Abstract
Klebsiella pneumoniae, a Gram-negative bacterium, is normally associated with pneumonia in patients with weakened immune systems. However, it is also a prevalent nosocomial infectious agent that can be found in infected surgical sites and combat wounds. Many of these clinical strains display multidrug resistance. We have worked with a clinical strain of K. pneumoniae that was initially isolated from a wound of an injured soldier. This strain demonstrated resistance to many commonly used antibiotics but sensitivity to carbapenems. This isolate was capable of forming biofilms in vitro, contributing to its increased antibiotic resistance and impaired clearance. We were interested in determining how sublethal concentrations of carbapenem treatment specifically affect K. pneumoniae biofilms both in morphology and in genomic expression. Scanning electron microscopy showed striking morphological differences between untreated and treated biofilms, including rounding, blebbing, and dimpling of treated cells. Comparative transcriptome analysis using RNA sequencing (RNA-Seq) technology identified a large number of open reading frames (ORFs) differentially regulated in response to carbapenem treatment at 2 and 24 h. ORFs upregulated with carbapenem treatment included genes involved in resistance, as well as those coding for antiporters and autoinducers. ORFs downregulated included those coding for metal transporters, membrane biosynthesis proteins, and motility proteins. Quantitative real-time PCR validated the general trend of some of these differentially regulated ORFs. Treatment of K. pneumoniae biofilms with sublethal concentrations of carbapenems induced a wide range of phenotypic and gene expression changes. This study reveals some of the mechanisms underlying how sublethal amounts of carbapenems could affect the overall fitness and pathogenic potential of K. pneumoniae biofilm cells.
INTRODUCTION
Antibiotic resistance and tolerance are common impediments to healing in cases of chronic infections (1). The bacteria implicated in causing chronic infections tend to be those able to persist in the patient in the form of biofilms (2–4). Biofilms are communities of microorganisms (fungal and/or bacterial) that are attached to surfaces and exist within self-produced extracellular polymeric substances (EPSs) (5, 6). Biofilms can form on a multitude of biotic and abiotic surfaces within the body, including implanted devices, such as artificial heart valves and catheters (5, 7). As biofilms are sessile, the microorganisms housed within are phenotypically different from their planktonic counterparts and are more resistant to antibiotics and clearance than those found in a free-living state (7–10). Biofilms are involved in at least 65% of all chronic bacterial infections, causing a wide variety of disease manifestations, including urinary tract infections (UTIs), endocarditis, and catheter infections (7, 8, 11, 12). Thus, identification of compounds that are effective against biofilms is of the utmost importance.
One challenge in treating biofilms is their increased antimicrobial resistance or tolerance. The structure of the biofilm itself can prevent antimicrobial penetration into the matrix, thus preventing contact with the cells (5, 6). The cells within a biofilm are generally less metabolically active than planktonic cells and therefore significantly less sensitive to mechanisms of action by many antimicrobials targeting synthesis of macromolecules or metabolic pathways (13). A percentage of cells within a biofilm may be persister cells, which are transiently antibiotic tolerant without the concomitant genetic changes seen in antimicrobial resistance (14). Furthermore, there is an increasing amount of antibiotic resistance seen in many strains of Gram-positive and Gram-negative bacteria as the results of increased expression of efflux pumps, mutations to the target site, and production of enzymes to inactivate the drug.
Klebsiella pneumoniae is a Gram-negative bacterium commonly found in soil and as normal human flora (15). However, K. pneumoniae biofilms can be associated with respiratory and urinary tract infections, especially in individuals with a weakened immune system (7, 8). K. pneumoniae also has emerging importance as a pathogen of surgical sites and chronic wounds (8, 11). Increasingly, multidrug-resistant (MDR) strains of K. pneumoniae are being isolated from wounded soldiers returning from combat (1, 7, 16, 17). MDR K. pneumoniae is also found in conjunction with other Gram-negative bacteria in nosocomial wound infections, especially those associated with burns (18).
The increasing antibiotic resistance seen in K. pneumoniae, including extended-spectrum β-lactamase (ESBL)- and, more recently, carbapenemase-producing strains is a source of concern in the medical community (19–21). Although carbapenem resistance is emerging, this antibiotic is one of the more effective antibiotics to combat bacterial infections. As such, we wanted to determine the effects of various clinically relevant carbapenems on biofilm cultures of K. pneumoniae in order to understand how antibiotic therapy might alter the gene expression of K. pneumoniae and whether these changes could decrease the pathogenicity or fitness of nosocomial pathogens. Previous work from our laboratory has shown that treatment of in vitro K. pneumoniae biofilms with low concentrations of imipenem (≥3.12 μg/ml) caused dramatic changes in morphology without a significant decrease in cell viability (7). Higher concentrations of imipenem (200 to 1,000 μg/ml) significantly reduced biofilm viability and therefore led to improved healing in the rabbit ear biofilm-infected wound model (7).
For this study, we wanted to elucidate the effects of carbapenems on K. pneumoniae biofilms independent of significant killing. To that end, we treated K. pneumoniae biofilms with sublethal concentrations of various carbapenems, resulting in remarkable morphological changes in addition to highly altered gene expression profiles. A greater understanding of mechanisms underlying sublethal effects of carbapenem treatment of K. pneumoniae could aid in determining alternative treatment modalities targeting reduction of overall fitness or attenuation of pathogenicity of this important nosocomial human pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A clinical isolate of K. pneumoniae, BAMC 07-18, was used for all experiments (22). This strain is multidrug resistant, with demonstrated resistance to a number of drugs, including ampicillin, azithromycin, ceftazidime, chloramphenicol, clarithromycin, gentamicin, linezolid, and tetracycline (7). K. pneumoniae cultures were grown in Luria-Bertani broth (LB) at 37°C. For cell enumeration experiments, K. pneumoniae was plated on Trypticase soy agar plates supplemented with 5% sheep's blood (TSA+). Cells were enumerated using the ProtoCOL automated colony counter (Microbiology International, Frederick, MD).
Determination of MICs.
To determine the MICs of BAMC 07-18 for imipenem, meropenem, and doripenem, these antibiotics were serially diluted in LB in a 96-well plate. To prepare the inoculum, an overnight culture of K. pneumoniae cells was diluted 1:100 in fresh LB, grown to an optical density at a wavelength of 600 nm (OD600) of 0.5, diluted to an OD600 of 0.05 in fresh LB, and added to each well of the plate containing the test carbapenems. The plate was incubated at 37°C overnight with shaking. The lowest concentration of antibiotic with no visible cell growth as determined by optical density spectrophotometrically was defined as the MIC of the test antibiotic. Each antibiotic was tested in triplicate.
Biofilm formation.
Borosilicate glass discs with a diameter of 1.75 mm (Ace Glass, Inc., Vineland, NJ) were coated with 10 μg/ml type I collagen in phosphate-buffered saline (PBS) overnight at 4°C. The K. pneumoniae inoculum, prepared as described above, was suspended in PBS (OD600 of 0.05) and was incubated with the glass discs in a 24-well plate for 2 h at room temperature. Each well contained 1 ml of bacterial suspension and one glass disc. After attachment, the discs were rinsed, and 1 ml fresh LB was added to each well. To test the effect of carbapenems on inhibition of biofilm formation, the discs were incubated for 24 h at 37°C with imipenem (2 μg/ml), meropenem (2 μg/ml), or doripenem (1 μg/ml) in LB. The discs were rinsed and placed in 1 ml of PBS. Biofilms were detached by sonication using the Microson XL ultrasonic cell disruptor (Qsonica, LLC, Newtown, CT) at 7 W for 2 min, and the cells were plated on TSA+ for enumeration after overnight incubation at 37°C. To test the effect of carbapenems on preformed biofilms, the discs were first incubated at 37°C for 24 h after attachment to allow optimal biofilm formation. The spent medium was removed, and the discs were incubated for 2 or 24 h with the aforementioned antibiotics. Plating for enumeration expressed as CFU was performed as described above.
Scanning electron microscopy.
Biofilms on glass discs were fixed in 2.5% phosphate-buffered glutaraldehyde for 1 h at 4°C and dehydrated in a graded series of cold ethanol-water mixtures (10, 20, 30, 50, 70, 80, 90, 95, and 100% ethanol) for 6 min each. Samples were dehydrated by critical point drying (EMS 850; Electron Microscopy Science, Hatfield, PA) and coated with palladium-gold using a Hummer 6.2 sputter coater (Anatech USA, Hayward, CA). After processing, samples were observed with a Sigma VP40 field emission scanning electron microscope (Carl Zeiss, Inc., Germany) in high-vacuum mode at 2 kV. Cell length was measured using ImageJ software (23).
RNA extraction.
To prepare K. pneumoniae biofilms for isolation of RNA, wells of a 6-well plate were coated with 10 μg/ml type I collagen in PBS overnight at 4°C. The bacterial suspension (OD600 of 0.05 in PBS), prepared as described above, was added to each well of the 6-well plate. After 2 h of incubation at room temperature, the wells were rinsed to remove unattached K. pneumoniae cells, and LB was added. After overnight incubation at 37°C, the wells were rinsed and treated with imipenem as described previously. Following 2 or 24 h of treatment, biofilms were removed from the plates and preserved in RNAprotect (QIAgen, Valencia, CA). RNA was extracted using the RNeasy minikit (QIAgen) according to the manufacturer's instructions. Samples were then treated twice with TURBO DNase (Ambion, Grand Island, NY) according to the manufacturer's instructions to remove contaminating genomic DNA. RNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and analyzed for quality using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA).
RNA sequencing and read mapping.
Total K. pneumoniae RNA from untreated and imipenem-treated biofilms was sequenced by the Genome Sequencing Facility at the University of Texas Health Science Center San Antonio (UTHSCSA). Briefly, 2 to 5 μg total RNA was used for preparation of the strand-specific RNA transcriptome sequencing (RNA-Seq) library. rRNA was depleted from total RNA using the Ribo-Zero Magnetic kit (Epicentre, Madison, WI). The directional RNA-Seq library was developed using the NEXTflex directional RNA-Seq (dUTP-based) kit (Bioo Scientific, Austin, TX). Following purification, mRNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers followed by second-strand cDNA synthesis using DNA polymerase I and RNase H. These cDNA fragments went through an end repair process, the addition of a single A base, and ligation of the adapters. The products were further purified and enriched with PCR to create the final RNA-Seq library. Directionality was retained by adding dUTP during the second-strand synthesis step and subsequent cleavage of the uridine-containing strand using uracil DNA glycosylase.
RNA-Seq libraries were subjected to the quantification process and pooled for cBot amplification and a subsequent sequencing run with HiSeq 2000 platform (Illumina, San Diego, CA). After the sequencing run, de-multiplexing with CASAVA was employed to generate a FASTQ file for each sample.
Single-end nucleotide reads were mapped to the annotated draft genomic sequence of K. pneumoniae BAMC 07-18 (GenBank accession no. JRQE00000000) using the software Bowtie (24). The mapped reads were separated into the forward and reverse complement directions using the software tool SAMtools (25). The mapped reads on each strand were visualized in the JBrowse genome viewer (26) for sequencing quality.
Differential gene expression analysis.
Raw read counts for the K. pneumoniae genes were determined with a Perl script based on the mapped read profiles determined above. The read counts were subjected to the Bioconductor software package “DESeq” (27) to evaluate the differential expression for the genes between experiments. Two sequencing runs derived from two independently conducted experiments were used in the DESeq analysis. Genes with significantly up- or downregulated expression levels were subject to the functional and pathway analysis using the DAVID software (28, 29).
qRT-PCR.
RNA extracted from K. pneumoniae biofilms was converted to cDNA using the iScript Select cDNA synthesis kit (Bio-Rad, Hercules, CA). cDNA was subjected to quantitative real-time PCR (qRT-PCR) with SYBR green PCR master mix (Bio-Rad) with a final concentration of 0.3 μM oligonucleotides (Table 1) using the ABI Prism 7300 system (Applied Biosystems, Grand Island, NY). Gene expression levels of treated and untreated biofilms were compared for selected hits generated from RNA sequencing results. Data were normalized to expression levels of gyrA.
TABLE 1.
Oligonucleotides used in this study
| Namea | Sequence (5′→3′) |
|---|---|
| csiDF | GCTTTTCTTTCACCCCTTCC |
| csiDR | CTGCTCGTCGATCTTCATCA |
| cspAF | CCGGTAAAATGACTGGTATCG |
| cspAR | GCTTTCGATGGTGAAGGATA |
| cysPF | GACAAGCTGACCATCAAGCA |
| cysPR | ACCCATCGTCGAATAGAACG |
| gyrAF | TACGCGGTATACGACACCAT |
| gyrAR | CGATGGAACCAAAGTTACCC |
| ibrAF | CTATGACTGGAAAACCGCCG |
| ibrAR | AGGTTCCAGCACGTGATACA |
| mreCF | ACGGTCAACGACCCTTACAG |
| mreCR | ATGCTCAAGCTGGAGATCGT |
| pmrJF | TACGGCTGGGATATTCTGCT |
| pmrJR | CAGCAGGTAGCGGTTAAAGC |
| sitAF | GCAATGGGCTTAATCTCGAA |
| sitAR | ATGGTGCTTTCGCTGAAGAT |
| tauBF | AGATGCTGAAGAAGGTGGGG |
| tauBR | CGGTCTCATGCCACAGTTTC |
In the primer names, the last letter indicates the orientation: F, forward; R, reverse.
Statistical analysis.
The unpaired Student's t test (two-tailed) was used to analyze cell viability and cell length data. All statistical data were calculated using GraphPad Prism version 4.0 for Macintosh (GraphPad Prism Software, San Diego, CA). Statistical significance was accepted when P values were less than 0.05.
Microarray data accession number.
The RNA-Seq reads and the DESeq results have been deposited in NCBI's Gene Expression Omnibus (30) and are accessible through GEO Series accession no. GSE62045.
RESULTS
MICs.
In order to determine the sublethal concentration for use in treating biofilms without drastic loss of cell viability, we used a broth dilution method to determine the MIC of each of the carbapenem antibiotics used. The MIC for both imipenem and meropenem was 0.4 μg/ml. The MIC for doripenem was 0.2 μg/ml. Five times the MIC of each test carbapenem was used as the sublethal (sub-antibiofilm) dose level to study its effect on each biofilm.
Meropenem and doripenem but not imipenem inhibited biofilm formation in K. pneumoniae.
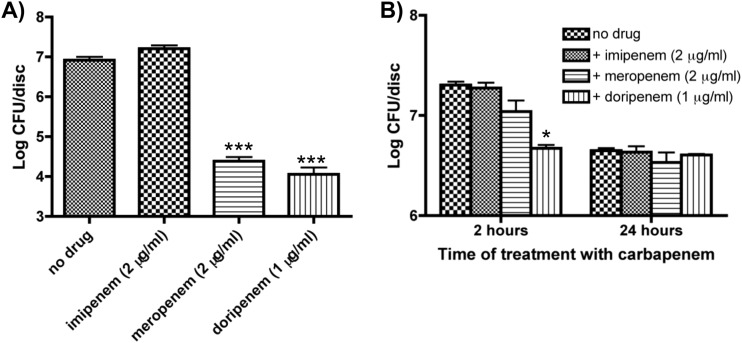
As shown in Fig. 1A, imipenem at 5× (2 μg/ml) its MIC had no significant effect on K. pneumoniae biofilm formation (untreated, 8.30 × 106 CFU/disc; imipenem, 9.26 × 106 CFU/disc; P = 0.76). In contrast, both meropenem (2.56 × 104 CFU/disc; P < 0.001) and doripenem (1.37 × 104 CFU/disc; P < 0.001) at 5× their MICs had negative effects on the formation of K. pneumoniae biofilms, reducing the viable cell count by over 99%.
FIG 1.
Effect of a sublethal concentration of carbapenems on biofilm formation and viability in K. pneumoniae. (A) To determine the effect of carbapenems on biofilm formation, the cells were grown for 24 h in Luria-Bertani liquid medium (LB) with 2 μg/ml imipenem/meropenem or 1 μg/ml doripenem. There was no significant effect seen on biofilm formation (P = 0.14) with imipenem; however, there was a significant effect seen with meropenem and doripenem. (B) To determine the effect of carbapenems on preformed biofilms, biofilms were grown for 24 h in LB. The discs were rinsed and placed in LB with the indicated carbapenems for 2 or 24 h. There was no significant effect on viability after 2 or 24 h of treatment (P = 0.31 and 0.59, respectively), with the exception of 2 h after treatment with doripenem. Data are representative of three independent experiments. *, P < 0.05; ***, P < 0.001.
High concentrations of carbapenems were effective in killing preformed 24-hour biofilms of K. pneumoniae.
Since we determined that 5× the MIC had a detrimental effect on biofilm formation, we decided to test the effect of the carbapenems on preformed biofilms. Preformed biofilms were treated with the aforementioned carbapenems at 5× their MICs for either 2 or 24 h (Fig. 1B). Following 2 h of treatment, doripenem caused a significant decrease in CFU/disc (P < 0.05), while imipenem and meropenem had no effect on cell viability within a preformed biofilm. Conversely, following 24 h of treatment with the various carbapenems, there were no significant decreases in the levels of CFU/disc.
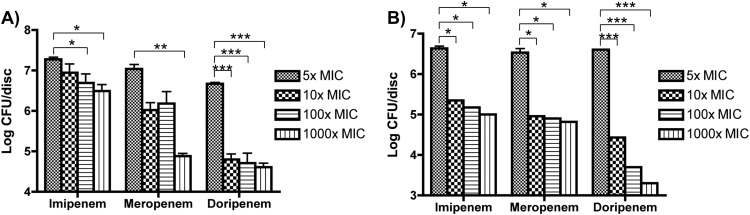
In order to determine if even higher levels of carbapenems could have a significant effect on the viability of K. pneumoniae in preformed biofilms, increased concentrations of carbapenems (10×, 100×, and 1,000× MIC) were used to treat biofilms for 2 and 24 h. We saw that using carbapenems at these higher levels caused significant reductions in biofilm viability after both 2 and 24 h of antibiotic treatment (Fig. 2); however, none of these antibiotics at even the highest levels was able to completely clear biofilms from the discs.
FIG 2.
Effect of increasing concentrations of carbapenems on biofilm viability in K. pneumoniae. Cells were grown for 24 h in LB and then incubated with the indicated fold increases of carbapenems over the MIC. (A) After 2 h of treatment, there were significant decreases in viability with 100× and 1,000× imipenem, 1,000× meropenem, and 10×, 100×, and 1,000× doripenem. (B) After 24 h of treatment, there were significant decreases in viability with all three drugs at all three increased concentrations. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Sub-antibiofilm levels of carbapenems caused transient changes of cell morphology in K. pneumoniae biofilms.
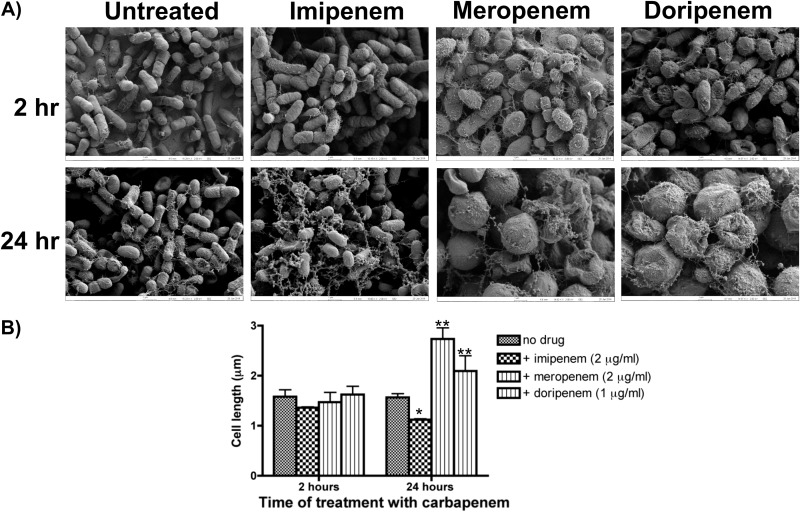
Although for the most part there were slight differences in the change of viability in response to treatment with carbapenems at 5× their MICs, there were striking differences in morphology when biofilms were visualized by scanning electron microscopy (SEM) after 2 or 24 h of treatment (Fig. 3A). Treated cells showed noticeable changes in cell morphology as early as 2 h after treatment. These changes included alterations in size and shape (most noticeable was rounding of biofilm cells) and the appearance of additional extracellular material in the treated versus untreated biofilms. We quantified the change in the size of treated cells by measuring cell length with ImageJ software (Fig. 3B). Despite visible changes in cell shape as revealed by SEM, there was no significant difference in cell length of biofilm cells treated for 2 h. In contrast, there were significant differences in cell length in biofilms treated with carbapenems for 24 h. Cells of imipenem-treated biofilms (1.12 μm) were significantly shorter than cells from untreated biofilms (1.57 μm; P < 0.05). Meropenem- and doripenem-treated cells were significantly larger and longer (2.73 and 2.09 μm, respectively) than cells in untreated biofilms (P < 0.05 for both treatments).
FIG 3.
A sublethal concentration of carbapenems alters the morphology and length of K. pneumoniae cells in a preformed biofilm. Biofilms were visualized using scanning electron microscopy. (A) Exposure of K. pneumoniae to a sublethal concentration of carbapenems for 24 h caused significant accumulation of material resembling cellular debris. Changes in cellular morphology started as early as 2 h after exposure to carbapenems. Images show 15,000× magnification and are representative of 3 independent experiments. (B) Following 24 h of imipenem treatment, there was significant cell shortening, while treatment with meropenem and doripenem led to significant cell lengthening compared to that in cells of an untreated biofilm. *, P < 0.05; **, P < 0.01.
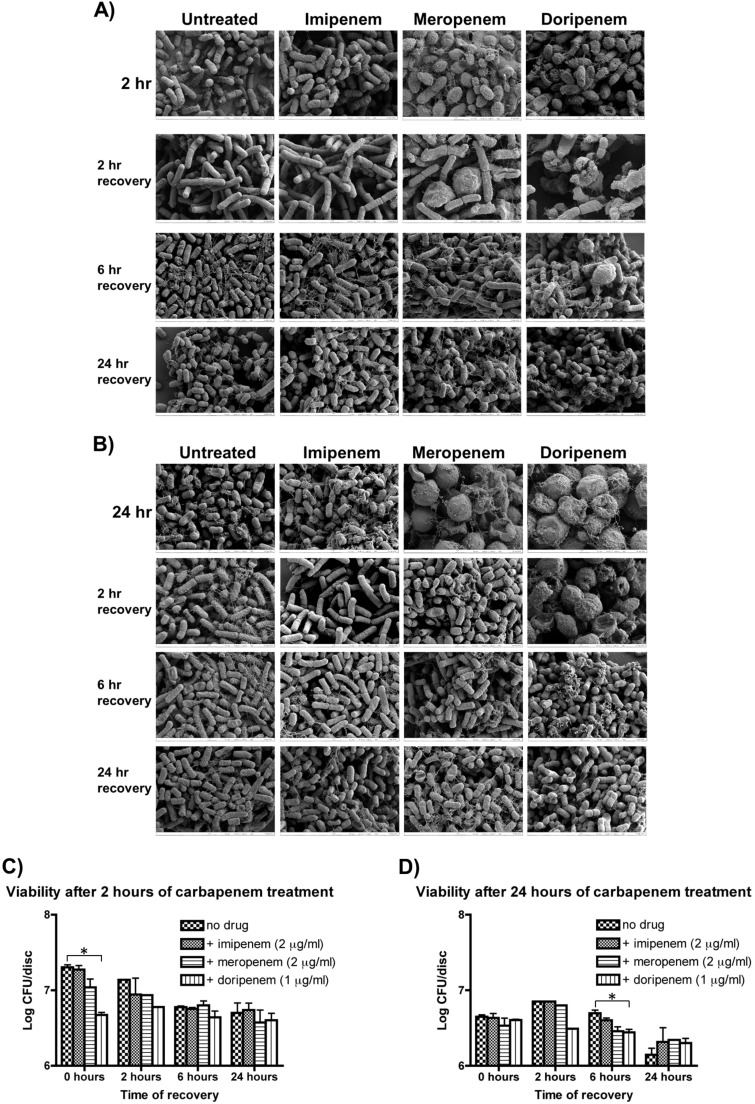
We also tested if the alteration in morphology was a transient effect by removing the antibiotics and allowing the cells to recover for periods of 2, 6, and 24 h. The cells were visualized using SEM and also plated for viability. Morphologically, the cells treated for 2 h with sublethal concentrations of carbapenems recovered after approximately 6 h in antibiotic-free medium (Fig. 4A), with no significant change in viability (Fig. 4C). Following 24 h of antibiotic treatment, a similar trend was seen, showing the overall morphology returning to normal after 6 h of recovery (Fig. 4B), with no significant change in viability (Fig. 4D).
FIG 4.
Recovery of K. pneumoniae following carbapenem treatment. Following 2 h (A and C) or 24 h (B and D) of treatment, biofilms were placed in fresh medium and allowed to recover for the indicated time periods. Biofilms were then visualized using SEM (A and B) or plated (C and D) to determine cell viability. SEM indicated a majority of cells had recovered normal cell morphology after 2 h in imipenem-treated biofilms, 6 h in meropenem-treated biofilms, and 24 h in doripenem-treated biofilms. In general, although there were morphological differences observed, there were not significant differences in viability in most treated biofilms, with the exception of doripenem-treated biofilm cells. *, P < 0.05.
Differential gene expression of K. pneumoniae biofilms in response to imipenem treatment.
RNA sequencing was performed on RNA extracted from K. pneumoniae biofilms treated for 2 h and 24 h with 2 μg/ml of imipenem. Two replicates of each experimental condition were performed, and the RNA was sequenced with the read numbers as follows: untreated, 15.4 and 10.6 million sequence reads; 2 h of treatment, 13.1 and 12.5 million sequence reads; and 24 h of treatment, 20.6 and 10.9 million reads. An average of 82% of the reads was successfully mapped to the K. pneumoniae strain BAMC 07-18 genome.
Comparison of 2 h of imipenem treatment to no treatment showed that there were 401 open reading frames (ORFs) upregulated (≥2-fold change; P < 0.05) and 257 ORFs downregulated (≥2-fold change; P < 0.05) out of the 5,409 ORFs predicted in the K. pneumoniae genome, representing 7.4% of ORFs upregulated and 4.7% downregulated (see Data Set S1 in the supplemental material). Following 24 h of imipenem treatment, there were 28 ORFs upregulated (0.5%) and 12 ORFs downregulated (0.2%) using the aforementioned criteria (see Data Set S2 in the supplemental material). When comparing cells treated for 24 h to those treated for 2 h, we found 217 ORFs (4.0%) upregulated and 308 ORFs (5.7%) downregulated (see Data Set S3 in the supplemental material). These results are summarized in Table 2. Considering treated versus untreated cells within the K. pneumoniae biofilm, the most dramatic changes in gene expression were seen following 2 h of treatment.
TABLE 2.
Differentially expressed ORFs organized by functional category
| ORF category by function(s) | No. (%) of ORFs with change in expression fora: |
|||||
|---|---|---|---|---|---|---|
| 2-h treatment vs no treatment |
24-h treatment vs no treatment |
24-h vs 2-h treatment |
||||
| Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | |
| Cellular processes and signaling | ||||||
| Cell cycle control, cell division, and chromosome partitioning | 1 (0.2) | 5 (1.9) | 2 (0.9) | 1 (0.3) | ||
| Cell wall, membrane, and envelope biogenesis | 14 (3.5) | 17 (6.6) | 1 (8.3) | 18 (8.3) | 10 (3.2) | |
| Cell motility | 3 (0.7) | 1 (0.4) | 1 (8.3) | 1 (0.5) | 3 (1.0) | |
| Posttranslational modification, protein turnover, and chaperones | 20 (5.0) | 10 (3.9) | 10 (4.6) | 14 (4.5) | ||
| Signal transduction mechanisms | 18 (4.5) | 8 (3.1) | 3 (25) | 2 (0.9) | 16 (5.2) | |
| Intracellular trafficking, secretion, and vesicular transport | 4 (1.0) | 4 (1.6) | 1 (8.3) | 3 (1.4) | 3 (1.0) | |
| Defense mechanisms | 4 (1.0) | 4 (1.6) | 3 (1.0) | |||
| Information storage and processing | ||||||
| Translation, ribosomal structure, and biogenesis | 3 (0.7) | 36 (14.0) | 29 (13.4) | 3 (1.0) | ||
| Transcription | 41 (10.0) | 11 (2.7) | 2 (7.1) | 1 (8.3) | 9 (4.1) | 33 (10.7) |
| Replication, recombination, and repair | 9 (2.2) | 9 (3.5) | 5 (2.3) | 3 (1.0) | ||
| Metabolism | ||||||
| Energy production and conversion | 47 (11.7) | 28 (10.9) | 6 (21.4) | 29 (13.4) | 41 (13.3) | |
| Amino acid transport and metabolism | 65 (16.2) | 21 (8.2) | 5 (17.9) | 2 (16.7) | 25 (11.5) | 43 (14.0) |
| Nucleotide transport and metabolism | 8 (2.0) | 18 (7.0) | 17 (7.8) | 7 (2.3) | ||
| Carbohydrate transport and metabolism | 55 (13.7) | 36 (14.0) | 3 (10.7) | 34 (15.7) | 40 (13.0) | |
| Coenzyme transport and metabolism | 7 (1.7) | 13 (5.1) | 1 (3.6) | 8 (3.7) | 7 (2.3) | |
| Lipid transport and metabolism | 24 (6.0) | 6 (2.3) | 4 (14.3) | 1 (8.3) | 3 (1.4) | 17 (5.5) |
| Inorganic ion transport and metabolism | 20 (5.0) | 18 (7.0) | 3 (10.7) | 1 (8.3) | 20 (9.2) | 20 (6.5) |
| Secondary metabolite biosynthesis, transport, and catabolism | 25 (6.2) | 2 (0.8) | 5 (17.9) | 1 (8.3) | 2 (0.9) | 17 (5.5) |
| Poorly characterized | ||||||
| General function prediction only | 19 (4.7) | 3 (1.2) | 1 (3.6) | 2 (0.9) | 11 (3.6) | |
| Function unknown | 65 (16.2) | 26 (10.1) | 6 (21.4) | 3 (25) | 21 (9.7) | 57 (18.5) |
Percentages total more than 100% due to ORFs belonging to multiple functional categories.
Imipenem treatment of K. pneumoniae biofilms caused upregulation of genes involved in transport and metabolism as well as protein turnover.
There was an overall trend of upregulation when looking at categories involving macromolecule transport and metabolism after cells were treated with imipenem. A large number of ORFs involved in amino acid transport and metabolism were upregulated, while comparatively fewer ORFs implicated in amino acid transport and metabolism were downregulated. ORFs involved in rRNA/tRNA processing and ribosome biosynthesis were generally downregulated. Although the protein biosynthetic machinery was downregulated, posttranslational modification, protein turnover, and chaperone ORFs were upregulated in response to imipenem treatment. There was also downregulation of genes involved in nucleotide and coenzyme biosynthesis. Lipid and fatty acid metabolism was commonly upregulated, as were the metabolism and transport of secondary metabolites.
There was a noticeable upregulation of genes involved in energy metabolism. Members and regulators of the glyoxylate cycle, pentose phosphate pathway, and the tricarboxylic acid (TCA) cycle were upregulated in response to imipenem treatment. ORFs involved in acetyl coenzyme A (acetyl-CoA) biosynthesis, formation of the succinate dehydrogenase complex, and cytochrome bo terminal oxidase also experienced significant upregulation in imipenem-treated biofilm cells. However, some ORFs involved in anaerobic respiration/fermentation were downregulated with imipenem treatment. These included genes essential for biosynthesis of fumarate reductase and dimethyl sulfoxide (DMSO) reductase. Also downregulated were cytochrome bd-I terminal oxidase ORFs and cytochrome c biogenesis ORFs.
Imipenem affected the expression of genes involved in cell wall/membrane biosynthesis of K. pneumoniae biofilm cells.
ORFs involved in peptidoglycan catabolism were upregulated, including bax and nlpD, with a concomitant downregulation of various ORFs involved in peptidoglycan biosynthesis, such as murDGP, mraYZ, uppP, and dacC, although two l,d-transpeptidase genes, erfK (31) and ynhG, responsible for peptidoglycan cross-linking were upregulated. Also upregulated were a number of ORFs implicated in altering morphology and responding to cell membrane and cell wall stress, such as the toxin gene vapC, bolA (coding for a morphogenic protein and member of the RpoS regulon), pspABCG, and spy. Conversely, genes coding for outer membrane (wbbM, wecFG, and wzyE) and lipid A (arcACT) biosynthetic proteins were downregulated. Associated with the decrease in peptidoglycan and outer membrane biosynthesis, cell division ORFs maf, mukF, and envC were downregulated in response to imipenem treatment.
Sublethal concentrations of imipenem activate the stress response in K. pneumoniae biofilm cells.
Of greatest interest were changes in expression levels of genes involved in the cellular stress response. ORFs representing heat shock (ibpAB and hspEQ) were upregulated, as were those involved in oxidative stress response (including fnr, sodAC, and sufABCDSE), phosphate starvation (psiF and phoH), carbon starvation (cstA and csiDE), and osmotic stress (osmBCEF). The master stress/stationary response regulator gene, rpoS, was upregulated, as was iraP, a gene coding for an RpoS stabilizer, and cspD, an rpoS-independent replication inhibitor gene induced in the stationary phase. Many ORFs implicated in protein repair or degradation of misfolded proteins (msrAB and cpxP) were upregulated in response to imipenem treatment, as were inhibitors of biofilm formation (bhsA, bssR, and ariR) and two autoinducers, lsrBF.
A number of stress response genes were also downregulated in response to imipenem treatment. The major chaperone gene hfq and a cold shock response DEAD box RNA helicase A gene, csdA/deaD, were downregulated, as were a few ORFs with roles in antibiotic resistance, including ampE (a repressor of the β-lactamase-encoding ampC gene), ermB, tetR, and a multidrug efflux pump gene, mdtC. A gene coding for a porin implicated in carbapenem sensitivity, oprD, was downregulated in imipenem-treated biofilm cells. Positive regulators of virulence, such as the sensor kinase genes baeS and barA, were also downregulated, as was comEC, a regulator of DNA uptake from the external environment. Finally, genes coding for a toxin (cvpA) and a holin (cidA) leading to cell lysis had decreased expression in biofilms treated with imipenem for 2 h.
Summary of differential gene expression following 24 h of imipenem treatment.
Much of the gene expression seems to have stabilized after 24 h of imipenem treatment when treated and untreated biofilms are compared. As there were very few ORFs differentially expressed in response to 24 h of imipenem treatment, the data do not show strong trends either way. Some of the energy metabolism genes were still upregulated, as were a few ORFs for amino acid, carbohydrate, and lipid metabolism and transport as well as acetyl-CoA biosynthesis. The two stress response genes showing continued significant upregulation after 24 h of imipenem treatment were the carbon starvation-induced gene, csiD, and the RpoS stabilizer gene, iraP. There were two stress response genes downregulated at 24 h, cpxP and a sigma factor gene generally induced in response to damaged or misfolded proteins, rpoE.
Comparison of gene expression levels between biofilms treated for 2 h and 24 h.
Although there was not an overall global alteration in gene expression levels when untreated biofilms were compared to those treated for 24 h, when we compared 24 h of treatment with 2 h of treatment, we observed that ORFs that were upregulated after 2 h showed a significant decrease in expression compared to a 24-h biofilm. Along those lines, we also noticed that ORFs that had been downregulated after 2 h compared to untreated biofilms experienced an upregulation following 24 h of imipenem treatment.
qRT-PCR analysis.
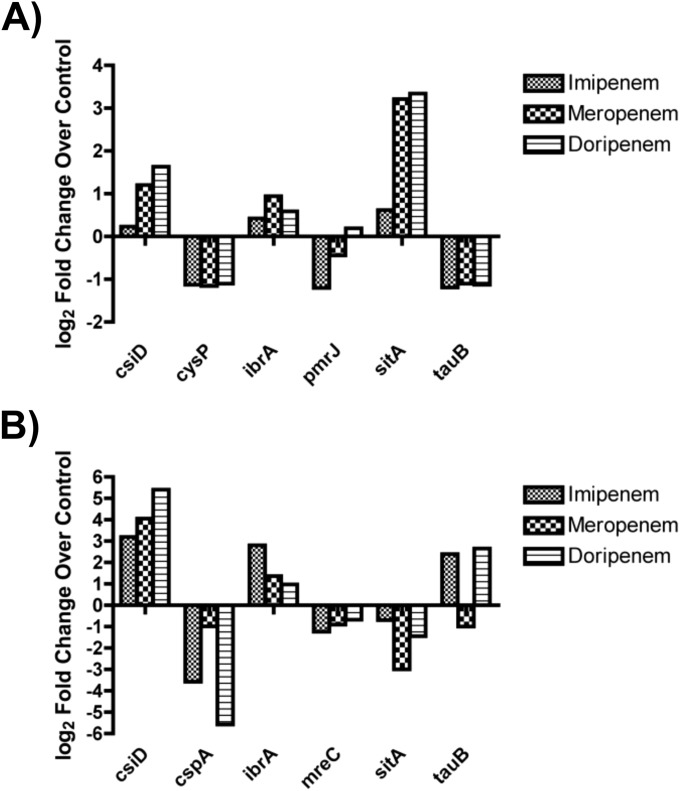
RNA converted to cDNA was used to perform quantitative real-time PCR (qRT-PCR) to verify some of the differentially expressed genes observed from RNA sequencing. Using RNA samples extracted from biofilms treated with carbapenems for 2 h, qRT-PCR using primers for csiD, ibrA, and sitA confirmed an upregulation of these ORFs compared to those in untreated biofilms. Similarly, qRT-PCR using primers for cysP, pmrJ (with the exception of doripenem-treated biofilms), and tauB showed a downregulation of the target ORFs. The results were consistent with the RNA-Seq data generated from imipenem-treated biofilms (Fig. 5A). Confirmation of expression trends was also seen when analyzing samples from biofilms treated for 24 h, with the exception of tauB in meropenem-treated biofilms (Fig. 5B).
FIG 5.
qRT-PCR confirms trends from RNA sequencing data. The remaining K. pneumoniae RNA from RNA-Seq and RNA isolated from meropenem- and doripenem-treated biofilms were converted to cDNA and amplified using qRT-PCR. qRT-PCR data confirmed expression trends from RNA-Seq data for a number of open reading frames following 2 h (A) and 24 h (B) of treatment with imipenem.
DISCUSSION
The emergence of increasingly multidrug-resistant (MDR) strains of K. pneumoniae necessitates the study of how currently effective therapies can be utilized to ameliorate virulence and eliminate infection without promoting the acquisition of a resistant phenotype. To that end, we studied the effects of sublethal concentrations of carbapenems on an ESBL-producing strain of K. pneumoniae, BAMC 07-18. For this study, we investigated three different carbapenems (imipenem, meropenem, and doripenem) to elucidate the pleiotropic effects of carbapenems at sublethal concentrations on a sensitive strain of K. pneumoniae. This study is the first of its kind to combine RNA-Seq analysis with SEM imaging of treated biofilms to examine the effects of sublethal concentrations of carbapenems on K. pneumoniae biofilms.
When K. pneumoniae biofilms were grown for 24 h and treated with increasing concentrations of carbapenems (up to 1,000× the MIC), we did not observe a significant decrease in viability until using 10× the MIC or higher (Fig. 2). For this reason, we chose to use 5× the MIC for all further experiments as the sublethal antibiofilm concentration for all test carbapenems since we could observe striking morphological changes without a killing effect (Fig. 3). Interestingly, even when treating biofilms with 1,000× the MIC of the most potent carbapenem available to us (doripenem), we did not observe total killing or clearance of the biofilm (Fig. 2). Since the levels of carbapenems used in this experiment would be much higher than in an in vivo situation (32–34), the results highlight the difficulty in treating established biofilm infections, regardless of whether or not the strain is resistant to the prescribed antibiotics.
Our previous studies showed that treatment of a preformed biofilm with imipenem in excess of the MIC did not significantly affect viability of the biofilm (7), we wanted to determine the effect of carbapenems on biofilm formation, mimicking a prophylaxis situation. We found that treatment with imipenem for 24 h had no effect on biofilm maturation of K. pneumoniae (Fig. 1A). In contrast, meropenem and doripenem did significantly decrease biofilm maturation; however, they did not completely kill cells that had already adhered to the disc surface (Fig. 1A). This is suggestive of the fact that even an immature biofilm (<2 h) is able to provide antibiotic tolerance to the cells within, again indicating the importance of prophylactic antibiotic therapy to prevent a biofilm from forming and the extraordinarily difficult task of clearance of recalcitrant biofilm infections once established.
Though we did not observe killing of K. pneumoniae biofilm cells with carbapenem treatment, we did see striking changes in cell morphology, including rounding, blebbing, and alteration of cell size (Fig. 3). We were curious as to whether this was a transient morphology, and if so, at what point the cells within the biofilm recovered. With both 2 h and 24 h of carbapenem treatment, after 2 h of recovery, the majority of imipenem- and meropenem-treated biofilms had returned to the untreated morphology. Near complete recovery of the cells was observed within 6 h of removal of the carbapenems. These observations via SEM were supported by the finding of no differences in CFU counts within the treated biofilm (Fig. 4). The data indicate that carbapenems lead to a transient change in morphology, and their removal allows the biofilms to quickly return to normal. In terms of an in vivo correlation, these data emphasize the importance of having consistent levels of antibiotic therapy, as the cells are able to recover rapidly, negating any benefit the antibiotics may have been providing.
Because we knew that carbapenems were having an effect on the cells without killing, we decided to use RNA-Seq to compare whole transcriptome profiles of K. pneumoniae biofilms treated with sublethal concentrations of imipenem for 2 and 24 h to those of untreated K. pneumoniae biofilms. RNA-Seq results gave hundreds of differentially expressed ORFs, many of which gave molecular insights into the morphological changes observed by SEM.
The upregulation of genes involved in peptidoglycan catabolism, such as bax, which is homologous to mannosyl-glycoprotein endo-β-N-acetylglucosamidase genes from other bacteria, and nlpD, which activates amidases at the septal ring (35), is indicative of how carbapenems could affect gene expression resulting in changes of cell morphology. Two l,d-transpeptidase genes (erfK and ynhG) generally responsible for peptidoglycan cross-linking were also upregulated (31, 36, 37). As carbapenems are able to inhibit l,d-transpeptidases (36), it follows that a treated bacterium could upregulate expression of the genes coding for these proteins in order to compensate for the deleterious effect of treatment.
A homolog of the morphogene bolA was significantly upregulated in response to imipenem treatment. In Escherichia coli, overexpression of bolA leads to a rounded morphology and induction of biofilm formation (38, 39). bolA is normally expressed in the stationary phase but can be induced under stress conditions; therefore, the increased expression of bolA and its regulator, rpoS, under the stress condition of antibiotic therapy is consistent with previous studies (38, 39). Also potentially affecting cell morphology could be the downregulation of genes involved in the biosynthesis of peptidoglycan, including murDGP, mraYZ, uppP, and dacC.
It has been previously noted that K. pneumoniae can form spheroplasts following treatment with antibiotics (40, 41). Our morphological analysis is consistent with these findings, and our RNA-Seq results suggest a molecular mechanism for this phenomenon. Furthermore, two proteins generally implicated as being upregulated in spheroplasts, CpxP and spheroplast protein y (Spy), are upregulated in imipenem-treated K. pneumoniae biofilms. The Cpx pathway is turned on under spheroplast conditions and leads to upregulation of spy (42, 43). Spy is a homolog of CpxP, which is hypothesized to be involved in the biogenesis of outer membrane proteins (OMP) (44).
In the analysis of our RNA sequencing data, we also noticed some significant changes in gene expression in the areas of generalized stress response, virulence, and the antibiotic resistance/tolerance in cells from an imipenem-treated biofilm. A large number of ORFs, including rpoS, were differentially expressed, contributing to the stress response observed. In E. coli, the Crl-RpoS regulon is shown to regulate metabolism, response to the environment, and DNA metabolism (45). Our study supports many of these results, with the upregulation of known RpoS-dependent ORFs. RpoS is the central regulator of general stress response in bacteria. Some of these upregulated ORFs are necessary for the oxidative (dps and sod family) and osmotic (osm family) stress responses, fatty acid biosynthesis (fad family), persister cell formation (hip family), and morphological changes (bolA), indicating the importance of the RpoS stress response to tolerance of antibiotic therapy. Also upregulated was the gene coding for cold shock protein D (cspD), which is induced in the stationary phase in an rpoS-independent manner. CspD is responsible for inhibiting replication by binding to single-stranded DNA (ssDNA), and its overexpression is implicated in increased persister cell formation (46). With upregulation of both hipB and cspD, it is possible that there are an increased number of persister cells found in imipenem-treated biofilms, potentially giving insight into the ability of these biofilms to remain viable even with a carbapenem treatment of 1,000× the MIC. As persister cells are dormant and transiently antibiotic tolerant, it may be necessary to develop novel therapies that are able to target these metabolically inactive cells to effectively clear biofilm infections.
As previously mentioned, this strain of K. pneumoniae is multidrug resistant, with ORFs encoding resistance to numerous classes of antibiotics. Therefore, it was not surprising to see differential expression of some of these ORFs in response to 2 h of imipenem treatment. We found upregulation of genes responsible for chloramphenicol and bleomycin resistance, as well as a predicted multidrug resistance gene. We also noted the downregulation of oprD, cidA, and ampDE (although only ampE was significantly downregulated). OprD is a porin, and decreased levels of this protein are implicated in increased carbapenem resistance (47). The ampDE genes are negative regulators of ampC, a β-lactamase-encoding gene (48). Finally, cidA mutants have increased vancomycin and rifampin tolerance (49). Taken together, these data suggest a generalized antibiotic-induced stress response instead of a specific response to the therapy being applied.
Interestingly, in response to 2 h of imipenem treatment, there was a downregulation of ORFs responsible for resistance to other antibiotics. Both tetR, which drives resistance to tetracycline, and ermC, which drives resistance to macrolides, were significantly downregulated in response to imipenem treatment. This finding provides a molecular basis for the potential of dual antibiotic therapy to more effectively target K. pneumoniae biofilms.
After 24 h of imipenem treatment, only two stress response genes were significantly upregulated, csiD and iraP. CsiD is expressed only under carbon starvation conditions and is positively regulated by rpoS. IraP functions to stabilize RpoS in the stationary phase and during starvation conditions. Other ORFs previously mentioned, including rpoS, the ibr family, msrAB, and bolA, are still upregulated compared to those in untreated biofilms, although the levels are insignificant.
A similar trend is seen with downregulated ORFs, as only two are significantly downregulated after 24 h of imipenem treatment—cpxP and rpoE. CpxP was shown to be upregulated after 2 h of imipenem treatment. CpxP can act as a negative regulator of its own expression. Therefore, extended production of CpxP could activate the feedback mechanism leading to decreased transcription of the cpxP gene, as shown here after 24 h of treatment. rpoE has a large regulon and varied responsibilities in the cell. A number of ORFs regulated by RpoE are involved in the synthesis and assembly of lipopolysaccharide (LPS) and OMPs (50). Additionally, the RpoE regulon consists of genes necessary for cell division and virulence. rpoE is downregulated following 2 h of exposure, although not significantly. It is interesting to speculate that a downregulation of rpoE following imipenem treatment may contribute to a decreased pathogenicity of K. pneumoniae without affecting viability of the cells.
Decreased virulence in the presence of sublethal concentrations of antibiotics has been shown in other organisms, such as Pseudomonas aeruginosa (51) and Staphylococcus aureus (52), among others. We found that a number of ORFs previously implicated in virulence of K. pneumoniae and other species were downregulated, indicating that treatment with sublethal levels of carbapenems may be altering the virulence potential of K. pneumoniae biofilms. The ORFs include genes required for adhesion (fimC), iron sequestration (feo and fec family members), nickel transport (nikR), two-component systems, anaerobic growth (dmsABC, frdC, and adhE), and lipid A (arnACT) and outer membrane production (wecFG, wbbM, and wzyE) (53–59). Sublethal antibiotics have also been implicated in altered biofilm formation in P. aeruginosa, E. coli, and S. aureus (60, 61) and we observed the differential gene expression of a number of biofilm-related ORFs, including cidA, ariR, mlrA, bssR, and cysB (62–66). As biofilm formation is yet another important virulence factor for K. pneumoniae and other bacteria, altering biofilms with sublethal concentrations of carbapenems may have positive effects on decreasing virulence potential.
While it is clear that the ability of sublethal concentrations of antibiotics to ameliorate the virulence of K. pneumoniae provides an opportunity to treat infections in novel ways, it is important to consider what, if any, deleterious effects could result from sublethal levels of antibiotic therapy. It has been shown that exposure to sublethal antibiotic concentrations in other species can promote the growth of resistant variants by both selecting for preexisting mutants and promoting new mutations (67). Sublethal antibiotic levels have also been shown to increase persister cell formation (67). Therefore, it would be interesting to generate mutations in genes we have classified as important for cell shape or virulence, for example, to determine the gene function. Once function has been confirmed, we could attempt to develop antipathogenic therapies by screening inhibitors against our candidate genes. Antipathogenic therapy is one of the most effect means to counteract microbial resistance as the end goal is not killing the bacteria, simply a reduction in virulence potential.
The increasing antibiotic resistance seen in pathogenic bacteria is a worrying trend necessitating the understanding of how bacteria are able to adapt to antibiotic insults. Our RNA-Seq results from imipenem-treated K. pneumoniae biofilms give a glimpse of the complex regulatory pathways being activated by antibiotic therapy, even at sublethal concentrations. These data hint at alternative targets for antimicrobial/antipathogenic therapies and provide suggestions for pathways that can be inhibited to promote elimination of biofilm infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Larry D. Swain for critical reading of the manuscript.
This work was supported by the U.S. Army Medical Research and Materiel Command, Combat Casualty Care Research Directorate and the Research Associateship Program from the National Research Council (T.A.V.L.).
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04581-14.
REFERENCES
- 1.Bennett JW, Robertson JL, Hospenthal DR, Wolf SE, Chung KK, Mende K, Murray CK. 2010. Impact of extended spectrum beta-lactamase producing Klebsiella pneumoniae infections in severely burned patients. J Am Coll Surg 211:391–399. doi: 10.1016/j.jamcollsurg.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 2.Meyle E, Stroh P, Gunther F, Hoppy-Tichy T, Wagner C, Hansch GM. 2010. Destruction of bacterial biofilms by polymorphonuclear neutrophils: relative contribution of phagocytosis, DNA release, and degranulation. Int J Artif Organs 33:608–620. [DOI] [PubMed] [Google Scholar]
- 3.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 4.Yao Y, Vuong C, Kocianova S, Villaruz AE, Lai Y, Sturdevant DE, Otto M. 2006. Characterization of the Staphylococcus epidermidis accessory-gene regulator response: quorum-sensing regulation of resistance to human innate host defense. J Infect Dis 193:841–848. doi: 10.1086/500246. [DOI] [PubMed] [Google Scholar]
- 5.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 6.Tsuneda S, Aikawa H, Hayashi H, Yuasa A, Hirata A. 2003. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS Microbiol Lett 223:287–292. doi: 10.1016/S0378-1097(03)00399-9. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Seth AK, Abercrombie JJ, Mustoe TA, Leung KP. 2014. Activity of imipenem against Klebsiella pneumoniae biofilms in vitro and in vivo. Antimicrob Agents Chemother 58:1208–1213. doi: 10.1128/AAC.01353-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Childers BM, Van Laar TA, You T, Clegg S, Leung KP. 2013. MrkD1P from Klebsiella pneumoniae strain IA565 allows for coexistence with Pseudomonas aeruginosa and protection from protease-mediated biofilm detachment. Infect Immun 81:4112–4120. doi: 10.1128/IAI.00521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 10.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 11.Rezaei E, Safari H, Naderinasab M, Aliakbarian H. 2011. Common pathogens in burn wound and changes in their drug sensitivity. Burns 37:805–807. doi: 10.1016/j.burns.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb Perspect Biol 2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fasani RA, Savageau MA. 2013. Molecular mechanisms of multiple toxin-antitoxin systems are coordinated to govern the persister phenotype. Proc Natl Acad Sci U S A 110:E2528–E2537. doi: 10.1073/pnas.1301023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagley ST. 1985. Habitat association of Klebsiella species. Infect Control 6:52–58. [DOI] [PubMed] [Google Scholar]
- 16.Murray CK, Yun HC, Griffith ME, Thompson B, Crouch HK, Monson LS, Aldous WK, Mende K, Hospenthal DR. 2009. Recovery of multidrug-resistant bacteria from combat personnel evacuated from Iraq and Afghanistan at a single military treatment facility. Mil Med 174:598–604. doi: 10.7205/MILMED-D-03-8008. [DOI] [PubMed] [Google Scholar]
- 17.Ressner RA, Murray CK, Griffith ME, Rasnake MS, Hospenthal DR, Wolf SE. 2008. Outcomes of bacteremia in burn patients involved in combat operations overseas. J Am Coll Surg 206:439–444. doi: 10.1016/j.jamcollsurg.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Lockhart SR, Abramson MA, Beekmann SE, Gallagher G, Riedel S, Diekema DJ, Quinn JP, Doern GV. 2007. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol 45:3352–3359. doi: 10.1128/JCM.01284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong JH, Clancy CJ, Cheng S, Shields RK, Chen L, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Characterization of porin expression in Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae identifies isolates most susceptible to the combination of colistin and carbapenems. Antimicrob Agents Chemother 57:2147–2153. doi: 10.1128/AAC.02411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Camacho E, Gomez-Gil R, Tobes R, Manrique M, Lorenzo M, Galvan B, Salvarelli E, Moatassim Y, Salanueva IJ, Pareja E, Codoner FM, Alvarez-Tejado M, Garcillan-Barcia MP, De la Cruz F, Mingorance J. 2014. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J Antimicrob Chemother 69:632–636. doi: 10.1093/jac/dkt419. [DOI] [PubMed] [Google Scholar]
- 21.Villa L, Capone A, Fortini D, Dolejska M, Rodriguez I, Taglietti F, De Paolis P, Petrosillo N, Carattoli A. 2013. Reversion to susceptibility of a carbapenem-resistant clinical isolate of Klebsiella pneumoniae producing KPC-3. J Antimicrob Chemother 68:2482–2486. doi: 10.1093/jac/dkt235. [DOI] [PubMed] [Google Scholar]
- 22.Van Laar TA, Chen T, Childers BM, Chen P, Abercrombie JJ, Leung KP. 2014. Genome sequence of a multidrug-resistant strain of Klebsiella pneumoniae, BAMC 07-18, isolated from a combat injury wound. Genome Announc 2(6):e01230-14. doi: 10.1128/genomeA.01230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. 2009. JBrowse: a next-generation genome browser. Genome Res 19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magnet S, Bellais S, Dubost L, Fourgeaud M, Mainardi JL, Petit-Frere S, Marie A, Mengin-Lecreulx D, Arthur M, Gutmann L. 2007. Identification of the l,d-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J Bacteriol 189:3927–3931. doi: 10.1128/JB.00084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craig WA. 1997. The pharmacology of meropenem, a new carbapenem antibiotic. Clin Infect Dis 24(Suppl 2):S266–S275. doi: 10.1093/clinids/24.Supplement_2.S266. [DOI] [PubMed] [Google Scholar]
- 33.Nandy P, Samtani MN, Lin R. 2010. Population pharmacokinetics of doripenem based on data from phase 1 studies with healthy volunteers and phase 2 and 3 studies with critically ill patients. Antimicrob Agents Chemother 54:2354–2359. doi: 10.1128/AAC.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed MD, Stern RC, O'Brien CA, Yamashita TS, Myers CM, Blumer JL. 1985. Pharmacokinetics of imipenem and cilastatin in patients with cystic fibrosis. Antimicrob Agents Chemother 27:583–588. doi: 10.1128/AAC.27.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters NT, Dinh T, Bernhardt TG. 2011. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J Bacteriol 193:4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mainardi JL, Hugonnet JE, Rusconi F, Fourgeaud M, Dubost L, Moumi AN, Delfosse V, Mayer C, Gutmann L, Rice LB, Arthur M. 2007. Unexpected inhibition of peptidoglycan ld-transpeptidase from Enterococcus faecium by the beta-lactam imipenem. J Biol Chem 282:30414–30422. doi: 10.1074/jbc.M704286200. [DOI] [PubMed] [Google Scholar]
- 37.Sanders AN, Pavelka MS. 2013. Phenotypic analysis of Escherichia coli mutants lacking l,d-transpeptidases. Microbiology 159:1842–1852. doi: 10.1099/mic.0.069211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guinote IB, Moreira RN, Barahona S, Freire P, Vicente M, Arraiano CM. 2014. Breaking through the stress barrier: the role of BolA in Gram-negative survival. World J Microbiol Biotechnol 30:2559–2566. doi: 10.1007/s11274-014-1702-4. [DOI] [PubMed] [Google Scholar]
- 39.Vieira HL, Freire P, Arraiano CM. 2004. Effect of Escherichia coli morphogene bolA on biofilms. Appl Environ Microbiol 70:5682–5684. doi: 10.1128/AEM.70.9.5682-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakao M, Nishi T, Tsuchiya K. 1981. In vitro and in vivo morphological response of Klebsiella pneumoniae to cefotiam and cefazolin. Antimicrob Agents Chemother 19:901–910. doi: 10.1128/AAC.19.5.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanberger H, Nilsson LE, Nilsson M, Maller R. 1991. Post-antibiotic effect of beta-lactam antibiotics on Gram-negative bacteria in relation to morphology, initial killing and MIC. Eur J Clin Invest 10:927–934. [DOI] [PubMed] [Google Scholar]
- 42.Hagenmaier S, Stierhof YD, Henning U. 1997. A new periplasmic protein of Escherichia coli which is synthesized in spheroplasts but not in intact cells. J Bacteriol 179:2073–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon E, Kim DY, Gross CA, Gross JD, Kim KK. 2010. The crystal structure Escherichia coli Spy. Protein Sci 19:2252–2259. doi: 10.1002/pro.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raivio TL, Laird MW, Joly JC, Silhavy TJ. 2000. Tethering of CpxP to the inner membrane prevents spheroplast induction of the cpx envelope stress response. Mol Microbiol 37:1186–1197. doi: 10.1046/j.1365-2958.2000.02074.x. [DOI] [PubMed] [Google Scholar]
- 45.Lelong C, Aguiluz K, Luche S, Kuhn L, Garin J, Rabilloud T, Geiselmann J. 2007. The Crl-RpoS regulon of Escherichia coli. Mol Cell Proteomics 6:648–659. doi: 10.1074/mcp.M600191-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Langklotz S, Narberhaus F. 2011. The Escherichia coli replication inhibitor CspD is subject to growth-regulated degradation by the Lon protease. Mol Microbiol 80:1313–1325. doi: 10.1111/j.1365-2958.2011.07646.x. [DOI] [PubMed] [Google Scholar]
- 47.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juan C, Macia MD, Gutierrez O, Vidal C, Perez JL, Oliver A. 2005. Molecular mechanisms of beta-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 49:4733–4738. doi: 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice KC, Nelson JB, Patton TG, Yang SJ, Bayles KW. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J Bacteriol 187:813–821. doi: 10.1128/JB.187.3.813-821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moreau PL. 2014. Protective role of the RpoE (sigma(E)) and Cpx envelope stress responses against gentamicin killing of nongrowing Escherichia coli incubated under aerobic, phosphate starvation conditions. FEMS Microbiol Lett 357:151–156. doi: 10.1111/1574-6968.12534. [DOI] [PubMed] [Google Scholar]
- 51.Molinari G, Guzman CA, Pesce A, Schito GC. 1993. Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. J Antimicrob Chemother 31:681–688. doi: 10.1093/jac/31.5.681. [DOI] [PubMed] [Google Scholar]
- 52.Otto MP, Martin E, Badiou C, Lebrun S, Bes M, Vandenesch F, Etienne J, Lina G, Dumitrescu O. 2013. Effects of subinhibitory concentrations of antibiotics on virulence factor expression by community-acquired methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 68:1524–1532. doi: 10.1093/jac/dkt073. [DOI] [PubMed] [Google Scholar]
- 53.Baltes N, Hennig-Pauka I, Jacobsen I, Gruber AD, Gerlach GF. 2003. Identification of dimethyl sulfoxide reductase in Actinobacillus pleuropneumoniae and its role in infection. Infect Immun 71:6784–6792. doi: 10.1128/IAI.71.12.6784-6792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge RG, Wang DX, Hao MC, Sun XS. 2013. Nickel trafficking system responsible for urease maturation in Helicobacter pylori. World J Gastroenterol 19:8211–8218. doi: 10.3748/wjg.v19.i45.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naylor J, Cianciotto NP. 2004. Cytochrome c maturation proteins are critical for in vivo growth of Legionella pneumophila. FEMS Microbiol Lett 241:249–256. doi: 10.1016/j.femsle.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 56.Nishiyama S, Murakami Y, Nagata H, Shizukuishi S, Kawagishi I, Yoshimura F. 2007. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology 153:1916–1925. doi: 10.1099/mic.0.2006/005561-0. [DOI] [PubMed] [Google Scholar]
- 57.Pullinger GD, van Diemen PM, Dziva F, Stevens MP. 2010. Role of two-component sensory systems of Salmonella enterica serovar Dublin in the pathogenesis of systemic salmonellosis in cattle. Microbiology 156:3108–3122. doi: 10.1099/mic.0.041830-0. [DOI] [PubMed] [Google Scholar]
- 58.Seyedmohammad S, Born D, Venter H. 2014. Expression, purification and functional reconstitution of FeoB, the ferrous iron transporter from Pseudomonas aeruginosa. Protein Expr Purif 101:138–145. doi: 10.1016/j.pep.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 59.Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5(3):e01163-14. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 61.Li D, Renzoni A, Estoppey T, Bisognano C, Francois P, Kelley WL, Lew DP, Schrenzel J, Vaudaux P. 2005. Induction of fibronectin adhesins in quinolone-resistant Staphylococcus aureus by subinhibitory levels of ciprofloxacin or by sigma B transcription factor activity is mediated by two separate pathways. Antimicrob Agents Chemother 49:916–924. doi: 10.1128/AAC.49.3.916-924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Attila C, Ueda A, Wood TK. 2009. 5-Fluorouracil reduces biofilm formation in Escherichia coli K-12 through global regulator AriR as an antivirulence compound. Appl Microbiol Biotechnol 82:525–533. doi: 10.1007/s00253-009-1860-8. [DOI] [PubMed] [Google Scholar]
- 63.Domka J, Lee J, Wood TK. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol 72:2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garo E, Eldridge GR, Goering MG, DeLancey Pulcini E, Hamilton MA, Costerton JW, James GA. 2007. Asiatic acid and corosolic acid enhance the susceptibility of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob Agents Chemother 51:1813–1817. doi: 10.1128/AAC.01037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grande R, Nistico L, Sambanthamoorthy K, Longwell M, Iannitelli A, Cellini L, Di Stefano A, Hall Stoodley L, Stoodley P. 2014. Temporal expression of agrB, cidA, and alsS in the early development of Staphylococcus aureus UAMS-1 biofilm formation and the structural role of extracellular DNA and carbohydrates. Pathog Dis 70:414–422. doi: 10.1111/2049-632X.12158. [DOI] [PubMed] [Google Scholar]
- 66.Lonn-Stensrud J, Naemi AO, Benneche T, Petersen FC, Scheie AA. 2012. Thiophenones inhibit Staphylococcus epidermidis biofilm formation at nontoxic concentrations. FEMS Immunol Med Microbiol 65:326–334. doi: 10.1111/j.1574-695X.2012.00964.x. [DOI] [PubMed] [Google Scholar]
- 67.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.