Abstract
The mechanisms of drug resistance development in the Plasmodium falciparum parasite to lumefantrine (LUM), commonly used in combination with artemisinin, are still unclear. We assessed the polymorphisms of Pfmspdbl2 for associations with LUM activity in a Kenyan population. MSPDBL2 codon 591S was associated with reduced susceptibility to LUM (P = 0.04). The high frequency of Pfmspdbl2 codon 591S in Kenya may be driven by the widespread use of lumefantrine in artemisinin combination therapy (Coartem).
TEXT
Chemotherapy is central to the treatment and control of Plasmodium falciparum malaria but faces the parasite's intrinsic ability to quickly develop resistance to antimalarials. The combination of artemisinin and lumefantrine (LUM) (Coartem) is the treatment of choice for uncomplicated malaria in much of Africa (1). However, parasites showing reduced LUM susceptibility have been reported in some countries (2–6). This has been associated with the selection of wild-type parasites at the P. falciparum chloroquine resistance transporter (crt) 76 locus (7, 8) and with at least a 2-fold increase in the frequency of the multidrug resistance 1 (MDR1) 86N mutants following treatment (2, 4, 5, 9). Recently, a single nucleotide polymorphism (SNP) in merozoite surface protein Duffy binding-like 2 (MSPDBL2) codon 591 (C591S) was shown to be associated with increased resistance to halofantrine, mefloquine, and LUM (10). Since LUM is now widely used in the treatment of malaria, it is important to understand the mechanisms of resistance to this drug. We therefore investigated the role of Pfmspdbl2 in the response to LUM in vitro using Kenyan isolates and chloroquine (CQ) as a reference drug. Pfmspdbl2 is a member of the MSP3 multigene family, including mspdbl1, msp3, and msp6 (11). The associated proteins are expressed simultaneously (11) and potentially interact with other proteins on the merozoite parasite membrane in the invasion of the erythrocyte (12, 13). Thus, all 4 genes were included in our investigation.
Parasite genomic DNA was extracted using a QIAmp DNA blood minikit (Qiagen, United Kingdom). We amplified the full-length Pfmspdbl1, Pfmspdbl2, Pfmsp3, and Pfmsp6 from 65 in vitro culture-adapted isolates with chemosensitivity data for CQ and LUM (7, 14) using the primers and PCR cycling conditions described in Table S1 in the supplemental material. PCR products were sequenced using BigDye Terminator v3.1 chemistry (Applied Biosystems, United Kingdom), and the resultant sequences were assembled and edited using SeqMan and aligned using MegAlign (Lasergene 7; DNASTAR, Madison, WI).
Pfmsp3, Pfmsp6, Pfmspdbl1, and Pfmspdbl2 alleles were defined on the basis of their haplotype structure and associations with in vitro drug responses assessed using the Kruskal-Wallis test and the Bonferroni correction for multiple comparisons (15). Thus, P values of <0.001 remained significant. We also determined the median 50% inhibitory concentrations (IC50s) and the 95% confidence intervals (CIs) for each SNP that showed a significant association (P < 0.05) with the drugs tested. Only SNPs with a >5% minor allele frequency were included, and all analyses were conducted using Stata v.11 (StataCorp, College Station, TX).
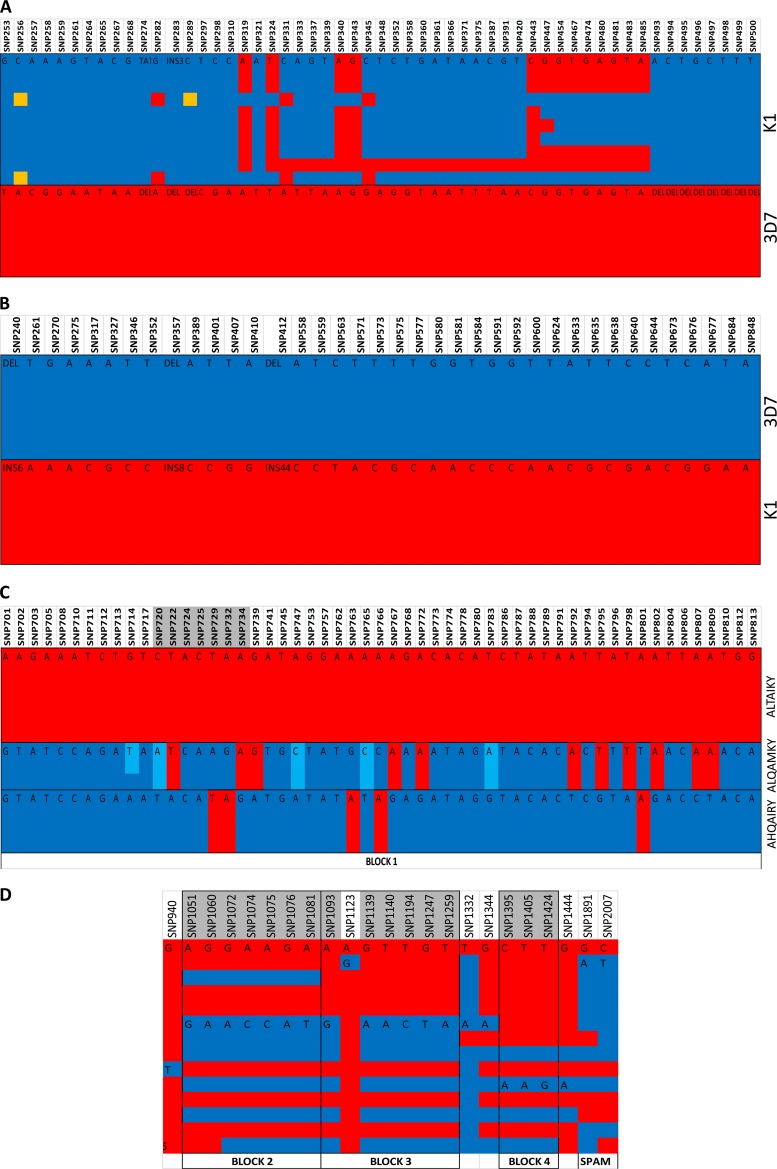
All msp3 and msp6 SNPs analyzed were in linkage disequilibrium, representing two previously defined alleles, K1 and 3D7 (Fig. 1A and B, respectively) (16, 17), therefore precluding individual SNP analysis. The activity of all the drugs tested did not differ in parasites harboring 3D7 or K1 alleles in the msp3 and msp6 genes (Table 1). The SNPs of Pfmspdbl1 and Pfmspdbl2 were used to determine the actual loci within the haplotypes associated with changes in drug responses.
FIG 1.
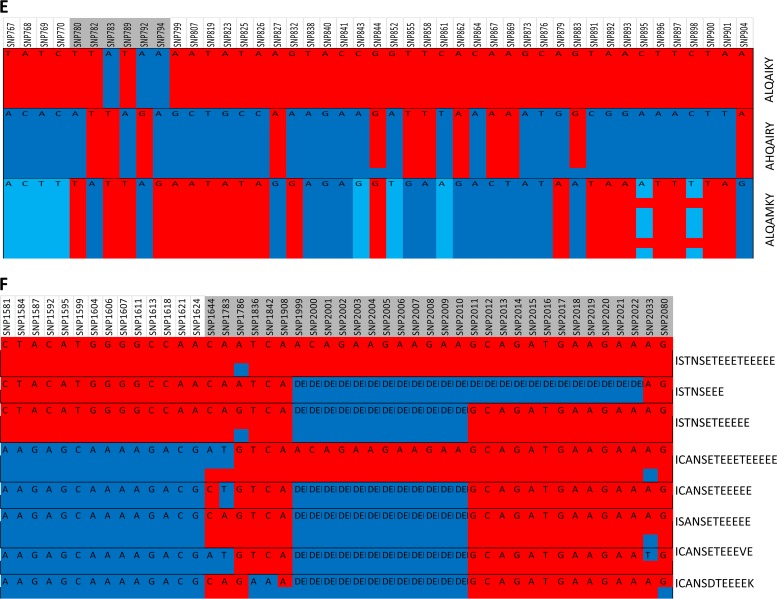
Linkage disequilibrium in the Pfmsp3 multigene family. The haplotypes were generated from an analysis of sequenced SNPs. Haplotypes shown are Pfmsp3 K1 and 3D7 (A), Pfmsp6 K1 and 3D7 (B), Pfmspdbl1 DBL domain AHQAIRY, ALTAIKY, and ALQAMKY (C), Pfmspdbl1 3′ DBL domain NEVRI, DKIQF, and NEIQF block 2, NGGRI and DEGIK block 3, TSV and TTG block 4, and the SPAM domain KN and EN (D), Pfmspdbl2 DBL domain AHQAIRY, ALQAIKY, and ALQAMKY (E), and Pfmspdbl2 SPAM domain 8 (F). Each column represents an SNP, each color in the column represents a different nucleotide, and each row represents an isolate sequence. The black outline depicts the allelic blocks. The columns shaded in gray are in linkage disequilibrium, and the amino acids from these polymorphisms were used to define the haplotypes. INS, number of nucleotides inserted (e.g., INS3 means 3 nucleotides); DEL, absence of sequence (i.e., a deletion).
TABLE 1.
Results of the drug sensitivity assays for chloroquine and lumefantrine compared to the Pfmsp3, Pfmsp6, Pfmspdbl1, and Pfmspdbl2 haplotypes
| Gene | Haplotypec | Chloroquine |
Lumefantrine |
||||
|---|---|---|---|---|---|---|---|
| n (%) | Median IC50 (95% CI) | P value | n (%) | Median IC50 (95% CI) | P value | ||
| MSP3 | K1 | 14 (64) | 36.27 (14.59–98.06) | 14 (64) | 67.34 (47.26–143.97) | ||
| 3D7 | 8 (36) | 23.29 (7.59–183.16) | 0.78 | 8 (36) | 82.7 (45.09–260.0) | 0.68 | |
| MSP6 | K1 | 11 (31) | 80.05 (25.46–181.35) | 11 (31) | 67.56 (40.04–236.42) | ||
| 3D7 | 25 (69) | 54.94 (18.22–87.09) | 0.21 | 25 (69) | 97.85 (66.25–167.88) | 0.36 | |
| MSPDBL1a | |||||||
| BLOCK 1 | AHQAIRY | 6 (20.7) | 83.56 (32.5–108.58) | 6 (20.7) | 74.55 (32.81–339.98) | ||
| ALTAIKY | 17 (58.6) | 31.62 (14.84–109.39) | 17 (58.6) | 124.32 (67.13–274.75) | |||
| ALQAMKY | 6 (20.7) | 56.32 (2.28–107.37) | 0.55 | 6 (20.7) | 197.62 (39.11–357.11) | 0.43 | |
| BLOCK 2 | NEVRI | 20 (56) | 85.7 (58.35–104.22) | 20 (56) | 75.07 (48.39–102.84) | ||
| DKIQF | 13 (36) | 14.99 (13.28–67.4) | 13 (36) | 169.29 (69.42–340.89) | |||
| NEIQF | 3 (8) | 109.46 (54.94–298.1) | 0.0093b | 3 (8) | 75.1 (67.56–104.41) | 0.06 | |
| BLOCK 3 | NGGRI | 23 (64) | 85.23 (55.94–99.74) | 23 (64) | 84.60 (53.17–102.04) | ||
| DEGIK | 13 (36) | 16.77 (13.28–102.07) | 0.068 | 13 (36) | 104.41 (62.7–340.89) | 0.054b | |
| BLOCK 4 | TSV | 31 (84) | 80.14 (30.46–91.55) | 31 (84) | 97.64 (68.23–140.13) | ||
| TTG | 6 (16) | 70.54 (10.53–279.59) | 0.77 | 6 (16) | 85.98 (52.36–402.73) | 0.21 | |
| SPAM domain | KN | 21 (64) | 30.7 (15.3–89.1) | 21 (64) | 97.6 (68.4–115.3) | ||
| EN | 12 (36) | 103.3 (59.2–114.9) | 0.007b | 12 (36) | 61.3 (36.9–270.6) | 0.28 | |
| MSPDBL2 | |||||||
| DBL domain | AHQAIRY | 10 (34.4) | 81.55 (30.42–112.89) | 10 (34.4) | 72.07 (37.26–229.12) | ||
| ALQAIKY | 11 (31.3) | 54.94 (11.08–86.81) | 11 (31.3) | 104.16 (72.53–252.08) | |||
| ALQAMKY | 11 (31.3) | 41.58 (12.74–92.17) | 0.25 | 11 (31.3) | 50.19 (37.91–142.39) | 0.17 | |
| SPAM domain | ISTNSETEEETEEEEE | 2 (6) | 8.41 (2.28–14.55) | 2 (6) | 248.05 (124.32–371.77) | ||
| ISTNSEEE | 2 (6) | 203.78 (109.46–298.1) | 2 (6) | 85.98 (67.56–104.41) | |||
| ISTNSETEEEEE | 8 (29) | 67.91 (16.16–98.52) | 8 (29) | 91.12 (66.47–238.47) | |||
| ICANSETEEETEEEEE | 3 (9) | 86.17 (83.06–104.5) | 3 (9) | 28.47 (22.95–36.76) | |||
| ICANSETEEEEE | 3 (9) | 92.96 (12.79–215.59) | 3 (9) | 48.59 (48.37–97.85) | |||
| ISANSETEEEEE | 6 (19) | 43.13 (11.51–109.58) | 6 (19) | 92.1 (59.18–227.23) | |||
| ICANSETEEEVE | 5 (16) | 30.73 (14.31–167.55) | 5 (16) | 59.54 (38.3–318.93) | |||
| ICANSDTEEEEK | 2 (6) | 15.8 (14.84–16.77) | 0.14 | 2 (6) | 294.57 (242.75–346.39) | 0.034b | |
Blocks 1, 2, and 3 are within the MSPDBL1 DBL domain, while block 4 is between the DBL and SPAM domains.
Significant result (P ≤ 0.05).
Boldface represents the 3D7 reference sequence.
Pfmspdbl1 contained a single DBL domain (11, 13), defined by 3 haplotype blocks (Fig. 1C and D), a secreted polymorphic antigen associated with merozoites (SPAM) domain (codons 631 and 669), and a haplotype block between the DBL and SPAM domains (Fig. 1D). Pfmspdbl2 also contained a single DBL domain (11, 13) and a SPAM domain (13), and we obtained sequence data from codons 127 to 520 (Fig. 1E) and 527 to 694 (Fig. 1F), respectively.
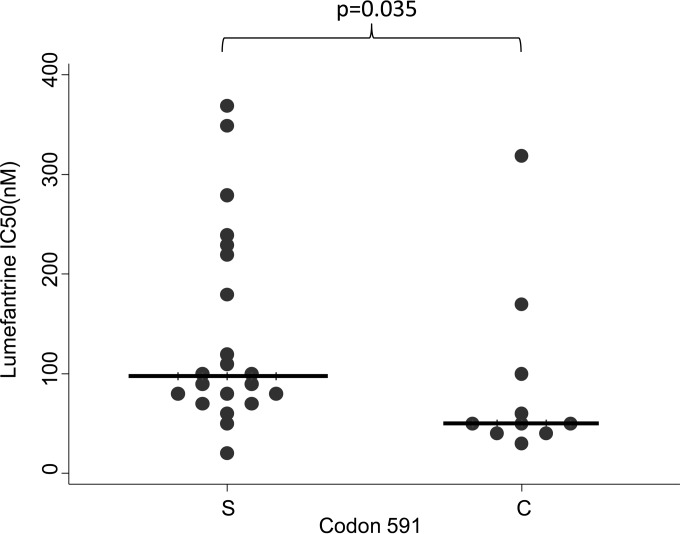
The MSPDBL1 haplotypes (n = 36) showed an association with CQ (P < 0.01) and LUM (P = 0.05) drug activity, while the haplotypes of MSPDBL2 (n = 31) showed an association with LUM (P = 0.03) (Table 1). Twelve Pfmspdbl1 SNPs (Table 2; see also Table S2 in the supplemental material) and 4 SNPs of Pfmspdbl2 (Table 3; see also Table S3 in the supplemental material) were associated with both CQ and LUM. Notably, Pfmspdbl2 SNP1783 (n = 31) codes for codon 591 (since the SNP followed 3 indels, 12 bp long), of which parasites containing serine were associated with reduced susceptibility to LUM (IC50, 97.6 nM; 95% CI, 77.7 to 199.8 nM; P = 0.04) (Fig. 2; Table 3). Codon 591S was also found at a high frequency (68%) in our population, similar to findings in Senegal (80% frequency) (10). This association of Pfmspdbl2 codon 591S with reduced susceptibility to LUM in a different African population adds support to the findings of the study in Senegal and suggests that codon 591 may be a marker for the surveillance of LUM resistance. Importantly, though, codon 591 is not likely to be the causal variant conferring resistance to LUM. Van Tyne et al. (10) demonstrated that stable integrants containing PfMSPDBL2 C591 were more sensitive to mefloquine, halofantrine, and LUM than those with the 591S parasite. The extensive use of LUM in Africa may be the major driving force favoring the high frequency of the 591S mutation.
TABLE 2.
Pfmspdbl1 SNPs significantly associated with drug responses to chloroquine and lumefantrine
| Codon(s) | SNP(s) | Nucleotide(s) (amino acid[s])a | Chloroquine |
Lumefantrine |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | Median IC50 (nM) | 95% CI | P value | n (%) | Median IC50 (nM) | 95% CI | P value | |||
| 351, 354 | 1051, 1060 | G, A (DK) | 12 (32) | 14.7 | 12.6–31.5 | 12 (32) | 206 | 52.4–344.9 | ||
| A, G (NE) | 25 (68) | 86.2 | 58.1–104.3 | 0.0013b | 25 (68) | 80.8 | 66.3–97.8 | 0.04b | ||
| 358, 359 | 1072, 1074, 1075, 1076 | A, C, C, A (IQ) | 14 (38) | 23.8 | 14.0–93.8 | 14 (38) | 136.8 | 66.2–334.8 | ||
| G, A, A, G (VR) | 23 (62) | 85.2 | 55.9–99.7 | 0.05b | 23 (62) | 84.6 | 53.2–102.0 | 0.04b | ||
| 361 | 1081 | T (F) | 15 (41) | 30.7 | 14.4–95.7 | 15 (41) | 111.6 | 68.9–330.0 | ||
| A (I) | 22 (59) | 85.7 | 56.4–102.3 | 0.08 | 22 (59) | 82.7 | 48.5–98.2 | 0.03b | ||
| 365 | 1093 | G (D) | 13 (36) | 16.8 | 13.3–102.1 | 13 (36) | 104.4 | 62.7–340.9 | ||
| A (N) | 23 (64) | 85.2 | 55.9–99.7 | 0.07 | 23 (64) | 84.6 | 53.2–102.0 | 0.05b | ||
| 380 | 1139 | A (E) | 14 (37) | 24.2 | 14.0–98.9 | 14 (37) | 108 | 66.2–334.8 | ||
| G (G) | 24 (63) | 84.1 | 48.2–96.1 | 0.12 | 24 (63) | 87.7 | 60.2–110.5 | 0.06 | ||
| 380, 398, 416 | 1140, 1194, 1247 | A, C, T (EGI) | 14 (38) | 24.2 | 14.0–98.9 | 14 (38) | 108 | 66.2–334.8 | ||
| T, T, G (GGR) | 23 (62) | 85.2 | 55.9–99.7 | 0.11 | 23 (62) | 84.6 | 53.2–102.0 | 0.045b | ||
| 420 | 1259 | A (K) | 15 (39) | 31.6 | 14.4–95.7 | 15 (39.5) | 111.6 | 68.9–330.0 | ||
| T (I) | 23 (61) | 85.2 | 55.9–99.7 | 0.09 | 23 (60.5) | 84.6 | 53.2–102.0 | 0.035b | ||
| 444 | 1332 | A (K) | 26 (74) | 84.1 | 31.1–99.7 | 26 (74) | 104.3 | 77.7–206.6 | ||
| T (N) | 9 (26) | 56.4 | 12.8–103.6 | 0.5 | 9 (26) | 48.6 | 36.9–93.8 | 0.03b | ||
| 631 | 1891 | A (K) | 24 (67) | 30.5 | 14.9–82.5 | 24 (67) | 100.7 | 76.9–160.3 | ||
| G (E) | 12 (33) | 103.3 | 59.2–114.9 | 0.0025b | 12 (33) | 61.8 | 36.9–270.6 | 0.17 | ||
| 669 | 2007 | T (N) | 19 (58) | 30.7 | 15.5–82.3 | 19 (58) | 97.6 | 73.3–138.1 | ||
| C (N) | 14 (42) | 106.4 | 78.6–123.8 | 0.0015b | 14 (42) | 58.1 | 38.0–240.1 | 0.19 | ||
The bold letters highlight the 3D7 reference alleles.
Significant result (P ≤ 0.05).
TABLE 3.
Pfmspdbl2 SNPs significantly associated with drug responses to chloroquine and lumefantrine
| Codon(s) | SNP | Nucleotide(s)a (amino acid[s]) | Chloroquine |
Lumefantrine |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | Median IC50 (nM) | 95% CI | P value | n (%) | Median IC50 (nM) | 95% CI | P value | |||
| 591 | 1783 | T (C) | 10 (32) | 58.9 | 14.4–147.1 | 10 (32) | 50.1 | 37.3–146.1 | ||
| A (S) | 21 (68) | 55.8 | 16.2–90.6 | 0.77 | 21 (68) | 97.6 | 77.7–199.8 | 0.035b | ||
| 608, 610 | 1836, 1842 | TC (NS) | 28 (93) | 68.2 | 30.1–95.7 | 28 (93) | 87.7 | 61.6–109.6 | ||
| AA (KR) | 2 (7) | 15.8 | 14.8–16.8 | 0.23 | 2 (7) | 294.6 | 242.8–346.4 | 0.046b | ||
| 667 | 2011–2010 | ACAGAAGAAGAA (TEEE) | 29 (94) | 54.9 | 16.4–84.0 | 2 (6) | 86 | 67.6–104.4 | ||
| DEL | 2 (6) | 203.8 | 109.5–298.1 | 0.04b | 29 (94) | 90.8 | 59.1–138.1 | 0.94 | ||
The bold letters highlight the 3D7 reference alleles.
Significant result (P ≤ 0.05).
FIG 2.
Pfmspdbl2 codon 591 (n = 46) S allele is associated with reduced susceptibility to LUM (P = 0.035). The horizontal lines indicate the median drug IC50s.
The observed inverse drug relationship of 4 Pfmspdbl1 codons (Table 2) associated with resistance to CQ, for instance, codons 351 and 354 (NE, IC50, 86.2 nM; 95% CI 58.1 to 104.3 nM; P = 0.001), and susceptibility to LUM (NE, IC50 = 80.8 nM, 95% CI, 66.3 to 97.8 nM; P = 0.04) is reminiscent of the previously described inverse relationship of wild-type CQ parasites showing resistance to LUM (8, 18, 19). This inverse relationship between drugs shown by SNPs of Pfmspdbl1 most likely occurs on a backdrop of the underlying inverse relationship driven by CQ and LUM, since they are drugs that have been widely used for malaria treatment. Consequently, LUM-artemisinin may select for wild-type crt, associated with CQ sensitivity (20) and implicated as a marker of LUM tolerance (2, 7), suggesting that LUM is likely to confer resistance via a different mechanism.
Not surprisingly, none of the Pfmsp3, Pfmsp6, Pfmspdbl1, or Pfmspdbl2 SNPs were in linkage disequilibrium with the K76T crt locus (data not shown). Additionally, the MSP3 gene family contains multiple high-frequency polymorphisms, which would increase the probability of random associations with drug activity. Furthermore, since Pfmspdbl2 has shown evidence of being under balancing selection and is likely to be under immune pressure (21), its role in immunity cannot be ignored. However, it remains to be determined if the C591S mutation can be used as a surveillance marker of LUM resistance in the field.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Malaria Capacity Development Consortium (MCDC) reentry grant to L.I.O.-O.
We thank Abdirahman Abdi for his comments on the manuscript, and we thank the director of the Kenya Medical Research Institute for permission to publish the article.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03522-14.
REFERENCES
- 1.WHO. 2013. Malaria: country antimalarial drug policies: by region. WHO, Geneva, Switzerland: http://www.who.int/malaria/am_drug_policies_by_region_searo/en/index.html. [Google Scholar]
- 2.Sisowath C, Strömberg J, Mårtensson A, Msellem M, Obondo C, Björkman A, Gil JP. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis 191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 3.Sisowath C, Ferreira PE, Bustamante LY, Dahlström S, Mårtensson A, Björkman A, Krishna S, Gil JP. 2007. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health 12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- 4.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. 2006. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother 50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJM, Mutabingwa TK, Sutherland CJ, Hallet RL. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother 51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Happi CT, Gbotosho GO, Folarin OA, Sowunmi A, Hudson T, O'Neil M, Milhous W, Wirth DF, Oduola AM. 2009. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob Agents Chemother 53:888–895. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, Bull P, Marsh K, Borrmann S, Nzila A. 2009. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother 53:5069–5073. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sisowath C, Petersen I, Veiga MI, Mårtensson A, Premji Z, Björkman A, Fidock DA, Gil JP. 2009. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis 199:750–757. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mårtensson A, Strömberg J, Sisowath C, Msellem MI, Gil JP, Montgomery SM, Olliaro P, Ali AS, Björkman A. 2005. Efficacy of artesunate plus amodiaquine versus that of artemether-lumefantrine for the treatment of uncomplicated childhood Plasmodium falciparum malaria in Zanzibar, Tanzania. Clin Infect Dis 41:1079–1086. doi: 10.1086/444460. [DOI] [PubMed] [Google Scholar]
- 10.Van Tyne D, Uboldi AD, Healer J, Cowman AF, Wirth DF. 2013. Modulation of PF10_0355 (MSPDBL2) alters Plasmodium falciparum response to antimalarial drugs. Antimicrob Agents Chemother 57:2937–2941. doi: 10.1128/AAC.02574-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh S, Soe S, Weisman S, Barnwell JW, Pérignon JL, Druilhe P. 2009. A conserved multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum. PLoS One 4:e5410. doi: 10.1371/journal.pone.0005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills KE, Pearce JA, Crabb BS, Cowman AF. 2002. Truncation of merozoite surface protein 3 disrupts its trafficking and that of acidic-basic repeat protein to the surface of Plasmodium falciparum merozoites. Mol Microbiol 43:1401–1411. doi: 10.1046/j.1365-2958.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 13.Hodder AN, Czabotar PE, Uboldi AD, Clarke OB, Lin CS, Healer J, Smith BJ, Cowman AF. 2012. Insights into Duffy binding-like domains through the crystal structure and function of the merozoite surface protein MSPDBL2 from Plasmodium falciparum. J Biol Chem 287:32922–32939. doi: 10.1074/jbc.M112.350504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okombo J, Kiara SM, Rono J, Mwai L, Pole L, Ohuma E, Borrmann S, Ochola LI, Nzila A. 2010. In vitro activities of quinine and other antimalarials and pfnhe polymorphisms in Plasmodium isolates from Kenya. Antimicrob Agents Chemother 54:3302–3307. doi: 10.1128/AAC.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bland JM, Altman DG. 1995. Multiple significance tests: the Bonferroni method. BMJ 310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber W, Felger I, Matile H, Lipps HJ, Steiger S, Beck HP. 1997. Limited sequence polymorphism in the Plasmodium falciparum merozoite surface protein 3. Mol Biochem Parasitol 87:231–234. doi: 10.1016/S0166-6851(97)00067-4. [DOI] [PubMed] [Google Scholar]
- 17.Pearce JA, Triglia T, Hodder AN, Jackson DC, Cowman AF, Anders RF. 2004. Plasmodium falciparum merozoite surface protein 6 is a dimorphic antigen. Infect Immun 72:2321–2328. doi: 10.1128/IAI.72.4.2321-2328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pradines B, Tall A, Fusai T, Spiegel A, Hienne R, Rogier C, Trape JF, Le Bras J, Parzy D. 1999. In vitro activities of benflumetol against 158 Senegalese isolates of Plasmodium falciparum in comparison with those of standard antimalarial drugs. Antimicrob Agents Chemother 43:418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price RN, Uhlemann A-C, van Vugt M, Brockman A, Hutagalung R, Nair S, Nash D, Singhasivanon P, Anderson TJ, Krishna S, White NJ, Nosten F. 2006. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis 42:1570–1577. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nzila A, Okombo J, Ohuma E, Al-Thukair A. 2012. Update on the in vivo tolerance and in vitro reduced susceptibility to the antimalarial lumefantrine. J Antimicrob Chemother 67:2309–2315. doi: 10.1093/jac/dks252. [DOI] [PubMed] [Google Scholar]
- 21.Ochola LI, Tetteh KKA, Stewart LB, Riitho V, Marsh K, Conway DJ. 2010. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol Biol Evol 27:2344–2351. doi: 10.1093/molbev/msq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.