Abstract
Dasabuvir (ABT-333) is a nonnucleoside inhibitor of the RNA-dependent RNA polymerase encoded by the hepatitis C virus (HCV) NS5B gene. Dasabuvir inhibited recombinant NS5B polymerases derived from HCV genotype 1a and 1b clinical isolates, with 50% inhibitory concentration (IC50) values between 2.2 and 10.7 nM, and was at least 7,000-fold selective for the inhibition of HCV genotype 1 polymerases over human/mammalian polymerases. In the HCV subgenomic replicon system, dasabuvir inhibited genotype 1a (strain H77) and 1b (strain Con1) replicons with 50% effective concentration (EC50) values of 7.7 and 1.8 nM, respectively, with a 13-fold decrease in inhibitory activity in the presence of 40% human plasma. This level of activity was retained against a panel of chimeric subgenomic replicons that contained HCV NS5B genes from 22 genotype 1 clinical isolates from treatment-naive patients, with EC50s ranging between 0.15 and 8.57 nM. Maintenance of replicon-containing cells in medium containing dasabuvir at concentrations 10-fold or 100-fold greater than the EC50 resulted in selection of resistant replicon clones. Sequencing of the NS5B coding regions from these clones revealed the presence of variants, including C316Y, M414T, Y448C, Y448H, and S556G, that are consistent with binding to the palm I site of HCV polymerase. Consequently, dasabuvir retained full activity against replicons known to confer resistance to other polymerase inhibitors, including the S282T variant in the nucleoside binding site and the M423T, P495A, P495S, and V499A single variants in the thumb domain. The use of dasabuvir in combination with inhibitors targeting HCV NS3/NS4A protease (ABT-450 with ritonavir) and NS5A (ombitasvir) is in development for the treatment of HCV genotype 1 infections.
INTRODUCTION
The hepatitis C virus (HCV) is an enveloped, single-strand, positive-sense RNA virus in the Flaviviridae family. HCV is a leading cause of cirrhosis and end-stage disease globally, with an estimated 170 million to 200 million people infected worldwide (1). Seven distinct HCV genotypes and numerous subtypes, with significant variability in their geographic distributions, have been characterized (2). HCV genotype 1 is the most common, accounting for 46 to 60% of the global infections, and is predominant in North America, Europe, and Japan (3–6).
HCV-infected patients typically have high-level viremia characterized by a vast heterogeneous pool of viral genome sequences (7). This quasispecies pool is produced by the error-prone HCV RNA-dependent RNA polymerase, which does not have an editing function to remove base incorporation errors (8). In some cases, these sequence mutations result in amino acid substitutions in nonstructural viral proteins such as NS3/NS4A protease, NS5A, or the RNA polymerase itself. Substitutions in these nonstructural proteins can potentially lead to drug resistance and treatment failure.
The HCV NS5B polymerase has multiple binding sites that can be targeted for inhibition of HCV replication. The most phylogenetically conserved of these is the catalytic site of the enzyme, which is inhibited by nucleoside/nucleotide inhibitors such as sofosbuvir, VX-135, IDX21437, IDX21459, and ACH-3422 (9, 10). Most of these nucleoside/nucleotide analogs select the S282T resistant variant in vitro in HCV subgenomic replicon assays (11). In addition to the active site, the HCV polymerase enzyme has 4 allosteric inhibitor binding sites, which are located within the canonical thumb and palm domains of the protein (12–14). The thumb domain contains two binding pockets, which are characterized by distinct sets of resistant variants. Compounds that bind to the lower thumb site (thumb II) include GS-9669, filibuvir, and lomibuvir and frequently select R422K, L419M, M423T, and I482L as major resistant variants (14, 15). Upper thumb (thumb I) binders, such as BI-207127 and BMS-791325, are associated with resistant variants P495A/S/L/T and V499A (16). The palm I and II sites are partially overlapping; palm II site inhibitors include HCV-796 and GSK5852 (17, 18). HCV-796 selects a highly resistant variant, C316Y, whereas GSK5852 retains activity against C316Y but is less active against variants C316F, S365F/L/T, and C445F. These palm II site inhibitors are differentiated from other nonnucleoside inhibitors in that they possess potent activity across genotype 1 to 4 polymerases (17, 18). Palm I site inhibitors include multiple series of benzothiadiazine-containing compounds, which select resistant variants C316Y, M414T, Y448H, and G554D (19, 20). The resistance profiles of the palm I site inhibitors and their overlap with the palm II site inhibitors are entirely consistent with published ligand-bound crystal structures of palm I and II site inhibitors (17, 21).
A high-throughput screening campaign identified an aryl dihydrouracil fragment that was subsequently modified to produce the potent nonnucleoside inhibitor known as dasabuvir (ABT-333) (22). In this paper, we report the in vitro potency and resistance profiles of dasabuvir. Dasabuvir is being developed for use in combination with ABT-450, a potent macrocyclic noncovalent peptidomimetic inhibitor of HCV NS3/NS4A protease, and the NS5A inhibitor ombitasvir (ABT-267), with or without ribavirin (RBV), for the treatment of HCV genotype 1 infections in patients with or without cirrhosis (23–27).
MATERIALS AND METHODS
Compound.
Dasabuvir, sodium N-[6-[3-tert-butyl-5-[2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl]-2-methoxyphenyl]naphthalen-2-yl]methanesulfonamide (Fig. 1), was synthesized at AbbVie.
FIG 1.
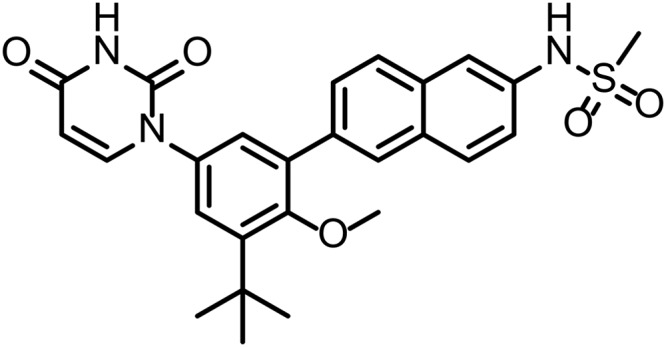
Structure of dasabuvir.
Inhibition of HCV polymerases in biochemical assays.
The recombinant HCV polymerases used in this study contained the first 570 amino acids of the 591-amino acid native protein sequence, with a six-histidine tag at the N terminus to facilitate purification by affinity chromatography. The polymerase enzymatic inhibition assay was described previously by Liu et al. (28). Briefly, dasabuvir was incubated with 5 to 50 nM polymerase for 15 min at room temperature, followed by the addition of nucleoside triphosphates (NTPs) and [3H]UTP for 3 h at 30°C. After termination of the reaction, the precipitated RNA was captured by filtration through a GF/B filter (Millipore). The amount of incorporated [3H]UTP was measured by scintillation counting with a Wallac 1450 MicroBeta counter. The percent inhibition was calculated from the initial rates of inhibited reactions relative to that of the uninhibited control reaction. The mean 50% inhibitory concentration (IC50) and the standard error of the mean (SEM) were calculated via nonlinear regression.
Inhibition of mammalian polymerases in biochemical assays.
The inhibition of human and mammalian DNA polymerases was evaluated by Replizyme Ltd. (Heslington, United Kingdom). The DNA-dependent RNA polymerase activity for human RNA polymerases II and III was measured using polymerases present in a HeLa cell extract and DNA templates containing promoters specific for either polymerase II or polymerase III. α-Amanitin, a potent inhibitor of human polymerase II and a modest inhibitor of polymerase III, was used as a control. The reaction mixtures contained 20 mM Tris-HCl (pH 8.0), 20% glycerol, 100 mM KCl, 1 mM dithiothreitol (DTT), 0.2 mM EDTA, 6 mM MgCl2, and either 1 μg/μl pAdVAntage plasmid (Promega, Madison, WI) (for the polymerase III assay) or 25 ng/μl cytomegalovirus (CMV) promoter DNA (Promega, Madison, WI) (for the polymerase II assay). The reaction mixtures also contained HeLa cell nuclear extract (ProteinOne, Rockville, MD), 400 μM ATP, CTP, and UTP, and 16 μM GTP and [α-33P]GTP. The reaction mixtures were incubated for 1 h at 30°C, quenched with the addition of proteinase K, SDS, and EDTA, incubated for 30 min at 56°C, and then analyzed on a Criterion Bio-Rad 5% acrylamide Tris-borate-EDTA (TBE)-urea gel. The gel was dried and placed on a PhosphorImager screen for overnight exposure. The volumes of the product bands were measured, and the percent inhibition was calculated; IC50 values were calculated using the following equation: % inhibition = 100[I]/([I] + IC50).
Activity in HCV replicons.
The genotype 1a replicon construct contained the 5′ nontranslated region (NTR) from genotype 1a (strain H77) (GenBank accession number NC004102) followed by the firefly luciferase reporter gene and the neomycin phosphotransferase gene (neo), which together constituted the first cistron of the bicistronic replicon construct. This was followed by the encephalomyocarditis virus (EMCV) internal ribosomal entry site (IRES), the second cistron containing the genotype 1a (H77) NS3-NS5B coding region with the adaptive substitutions encoding E1202G, K1691R, K2040R, and S2204I, and finally the 1a (H77) 3′ NTR. The genotype 1b (strain Con1) replicon construct contained the 5′ NTR from 1b (Con1) (GenBank accession number AJ238799) followed by the firefly luciferase reporter gene and the neo gene, which together constituted the first cistron of the bicistronic replicon construct. This was followed by the EMCV IRES, the second cistron containing the genotype 1b (Con1) NS3-NS5B coding region with the adaptive substitutions encoding E1202G, T1280I, and S2204I, and finally the 1b (Con1) 3′ NTR. Both replicon-containing cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 100 IU/ml penicillin, 100 μg/ml streptomycin, and 200 μg/ml G418 (all from Invitrogen, Carlsbad, CA) with 10% (vol/vol) fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA). The inhibitory effects of dasabuvir on HCV RNA replication were determined in DMEM containing 5% FBS, with or without 40% human plasma (Bioreclamation, Westbury, NY), by measuring the activity of the luciferase reporter gene. The cells were incubated for 3 days in the presence of dasabuvir and subsequently were lysed and processed according to the manufacturer's instructions (Promega, Madison, WI). The luciferase activity in the cells was measured using a Victor II luminometer (Perkin-Elmer, Waltham, MA); 50% effective concentration (EC50) values were calculated using nonlinear regression curve fitting to a 4-parameter logistic equation with GraphPad Prism 4 software. EC50s and standard deviations (SDs) were calculated from at least 3 experiments conducted in duplicate. The cytotoxicity of dasabuvir was determined with the colorimetric assay using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (5 mg/ml; Sigma-Aldrich, St. Louis, MO) (29); 50% cytotoxic concentration (CC50) values were calculated from the optical density data using nonlinear regression curve fitting to a 4-parameter logistic equation with GraphPad Prism 4 software.
Activity against panel of genotype 1 clinical isolates.
Methods for the measurement of activity against HCV replicons containing the NS5B gene from HCV-infected samples were described previously (30). Briefly, viral RNA was isolated from serum samples obtained from HCV-infected patients by using the QIAamp viral RNA isolation kit (Qiagen, Valencia, CA), according to the manufacturer's instructions. Reverse transcription (RT)-PCR using SuperScript III with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) and gene-specific primers was used to generate a DNA fragment containing the NS5B gene. This fragment was ligated into a genotype 1a or 1b replicon shuttle vector plasmid and transformed into competent Escherichia coli cells. After overnight growth in liquid culture, the plasmid DNA from the entire population was isolated, purified, and then linearized by restriction enzyme digestion. The NS5B gene was sequenced using the ABI Prism dye terminator cycle sequencing ready reaction kit, with an Applied Biosystems 3100 genetic analyzer. In transient expression assays, the chimeric replicon containing the gene of interest from an HCV-containing plasma or serum sample was transfected into a Huh-7-derived cell line via electroporation (30, 31). The EC50s were calculated as described above.
Resistance selection.
In order to characterize replicon variants with reduced susceptibility to dasabuvir, resistance selection was conducted with the HCV replicon-containing stable cell lines 1a (H77) and 1b (Con1), as described previously (19). Replicon variant cells (104 to 105, depending on the concentration of inhibitor) were plated in 150-mm cell culture plates and grown in the presence of G418 (400 μg/ml) and dasabuvir, at a concentration that was either 10-fold or 100-fold above the EC50 for the respective cell line. After 3 weeks of treatment, the majority of replicon cells were cleared of replicon RNA and therefore were unable to survive in the G418-containing medium. The cells containing resistant replicon variants survived and formed colonies; these colonies were picked and further expanded. To characterize the genotypes of the resistant replicon variants, total RNA was extracted from the expanded colonies using a Qiagen RNeasy spin kit (Qiagen, Valencia, CA). The NS5B coding region was amplified by RT-PCR (with SuperScript III and Platinum Pfx DNA polymerase; Invitrogen, Carlsbad, CA) using specific primers. The amplified samples were purified using a PCR purification kit (Qiagen, Valencia, CA) and sequenced using the ABI Prism dye terminator cycle sequencing ready reaction kit, with an Applied Biosystems 3100 genetic analyzer.
Antireplicon activity against a panel of resistant variants.
Methods for measurement of the effects of individual amino acid substitutions on the activity of an inhibitor in HCV replicon cell culture assays were described previously (19). Briefly, the resistance-associated NS5B mutations were each introduced into the genotype 1a (H77) or 1b (Con1) replicons described above but without the neo gene, using a Change-IT multiple mutation site-directed mutagenesis kit (Affymetrix, Santa Clara, CA). After the mutagenesis had been confirmed by sequence analysis, the plasmids were linearized and a TranscriptAid T7 high yield transcription kit (Fermentas, Glen Burnie, MD) was used to transcribe the HCV subgenomic RNA from the plasmids. In transient expression assays, the replicon containing the substitution was transfected into a Huh-7 cell line via electroporation. EC50s and standard deviations were calculated from at least 3 experiments conducted in duplicate, as described above. Replication efficiency was calculated as a percentage of wild-type replication using the following equation: 100 × [(variant 4-day luciferase activity/wild-type 4-day luciferase activity)/(variant 4-h luciferase activity/wild-type 4-h luciferase activity)].
RESULTS
Activity in polymerase enzymatic assays.
Dasabuvir inhibited the polymerase enzymatic activity of genotype 1 laboratory strain enzymes (H77, BK, N, and Con1 strains), as well as enzymes produced from polymerase genes from HCV genotype 1-infected subjects, with IC50s between 2.2 and 10.7 nM (Table 1). Dasabuvir was also tested for inhibition of a variety of human and mammalian DNA-dependent DNA polymerases (polymerases alpha, beta, and gamma) and DNA-dependent RNA polymerases (e.g., RNA polymerases II and III) and one RNA-dependent DNA polymerase (polymerase gamma-RT function). Dasabuvir was highly selective for the inhibition of HCV genotype 1 polymerases over representative human/mammalian polymerases, as judged by selectivity ratios that were at least 7,000:1 (Table 1).
TABLE 1.
Inhibition of HCV polymerases and mammalian polymerases by dasabuvir
| Polymerase | IC50 (mean ± SEM) (nM) | Selectivitya |
|---|---|---|
| HCV genotype 1a (H77) | 2.8 ± 0.2 | |
| HCV genotype 1a no. 1b | 2.4 ± 0.2 | |
| HCV genotype 1a no. 2b | 4.2 ± 0.4 | |
| HCV genotype 1b (Con1) | 10.7 ± 1.4 | |
| HCV genotype 1b (N) | 2.2 ± 0.3 | |
| HCV genotype 1b (BK) | 3.1 ± 0.2 | |
| HCV genotype 1b no. 1b | 4.3 ± 0.5 | |
| HCV genotype 2a no. 1b | 16,400 ± 2,100 | |
| HCV genotype 2b no. 1b | >20,000 | |
| HCV genotype 2b no. 2b | 16,100 ± 1,700 | |
| HCV genotype 3a no. 1b | 5,600 ± 800 | |
| HCV genotype 3a no. 2b | 11,900 ± 1,300 | |
| HCV genotype 3a no. 3b | 3,700 ± 1,000 | |
| HCV genotype 4a no. 1b | 900 ± 200 | |
| HCV genotype 4a no. 2b | 3,700 ± 1,000 | |
| Bovine polymerase alpha | 76,000 ± 9,200 | >7,102 |
| Human polymerase beta | >100,000 | >9,345 |
| Human polymerase gamma, polymerase function | >100,000 | >9,345 |
| Human polymerase gamma, reverse transcriptase function | >100,000 | >9,345 |
| Bovine terminal deoxynucleotidyl transferase | >100,000 | >9,345 |
| Human DNA-dependent RNA polymerase II | >100,000 | >9,345 |
| Human DNA-dependent RNA polymerase II | >100,000 | >9,345 |
Calculated from the ratio of the IC50 for the mammalian polymerase to the IC50 of 10.7 nM for HCV genotype 1b (Con1) polymerase.
NS5B derived from a clinical isolate.
Activity in HCV replicons.
Dasabuvir inhibited replication of HCV subgenomic replicons in cell culture assays, with EC50 values of 7.7 and 1.8 nM against genotype 1a (H77) and 1b (Con1), respectively (Table 2). In the presence of 40% human plasma, there was a 12- to 13-fold decrease in inhibitory potency, yielding EC50s of 99 and 21 nM for HCV genotype 1a (H77) and 1b (Con1) replicons, respectively. The CC50 for dasabuvir was 10,360 nM, resulting in a therapeutic index that exceeded 1,345.
TABLE 2.
Dasabuvir EC50 values against stable subgenomic replicon cell lines
| HCV replicon subtype | EC50 (mean ± SD) (nM)a |
|
|---|---|---|
| With 0% human plasma | With 40% human plasma | |
| Genotype 1a (H77) | 7.7 ± 3.8 | 99 ± 13 |
| Genotype 1b (Con1) | 1.8 ± 0.98 | 21 ± 10 |
The EC50s and SDs were calculated from at least 3 experiments conducted in duplicate. Both the 0% and 40% human plasma assays also contained 5% fetal bovine serum.
Resistance in replicon cell culture assays.
Subsequent to passaging of HCV genotype 1 replicon cell lines in the presence of dasabuvir and G418, the NS5B sequence from surviving colonies was analyzed (Table 3). Using concentrations of dasabuvir that were 10-fold greater than the wild-type genotype 1a (H77) EC50, the predominant variant selected was S556G, which was observed in 43% of the colonies. Variants at amino acid positions 395, 414, 444, 448, 559, and 565 were also observed, but each substitution was found in only a single colony. Using dasabuvir concentrations that were 100-fold greater than the EC50 resulted in selection of C316Y as the predominant variant, with a prevalence of 40%, whereas the Y448C, C451R, and S556G variants were each detected at a prevalence of 20%. The impact of these variants on the EC50 of dasabuvir was evaluated by introduction of each amino acid substitution into the 1a (H77) replicon through mutagenesis or by evaluation of activity in the selected replicon colony harboring the variant. Variants A395G, M414T, N444K, S556G, S556N, and S565F conferred 10- to 32-fold resistance to dasabuvir. This is consistent with the observation that most of these variants were observed following selection with concentrations 10-fold greater than the EC50 of dasabuvir. Variants C316Y, Y448C, and Y448H conferred over 940-fold resistance, which is consistent with the observation that these variants were generally selected with concentrations 100-fold greater than the EC50 of dasabuvir. Variants C451R and D559G had poor replication capacity and could not be evaluated in replicon assays.
TABLE 3.
Selection of resistant variants in NS5B by dasabuvir in genotype 1 replicon cell lines and resistance in transient expression assays
| Genotype | Variant | Prevalencea |
EC50 (mean ± SD) (nM)b | Fold resistance | Replication efficiency (%)c | |
|---|---|---|---|---|---|---|
| 10× | 100× | |||||
| 1a (H77) | Wild-type | 0.96 ± 0.2 | 100 | |||
| C316Y | 4/10 | 1,413 ± 415 | 1,472 | 82 | ||
| A395G | 1/14 | 10d | ||||
| M414T | 1/14 | 31 ± 9 | 32 | 110 | ||
| N444K | 1/14 | 23d | ||||
| Y448C | 1/14 | 2/10 | 902 ± 270 | 940 | 19 | |
| Y448H | 1/14 | 936 ± 294 | 975 | 41 | ||
| C451R | 2/10 | NA | <0.5 | |||
| S556G | 6/14 | 2/10 | 29 ± 8 | 30 | 59 | |
| S556N | 1/14 | 29d | ||||
| D559G | 1/14 | NA | <0.5 | |||
| S565F | 1/14 | 17d | ||||
| 1b (Con1) | Wild-type | 0.59 ± 0.14 | 100 | |||
| C316Ne | 3.2 ± 0.14 | 5 | 154 | |||
| C316Y | 3/12 | 12/12 | 926 ± 196 | 1,569 | 96 | |
| S368T | 1/12 | 82 ± 10 | 139 | 11 | ||
| N411S | 1/12 | 50 ± 5 | 84 | 13 | ||
| M414T | 4/12 | 28 ± 3 | 47 | 31 | ||
| C445F, C451S, I585V | 1/12 | 16d | ||||
| Y448Ce | 244 ± 28 | 414 | 7.2 | |||
| Y448He | 27 ± 4 | 46 | 58 | |||
| A553V | 1/12 | 71 ± 6 | 120 | 14 | ||
| S556G | 1/12 | 6.5 ± 0.2 | 11 | 62 | ||
Number of times the variant was found out of the total number of colonies analyzed.
The EC50s and SDs were calculated from at least 3 experiments conducted in duplicate with replicons containing the variants in NS5B, using transient expression luciferase assays. NA, not assessed, as resistance could not be evaluated due to poor replication capacity of the variant.
Replication efficiency was calculated as a percentage of replication relative to the wild-type replicon.
Fold resistance was evaluated in a stable chimeric replicon cell line harboring the amino acid substitution; therefore, EC50 in a transient expression assay is not shown.
Substitutions at resistance-associated amino acid positions but not selected by dasabuvir in vitro.
In experiments conducted with genotype 1b (Con1) at a concentration that was 10-fold greater than the EC50 of dasabuvir, the predominant variants selected were C316Y and M414T. Variants at amino acid positions 368, 411, 445, 451, 553, and 556 were each observed in one colony. Variant S556G in genotype 1b (Con1) conferred 11-fold resistance, while the S368T, N411S, M414T, and A553V substitutions each conferred 47- to 139-fold resistance to dasabuvir. A replicon colony containing variants C445F, C451S, and I585V demonstrated 16-fold resistance to dasabuvir. Variant C316Y in the genotype 1b (Con1) replicon was found to confer 1,569-fold resistance to dasabuvir. The C316Y substitution was observed in all of the surviving colonies in studies conducted with concentrations 100-fold greater than the EC50 of dasabuvir.
The activity of dasabuvir was evaluated against variants that confer resistance to compounds binding to the nucleoside binding site (S282T), the thumb I site (P495A, P495S, and V499A), and the thumb II site (M423T). Dasabuvir retained wild-type levels of activity against all of these variants (Table 4).
TABLE 4.
Activity of dasabuvir against variants in the nucleoside binding site, thumb I site, or thumb II site
| Genotype 1b variant | EC50 (mean ± SD) (nM)a | Fold resistance |
|---|---|---|
| S282T | 1.1 ± 0.22 | 0.6 |
| M423T | 0.64 ± 0.18 | 0.4 |
| P495A | 4.3 ± 1.2 | 2.4 |
| P495S | 1.9 ± 0.7 | 1.1 |
| V499A | 2.5 ± 0.5 | 1.4 |
The EC50s and SDs were calculated from at least 3 experiments conducted in duplicate with replicons containing the variants in NS5B, using transient luciferase assays.
Activity of dasabuvir against a panel of genotype 1 clinical isolates.
Given the genetic diversity of HCV, the activity of dasabuvir was evaluated against a panel of clinical isolates derived from treatment-naive individuals infected with genotype 1a or genotype 1b virus (Table 5). The variability at key amino acid positions in the nucleoside, thumb I, thumb II, and palm sites of NS5B, including amino acid positions 96, 282, 316, 368, 395, 411, 414, 423, 445, 448, 451, 495, 496, 499, 553, 556, 559, and 565, was analyzed by population sequencing. In the European HCV database (32), amino acids at the positions listed above are conserved among the genotype 1a sequences. The EC50 of dasabuvir against 11 genotype 1a clinical isolates ranged between 0.18 and 8.57 nM. Variants S556G and M414I were each observed in one subject, and the EC50 values of dasabuvir were 5.17 and 8.57 nM, respectively, in those samples. Among the genotype 1b sequences in the European database, the prevalence of variants at amino acid positions 316, 451, 499, and 566 ranged between 11% to 37%, while the residues at all of the other positions listed above were conserved. The EC50 of dasabuvir against 11 genotype 1b clinical isolates ranged between 0.15 and 2.98 nM. Variants C451G, C451T, V499A, and S556G were observed in isolates from two individuals; however, dasabuvir remained active against both samples. As none of the clinical isolates in this panel contained C316N, a common polymorphism in genotype 1b NS5B according to the sequence database, this variant was introduced into the 1b (Con1) replicon and was found to confer 5-fold resistance to dasabuvir (Table 3).
TABLE 5.
Antiviral activity of dasabuvir in transient expression assays with HCV replicons containing NS5B genes from treatment-naive HCV-infected subjects
| Sample no. | Varianta | EC50 (mean ± SD) (nM) |
|---|---|---|
| Genotype 1a | ||
| 1 | None | 0.99 ± 0.02 |
| 2 | None | 0.18 ± 0.02 |
| 3 | None | 0.40 ± 0.06 |
| 4 | S556G | 5.17 ± 0.58 |
| 5 | None | 0.48 ± 0.07 |
| 6 | M414I | 8.57 ± 1.12 |
| 7 | None | 0.77 ± 0.06 |
| 8 | None | 0.71 ± 0.07 |
| 9 | None | 1.98 ± 0.45 |
| 10 | None | 1.87 ± 0.16 |
| 11 | None | 2.00 ± 0.52 |
| Genotype 1b | ||
| 1 | None | 2.98 ± 0.44 |
| 2 | C451T, V499A | 0.30 ± 0.09 |
| 3 | None | 0.17 ± 0.01 |
| 4 | None | 0.27 ± 0.09 |
| 5 | C451G/C, S556G/S | 0.85 ± 0.09 |
| 6 | None | 0.61 ± 0.08 |
| 7 | None | 0.15 ± 0.05 |
| 8 | None | 0.50 ± 0.05 |
| 9 | None | 0.44 ± 0.14 |
| 10 | None | 0.23 ± 0.12 |
| 11 | None | 0.47 ± 0.10 |
None, variants relative to the genotype 1a (H77) or 1b (Con1) sequence at amino acid positions 96, 282, 316, 368, 395, 411, 414, 423, 445, 448, 451, 495, 496, 499, 553, 556, 559, and 565 were not observed.
DISCUSSION
Dasabuvir was developed to be an inhibitor of the HCV RNA polymerase and to contribute to a multitarget strategy for treatment of HCV genotype 1 infections. The in vitro characterization of dasabuvir consisted of profiling the inhibitory activity of the compound in biochemical enzyme assays and cellular replicon assays, as well as establishing its resistance profile. In biochemical assays, dasabuvir inhibited HCV genotype 1 polymerases with IC50s between 2.2 and 10.7 nM and was largely inactive toward polymerases from HCV genotypes 2, 3, and 4. These results suggest that dasabuvir will be most useful for treating HCV genotype 1 infections. There was no significant inhibitory activity against endogenous human/mammalian polymerases, leading to selectivity factors exceeding 7000:1. This high degree of selectivity likely results from the fact that there is no human counterpart for the RNA-dependent RNA polymerase enzymatic activity possessed by HCV polymerase. In the genotype 1 cell culture system, dasabuvir inhibited replication of HCV subgenomic replicons, with EC50s of 7.7 and 1.8 nM against genotype 1a (H77) and 1b (Con1), respectively. Importantly, dasabuvir was also quite potent (EC50 values of <10 nM) against a panel of 22 genotype 1 treatment-naive clinical isolates, thereby demonstrating that dasabuvir possesses broad activity toward HCV genotype 1a and 1b clinical strains. In the European HCV database, NS5B sequences containing C316N and S556G occur as genotype 1b variants, with prevalences of approximately 37% and 11%, respectively (32). However, C316N and S556G variants in the genotype 1b (Con1) replicon conferred only 5- and 11-fold resistance to dasabuvir, respectively.
The evaluation of the susceptibility of viral sequence variants to dasabuvir was accomplished via resistance selection with genotype 1 replicons. These experiments selected variants at several amino acid positions in the NS5B protein. The predominant variants selected in the genotype 1a replicon were S556G and C316Y, while C316Y and M414T were predominant in the genotype 1b replicon. The C316Y variant in both genotype 1a and 1b replicons and Y448C/H in the genotype 1a replicon conferred >900-fold resistance to dasabuvir. Other variants at amino acid positions 368, 411, 414, 553, 556, and 559 conferred lower levels of resistance. Variants C316Y, M414T, Y448H, and S556G in both genotype 1a and 1b replicons had >30% replication capacity, while the other variants tested at amino acid positions 368, 411, 445, 451, 553, and 559 had <20% replication capacity relative to the wild-type replicon (Table 3). The high replication capacities of the C316Y, M414T, Y448H, and S556G variants combined with high levels of resistance strongly suggest that these viral variants might be expected to arise as variants resistant to dasabuvir monotherapy in human subjects. Indeed, dasabuvir monotherapy has been studied in human subjects as part of clinical studies M10-380 (registered at ClinicalTrials.gov under registration number NCT00851890) and M11-602 (registered at ClinicalTrials.gov under registration number NCT01074008). Both studies were randomized, placebo-controlled, dose-ranging studies evaluating the pharmacokinetics, safety, tolerability, antiviral activity, and resistance of dasabuvir in HCV-infected treatment-naive adults (33, 34). Both studies contained a 3-day monotherapy period followed by a treatment regimen that included pegylated interferon and RBV, which allowed for early on-treatment evaluation of resistant variants. Analyses conducted at postbaseline time points showed the presence of variants C316Y, M414T/I, Y448H, S556G, and D559G/N (33, 34). Thus, the in vitro resistance selection results correlated well with the resistant variants that arose in study participants treated with dasabuvir as monotherapy. Moreover, the selection of significant resistant variants in vitro and in vivo after exposure to dasabuvir emphasizes that the treatment of HCV will require a multitarget approach using a combination of potent direct-acting antiviral agents with nonoverlapping viral targets.
Insights regarding the potential binding sites and inhibitory mechanisms of dasabuvir with the HCV polymerase can also be gleaned from an analysis of resistant variants. The variants selected by dasabuvir were similar to those selected by the benzothiadiazine class of inhibitors, compounds that bind to the palm I site of NS5B (19–21). The palm I site overlaps with the palm II site near the C316 residue, as indicated by the fact that the C316Y variant is resistant to the known palm II inhibitor HCV-796. However, HCV-796 possesses significant inhibitory activity toward genotype 2a, 2b, and 3 polymerases (18). The observation that dasabuvir is not active against genotype 2 and 3 polymerases suggests that it is likely a palm I site binder. Sequence alignments of the palm I site reveal that the genotype 1 Met414 residue is Gln in genotype 2 and Val in genotype 4a. In addition, the genotype 1 Ile447 residue is Met in genotypes 2 to 5. HCV genotype 1 polymerases containing substitutions at positions 414 and 448 are resistant to dasabuvir, thereby demonstrating the importance of the amino acids in this region for the binding of dasabuvir and perhaps providing a rationale for the genotype 1 selectivity of dasabuvir. Thus, the available data suggest that dasabuvir inhibits HCV polymerase through an allosteric mechanism that is similar to that of the benzothiadiazines, blocking the RNA chain initiation steps of the polymerase (Y. Liu, unpublished data). Importantly, the results in Table 4 demonstrate that variants that are typically selected by nucleoside/nucleotide, thumb I, or thumb II site inhibitors are not resistant to dasabuvir. The absence of overlapping resistance profiles suggests that dasabuvir could be effective as part of combination therapy that includes other types of polymerase inhibitors.
In summary, dasabuvir is a selective nonnucleoside inhibitor of HCV genotype 1 RNA polymerases that likely binds to the palm I allosteric inhibitory site of the protein. The resistance profile of dasabuvir supports its use in combination with other direct-acting anti-HCV drugs for the treatment of genotype 1 infections. This in vitro profile provided one of the bases for investigating the combination of dasabuvir with the NS3/NS4A protease inhibitor ABT-450 (plus ritonavir) and the NS5A inhibitor ombitasvir (ABT-267) in human subjects infected with HCV of genotype 1. The combination of these three direct-acting anti-HCV agents provides a high barrier to resistance, as evidenced by the high sustained virological response rates in treatment-naive and treatment-experienced patients, with or without cirrhosis, treated to date in the six phase 3 clinical trials conducted with or without ribavirin (23–27). Therefore, dasabuvir has the potential to play a key role in the treatment of HCV genotype 1 infections worldwide.
ACKNOWLEDGMENTS
The design, study conduct, and financial support for this study were provided by AbbVie. AbbVie participated in interpretation of the data and review and approval of the publication.
Y.L. and R.M. are former employees of AbbVie and may own AbbVie stock. All other authors are employees of AbbVie and may own AbbVie stock.
We thank Hock Ben Lim for assistance with biochemical assays and Barbara McGovern for critical review of the manuscript.
REFERENCES
- 1.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment Web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blatt LM, Mutchnick MG, Tong MJ, Klion FM, Lebovics E, Freilich B, Bach N, Smith C, Herrera J, Tobias H, Conrad A, Schmid P, McHutchison JG. 2000. Assessment of hepatitis C virus RNA and genotype from 6807 patients with chronic hepatitis C in the United States. J Viral Hepat 7:196–202. doi: 10.1046/j.1365-2893.2000.00221.x. [DOI] [PubMed] [Google Scholar]
- 4.Zein NN. 2000. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev 13:223–235. doi: 10.1128/CMR.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyoda H, Kumada T, Takaguchi K, Shimada N, Tanaka J. 2014. Changes in hepatitis C virus genotype distribution in Japan. Epidemiol Infect 142:2624–2628. doi: 10.1017/S0950268814000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. 2014. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus: 15 years on. J Gen Virol 85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 8.Soriano V, Perelson AS, Zoulim F. 2008. Why are there different dynamics in the selection of drug resistance in HIV and hepatitis B and C viruses? J Antimicrob Chemother 62:1–4. doi: 10.1093/jac/dkn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pawlotsky JM. 2014. New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology 146:1176–1192. doi: 10.1053/j.gastro.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Coats SJ, Garnier-Amblard EC, Amblard F, Ehteshami M, Amiralaei S, Zhang H, Zhou L, Boucle SR, Lu X, Bondada L, Shelton JR, Li H, Liu P, Li C, Cho JH, Chavre SN, Zhou S, Mathew J, Schinazi RF. 2014. Chutes and ladders in hepatitis C nucleoside drug development. Antiviral Res 102:119–147. doi: 10.1016/j.antiviral.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Pogam S, Jiang WR, Leveque V, Rajyaguru S, Ma H, Kang H, Jiang S, Singer M, Ali S, Klumpp K, Smith D, Symons J, Cammack N, Najera I. 2006. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349–359. doi: 10.1016/j.virol.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 12.Le Pogam S, Kang H, Harris SF, Leveque V, Giannetti AM, Ali S, Jiang WR, Rajyaguru S, Tavares G, Oshiro C, Hendricks T, Klumpp K, Symons J, Browner MF, Cammack N, Najera I. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J Virol 80:6146–6154. doi: 10.1128/JVI.02628-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomei L, Altamura S, Bartholomew L, Biroccio A, Ceccacci A, Pacini L, Narjes F, Gennari N, Bisbocci M, Incitti I, Orsatti L, Harper S, Stansfield I, Rowley M, De Francesco R, Migliaccio G. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J Virol 77:13225–13231. doi: 10.1128/JVI.77.24.13225-13231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kukolj G, McGibbon GA, McKercher G, Marquis M, Lefebvre S, Thauvette L, Gauthier J, Goulet S, Poupart MA, Beaulieu PL. 2005. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J Biol Chem 280:39260–39267. doi: 10.1074/jbc.M506407200. [DOI] [PubMed] [Google Scholar]
- 15.Fenaux M, Eng S, Leavitt SA, Lee YJ, Mabery EM, Tian Y, Byun D, Canales E, Clarke MO, Doerffler E, Lazerwith SE, Lew W, Liu Q, Mertzman M, Morganelli P, Xu L, Ye H, Zhang J, Matles M, Murray BP, Mwangi J, Zhang J, Hashash A, Krawczyk SH, Bidgood AM, Appleby TC, Watkins WJ. 2013. Preclinical characterization of GS-9669, a thumb site II inhibitor of the hepatitis C virus NS5B polymerase. Antimicrob Agents Chemother 57:804–810. doi: 10.1128/AAC.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemm JA, Liu M, Gentles RG, Ding M, Voss S, Pelosi LA, Wang YK, Rigat KL, Mosure KW, Bender JA, Knipe JO, Colonno R, Meanwell NA, Kadow JF, Santone KS, Roberts SB, Gao M. 2014. Preclinical characterization of BMS-791325, an allosteric inhibitor of hepatitis C virus NS5B polymerase. Antimicrob Agents Chemother 58:3485–3495. doi: 10.1128/AAC.02495-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howe AY, Cheng H, Johann S, Mullen S, Chunduru SK, Young DC, Bard J, Chopra R, Krishnamurthy G, Mansour T, O'Connell J. 2008. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob Agents Chemother 52:3327–3338. doi: 10.1128/AAC.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voitenleitner C, Crosby R, Walker J, Remlinger K, Vamathevan J, Wang A, You S, Johnson J III, Woldu E, Van Horn S, Horton J, Creech K, Shotwell JB, Hong Z, Hamatake R. 2013. In vitro characterization of GSK2485852, a novel hepatitis C virus polymerase inhibitor. Antimicrob Agents Chemother 57:5216–5224. doi: 10.1128/AAC.00874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo H, Lu L, Pilot-Matias T, Pithawalla R, Mondal R, Masse S, Dekhtyar T, Ng T, Koev G, Stoll V, Stewart KD, Pratt J, Donner P, Rockway T, Maring C, Molla A. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob Agents Chemother 49:4305–4314. doi: 10.1128/AAC.49.10.4305-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Dekhtyar T, Masse S, Pithawalla R, Krishnan P, He W, Ng T, Koev G, Stewart K, Larson D, Bosse T, Wagner R, Pilot-Matias T, Mo H, Molla A. 2007. Identification and characterization of mutations conferring resistance to an HCV RNA-dependent RNA polymerase inhibitor in vitro. Antiviral Res 76:93–97. doi: 10.1016/j.antiviral.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Webber SE, Murphy DE, Li L-S, Dragovich PS, Tran CV, Zhongxiang S, Ruebsam F, Shah AM, Tsan M, Showalter RE, Patel R, Li B, Zhao Q, Han Q, Hermann T, Kissinger CR, LeBrun L, Sergeeva MV, Kirkovsky L. 2008. Novel HCV NS5B polymerase inhibitors derived from 4-(1′,1′-dioxo-1′,4′-dihydro-1′λ6-benzo[1′,2′,4′]thiadiazin-3′-yl)-5-hydroxy-2H-pyridazin-3-ones. 1. Exploration of 7′-substitution of benzothiadiazine. Bioorg Med Chem Lett 18:1413–1418. doi: 10.1016/j.bmcl.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Lim BH, Jiang WW, Flentge CA, Hutchinson DK, Madigan DL, Randolph JT, Wagner R, Maring CJ, Kati WM, Molla A. 2012. Identification of aryl dihydrouracil derivatives as palm initiation site inhibitors of HCV NS5B polymerase. Bioorg Med Chem Lett 22:3747–3750. doi: 10.1016/j.bmcl.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Feld JJ, Kowdley KV, Coakley E, Sigal S, Nelson DR, Crawford D, Weiland O, Aguilar H, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 24.Ferenci P, Bernstein D, Lalezari J, Cohen D, Luo Y, Cooper C, Tam E, Marinho RT, Tsai N, Nyberg A, Box TD, Younes Z, Enayati P, Green S, Baruch Y, Bhandari BR, Caruntu FA, Sepe T, Chulanov V, Janczewska E, Rizzardini G, Gervain J, Planas R, Moreno C, Hassanein T, Xie W, King M, Podsadecki T, Reddy KR. 2014. ABT-450/r-ombitasvir and dasabuvir with or without ribavirin for HCV. N Engl J Med 370:1983–1992. doi: 10.1056/NEJMoa1402338. [DOI] [PubMed] [Google Scholar]
- 25.Poordad F, Hezode C, Trinh R, Kowdley KV, Zeuzem S, Agarwal K, Shiffman ML, Wedemeyer H, Berg T, Yoshida EM, Forns X, Lovell SS, Da Silva-Tillmann B, Collins CA, Campbell AL, Podsadecki T, Bernstein B. 2014. ABT-450/r-ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N Engl J Med 370:1973–1982. doi: 10.1056/NEJMoa1402869. [DOI] [PubMed] [Google Scholar]
- 26.Zeuzem S, Jacobson IM, Baykal T, Marinho RT, Poordad F, Bourliere M, Sulkowski MS, Wedemeyer H, Tam E, Desmond P, Jensen DM, Di Bisceglie AM, Varunok P, Hassanein T, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Retreatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1604–1614. doi: 10.1056/NEJMoa1401561. [DOI] [PubMed] [Google Scholar]
- 27.Andreone P, Colombo MG, Enejosa JV, Koksal I, Ferenci P, Maieron A, Müllhaupt B, Horsmans Y, Weiland O, Reesink HW, Rodrigues L, Hu YB, Podsadecki T, Bernstein B. 2014. ABT-450, ritonavir, ombitasvir, and dasabuvir achieves 97% and 100% sustained virologic response with or without ribavirin in treatment-experienced patients with HCV genotype 1b infection. Gastroenterology 147:359–365.e1. doi: 10.1053/j.gastro.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Jiang WW, Pratt J, Rockway T, Harris K, Vasavanonda S, Tripathi R, Pithawalla R, Kati WM. 2006. Mechanistic study of HCV polymerase inhibitors at individual steps of the polymerization reaction. Biochemistry 45:11312–11323. doi: 10.1021/bi060511j. [DOI] [PubMed] [Google Scholar]
- 29.Halfman CJ. 1981. Concentrations of binding protein and labeled analyte that are appropriate for measuring at any analyte concentration range in radioimmunoassays. Methods Enzymol 74C:481–497. [DOI] [PubMed] [Google Scholar]
- 30.Middleton T, He Y, Pilot-Matias T, Tripathi R, Lim BH, Roth A, Chen CM, Koev G, Ng TI, Krishnan P, Pithawalla R, Mondal R, Dekhtyar T, Lu L, Mo H, Kati WM, Molla A. 2007. A replicon-based shuttle vector system for assessing the phenotype of HCV NS5B polymerase genes isolated from patient populations. J Virol Methods 145:137–145. doi: 10.1016/j.jviromet.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 31.Tripathi RL, Krishnan P, He Y, Middleton T, Pilot-Matias T, Chen CM, Lau DT, Lemon SM, Mo H, Kati W, Molla A. 2007. Replication efficiency of chimeric replicon containing NS5A-5B genes derived from HCV-infected patient sera. Antiviral Res 73:40–49. doi: 10.1016/j.antiviral.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Le Mercier P, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky JM, Rice CM, Trepo C, Penin F, Deleage G. 2007. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res 35:D363–D366. doi: 10.1093/nar/gkl970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Middleton T, He Y, Beyer J, Menon R, Klein C, Cohen D, Collins C. 2010. Resistance profile of ABT-333 and relationship to viral load decrease in patients treated in combination with peg-interferon and ribavirin for 28 days. J Hepatol 52:S296–S297. doi: 10.1016/S0168-8278(10)60764-7. [DOI] [Google Scholar]
- 34.Middleton T, He Y, Beyer J, Gaultier IA, Cohen DE, Podsadecki TJ, Bernstein B, Collins C. 2011. Factors affecting HCV viral load response to the nonnucleoside polymerase inhibitors ABT-072 and ABT-333. J Hepatol 54:S483–S484. doi: 10.1016/S0168-8278(11)61226-9. [DOI] [Google Scholar]


