Abstract
Sulbactam is a class A β-lactamase inhibitor with intrinsic whole-cell activity against certain bacterial species, including Acinetobacter baumannii. The clinical use of sulbactam for A. baumannii infections is of interest due to increasing multidrug resistance in this pathogen. However, the molecular drivers of its antibacterial activity and resistance determinants have yet to be precisely defined. Here we show that the antibacterial activities of sulbactam vary widely across contemporary A. baumannii clinical isolates and are mediated through inhibition of the penicillin-binding proteins (PBPs) PBP1 and PBP3, with very low frequency of resistance; the rare pbp3 mutants with high levels of resistance to sulbactam are attenuated in fitness. These results support further investigation of the potential clinical utility of sulbactam.
INTRODUCTION
Acinetobacter baumannii is an opportunistic pathogen that has emerged globally as an important cause of nosocomial infections in immunocompromised patients (1–3). A. baumannii can cause a multitude of diseases, including urinary tract, wound, and surgical site infections, bacteremia, meningitis, and ventilator-associated pneumonia (VAP) (4–8). VAP is the most frequently reported A. baumannii infection among patients in the intensive care unit (ICU), with mortality rates ranging between 25% and 75% (1, 8–10). A. baumannii has also been associated with infections among military personnel who sustained severe combat wounds in Afghanistan and Iraq (11, 12).
Over the past decade, emerging A. baumannii infections have become of great concern clinically, due to the limited number of antibiotics that are effective treatments for infections (7, 10, 13, 14). This inadequate availability of therapeutic options is largely due to the organism's profound ability to acquire and to maintain resistance determinants for multiple classes of antibiotics (7, 10, 13). Even more alarming is the increasing prevalence of carbapenem-resistant A. baumannii (CRAB) (13, 15). Most antibiotics are not effective against CRAB infections, leaving reliable treatment options reduced to only a few drugs, such as colistin, tigecycline, or combinations thereof (13, 15).
Sulbactam is a β-lactamase inhibitor of many Ambler class A enzymes that is commercially available in combination with ampicillin. This β-lactam/β-lactamase inhibitor combination has been approved by the FDA for treatment of skin, gynecological, and intra-abdominal infections (16). Although sulbactam is used clinically as a β-lactamase inhibitor, it also has inherent antibacterial activity against a limited number of bacterial species, including Neisseria gonorrhoeae, Bacteroides fragilis, and, fortuitously, Acinetobacter spp. (17). Preliminary in vitro experiments have demonstrated that sulbactam binds to penicillin-binding proteins (PBPs) of Acinetobacter spp., and it is presumed that this activity is responsible for the observed bacterial killing (18, 19). Although historically ampicillin-sulbactam has been effective in treating VAP, bacteremia, and other nosocomial infections caused by A. baumannii (20–23), clinical resistance is emerging (24). Several recent clinical studies evaluated the activity of sulbactam combined with other antibiotics, such as fosfomycin (25), cefoperazone (26), minocycline (26), aminoglycosides (27), and colistin (28), for improved efficacy against multidrug-resistant (MDR) A. baumannii. In most cases, the addition of sulbactam resulted in increased efficacy in treating these infections, most likely due to the intrinsic activity of sulbactam (23, 26–29).
Despite the potential clinical utility of these combinations, our understanding of the molecular mechanisms driving sulbactam activity and resistance is still in its infancy. Some evidence suggests that loss of PBP2 activity (30) or upregulated efflux (31) may play a role in resistance. A clear link between TEM-1 expression and diminished sulbactam activity has recently been established (32). In this study, we expand on the findings described above by determining (i) the relative binding affinities and acylation rates of sulbactam with the major PBPs in A. baumannii, (ii) the activity of sulbactam versus comparator agents against a panel of recent clinical isolates of MDR A. baumannii that have been characterized by whole-genome sequencing, and (iii) the frequency and mechanism of spontaneous resistance in A. baumannii. The antibacterial activity of sulbactam was found to vary widely across contemporary A. baumannii clinical isolates, was mediated through inhibition of PBP1 and PBP3 but not PBP2, and was associated with a very low frequency of resistance. High-level resistance mapped to mutations in pbp3 and was accompanied by a fitness penalty, whereas low-level resistance arose from mutations in cell wall biosynthesis or stress response genes.
MATERIALS AND METHODS
Strains and media.
A. baumannii ATCC 19606T was obtained from the American Type Culture Collection (Manassas, VA). A. baumannii ATCC 17978 and ARC5468 (98-37-09) were generous gifts from Paul Dunman (University of Rochester, Rochester, NY). A. baumannii strains ARC2058 and ARC2461 were obtained from the AstraZeneca culture collection. A total of 60 Acinetobacter isolates that were characterized for β-lactamase genes were included in the MIC90 test panel; the majority of these were A. baumannii, but several were found to belong to the A. baumannii/A. calcoaceticus complex upon whole-genome sequencing analysis, as indicated in Table 2. The panel included isolates from the AstraZeneca strain collection as well as recently obtained worldwide surveillance isolates from International Health Management Associates Inc. (Schaumburg, IL). These isolates were deliberately selected for characterization because of their wide range of susceptibility to sulbactam and their global origins. All bacterial strains were routinely grown from frozen glycerol stocks on blood agar plates. Susceptibility testing was performed and growth rates were assessed in cation-adjusted Mueller-Hinton broth (MHB-II) according to Clinical and Laboratory Standards Institute (CLSI) guidelines (33). The relative growth of wild-type versus sulbactam-resistant A. baumannii strains was determined by monitoring the change in optical density at 595 nm (OD595) over time at 37°C in cation-adjusted Mueller-Hinton broth (MHB-II) in a 96-well format, using a SpectraMax Plus spectrophotometer.
Table 2.
Sulbactam activities against recent clinical strains of A. baumannii
| Strain | β-Lactamase content | MIC (μg/ml)a |
||||||
|---|---|---|---|---|---|---|---|---|
| SUL | UNA | MEM | CST | LVX | GEN | TET | ||
| ARC3491b | OXA-215 | 0.5 | 1 | 2 | 0.5 | 1 | 2 | 4 |
| ATCC 19606T | OXA-98 | 1 | 2 | 1 | 0.5 | 0.25 | 8 | 2 |
| ARC2582 | OXAc | 1 | 2 | 0.25 | 0.125 | 0.125 | 4 | 2 |
| ARC2597b | OXAc | 1 | 2 | 0.125 | 0.125 | <0.03 | 0.25 | 1 |
| ARC2058 | OXA-95 | 2 | 2 | 0.25 | 0.5 | 0.125 | 1 | 1 |
| ARC2728b | OXAc | 2 | 2 | 0.25 | 0.25 | 0.125 | 0.25 | 1 |
| ARC5090 | OXAc | 2 | 2 | 0.25 | 0.25 | 0.125 | 0.25 | 2 |
| ARC2719 | OXAc | 2 | 4 | 0.25 | 0.125 | 0.125 | 0.5 | 2 |
| ARC2720b | OXAc | 2 | 4 | 1 | 0.125 | 2 | 0.5 | 2 |
| ARC3489 | OXA,c OXA-68 | 2 | 8 | 4 | 0.25 | 16 | >32 | >32 |
| ARC3494 | OXA-65 | 4 | 2 | 0.25 | 0.5 | 0.25 | 0.25 | 2 |
| ARC2780b | OXA,c OXA-2, IMP-1 | 4 | 4 | 32 | 4 | 4 | >32 | 2 |
| ARC3487 | OXA-20, OXA-58, OXA-66 | 4 | 8 | 8 | 0.25 | 8 | 8 | 16 |
| ARC3659 | OXA-23, OXAc | 4 | 8 | 8 | 0.25 | 8 | >32 | 8 |
| ARC5084 | IMP-4(B), OXA-58, OXA-65 | 4 | 8 | >32 | 0.125 | 4 | >32 | 2 |
| ARC2682 | SHV-5, OXA-113 | 4 | 16 | 32 | 0.25 | 16 | >32 | 8 |
| ARC2059 | PSE-2, PSE-1 | 8 | 16 | 0.5 | 0.25 | >32 | 1 | 4 |
| ARC5092 | OXA-23, OXA-64 | 8 | 16 | 16 | >32 | 8 | >32 | 32 |
| ARC3485 | OXA-82 | 8 | 32 | 16 | 0.25 | 16 | 1 | 32 |
| ARC2674 | SHV-5, OXA-113 | 8 | 32 | 8 | 0.25 | 16 | 8 | >32 |
| ARC2788 | OXA-65, TEM-1 | 8 | 32 | 1 | 0.125 | 32 | 16 | 4 |
| ARC3515 | OXA-64, OXA-58 | 8 | 32 | 4 | 0.25 | 8 | 4 | >32 |
| ARC5081 | OXA-94, OXA-23 | 8 | 32 | 16 | 0.125 | 8 | 0.25 | 2 |
| ARC5091 | OXA-82, OXA-23 | 8 | 32 | 32 | 8 | >32 | >32 | 32 |
| ARC2777 | OXA-172, TEM-1 | 8 | >32 | 32 | 0.5 | 32 | 16 | 32 |
| ARC3488 | OXA,c OXA-68 | 16 | 16 | 4 | 2 | 32 | >32 | >32 |
| ARC5075 | SHV-5, OXA-113 | 16 | 16 | 32 | 0.25 | 32 | >32 | >32 |
| ARC5088 | OXA-20, OXA-58, OXA-66 | 16 | 16 | 8 | 0.125 | 8 | 8 | 16 |
| ARC2675 | SHV-5, OXA-113 | 16 | 32 | >32 | 0.125 | 32 | >32 | 16 |
| ARC2681 | OXA-40, TEM-1, OXA-132 | 16 | 32 | 32 | 0.25 | 16 | >32 | >32 |
| ARC2778 | OXA-40, TEM-1, OXA-65 | 16 | 32 | >32 | 0.25 | 32 | >32 | >32 |
| ARC2779b | OXA-2, VIM-2 | 16 | 32 | 16 | 0.25 | 0.125 | >32 | 2 |
| ARC3484 | TEM-1, OXA-23, OXA-64 | 16 | 32 | 32 | 0.125 | 8 | >32 | >32 |
| ARC3492 | OXA-40, OXA-132, TEM-1 | 16 | 32 | >32 | 0.25 | 8 | >32 | >32 |
| ARC3513 | TEM-1, OXA-23, OXA-65 | 16 | 32 | 32 | 1 | 16 | >32 | 8 |
| ARC5073 | OXA-23, TEM-1, OXA-64, PER-1 | 16 | 32 | >32 | 0.125 | 8 | 0.5 | >32 |
| ARC5083 | OXA-66, OXA-23 | 16 | 32 | 16 | 0.125 | >32 | >32 | >32 |
| ARC2461 | OXA-66, TEM-1 | 16 | >32 | 2 | 0.125 | 16 | >32 | >32 |
| ARC2462 | TEM-1, OXA-66 | 16 | >32 | 4 | 0.125 | 16 | >32 | >32 |
| ARC2598 | OXA,c TEM-1, OXA-113 | 16 | >32 | 8 | 0.25 | 4 | 2 | >32 |
| ARC2635 | OXA-65, OXA-40, TEM-1 | 32 | 32 | >32 | 0.25 | 16 | >32 | 8 |
| ARC5085 | OXA,c TEM-1 | 32 | 32 | 8 | 1 | 32 | >32 | >32 |
| ARC3657 | OXA-130 | 32 | >32 | 2 | 0.5 | 16 | 0.25 | 8 |
| ARC2636 | OXA-65, OXA-40, TEM-1 | 32 | >32 | >32 | 0.125 | 16 | >32 | 16 |
| ARC2782 | OXA-66, OXA-23, TEM-1, PER-1 | 32 | >32 | 16 | 0.125 | 4 | >32 | >32 |
| ARC3486 | OXA-72, OXA-66, TEM-1 | 32 | >32 | >32 | 0.25 | 8 | >32 | >32 |
| ARC3490 | TEM-1+, PSE-2, OXA-69 | 32 | >32 | 0.5 | 0.5 | 16 | 16 | 8 |
| ARC3495 | OXA-40, OXA-109 | 32 | >32 | >32 | 0.25 | 4 | >32 | >32 |
| ARC3658 | OXA-66, PER-1, TEM-1, OXA-23 | 32 | >32 | 32 | 0.25 | 8 | >32 | >32 |
| ARC5076 | TEM-1, OXA-23, OXA-66 | 32 | >32 | 32 | 0.25 | 8 | 8 | >32 |
| ARC5077 | OXA,c OXA-72 | 32 | >32 | >32 | 0.5 | 16 | >32 | >32 |
| ARC5079 | OXA-72, OXA-65 | 32 | >32 | >32 | 0.125 | 16 | 8 | 8 |
| ARC5080 | OXA-71, OXA-40 | 32 | >32 | >32 | 0.25 | 16 | >32 | 16 |
| ARC5086 | OXA,c TEM-1, OXA-72, OXA-66 | 32 | >32 | >32 | 0.125 | 16 | >32 | >32 |
| ARC5087 | OXA-66, OXA-23 | 32 | >32 | 16 | 0.25 | >32 | >32 | >32 |
| ARC5089 | PER,c TEM-1, OXA-23, OXA-66 | 32 | >32 | 32 | 0.125 | 16 | >32 | 4 |
| ARC3493 | OXA-40, OXA-66 | 64 | >32 | >32 | 32 | 4 | >32 | >32 |
| ARC5074 | GES,c TEM-1, OXA-51 | 64 | >32 | 8 | 0.125 | 4 | 0.125 | 1 |
| ARC5082 | OXA-66, OXA-23 | 64 | >32 | >32 | 0.5 | 8 | 0.5 | >32 |
| ARC3882 | OXA-23, NDM-1, OXA-10c | >64 | >32 | >32 | 0.125 | 4 | >32 | 8 |
| Range | 0.5 to >64 | 1 to >32 | <0.03 to >32 | 0.125 to >32 | <0.03 to >32 | 0.06 to >32 | 0.5 to >32 | |
SUL, sulbactam; UNA, Unasyn (2:1 combination of ampicillin and sulbactam); MEM, meropenem; CST, colistin; LVX, levofloxacin; GEN, gentamicin; TET, tetracycline.
Member of the A. baumannii-A. calcoaceticus complex family.
The gene encodes a closely related variant of the indicated β-lactamase family.
Isolation of membrane-bound penicillin-binding proteins from A. baumannii.
A. baumannii bacteria were grown in 1 liter of Luria-Bertani (LB) medium, and cells were harvested at the log phase of growth, at an OD595 of 0.4. Cells were centrifuged at 6,000 rpm, and pellets were frozen at −20°C for future use. The cell pellets were thawed on ice and washed once with 50 ml of ice-cold 100 mM sodium phosphate buffer (pH 7.0) containing 100 mM NaCl. The cells were resuspended in 10 ml of the same buffer. After 200 μl of 50 mg/ml lysozyme (Pierce) and one tablet of mini cOmplete EDTA-free protease inhibitor cocktail (Roche) were added, the cells were incubated at 37°C for 45 min; 200 μl of DNase I (Pierce) in 10 mM MgSO4 (final concentration) was then added, and the cells were treated with three cycles of freezing and thawing (10 min in a dry ice-ethanol bath followed by 20 min in a 37°C water bath). The cells were transferred to 50-ml glass tubes and sonicated (Branson Sonifier 250, output control 6) on ice four times, for 30 s each time, with 1 min of cooling between the sonication steps. Cell debris was removed by centrifugation at 12,000 × g for 10 min at 4°C. The supernatant was ultracentrifuged at 100,000 × g for 45 min at 4°C to collect the crude membranes. The membranes were washed three times with 5 ml of cold 100 mM sodium phosphate buffer (pH 7.0), suspended in 1.5 ml of 100 mM sodium phosphate buffer (pH 7.0), and then homogenized with a 10-ml tissue grinder (Wheaton). The membrane preparations were divided into aliquots and stored at −80°C for future use. The concentrations of membrane proteins were determined using the Bradford assay (Bio-Rad), with bovine serum albumin (BSA) (Sigma) as the standard. β-Lactamase activity present in the membranes was determined with a standard nitrocefin hydrolysis assay (BioVision), according to the manufacturer's instructions. Several strains of A. baumannii were tested; because ATCC 17978 demonstrated the least β-lactamase activity and the best PBP profiles, it was selected for use in the compound affinity test.
Determination of apparent compound affinities for membrane-bound penicillin-binding proteins.
A total of 150 μg of membrane proteins from the ATCC 17978 strain was added to 100 mM sodium phosphate (pH 7.0) to bring the mixture to a total volume of 10 μl. The solution was mixed with 2 μl of buffer or test compound at various concentrations and incubated at 30°C for 30 min. Then, 0.5 μl of 3 mM Bocillin FL (Life Technologies) in water was added, and the mixture was incubated at 30°C for another 30 min before the addition of 0.6 μl of 45% sarcosyl (Sigma) and 45% penicillin G (Sigma). After 30 min of incubation at 30°C, the mixture was centrifuged at 20,000 × g for 10 min at 4°C, and 11 μl of the supernatant was mixed with 4 μl of 5× SDS sample loading buffer and 5 μl of water. After being heated at 95°C for 4 min, the sample was loaded onto an 8% acrylamide–Tris-glycine gel (Life Technologies). PBPs were separated at 140 V for 1.5 h. The gel was washed briefly with water, fixed for 15 min in destaining solution containing 10% acetic acid and 40% methanol in water, and washed with water for 1 h before scanning with an LAS-4000 FluorImager (Fuji) with 520-nm green EPI light-emitting diode (LED) light and a 575DF20 filter.
Cloning, expression, and purification of A. baumannii and Pseudomonas aeruginosa PBPs.
The cloning, expression, and purification of P. aeruginosa PBP2 and PBP3 and A. baumannii PBP3 were performed as described previously (34, 35). All protein concentrations were determined by the Bradford method, and proteins were characterized by SDS-PAGE analysis and analytical liquid chromatography-mass spectrometry (LC-MS). Purified PBPs were stored at −80°C. A. baumannii PBP1a and PBP2 and P. aeruginosa PBP1a were cloned, expressed, and purified as follows. Genomic DNA isolation, plasmid DNA purification, and PCR product purification were performed using a Wizard genomic DNA purification kit (Promega, Madison, WI), PureYield Plasmid Midiprep system (Promega), and QuickStep2 PCR purification kit (EdgeBio), respectively. Primers for PCR DNA amplification were purchased from Eurofins MWG Operon (Huntsville, AL). All PCRs were performed with High Fidelity PCR Master Mix (Roche Applied Science), using reaction conditions specified by the manufacturer. All ligation-independent cloning reactions were performed using the pET-43.1 Ek/LIC vector kit (EMD Millipore), according to the manufacturer's instructions. DNA sequences of the cloned genes were confirmed by sequencing on an ABI Prism 3100 DNA sequencer (Applied Biosystems), using the BigDye terminator cycle sequencing kit (Applied Biosystems). Computer analysis of DNA sequences was performed with Sequencher (Gene Codes Corp., Ann Arbor, MI).
A. baumannii pbp1a.
The ponA (pbp1a) gene from A. baumannii was codon-optimized for expression in Escherichia coli and custom synthesized with the N-terminal His6 purification tag and a tobacco etch virus (TEV) protease cleavage site (Blue Sky Bioservices). The 25-amino-acid N-terminal secretion signal was deleted. The optimized gene was cloned into pET-28a(+) (Novagen Biosciences) using NcoI and XhoI restriction sites, to create the plasmid pNG141. For protein expression, the plasmid was transformed into BL21-Gold(DE3) (Agilent Technologies), plated on Luria-Bertani (LB) medium containing 25 μg/ml kanamycin, and incubated overnight at 37°C. A single colony of BL21-Gold(DE3)/pNG141 was inoculated into a 100-ml culture of LB medium containing 25 μg/ml kanamycin and was grown overnight at 37°C. The overnight culture medium was diluted to an OD600 of 0.1 in 2 liters of LB medium containing 25 μg/ml kanamycin and was grown to mid-logarithmic phase (OD600 = 0.6) at 30°C, with aeration. The culture was incubated on ice for 30 min and transferred to 18°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to a final concentration in each culture of 0.1 mM. After overnight induction at 18°C, the cells were harvested by centrifugation at 5,000 × g for 15 min at 25°C. Cell pastes were stored at −20°C. The frozen cell paste from 2 liters of cell culture medium was suspended in 40 ml of buffer A, consisting of 25 mM Tris-HCl (pH 7.5), 0.5 M NaCl, and 5% (vol/vol) glycerol, supplemented with one EDTA-free protease inhibitor cocktail tablet (Roche Molecular Biochemical). Cells were disrupted twice with a French press at 18,000 lb/in2 at 4°C, and the crude extract was centrifuged at 150,000 × g (45 Ti rotor; Beckman-Coulter) for 30 min at 4°C. The supernatant was applied, at a flow rate of 2.0 ml/min, to a 5-ml HiTrap Ni2+-chelating column (GE Healthcare Life Sciences) preequilibrated with buffer A. The column was washed with buffer A, and PBP1a was eluted with a linear gradient of 0 M to 0.5 M imidazole in buffer A. Fractions containing PBP1a were pooled and concentrated to 5 ml with an Amicon Ultracel-10K filter unit (Millipore). The 5-ml sample was applied, at a flow rate of 1.5 ml/min, to a 120-ml Superdex 200 (HR 16/60) column (GE Healthcare Life Sciences) preequilibrated with buffer B, consisting of 25 mM Tris-HCl (pH 7.5), 0.5 M NaCl, 1 mM EDTA, 2 mM dithiothreitol, and 5% (vol/vol) glycerol. The fractions containing PBP1a were pooled and concentrated with an Amicon Ultracel-10K filter unit (Millipore). The final yield of purified A. baumannii PBP1a was 57 mg from 2 liters of cell culture medium.
A. baumannii pbp2.
A bacterial plasmid for expression of NusA-tagged Acinetobacter baumannii PBP2 (residues 40 to 672) with an enterokinase cleavage site was made by amplifying the gene encoding Acinetobacter baumannii ARC2058 PBP2 (residues 40 to 672) from genomic DNA. The resulting PCR product was spin column purified and cloned into the plasmid pET-43.1 by ligation-independent cloning. The cloning reaction product was transformed into NovaBlue GigaSingles competent cells (EMD Millipore). Transformants were selected on LB medium with 100 μg/ml ampicillin, plasmids were isolated, and inserts were verified by PCR and sequencing. For protein expression, the plasmid was transformed into BL21(DE3) (EMD Chemicals, Gibbstown, NJ), plated on Luria-Bertani (LB) medium containing 100 μg/ml ampicillin, and incubated overnight at 37°C. A single colony of BL21(DE3)/pJT1294 was inoculated into a 100-ml culture of LB medium containing 100 μg/ml ampicillin and was grown overnight at 37°C. The overnight culture medium was diluted to an OD600 of 0.1 in 2 liters of LB medium containing 100 μg/ml ampicillin and was grown to mid-logarithmic phase (OD600 = 0.6) at 30°C, with aeration. The culture was incubated on ice for 30 min and transferred to 18°C. IPTG was then added to a final concentration in each culture of 0.5 mM. After overnight induction at 18°C, the cells were harvested by centrifugation at 5,000 × g for 15 min at 25°C. Cell pastes were stored at −20°C. The frozen cell paste from 2 liters of cell culture medium was suspended in 40 ml of buffer A, consisting of 25 mM Tris-HCl (pH 8.0), 0.5 M NaCl, and 5% (vol/vol) glycerol, supplemented with one EDTA-free protease inhibitor cocktail tablet (Roche Molecular Biochemical). Cells were disrupted twice with a French press at 18,000 lb/in2 at 4°C, and the crude extract was centrifuged at 150,000 × g (45 Ti rotor; Beckman-Coulter) for 30 min at 4°C. The supernatant was applied, at a flow rate of 2.0 ml/min, to a 5-ml HiTrap Ni2+-chelating column (GE Healthcare Life Sciences) preequilibrated with buffer A. The column was then washed with buffer A, and NusA-PBP2 was eluted with a linear gradient of 0 M to 0.5 M imidazole in buffer A. Fractions containing NusA-PBP2 were pooled and concentrated to 5 ml with an Amicon Ultracel-10K filter unit (Millipore). The 5-ml sample was applied, at a flow rate of 1.5 ml/min, to a 120-ml Superdex 200 (HR 16/60) column (GE Healthcare Life Sciences) preequilibrated with buffer B, consisting of 25 mM HEPES (pH 7.3), 0.15 M NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 10% (vol/vol) glycerol. The fractions containing NusA-PBP2 were pooled and concentrated with an Amicon Ultracel-10K filter unit (Millipore). The final yield of purified NusA-PBP2 was 15 mg from 2 liters of cell culture medium.
P. aeruginosa pbp1a.
The ponA (pbp1a) gene from P. aeruginosa was codon-optimized for expression in E. coli and custom synthesized with the N-terminal His10 purification tag and a TEV protease cleavage site (Blue Sky Bioservices). The 26-amino-acid N-terminal secretion signal was deleted. The optimized gene was cloned into pET-28a(+) (Novagen Biosciences) using NheI and XhoI restriction sites, to create the plasmid pNG140. For protein expression, the plasmid was transformed into BL21-Gold(DE3) (Agilent Technologies), plated on Luria-Bertani (LB) medium containing 25 μg/ml kanamycin, and incubated overnight at 37°C. A single colony of BL21-Gold(DE3)/pNG141 was inoculated into a 100-ml culture of LB medium containing 25 μg/ml kanamycin and was grown overnight at 37°C. The overnight culture was diluted to an OD600 of 0.1 in 2 liters of LB medium containing 25 μg/ml kanamycin and was grown to mid-logarithmic phase (OD600 = 0.6) at 30°C, with aeration. The culture was incubated on ice for 30 min and transferred to 18°C. IPTG was then added to a final concentration in each culture of 0.1 mM. After overnight induction at 18°C, the cells were harvested by centrifugation at 5,000 × g for 15 min at 25°C. Cell pastes were stored at −20°C. The frozen cell paste from 2 liters of cell culture medium was suspended in 40 ml of buffer A, consisting of 25 mM Tris-HCl (pH 7.5), 0.5 M NaCl, and 5% (vol/vol) glycerol, supplemented with one EDTA-free protease inhibitor cocktail tablet (Roche Molecular Biochemical). Cells were disrupted twice with a French press at 18,000 lb/in2 at 4°C, and the crude extract was centrifuged at 150,000 × g (45 Ti rotor, Beckman-Coulter) for 30 min at 4°C. The supernatant was applied, at a flow rate of 2.0 ml/min, to a 5-ml HiTrap Ni2+-chelating column (GE Healthcare Life Sciences) preequilibrated with buffer A. The column was washed with buffer A, and A. baumannii PBP1a was eluted with a linear gradient of 0 M to 0.5 M imidazole in buffer A. Fractions containing PBP1a were pooled and concentrated to 5 ml with an Amicon Ultracel-10K filter unit (Millipore). The 5-ml sample was applied, at a flow rate of 1.5 ml/min, to a 120-ml Superdex 200 (HR 16/60) column (GE Healthcare Life Sciences) preequilibrated with buffer B, consisting of 25 mM Tris-HCl (pH 7.5), 0.5 M NaCl, 1 mM EDTA, 2 mM dithiothreitol, and 5% (vol/vol) glycerol. The fractions containing PBP1a were pooled and concentrated with an Amicon Ultracel-10K filter unit (Millipore). The final yield of purified A. baumannii PBP1a was 57 mg from 2 liters of cell culture medium.
Fluorescence anisotropy assay.
The fluorescence anisotropy assay, which measures the ability of each test compound to compete with fluorescent Bocillin FL for time-dependent PBP acylation, was performed as described previously (34). Briefly, the assay was performed in 0.1 M sodium phosphate buffer at pH 7.0 (except for P. aeruginosa PBP2, for which the assay was performed at pH 6.2) containing 0.01% Triton X-100 detergent (Surfact-Amps 100; Thermo Fisher Scientific). The final concentration of each component was as follows: Bocillin FL, 30 nM; all PBPs, 60 nM (except for A. baumannii PBP2 and P. aeruginosa PBP2, which were used at 48 and 300 nM, respectively).
Cell morphology.
Bacterial strains (ARC2058 and ATCC 17978) were cultured in MHB-II overnight at 37°C. A 40-μl sample of the overnight culture was used to inoculate 4 ml of fresh MHB-II. After 30 min of shaking at 37°C, compounds (sulbactam, aztreonam, ceftazidime, or meropenem) were added to a final concentration of 0.5× MIC. After incubation for 3 h, 20 μl of the culture medium was removed and heat fixed onto a glass microscope slide. After fixation, the slide was stained with acridine orange (BD Diagnostic Systems, Sparks, MD) for 3 min and rinsed with distilled water. Cell morphology was examined with an Olympus BX50 fluorescence microscope (Olympus America Inc., Melville, NY).
Whole-genome sequencing and analysis.
Genomic DNA was purified using the Wizard genomic purification kit (Promega Corp.) and was used for library construction. DNA libraries were prepared using the Nextera library construction protocol (Illumina), following the manufacturer's instructions, and were sequenced with a MiSeq sequencer (Illumina). For each isolate, approximately 2.5 million 150-bp paired-end sequence reads were assembled de novo and analyzed using the CLC bio suite of software tools (CLC bio, Cambridge, MA).
Determination of frequency of resistance.
A. baumannii strains were grown overnight at 37°C on blood agar plates. After 24 h of incubation, a suspension of 109 to 1010 cells/ml (OD600 of ∼3) was made in MHB-II and then spread on agar plates containing concentrations of sulbactam 2 to 32 times the agar microdilution MIC. These plates were incubated for 48 h at 37°C, and the numbers of colonies that grew were counted for each strain. In addition, the CFU/ml on drug-free plates was determined for each strain to allow calculation of the frequency of resistance. Several sulbactam-resistant colonies were selected, passaged twice on blood agar plates in the absence of compound, and retested for susceptibility. DNA was isolated from strains that maintained decreased sulbactam susceptibility and was compared with DNA isolated from the parental strain through whole-genome sequencing, as described above.
Proliferation of A. baumannii in murine blood.
Bacterial strains of interest were struck out on blood agar plates and incubated overnight at 37°C. Bacteria from these plates were resuspended in phosphate-buffered saline (PBS) at a concentration of 2 × 107 CFU/ml; 2 μl was added to 200 μl of freshly harvested, heparinized, whole mouse blood (pooled from at least 5 mice), resulting in a final inoculum of 2 × 105 CFU/ml, and the mixture was incubated at 37°C in a 5% CO2 incubator, with shaking at 120 rpm. Ten-microliter samples were removed at the indicated times, and bacterial proliferation was measured by performing colony counts of 2-fold serial dilutions of these samples on blood agar, after an overnight incubation at 37°C.
RESULTS
Sulbactam inhibits PBP1 and PBP3 but not PBP2 in A. baumannii.
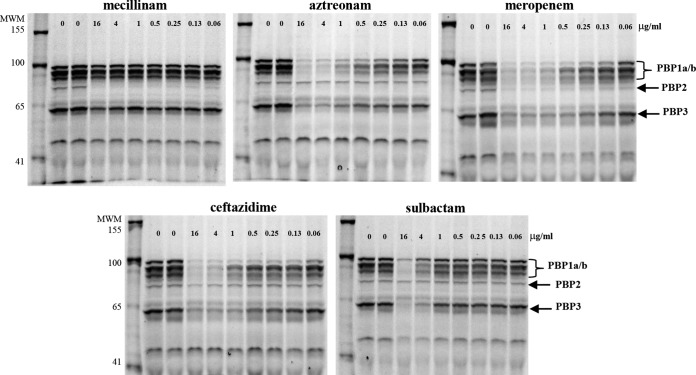
It was first suggested that the mechanism of the antibacterial activity of sulbactam against A. baumannii was primarily via inhibition of PBP1a and PBP2 (17). Although it has been reported that sulbactam preferentially binds PBP3 over PBP1a, with 50% inhibitory concentration (IC50) values of 4 μM and 55 μM, respectively (18), with a mode of binding to the active site similar to that of ampicillin (18), no evaluation of the binding affinity for PBP2 was made in that study. Therefore, to understand the relative affinity of sulbactam for PBP2 versus PBP1 orthologs or PBP3, membranes containing these proteins were purified from an A. baumannii clinical isolate, ATCC 17978, and subjected to a standard gel-based Bocillin FL competition assay (37). The activity of sulbactam was compared with that of several control compounds for which a PBP binding preference has been established. As shown in Fig. 1, sulbactam bound weakly to all PBP1 orthologs and to PBP3 but not to PBP2, a binding pattern that is quite similar to that observed with aztreonam. As expected, mecillinam bound exclusively to PBP2, whereas ceftazidime and meropenem interacted with all high-molecular-weight PBPs, albeit with different affinities, in agreement with previous reports (38–40) (Fig. 1).
FIG 1.
Sulbactam binding to PBP1 and PBP3 but not PBP2 in A. baumannii. Increasing amounts of compounds of interest were incubated with membrane proteins harvested from A. baumannii ATCC 17978 in a standard, gel-based, Bocillin FL competition assay. Arrows, PBPs 1a/b, PBP2, and PBP3. There is no pbp1c gene in any of the published A. baumannii genomes. It is likely that the band corresponding to this protein, as documented for other Gram-negative orthologs, is a proteolytic fragment or results from an alternate start site for PBP1a or PBP1b. MWM, molecular weight markers.
To quantify these binding activities, the relative abilities of the same set of compounds to compete with Bocillin FL for time-dependent acylation of purified PBPs were measured using fluorescence anisotropy, as described previously (34) (Table 1). Acylation rate constants generally corresponded well with the gel-based competition results shown in Fig. 1. Sulbactam preferentially inhibited PBP1a and PBP3 over PBP2, as did aztreonam and ceftazidime, although the latter two compounds were notably more reactive. Mecillinam reacted predominantly with PBP2, whereas meropenem was quite reactive with all three PBPs tested but with lower potency against PBP3 than against PBP1a or PBP2, as described previously (40). Some interesting differences in the relative reactivities of the same set of compounds were found when the compounds were tested against P. aeruginosa PBP1a, PBP2, and PBP3 (Table 1), some of which have been reported previously (34, 35). Most notable was the difference in the reactivities toward PBP3 orthologs, with aztreonam, ceftazidime, and meropenem exhibiting much greater potency for P. aeruginosa than A. baumannii. Another interesting difference was in the activity of meropenem, which showed higher reactivities with A. baumannii PBP1a and PBP2 than PBP3, whereas the pattern was reversed for P. aeruginosa. However, no such species-dependent reactivity was observed with sulbactam, which was equally ineffective in binding to either of the PBP2 orthologs (Table 1).
TABLE 1.
Acylation rate constants for acylation of A. baumannii and P. aeruginosa PBP1a, PBP2, and PBP3 by various inhibitors
| Compound |
kon/Ki (M−1 s−1) |
|||||
|---|---|---|---|---|---|---|
|
A. baumannii |
P. aeruginosa |
|||||
| PBP1a | PBP2 | PBP3 | PBP1a | PBP2 | PBP3 | |
| Bocillin FL | 5,500 | 13,000 | 32,000 | 9,270 | 1,030 | 18,600 |
| Aztreonam | 1,200 | 0.12 | 520 | 85 | <5 | 296,000 |
| Ceftazidime | 5,000 | 1.2 | 780 | 3,760 | <5 | 69,000 |
| Mecillinam | 1.6 | 6,200 | <15 | <7 | 1,500 | NTa |
| Meropenem | 28,000 | 25,000 | 1,600 | 5,040 | 1,200 | 49,000 |
| Sulbactam | 8.8 | 0.34 | 17 | 5.9 | 0.12 | 1.7 |
NT, not tested.
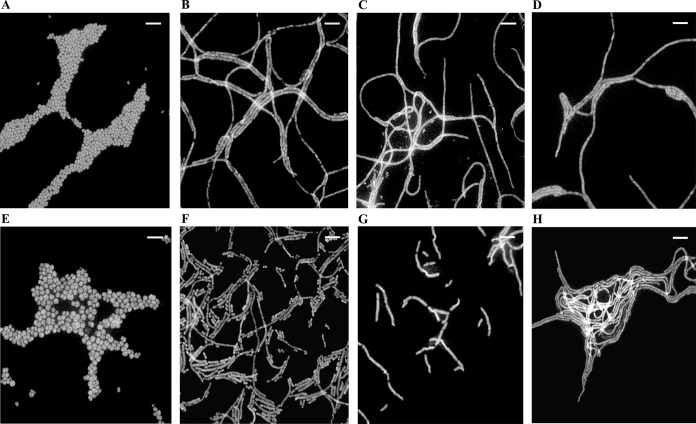
The morphology of A. baumannii treated with these compounds was then examined by fluorescence microscopy after 3-h exposure to 0.5× MIC of each compound (mecillinam was excluded because it lacks anti-Acinetobacter whole-cell activity, presumably due to the lack of membrane penetration). As shown in Fig. 2, unlike the coccobacillary morphology of untreated A. baumannii (Fig. 2A), sulbactam-treated cells (Fig. 2B) formed “spaghetti-like” filaments, quite similar to the well-established phenotype observed upon treatment with ceftazidime and aztreonam (Fig. 2C and D) (41) and consistent with their similar mechanisms of PBP inhibition. In contrast, treatment of A. baumannii with 0.5× MIC of meropenem resulted in spheroplasts that were larger and more rounded (Fig. 2E). These phenotypes were not strain specific, as they were also observed in ATCC 17978 (data not shown). It has been well established that meropenem (as well as other β-lactam) treatment of other Gram-negative bacterial species such as P. aeruginosa notably results in filamentous cells with distinct bulges at lower concentrations, with spheroplast formation occurring only at much higher concentrations (42). Attempts to evaluate morphological changes in A. baumannii at higher concentrations of meropenem were unsuccessful due to rapid lysis of the cells under these conditions. It is likely that this concentration-dependent difference in morphology between A. baumannii and other Gram-negative bacteria upon exposure to meropenem is due to the higher affinity of the drug for A. baumannii PBP2 than for orthologs in other species (as exemplified in Table 1).
FIG 2.
Fluorescence microscopic images of cells after 3-h exposure to 0.5× MIC levels of antimicrobials. (A to E) A. baumannii ARC2058. (A) No drug. (B) Sulbactam (0.5 μg/ml). (C) Ceftazidime (4 μg/ml). (D) Aztreonam (16 μg/ml). (E) Meropenem (0.125 μg/ml). (F) Sulbactam-resistant ATCC 17978 pbp3 S395F mutant, without treatment. (G and H) ARC2058 pbp3 S390T mutant, without treatment (G) and after exposure to sulbactam at 16 μg/ml (H). Bars, 5 μm. Similar results were obtained with ATCC 17978 (data not shown).
Antibacterial activities of sulbactam against contemporary clinical isolates.
Sulbactam used to be considered a therapeutic option for the treatment of infections caused by MDR strains of A. baumannii due to its intrinsic antibacterial activities against many of these strains (43). However, because (i) it is marketed only in combination with ampicillin in most countries and (ii) its clinical use occurs most frequently in combination with other agents, such as colistin and carbapenems (15), there are no reports of the activities of sulbactam alone against current clinical isolates. Therefore, 60 geographically diverse clinical isolates of A. baumannii from recent (post-2006) nosocomial infections were tested for their susceptibilities to sulbactam alone, ampicillin and sulbactam in a 2:1 ratio, and five control compounds. As shown in Table 2, the intrinsic antibacterial activities of sulbactam against these isolates ranged from 0.5 to >64 μg/ml. Ampicillin-sulbactam was about 2-fold less active, with activities ranging from 1 to >32 μg/ml. This finding demonstrates that the antibacterial activity of ampicillin-sulbactam is due to the sulbactam component, in agreement with previous studies (36, 44, 45).
Sulbactam activities against isolates with defined β-lactamase genes.
It has recently been shown that sulbactam activity is compromised by TEM-1 expression, both in resistant clinical isolates of A. baumannii and when overexpressed in a susceptible A. baumannii strain (32). In order to further explore the contributions of this and other β-lactamases to sulbactam activity against A. baumannii, the β-lactamase contents of the isolates listed in Table 2 were determined by sequencing analysis of their entire genomes. There was a notable correlation between decreased activity of sulbactam and the presence of TEM-1 (Table 2). In addition, sulbactam was much less active against strains bearing class B metallo-β-lactamases (MBLs), such as the VIM-2-positive strain ARC2779 and the NDM-1-positive strain ARC3882 (Table 2).
The frequency of resistance to sulbactam is low in A. baumannii and maps to pbp3.
To further define the activity of sulbactam in A. baumannii, the frequency of the emergence of resistance to increasing concentrations of this compound was determined for five unrelated clinical isolates of A. baumannii with different degrees of susceptibility. As shown in Table 3, the frequency of sulbactam resistance was quite low for all strains tested, occurring at ∼4 × 10−9 at only 2× and 4× MIC for two of the strains (ARC2058 and ATCC 17978) and at ∼2 × 10−9 at 8× MIC for ARC2058. The resistance frequency was below the limit of detection at higher concentrations for these two strains and at all concentrations tested for the remaining strains (ATCC 19606T, ARC5468, and ARC2461).
TABLE 3.
Frequencies of resistance to sulbactam among susceptible A. baumannii strains at the indicated concentrations
| Strain | Sulbactam MIC (μg/ml) | Frequency of resistance at: |
|||
|---|---|---|---|---|---|
| 2× MIC | 4× MIC | 8× MIC | 16× MIC | ||
| ARC2058 | 1 | 4.41E−09 | 1.39E−09 | 2.11E−09 | <4.15E−10 |
| ATCC 17978 | 1 | 8.50E−09 | 8.10E−09 | <4.05E−10 | 4.05E−10 |
| ATCC 19606T | 1 | <4.15E−10 | <4.15E−10 | <4.15E−10 | <4.15E−10 |
| ARC5468 | 4 | <4.15E−10 | <4.15E−10 | <4.15E−10 | <4.15E−10 |
| ARC2461 | 16 | <4.15E−10 | <4.15E−10 | <4.15E−10 | <4.15E−10 |
To determine the genetic basis of resistance, the entire genomes of several stably resistant colonies from ARC2058 and the only stably resistant colony isolated from ATCC 17978 were sequenced and compared with those of their parental strains. As shown in Table 4, sulbactam resistance at 4× MIC mapped to pbp3 in both strain backgrounds, resulting in a serine-to-threonine substitution at position 390 in the resistant ATCC 17978 mutant and a serine-to-phenylalanine substitution at position 395 in one of the resistant ARC2058 mutants. Both of these mutations are at or near the sulbactam binding site of PBP3 (18). The resistant ARC2058 pbp3 mutant strain also had a frameshift mutation in a hypothetical protein that has similarity to a protein that belongs to the peptidase C13 family. Both of these strains with pbp3 mutations showed high levels of resistance to sulbactam, with MICs increased 64-fold and 32-fold, respectively. Neither of these sulbactam-resistant mutants was cross-resistant to other classes of drugs tested (i.e., aztreonam, ceftazidime, meropenem, ciprofloxacin, and colistin). Examination by fluorescence microscopy revealed that both of the PBP3 mutant cell types were much more rod-shaped than the parent strains in both backgrounds (Fig. 2F and G). However, only the resistant ARC2058 pbp3 mutant formed “spaghetti-like” filaments (Fig. 2H) when treated with 0.5× MIC of sulbactam. The resistant ATCC 17978 pbp3 mutant morphology did not change upon exposure to sulbactam (data not shown). Despite multiple attempts, it was not possible to determine whether the difference in morphology between these two mutants upon exposure to sulbactam was due to the additional disrupted peptidase gene of ARC2058, because both types of mutations were found to be highly recalcitrant to backcrossing into isogenic backgrounds.
TABLE 4.
Genetic contents and antibiotic susceptibilities of A. baumannii sulbactam-resistant mutants
| Strain | Concn at which mutant was isolated | Mutation | MIC (μg/ml)a |
|||||
|---|---|---|---|---|---|---|---|---|
| SUL | ATM | CAZ | MEM | CIP | CST | |||
| ATCC 17978 | ||||||||
| Wild-type | NA | NA | 1 | 16 | 16 | 0.25 | 0.25 | 0.25 |
| 4-2 | 4× MIC | PBP3 (S390T) | 64 | 32 | 8 | 0.125 | 0.25 | 0.25 |
| ARC2058 | ||||||||
| Wild-type | NA | NA | 1 | 32 | 8 | 0.25 | 0.25 | 0.5 |
| 2-16 | 2× MIC | GalE (E192K) | 4 | 32 | 8 | 0.0.5 | 0.0.5 | 2 |
| 4-2 | 4× MIC | PBP3 (S395F)b | 32 | 32 | 8 | 0.5 | 0.5 | 0.5 |
| 4-28 | 4× MIC | l,d-Transpeptidasec | 4 | 32 | 8 | 0.25 | 0.25 | 1 |
| 4-29 | 4× MIC | MraY (T23I) | 8 | 32 | 16 | 0.5 | 0.5 | 1 |
| 8-18 | 8× MIC | RpoC (R803H) | 8 | 128 | 64 | 0.5 | 0.5 | 0.5 |
SUL, sulbactam; ATM, aztreonam; CAZ, ceftazidime; MEM, meropenem; CIP, ciprofloxacin; CST, colistin; NA, not applicable.
Also contains a frameshift mutation in a peptidase C13 family protein at Gly50.
L20fs:Δ82 bp.
Several other strains with lower levels of resistance to sulbactam in the ARC2058 background were found to have mutations in genes related to either cell wall metabolism or stress responses, including an E192K mutation in GalE (UDP-glucose 4-epimerase), a frameshift mutation in l,d-transpeptidase, a T23I mutation in MraY (phospho-N-acetylmuramoyl-pentapeptide transferase), and an R803H mutation in RpoC (the β′ subunit of RNA polymerase) (Table 4). None of these mutants had distinguishing morphological changes, compared to their wild-type parent strains, in examinations by fluorescence microscopy (data not shown). Of these non-pbp3 mutants, only the rpoC mutant conferred significant cross-resistance to aztreonam and ceftazidime, and not to the other compounds tested (Table 4).
Sulbactam resistance confers a fitness penalty in A. baumannii.
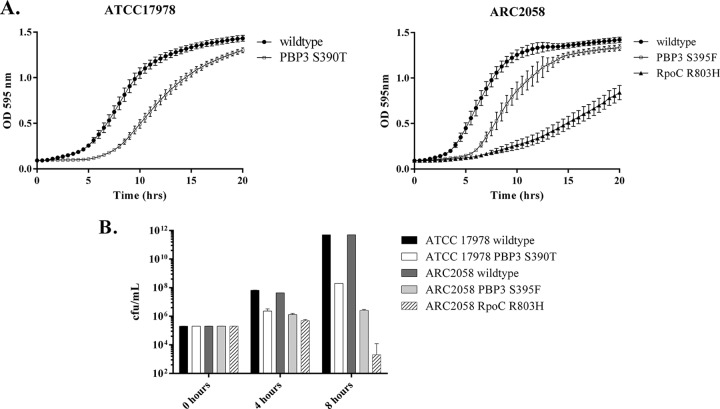
The proliferation rates of the mutants described above were evaluated in MHB-II, to determine whether sulbactam resistance affects the physiology of A. baumannii. Although the mutants with low-level resistance related to cell wall function did not show any growth deficiency (data not shown), both pbp3 and rpoC mutant strains had significantly longer doubling times than their wild-type parents during the lag phase (Fig. 3A). These strains were also more attenuated with respect to fitness when grown in freshly harvested murine blood (Fig. 3B), suggesting that their ability to cause infection may also be greatly reduced. Overall, these data suggest that sulbactam resistance mapping to pbp3 or rpoC may confer a fitness penalty in A. baumannii.
FIG 3.
Sulbactam-resistant A. baumannii attenuation in growth. Wild-type or sulbactam-resistant mutant strains were incubated at 37°C with shaking, either in MHB-II (A) or in freshly harvested, pooled, mouse blood in a 5% CO2 incubator (B). Samples were removed at the indicated times, and bacterial proliferation was measured by performing colony counts of 2-fold serial dilutions of these samples on blood agar, after overnight incubation at 37°C. Results shown are averages of at least three independent experiments.
DISCUSSION
What drives anti-Acinetobacter whole-cell activity?
The results presented above clearly show that sulbactam preferentially binds A. baumannii PBP1 orthologs and PBP3 over PBP2, using both gel-based and real-time fluorescence anisotropy competition assays (Fig. 1 and Table 1). However, both methods also reveal that sulbactam's rate of acylation of these proteins is considerably lower than that of the reference β-lactams and is similar to its low activity against P. aeruginosa orthologs. Therefore, the whole-cell antibacterial activity of sulbactam in A. baumannii is not due to greater affinity for target enzymes, compared to other strains. Although it is possible that differences in the outer membrane of A. baumannii allow for greater compound uptake in this organism than in species such as P. aeruginosa, it is worth noting that sulbactam has no activity against P. aeruginosa efflux-deficient strains and none of the resistance mutations that were sequenced mapped to outer membrane porins or transporters (data not shown). Another possible explanation is that sulbactam simultaneously inhibits other targets, such as low-molecular-weight PBPs (which were not reproducibly detected in the gel-based competition assays), to a sufficient extent that whole-cell activity is obtained. The latter hypothesis is supported by the results of the resistance studies described herein, in which resistance, although low, mapped to other genes related to cell wall biosynthesis besides pbp3, with concomitant slight increases in sulbactam MICs (Table 4).
While the morphological changes in A. baumannii upon exposure to sulbactam described above suggest that pbp3 inhibition does indeed occur, further investigation of the relative contribution of this enzyme inhibition to overall whole-cell killing could potentially be accomplished using more sophisticated microscopic approaches, such as direct detection of target binding in situ with fluorescently labeled derivatives, as described by Kocaoglu and colleagues (46). Alternatively, chemical proteomics, a recently developed platform that allows the identification of all proteins that specifically interact with small molecules of interest (47), may be a useful approach to further define the targets of inhibition for sulbactam.
Resistance studies.
The results presented above show that spontaneous resistance in susceptible strains is rare, with high-level resistance mapping to pbp3 at a fitness cost to the organism. Although these mutants were generated in a laboratory setting, no preexisting pbp3 mutations in the sulbactam-resistant clinical isolates listed in Table 2 were found, supporting a potential fitness penalty associated with these mutations. In contrast, low-level resistance mapped to genes associated with cell wall biosynthesis (galE, mraY, or l,d-transpeptidase) or stress responses (rpoC), with a fitness penalty associated with the latter mechanism (Fig. 3). Analysis by sequence alignment of the R805H mutation shows that it lies within the conserved, C-terminal, DNA binding region of the β′ subunit of RNA polymerase. This is consistent with a report describing a similar mutation that confers indirect resistance to cephalosporins in Bacillus subtilis (48). Taken together, these results suggest that sulbactam resistance in A. baumannii clinical isolates is multifactorial, as is the case for other β-lactams in nonfermenting Gram-negative pathogens (49). In summary, the broad range of anti-Acinetobacter activities of sulbactam against contemporary clinical isolates and the low frequency of spontaneous resistance as shown here, plus its previously demonstrated potentiation of other antibiotics (25–28), support further investigation into the potential of sulbactam for the treatment of serious MDR A. baumannii infections, as part of a multidrug regimen.
ACKNOWLEDGMENTS
This work was fully funded by AstraZeneca.
We acknowledge the following individuals for their help with this work: Michele Johnstone for help with resistance studies, Sarah Patey for help with MIC determinations, Jim Whiteaker for sample processing for whole-genome sequencing, Lisa Aschenbrenner for help with fluorescence microscopy, and Lakshmi Miller-Vedam for help with fitness evaluations. We are also grateful to Patricia Bradford for her helpful comments on the manuscript.
REFERENCES
- 1.Daniels TL, Deppen S, Arbogast PG, Griffin MR, Schaffner W, Talbot TR. 2008. Mortality rates associated with multidrug-resistant Acinetobacter baumannii infection in surgical intensive care units. Infect Control Hosp Epidemiol 29:1080–1083. doi: 10.1086/591456. [DOI] [PubMed] [Google Scholar]
- 2.Perez F, Ponce-Terashima R, Adams MD, Bonomo RA. 2011. Are we closing in on an “elusive enemy”? The current status of our battle with Acinetobacter baumannii. Virulence 2:86–90. doi: 10.4161/viru.2.2.15748. [DOI] [PubMed] [Google Scholar]
- 3.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y-T, Kuo S-C, Yang S-P, Lin Y-T, Tseng F-C, Chen T-L, Fung C-P. 2012. Impact of appropriate antimicrobial therapy on mortality associated with Acinetobacter baumannii bacteremia: relation to severity of infection. Clin Infect Dis 55:209–215. doi: 10.1093/cid/cis385. [DOI] [PubMed] [Google Scholar]
- 5.Bergogne-Bérézin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang M, Hu Z, Hu F. 2012. Nosocomial meningitis caused by Acinetobacter baumannii: risk factors and their impact on patient outcomes and treatments. Future Microbiol 7:787–793. doi: 10.2217/fmb.12.42. [DOI] [PubMed] [Google Scholar]
- 7.Peleg AY, de Breij A, Adams MD, Cerqueira GM, Mocali S, Galardini M, Nibbering PH, Earl AM, Ward DV, Paterson DL, Seifert H, Dijkshoorn L. 2012. The success of Acinetobacter species: genetic, metabolic and virulence attributes. PLoS One 7:e46984. doi: 10.1371/journal.pone.0046984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Özgür ES, Horasan ES, Karaca K, Ersöz G, Naycı Atış S, Kaya A. 2014. Ventilator-associated pneumonia due to extensive drug-resistant Acinetobacter baumannii: risk factors, clinical features, and outcome. Am J Infect Control 42:206–208. doi: 10.1016/j.ajic.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Chaari A, Mnif B, Bahloul M, Mahjoubi F, Chtara K, Turki O, Gharbi N, Chelly H, Hammami A, Bouaziz M. 2013. Acinetobacter baumannii ventilator-associated pneumonia: epidemiology, clinical characteristics and prognosis factors. Int J Infect Dis 17:e1225–e1228. doi: 10.1016/j.ijid.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Visca P, Seifert H, Towner KJ. 2011. Acinetobacter infection: an emerging threat to human health. IUBMB Life 63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 11.Davis KA, Moran KA, McAllister CK, Gray PJ. 2005. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg Infect Dis 11:1218–1224. doi: 10.3201/1108.050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Shea MK. 2012. Acinetobacter in modern warfare. Int J Antimicrob Agents 39:363–375. doi: 10.1016/j.ijantimicag.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Evans BA, Hamouda A, Amyes SGB. 2013. The rise of carbapenem-resistant Acinetobacter baumannii. Curr Pharm Des 19:223–238. doi: 10.2174/138161213804070285. [DOI] [PubMed] [Google Scholar]
- 14.Singh H, Thangaraj P, Chakrabarti A. 2013. Acinetobacter baumannii: a brief account of mechanisms of multidrug resistance and current and future therapeutic management. J Clin Diagn Res 7:2602–2605. doi: 10.7860/JCDR/2013/6337.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durante-Mangoni E, Utili R, Zarrilli R. 2014. Combination therapy in severe Acinetobacter baumannii infections: an update on the evidence to date. Future Microbiol 9:773–789. doi: 10.2217/fmb.14.34. [DOI] [PubMed] [Google Scholar]
- 16.Adnan S, Paterson DL, Lipman J, Roberts JA. 2013. Ampicillin/sulbactam: its potential use in treating infections in critically ill patients. Int J Antimicrob Agents 42:384–389. doi: 10.1016/j.ijantimicag.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi JA, Gill MA. 1988. Sulbactam: a beta-lactamase inhibitor. Clin Pharm 7:37–51. [PubMed] [Google Scholar]
- 18.Papp-Wallace KM, Senkfor B, Gatta J, Chai W, Taracila MA, Shanmugasundaram V, Han S, Zaniewski RP, Lacey BM, Tomaras AP, Skalweit MJ, Harris ME, Rice LB, Buynak JD, Bonomo RA. 2012. Early insights into the interactions of different β-lactam antibiotics and β-lactamase inhibitors against soluble forms of Acinetobacter baumannii PBP1 and Acinetobacter sp. PBP3. Antimicrob Agents Chemother 56:5687–5692. doi: 10.1128/AAC.01027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban C, Go E, Mariano N, Rahal JJ. 1995. Interaction of sulbactam, clavulanic acid and tazobactam with penicillin-binding proteins of imipenem-resistant and -susceptible Acinetobacter baumannii. FEMS Microbiol Lett 125:193–197. doi: 10.1111/j.1574-6968.1995.tb07357.x. [DOI] [Google Scholar]
- 20.Wood GC, Scott DH, Martin AC, Timothy CF, Bradley AB. 2002. Comparison of ampicillin-sulbactam and imipenem-cilastatin for the treatment of Acinetobacter ventilator-associated pneumonia. Clin Infect Dis 34:1425–1430. doi: 10.1086/340055. [DOI] [PubMed] [Google Scholar]
- 21.Jellison TK, McKinnon PS, Rybak MJ. 2001. Epidemiology, resistance, and outcomes of Acinetobacter baumannii bacteremia treated with imipenem-cilastatin or ampicillin-sulbactam. Pharmacotherapy 21:142–148. doi: 10.1592/phco.21.2.142.34114. [DOI] [PubMed] [Google Scholar]
- 22.Betrosian AP, Frantzeskaki F, Xanthaki A, Douzinas EE. 2008. Efficacy and safety of high-dose ampicillin/sulbactam vs. colistin as monotherapy for the treatment of multidrug resistant Acinetobacter baumannii ventilator-associated pneumonia. J Infect 56:432–436. doi: 10.1016/j.jinf.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Levin AS, Levy CE, Manrique AEI, Medeiros EAS, Costa SF. 2003. Severe nosocomial infections with imipenem-resistant Acinetobacter baumannii treated with ampicillin/sulbactam. Int J Antimicrob Agents 21:58–62. doi: 10.1016/S0924-8579(02)00276-5. [DOI] [PubMed] [Google Scholar]
- 24.Jones RN, Flonta M, Gurler N, Cepparulo M, Mendes RE, Castanheira M. 2014. Resistance surveillance program report for selected European nations (2011). Diagn Microbiol Infect Dis 78:429–436. doi: 10.1016/j.diagmicrobio.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Santimaleeworagun W, Wongpoowarak P, Chayakul P, Pattharachayakul S, Tansakul P, Garey KW. 2011. In vitro activity of colistin or sulbactam in combination with fosfomycin or imipenem against clinical isolates of carbapenem-resistant Acinetobacter baumannii producing OXA-23 carbapenemases. Southeast Asian J Trop Med Public Health 42:890–900. [PubMed] [Google Scholar]
- 26.Pei G, Mao Y, Sun Y. 2012. In vitro activity of minocycline alone and in combination with cefoperazone-sulbactam against carbapenem-resistant Acinetobacter baumannii. Microb Drug Resist 18:574–577. doi: 10.1089/mdr.2012.0076. [DOI] [PubMed] [Google Scholar]
- 27.Poulikakos P, Tansarli GS, Falagas ME. 2014. Combination antibiotic treatment versus monotherapy for multidrug-resistant, extensively drug-resistant, and pandrug-resistant Acinetobacter infections: a systematic review. Eur J Clin Microbiol Infect Dis 33:1675–1685. doi: 10.1007/s10096-014-2124-9. [DOI] [PubMed] [Google Scholar]
- 28.Kalin G, Alp E, Akin A, Coskun R, Doganay M. 2014. Comparison of colistin and colistin/sulbactam for the treatment of multidrug resistant Acinetobacter baumannii ventilator-associated pneumonia. Infection 42:37–42. doi: 10.1007/s15010-013-0495-y. [DOI] [PubMed] [Google Scholar]
- 29.Kempf M, Djouhri-Bouktab L, Brunel J-M, Raoult D, Rolain J-M. 2012. Synergistic activity of sulbactam combined with colistin against colistin-resistant Acinetobacter baumannii. Int J Antimicrob Agents 39:180–181. doi: 10.1016/j.ijantimicag.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Fernández-Cuenca F, Martínez-Martínez L, Conejo MC, Ayala JA, Perea EJ, Pascual A. 2003. Relationship between β-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J Antimicrob Chemother 51:565–574. doi: 10.1093/jac/dkg097. [DOI] [PubMed] [Google Scholar]
- 31.Chiu C-H, Lee H-Y, Tseng L-Y, Chen C-L, Chia J-H, Su L-H, Liu S-Y. 2010. Mechanisms of resistance to ciprofloxacin, ampicillin/sulbactam and imipenem in Acinetobacter baumannii clinical isolates in Taiwan. Int J Antimicrob Agents 35:382–386. doi: 10.1016/j.ijantimicag.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Krizova L, Poirel L, Nordmann P, Nemec A. 2013. TEM-1 β-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J Antimicrob Chemother 68:2786–2791. doi: 10.1093/jac/dkt275. [DOI] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—9th ed. CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Shapiro AB, Gu R-F, Gao N, Livchak S, Thresher J. 2013. Continuous fluorescence anisotropy-based assay of BOCILLIN FL penicillin reaction with penicillin binding protein 3. Anal Biochem 439:37–43. doi: 10.1016/j.ab.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro AB, Gao N, Gu R-F, Thresher J. 2014. Fluorescence anisotropy-based measurement of Pseudomonas aeruginosa penicillin-binding protein 2 transpeptidase inhibitor acylation rate constants. Anal Biochem 463:15–22. doi: 10.1016/j.ab.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Corbella X, Ariza J, Ardanuy C, Vuelta M, Tubau F, Sora M, Pujol M, Gudiol F. 1998. Efficacy of sulbactam alone and in combination with ampicillin in nosocomial infections caused by multiresistant Acinetobacter baumannii. J Antimicrob Chemother 42:793–802. doi: 10.1093/jac/42.6.793. [DOI] [PubMed] [Google Scholar]
- 37.Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. 1999. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother 43:1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spratt BG. 1977. The mechanism of action of mecillinam. J Antimicrob Chemother 3:13–19. [DOI] [PubMed] [Google Scholar]
- 39.Hayes MV, Orr DC. 1983. Mode of action of ceftazidime: affinity for the penicillin-binding proteins of Escherichia coli K12, Pseudomonas aeruginosa and Staphylococcus aureus. J Antimicrob Chemother 12:119–126. [DOI] [PubMed] [Google Scholar]
- 40.Hashizume T, Ishino F, Nakagawa J, Tamaki S, Matsuhashi M. 1984. Studies on the mechanism of action of imipenem (N-formimidoylthienamycin) in vitro: binding to the penicillin-binding proteins (PBPs) in Escherichia coli and Pseudomonas aeruginosa, and inhibition of enzyme activities due to the PBPs in E. coli. J Antibiot (Tokyo) 37:394–400. doi: 10.7164/antibiotics.37.394. [DOI] [PubMed] [Google Scholar]
- 41.Buijs J, Dofferhoff ASM, Mouton JW, Wagenvoort JHT, Van Der Meer JWM. 2008. Concentration-dependency of β-lactam-induced filament formation in Gram-negative bacteria. Clin Microbiol Infect 14:344–349. doi: 10.1111/j.1469-0691.2007.01940.x. [DOI] [PubMed] [Google Scholar]
- 42.Trautmann M, Heinemann M, Zick R, Möricke A, Seidelmann M, Berger D. 1998. Antibacterial activity of meropenem against Pseudomonas aeruginosa, including antibiotic-induced morphological changes and endotoxin-liberating effects. Eur J Clin Microbiol Infect Dis 17:754–760. doi: 10.1007/s100960050180. [DOI] [PubMed] [Google Scholar]
- 43.Michalopoulos A, Falagas ME. 2010. Treatment of Acinetobacter infections. Expert Opin Pharmacother 11:779–788. doi: 10.1517/14656561003596350. [DOI] [PubMed] [Google Scholar]
- 44.Levin AS. 2002. Multidrug resistant Acinetobacter infections: a role for sulbactam combinations in overcoming an emerging worldwide problem. Clin Microbiol Infect 8:144–153. doi: 10.1046/j.1469-0691.2002.00415.x. [DOI] [PubMed] [Google Scholar]
- 45.Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. 2004. In vitro activities of the β-lactamase inhibitors clavulanic acid, sulbactam, and tazobactam alone or in combination with β-lactams against epidemiologically characterized multidrug-resistant Acinetobacter baumannii strains. Antimicrob Agents Chemother 48:1586–1592. doi: 10.1128/AAC.48.5.1586-1592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kocaoglu O, Calvo RA, Sham L-T, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE. 2012. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem Biol 7:1746–1753. doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rix U, Superti-Furga G. 2009. Target profiling of small molecules by chemical proteomics. Nat Chem Biol 5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 48.Lee YH, Nam KH, Helmann JD. 2013. A mutation of the RNA polymerase β′ subunit (rpoC) confers cephalosporin resistance in Bacillus subtilis. Antimicrob Agents Chemother 57:56–65. doi: 10.1128/AAC.01449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock REW, Martínez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to β-lactam antibiotics. Antimicrob Agents Chemother 54:4159–4167. doi: 10.1128/AAC.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]