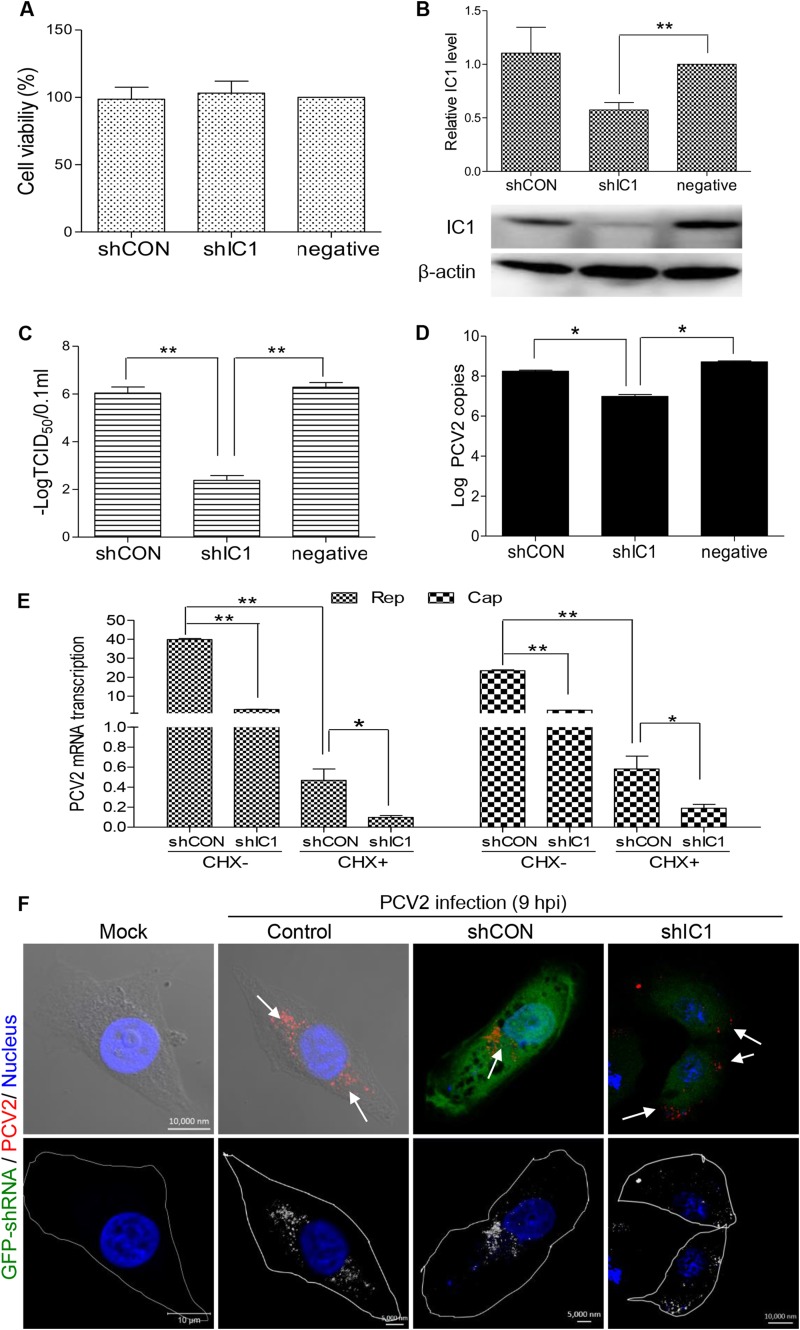
Knockdown of IC1 disrupts intracellular PCV2 trafficking.
The direct interaction between Cap and IC1 prompted us to study the critical role of IC1 transport in the life cycle of PCV2 infection. Therefore, we created a small hairpin RNA (shRNA)-mediated knockdown of IC1. IC1-specific shRNA (shIC1) or the nontargeting control shRNA (shCON) was transfected into PK15 cells by lentivirus-mediated shRNA transfer to construct cell lines stably expressing GFP-shRNAs. When the transfected cells were selected by BSD, flow cytometry analysis showed that the proportions of cells expressing GFP-shRNA were 92.3% for GFP-shCON and 93.7% for GFP-shIC1 (data not shown). To assess possible toxic effects of expression of GFP-shRNA in PK15 cells, cell viability was determined using the CKK-8 assay, which showed that there were no significant differences from mock-transfected cell results (P > 0.05) (Fig. 7A). Western blot analysis confirmed that IC1 expression was significantly decreased (P < 0.01) in PK15 cells stably expressing shIC1 (Fig. 7B). Subsequently, IC1-silenced PK15 cells were inoculated with PCV2 at an MOI of 1 and PCV2 replication was assessed by determining the virus titer (TCID50) and by real-time PCR. The data showed that the virus titer and viral genomic DNA levels were significantly decreased (P < 0.05) in IC1-silenced PK15 cells (Fig. 7C and D) compared with parental PK15 cells and shCON-transfected cells. This indicated that IC1 silencing significantly decreased nuclear trafficking of newly invading PCV2 virions and production of progeny PCV2 particles. Further transcriptional analysis of the viral Cap and Rep genes demonstrated that the mRNA of the PCV2 Cap and Rep genes was significantly downregulated in shIC1-silenced PK15 cells treated with or without cycloheximide (CHX) (P < 0.01) (Fig. 7E), indicating that IC1 affected PCV2 replication at the transcriptional level. Additionally, confocal analysis revealed that PCV2 virions failed to migrate to the perinuclear region and stopped in the cytoplasm, adjacent to the cell membrane of shIC1-silenced PK15 cells, at 9 hpi (Fig. 7F). In contrast, virus particles moved to the perinuclear region in shCON-transfected and normal PK15 cells at 9 hpi. This indicated that IC1 knockdown stopped the intracellular movement of PCV2 virions. Collectively, our results show that microtubule motor dynein-dependent transport of IC1 to the nucleus is critical for PCV2 replication. Furthermore, dynein may be recruited directly by the PCV2 Cap protein following release of the viral capsid from early endosomes (Fig. 8).
FIG 7.
Effective infection of PCV2 is IC1 dependent. (A) The viability of PK15 cells stably expressing shIC1 was analyzed with a CCK-8 assay. (B) PK15 cells were transduced with lentivirus containing shRNA. The cell lysates of control shRNA (shCON) and IC1 shRNA-transduced PK15 cells (shIC1) were analyzed by immunoblotting to examine protein levels of IC1 and β-actin upon lentivirus transduction. (C) PK15 cells (negative) and shIC1- or shCON-transduced PK15 cells were each infected with PCV2 at an MOI of 1 for 72 h. Viral titers were then determined to analyze the effects of IC1 knockdown on PCV2 infectivity. (D) Experiments were performed as described for panel B, and virus DNA levels were quantified by real-time PCR to analyze the effect of IC1 knockdown on virus gene replication. (E) PK15 cells transduced (or not) with lentivirus were infected with PCV2 at an MOI of 1 for 8 h and treated with CHX for another 8 h. Cap and Rep mRNA transcripts of the PCV2 genome were determined by real-time comparative quantitative PCR and analyzed using 2−ΔΔCT means. (F) PK15 cells transduced (or not) with lentivirus were infected with PCV2 at an MOI of 25 for 9 h. Cells were incubated with mouse anti-Cap IgG overnight at 4°C and then stained with anti-mouse Alexa Fluor 546. Green fluorescence represents cells transduced with lentivirus and expressing GFP-shRNA. Subcellular localization of viral particles was examined by immunofluorescence confocal microscopy to analyze the effect of IC1 knockdown on viral transport. To make viral particles easier to view, they were inverted into gray-scale images. Values represent means ± SD of the results of three independent experiments performed in triplicate. One asterisk (*) represents P < 0.05; two asterisks (**) represent P < 0.01.
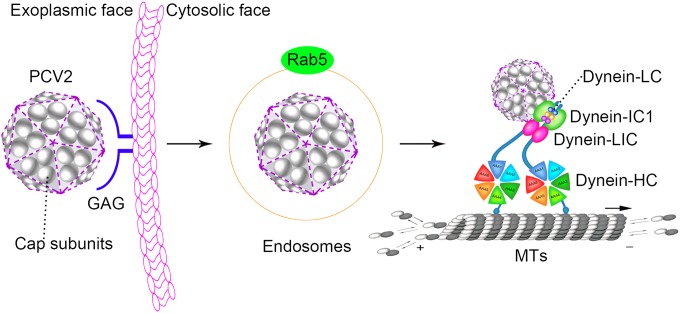
FIG 8.
Schematic model depicting the entry and nuclear targeting of PCV2. PCV2 particles were localized with endosomes during the early stages of infection. Following release of the capsid from the early endosome, the capsid protein (gray) directly binds to the IC1 subunit of the dynein complex in order to travel along microtubules.
DISCUSSION
Microtubule motor dynein-dependent nuclear trafficking of PCV2.
Upon entering cells, many viruses engage with microtubule motors to overcome the diffusion barrier and traverse the cytoplasm (63). Some viruses employ the host microtubule/dynein machinery to facilitate their intracellular transport (42). Research on cell receptors demonstrated that PCV2 particles enter target cells by binding to heparan sulfate on the cell surface (24–26) or via clathrin-mediated endocytosis in monocytic cells (27). However, there has been very little insight into how the circovirus arrives at its replication destination. Microtubules are highly dynamic and unstable polymers that undergo rapid cycles of polymerization and depolymerization (64). In the present study, PCV2 trafficking to the nucleus was inhibited by microtubule depolymerization, with nocodazole binding to β-tubulin in a dose-dependent fashion (Fig. 3). In addition, PCV2 progeny replication was significantly decreased (Fig. 1), suggesting that microtubule depolymerization disturbs PCV2 replication by inhibiting virus transport to the nucleus.
Dynein is the minus-end-directed microtubule motor that is responsible for the progressive retrograde transport of cytoplasmic cargoes in mammalian cells (65). The subunits, including ICs, LICs, and LCs, form the cytoplasmic dynein tail and participate in linking dynein to physiological forms of the cargo (66, 67). The mechanism of dynein recruitment is not well understood for any virus. Whether the intrinsic activities of virus proteins are specifically responsible for dynein-based transport during infection has proven difficult to determine in vivo. In addition, increasing evidence demonstrates an alternative mechanism of dynein recruitment, which has been documented in several viruses: the direct interaction of the adenovirus hexon subunit with dynein chains IC and LIC (48), the interaction of the papillomavirus L2 protein with dynein chain LC (68), the interaction of the enveloped pseudorabies virus VP1/2 protein with dynein/dynactin microtubule motor complex (69), and the interaction of African swine fever virus protein p54 with dynein light-chain 8 in the early stages of the viral life cycle (47). In this study, we found that dynein-dependent endosomes are involved in the nuclear trafficking of PCV2 particles (Fig. 3C). We hypothesize that PCV2 Cap protein subunits bind directly to dynein IC1 during the early stages of infection, following release of the capsid from the early endosomes. Dynein IC and LIC subunits have been shown to bind directly to the capsid protein hexon following release of capsids from the early endosome (48). Many viral infections have been found to be associated with Rab GTPases (70). Simian virus 40 enters endosomes to reach the endoplasmic reticulum, and viral infection is sensitive to perturbations that inhibit endosome acidification and maturation (71). Adenoassociated viruses are known to exploit microtubules and dynein for rapid cytoplasmic trafficking from endosomal compartments toward the perinuclear region for replication (72). Despite significant progress in this work, PCV2-containing vesicular transport from early to late endosomes and the process of virus uncoating require further study.
Our previous report identified the PCV2 Cap protein bound to α-tubulin in a coimmunoprecipitation assay (28, 73). In this study, we demonstrated that Cap also colocalized and interacted with the microtubule-associated dynein IC1 chain. However, in a pulldown assay, the viral protein Cap was shown to bind only to IC1 but not to α-tubulin (Fig. 6), indicating a direct interaction between Cap and IC1. Deletion of the N-terminal region (aa 42 to 100) of Cap showed that this region functions as the binding domain (Fig. 6). Abrogation of the Cap/IC1 interaction and knockdown of IC1 inhibited PCV2 transport to the nucleus (Fig. 7). Together, these findings show that the cytoplasmic dynein IC1 chain is the likely intracellular receptor for Cap and that the Cap/IC1 interaction plays a critical role in PCV2-dependent microtubule transport.
The abnormal dynamic location of IC1 and its potential role.
Cytoplasmic dynein was reported to mediate cargo transport only through the cytoplasm and to localize to the nuclear envelope or nuclear pores to force nuclear migration and mitotic entry (58, 59). Dynein IC1 can also interact with the Gβγ subunit, which plays a central role in G-protein signaling and is involved in cell division, vesicle trafficking, and signal integration (74). The Trk-ERK1/2-stimulated phosphorylation of dynein IC is required for regulation of dynein activity and affects certain cellular events, such as microtubule-dependent sliding and mitosis (75–77). The middle part of IC1 consists of repetitive sequences homologous to the sequence of the nucleoside diphosphate kinase (NDPK), indicating that IC1 may function as an NDPK that regulates RNA, DNA, and polysaccharide synthesis (78–80). The results show that dynein is a multifunctional protein. On the basis of these potential roles of IC1, we propose that dynein IC1 may regulate nuclear metabolism via an unidentified signal transduction pathway to promote viral genome replication and transcription, but this hypothesis requires further study.
As observed previously, in the present study, the microtubule motor dynein IC1 was present only in the cytoplasm of noninfected cells (Fig. 5). Surprisingly, IC1 was detected in the nuclei of infected cells and interacted with the viral Cap protein (Fig. 5). Whether the nuclear distribution of dynein IC1 is related to NDPK activity needs further investigation. How dynein IC1 enters the nucleus is still unknown. Moreover, we observed that the intranuclear Cap/IC1 complex aggregated in the cytoplasm during the latter stages of infection, suggesting that Cap can be exported from the nucleus, which implies that it has a nuclear export signal. Identification and characterization of such a signal will allow us to further understand the function of Cap and the dynamic location of the Cap/IC1 complex.
Acetylation of α-tubulin benefits PCV2 infection.
Acetylation of α-tubulin is involved in stabilizing microtubules and is regulated by HDAC6 (81–83). Acetylated stable microtubules promote cell fusion, cell differentiation, and actin-dependent ruffle formation (84, 85). During maturation of the osteoclast, Rho-GTPase influences the levels of acetylated microtubules and actin organization by HDAC6 and mDia2 (86). Moreover, acetylation of α-tubulin is involved in virus infection. For instance, binding of the gp120 protein of human immunodeficiency virus type 1 to CD4+-permissive cells increases the levels of acetylated α-tubulin, and HDAC6 plays a significant role in regulating human immunodeficiency virus type 1 infection and envelope-mediated syncytium formation (87). Infection with influenza A virus also induces acetylation of α-tubulin in epithelial cells, and enhanced microtubule acetylation increases the release of virions from infected cells (88). Kaposi's sarcoma-associated herpesvirus induces acetylation and aggregation of microtubules via RhoA (89). However, the mechanism of α-tubulin acetylation during viral infection is unclear. In the present study, the viral Cap protein significantly induced acetylation of α-tubulin (Fig. 2), suggesting that PCV2 has the ability to activate microtubule acetylation and enhance intracellular transport. Notably, the Cap protein was also able to induce acetylation of α-tubulin weakly in cells overexpressing HDAC6 (Fig. 2). This indicates that Cap induces α-tubulin acetylation by inhibiting HDAC6 activity, but it is now important to determine which pathway is utilized by the viral Cap protein to achieve this. HDACs are reportedly involved in herpesvirus replication and virus-stimulated host defenses (90, 91). However, our results showed that treatment with TSA or NaBut (an inhibitor of HDACs other than HDAC6) could induce the acetylation of histone 3 in both normal cells and cells overexpressing HDAC6 but not in PCV2-infected or Cap-containing cells. This suggests that histone 3 acetylation is not regulated by PCV2 infection, and HDAC6 activity does not appear to be related to histone 3 acetylation.
These results highlight the importance of the microtubule dynein motor; the nuclear targeting of PCV2 was found to be entirely dependent on dynein. The IC1 subunit of dynein-mediated PCV2 binding to the microtubule/dynein machinery and the viral Cap protein induced α-tubulin acetylation. In addition, intracellular translocation of IC1 during PCV2 replication suggests that IC1 may be involved in replication of PCV2. These findings deepen our understanding of the molecular mechanisms of PCV2 infection. Further investigations are required to examine in more detail the role of the microtubule/dynein machinery in nuclear trafficking of PCV2. Identifying a peptide that competes with IC1 for the binding domain may be a promising strategy for blocking the intracellular transport of circovirus.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grant number 31230072) and the National High-Tech Program of China (grant number 2011AA10A208).
We thank Yun-Qin Li for technical assistance in laser confocal microscopy.
REFERENCES
- 1.Biagini P, Bendinelli M, Hino S, Kakkola L, Mankertz A, Niel C, Okamoto H, Raidal S, Teo CG, Todd D. 2012. Circoviridae, p 343–349 InKing AMQ, Lefkowitz E, Adams MJ, Carstens EB (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom. [Google Scholar]
- 2.Tischer I, Gelderblom H, Vettermann W, Koch MA. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 3.Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM. 1998. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the U S A and Europe. J Vet Diagn Invest 10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 4.Allan GM, Ellis JA. 2000. Porcine circoviruses: a review. J Vet Diagn Invest 12:3–14. doi: 10.1177/104063870001200102. [DOI] [PubMed] [Google Scholar]
- 5.Segalés J, Allan GM, Domingo M. 2005. Porcine circovirus diseases. Anim Health Res Rev 6:119–142. doi: 10.1079/AHR2005106. [DOI] [PubMed] [Google Scholar]
- 6.Opriessnig T, Meng XJ, Halbur PG. 2007. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest 19:591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- 7.Baekbo P, Kristensen CS, Larsen LE. 17January2012, posting date Porcine circovirus diseases: a review of PMWS. Transbound Emerg Dis doi: 10.1111/j.1865-1682.2011.01288.x. [DOI] [PubMed] [Google Scholar]
- 8.Tischer I, Bode L, Apodaca J, Timm H, Peters D, Rasch R, Pociuli S, Gerike E. 1995. Presence of antibodies reacting with porcine circovirus in sera of humans, mice, and cattle. Arch Virol 140:1427–1439. doi: 10.1007/BF01322669. [DOI] [PubMed] [Google Scholar]
- 9.Gilliland SM, Forrest L, Carre H, Jenkins A, Berry N, Martin J, Minor P, Schepelmann S. 2012. Investigation of porcine circovirus contamination in human vaccines. Biologicals 40:270–277. doi: 10.1016/j.biologicals.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Dubin G, Toussaint JF, Cassart JP, Howe B, Boyce D, Friedland L, Abu-Elyazeed R, Poncelet S, Han HH, Debrus S. 2013. Investigation of a regulatory agency enquiry into potential porcine circovirus type 1 contamination of the human rotavirus vaccine, Rotarix: approach and outcome. Hum Vaccin Immunother 9:2398–2408. doi: 10.4161/hv.25973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beach NM, Cordoba L, Kenney SP, Meng XJ. 2011. Productive infection of human hepatocellular carcinoma cells by porcine circovirus type 1. Vaccine 29:7303–7306. doi: 10.1016/j.vaccine.2011.06.097. [DOI] [PubMed] [Google Scholar]
- 12.Hamel AL, Lin LL, Nayar GP. 1998. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol 72:5262–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou JY, Chen QX, Ye JX, Shen HG, Chen TF, Shang SB. 2006. Serological investigation and genomic characterization of PCV2 isolates from different geographic regions of Zhejiang province in China. Vet Res Commun 30:205–220. doi: 10.1007/s11259-006-3203-x. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Chen I, Kwang J. 2005. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol 79:8262–8274. doi: 10.1128/JVI.79.13.8262-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Cao J, Zhou N, Jin Y, Wu J, Zhou J. 2013. Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J Virol 87:1420–1429. doi: 10.1128/JVI.01443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mankertz A, Mankertz J, Wolf K, Buhk HJ. 1998. Identification of a protein essential for replication of porcine circovirus. J Gen Virol 79(Pt 2):381–384. [DOI] [PubMed] [Google Scholar]
- 17.Mankertz A, Hillenbrand B. 2001. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 279:429–438. doi: 10.1006/viro.2000.0730. [DOI] [PubMed] [Google Scholar]
- 18.Cheung AK. 2006. Rolling-circle replication of an animal circovirus genome in a theta-replicating bacterial plasmid in Escherichia coli. J Virol 80:8686–8694. doi: 10.1128/JVI.00655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J Gen Virol 81:2281–2287. [DOI] [PubMed] [Google Scholar]
- 20.Trible BR, Suddith AW, Kerrigan MA, Cino-Ozuna AG, Hesse RA, Rowland RR. 2012. Recognition of the different structural forms of the capsid protein determines the outcome following infection with porcine circovirus type 2. J Virol 86:13508–13514. doi: 10.1128/JVI.01763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahé D, Blanchard P, Truong C, Arnauld C, Le Cann P, Cariolet R, Madec F, Albina E, Jestin A. 2000. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J Gen Virol 81(Pt 7):1815–1824. [DOI] [PubMed] [Google Scholar]
- 22.Lekcharoensuk P, Morozov I, Paul PS, Thangthumniyom N, Wajjawalku W, Meng XJ. 2004. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J Virol 78:8135–8145. doi: 10.1128/JVI.78.15.8135-8145.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang SB, Jin YL, Jiang XT, Zhou JY, Zhang X, Xing G, He JL, Yan Y. 2009. Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol Immunol 46:327–334. doi: 10.1016/j.molimm.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Misinzo G, Meerts P, Bublot M, Mast J, Weingartl HM, Nauwynck HJ. 2005. Binding and entry characteristics of porcine circovirus 2 in cells of the porcine monocytic line 3D4/31. J Gen Virol 86:2057–2068. doi: 10.1099/vir.0.80652-0. [DOI] [PubMed] [Google Scholar]
- 25.Misinzo G, Delputte PL, Meerts P, Lefebvre DJ, Nauwynck HJ. 2006. Porcine circovirus 2 uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells. J Virol 80:3487–3494. doi: 10.1128/JVI.80.7.3487-3494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A, Johnson JE. 2011. The 2.3-angstrom structure of porcine circovirus 2. J Virol 85:7856–7862. doi: 10.1128/JVI.00737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misinzo G, Delputte PL, Lefebvre DJ, Nauwynck HJ. 2009. Porcine circovirus 2 infection of epithelial cells is clathrin-, caveolae- and dynamin-independent, actin and Rho-GTPase-mediated, and enhanced by cholesterol depletion. Virus Res 139:1–9. doi: 10.1016/j.virusres.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Zhou J, Wu Y, Zheng X, Ma G, Wang Z, Jin Y, He J, Yan Y. 2009. Differential proteome analysis of host cells infected with porcine circovirus type 2. J Proteome Res 8:5111–5119. doi: 10.1021/pr900488q. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Bai J, Zhang L, Jiang Z, Wang X, Li Y, Jiang P. 2013. Hsp70 positively regulates porcine circovirus type 2 replication in vitro. Virology 447:52–62. doi: 10.1016/j.virol.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 30.Radtke K, Dohner K, Sodeik B. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol 8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 31.Sieczkarski SB, Whittaker GR. 2002. Dissecting virus entry via endocytosis. J Gen Virol 83:1535–1545. [DOI] [PubMed] [Google Scholar]
- 32.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. 2012. GTPase networks in membrane traffic. Annu Rev Biochem 81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun X, Whittaker GR. 2007. Role of the actin cytoskeleton during influenza virus internalization into polarized epithelial cells. Cell Microbiol 9:1672–1682. doi: 10.1111/j.1462-5822.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. 1999. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol 1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- 35.Huotari J, Helenius A. 2011. Endosome maturation. EMBO J 30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greber UF, Puntener D. 2009. DNA-tumor virus entry–from plasma membrane to the nucleus. Semin Cell Dev Biol 20:631–642. doi: 10.1016/j.semcdb.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 37.Döhner K, Nagel CH, Sodeik B. 2005. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol 13:320–327. doi: 10.1016/j.tim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Sodeik B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol 8:465–472. doi: 10.1016/S0966-842X(00)01824-2. [DOI] [PubMed] [Google Scholar]
- 39.Hoogenraad CC, Bradke F. 2009. Control of neuronal polarity and plasticity–a renaissance for microtubules? Trends Cell Biol 19:669–676. doi: 10.1016/j.tcb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Buxton GA, Siedlak SL, Perry G, Smith MA. 2010. Mathematical modeling of microtubule dynamics: insights into physiology and disease. Prog Neurobiol 92:478–483. doi: 10.1016/j.pneurobio.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garnham CP, Roll-Mecak A. 2012. The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton (Hoboken) 69:442–463. doi: 10.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greber UF, Way M. 2006. A superhighway to virus infection. Cell 124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 43.King SJ, Schroer TA. 2000. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol 2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 44.Pfister KK, Fisher EM, Gibbons IR, Hays TS, Holzbaur EL, McIntosh JR, Porter ME, Schroer TA, Vaughan KT, Witman GB, King SM, Vallee RB. 2005. Cytoplasmic dynein nomenclature. J Cell Biol 171:411–413. doi: 10.1083/jcb.200508078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikkawa M. 2013. Big steps toward understanding dynein. J Cell Biol 202:15–23. doi: 10.1083/jcb.201304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J 30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hernáez B, Tarragó T, Giralt E, Escribano JM, Alonso C. 2010. Small peptide inhibitors disrupt a high-affinity interaction between cytoplasmic dynein and a viral cargo protein. J Virol 84:10792–10801. doi: 10.1128/JVI.01168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bremner KH, Scherer J, Yi J, Vershinin M, Gross SP, Vallee RB. 2009. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 6:523–535. doi: 10.1016/j.chom.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou JY, Shang SB, Gong H, Chen QX, Wu JX, Shen HG, Chen TF, Guo JQ. 2005. In vitro expression, monoclonal antibody and bioactivity for capsid protein of porcine circovirus type II without nuclear localization signal. J Biotechnol 118:201–211. doi: 10.1016/j.jbiotec.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Ma G, Li Y, Jiang X, He J, Zhou J. 2009. Characterization of monoclonal antibody against replication-associated protein of porcine circovirus. DNA Cell Biol 28:23–29. doi: 10.1089/dna.2008.0800. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Wu X, Zan J, Wu Y, Ye C, Ruan X, Zhou J. 2013. Cellular chaperonin CCTgamma contributes to rabies virus replication during infection. J Virol 87:7608–7621. doi: 10.1128/JVI.03186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hierholzer J, Killington R. 1996. Virus isolation and quantitation, p 25–46 InMahy BW, Kangro HO (ed), Virology methods manual. Academic Press Inc., San Diego, CA. [Google Scholar]
- 53.Shen HG, Zhou JY, Huang ZY, Guo JQ, Xing G, He JL, Yan Y, Gong LY. 2008. Protective immunity against porcine circovirus 2 by vaccination with ORF2-based DNA and subunit vaccines in mice. J Gen Virol 89:1857–1865. doi: 10.1099/vir.0.2008/000125-0. [DOI] [PubMed] [Google Scholar]
- 54.Wu X, Wang S, Yu Y, Zhang J, Sun Z, Yan Y, Zhou J. 2013. Subcellular proteomic analysis of human host cells infected with H3N2 swine influenza virus. Proteomics 13:3309–3326. doi: 10.1002/pmic.201300180. [DOI] [PubMed] [Google Scholar]
- 55.Aniento F, Emans N, Griffiths G, Gruenberg J. 1993. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol 123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Driskell OJ, Mironov A, Allan VJ, Woodman PG. 2007. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat Cell Biol 9:113–120. doi: 10.1038/ncb1525. [DOI] [PubMed] [Google Scholar]
- 57.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol 139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shekhar N, Wu J, Dickinson RB, Lele TP. 2013. Cytoplasmic dynein: tension generation on microtubules and the nucleus. Cell Mol Bioeng 6:74–81. doi: 10.1007/s12195-012-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu DJ, Baffet AD, Nayak T, Akhmanova A, Doye V, Vallee RB. 2013. Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell 154:1300–1313. doi: 10.1016/j.cell.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheung AK, Bolin SR. 2002. Kinetics of porcine circovirus type 2 replication. Arch Virol 147:43–58. doi: 10.1007/s705-002-8302-4. [DOI] [PubMed] [Google Scholar]
- 61.Liu Q, Tikoo SK, Babiuk LA. 2001. Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology 285:91–99. doi: 10.1006/viro.2001.0922. [DOI] [PubMed] [Google Scholar]
- 62.Finsterbusch T, Steinfeldt T, Caliskan R, Mankertz A. 2005. Analysis of the subcellular localization of the proteins Rep, Rep' and Cap of porcine circovirus type 1. Virology 343:36–46. doi: 10.1016/j.virol.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 63.Luby-Phelps K. 2000. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol 192:189–221. [DOI] [PubMed] [Google Scholar]
- 64.Desai A, Mitchison TJ. 1997. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 65.Vallee RB, Williams JC, Varma D, Barnhart LE. 2004. Dynein: an ancient motor protein involved in multiple modes of transport. J Neurobiol 58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 66.Goodman BS, Derr ND, Reck-Peterson SL. 2012. Engineered, harnessed, and hijacked: synthetic uses for cytoskeletal systems. Trends Cell Biol 22:644–652. doi: 10.1016/j.tcb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allan VJ. 2011. Cytoplasmic dynein. Biochem Soc Trans 39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 68.Schneider MA, Spoden GA, Florin L, Lambert C. 2011. Identification of the dynein light chains required for human papillomavirus infection. Cell Microbiol 13:32–46. doi: 10.1111/j.1462-5822.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 69.Zaichick SV, Bohannon KP, Hughes A, Sollars PJ, Pickard GE, Smith GA. 2013. The herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe 13:193–203. doi: 10.1016/j.chom.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Macovei A, Petrareanu C, Lazar C, Florian P, Branza-Nichita N. 2013. Regulation of hepatitis B virus infection by Rab5, Rab7, and the endolysosomal compartment. J Virol 87:6415–6427. doi: 10.1128/JVI.00393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Engel S, Heger T, Mancini R, Herzog F, Kartenbeck J, Hayer A, Helenius A. 2011. Role of endosomes in simian virus 40 entry and infection. J Virol 85:4198–4211. doi: 10.1128/JVI.02179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao PJ, Samulski RJ. 2012. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J Virol 86:10462–10473. doi: 10.1128/JVI.00935-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng S, Zhang M, Li W, Wang Y, Liu Y, He Q. 2012. Proteomic analysis of porcine alveolar macrophages infected with porcine circovirus type 2. J Proteomics 75:3258–3269. doi: 10.1016/j.jprot.2012.03.039. [DOI] [PubMed] [Google Scholar]
- 74.Chen S, Spiegelberg BD, Lin F, Dell EJ, Hamm HE. 2004. Interaction of Gbetagamma with RACK1 and other WD40 repeat proteins. J Mol Cell Cardiol 37:399–406. doi: 10.1016/j.yjmcc.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 75.Habermacher G, Sale WS. 1997. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J Cell Biol 136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang CY, Chang CP, Huang CL, Ferrell JE Jr. 1999. M phase phosphorylation of cytoplasmic dynein intermediate chain and p150(Glued). J Biol Chem 274:14262–14269. doi: 10.1074/jbc.274.20.14262. [DOI] [PubMed] [Google Scholar]
- 77.Mitchell DJ, Blasier KR, Jeffery ED, Ross MW, Pullikuth AK, Suo D, Park J, Smiley WR, Lo KWH, Shabanowitz J, Deppmann CD, Trinidad JC, Hunt DF, Catling AD, Pfister KK. 2012. Trk Activation of the ERK1/2 Kinase Pathway Stimulates Intermediate Chain Phosphorylation and Recruits Cytoplasmic Dynein to Signaling Endosomes for Retrograde Axonal Transport. J Neurosci 32:15495–15510. doi: 10.1523/JNEUROSCI.5599-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ogawa K, Takai H, Ogiwara A, Yokota E, Shimizu T, Inaba K, Mohri H. 1996. Is outer arm dynein intermediate chain 1 multifunctional? Mol Biol Cell 7:1895–1907. doi: 10.1091/mbc.7.12.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Padma P, Hozumi A, Ogawa K, Inaba K. 2001. Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene 275:177–183. doi: 10.1016/S0378-1119(01)00661-8. [DOI] [PubMed] [Google Scholar]
- 80.Chakrabarty AM. 1998. Nucleoside diphosphate kinase: role in bacterial growth, virulence, cell signalling and polysaccharide synthesis. Mol Microbiol 28:875–882. doi: 10.1046/j.1365-2958.1998.00846.x. [DOI] [PubMed] [Google Scholar]
- 81.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, Horinouchi S, Yoshida M. 2002. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J 21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piperno G, LeDizet M, Chang XJ. 1987. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol 104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maruta H, Greer K, Rosenbaum JL. 1986. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J Cell Biol 103:571–579. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Waterman-Storer CM, Salmon ED. 1999. Positive feedback interactions between microtubule and actin dynamics during cell motility. Curr Opin Cell Biol 11:61–67. doi: 10.1016/S0955-0674(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 85.Conacci-Sorrell M, Ngouenet C, Eisenman RN. 2010. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell 142:480–493. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, Jurdic P. 2005. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci 118:2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- 87.Valenzuela-Fernández A, Alvarez S, Gordon-Alonso M, Barrero M, Ursa A, Cabrero JR, Fernández G, Naranjo-Suárez S, Yáñez-Mo M, Serrador JM, Muñoz-Fernández MA, Sánchez-Madrid F. 2005. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol Biol Cell 16:5445–5454. doi: 10.1091/mbc.E05-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Husain M, Harrod KS. 2011. Enhanced acetylation of alpha-tubulin in influenza A virus infected epithelial cells. FEBS Lett 585:128–132. doi: 10.1016/j.febslet.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 89.Naranatt PP, Krishnan HH, Smith MS, Chandran B. 2005. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol 79:1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guise AJ, Budayeva HG, Diner BA, Cristea IM. 2013. Histone deacetylases in herpesvirus replication and virus-stimulated host defense. Viruses 5:1607–1632. doi: 10.3390/v5071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Opdenbosch N, Favoreel H, Van de Walle GR. 2012. Histone modifications in herpesvirus infections. Biol Cell 104:139–164. doi: 10.1111/boc.201100067. [DOI] [PubMed] [Google Scholar]