Abstract
Most Mycobacterium tuberculosis rifampin-resistant strains have been associated with mutations in an 81-bp rifampin resistance-determining region (RRDR) in the gene rpoB. However, if this region alone were targeted, rifampin-resistant strains with mutations outside the RRDR would not be detected. In this study, among 51 rifampin-resistant clinical isolates analyzed by sequencing 1,681-bp-long DNA fragments containing the RRDR, 47 isolates contained mutations within the RRDR, three isolates contained mutations both within and outside the RRDR, and only one isolate had a single missense mutation (Arg548His) located outside the RRDR. A drug susceptibility test of recombinant Mycobacterium smegmatis and M. tuberculosis isolates carrying mutated rpoB (Arg548His) showed an increased MIC for rifampin compared to that of the control strains. Modeling of the Arg548His mutant RpoB-DNA complex revealed that the His548 side chain formed a more stable hydrogen bond structure than did Arg548, reducing the flexibility of the rifampin-resistant cluster II region of RpoB, suggesting that the RpoB Arg548His mutant does not effectively interact with rifampin and results in bacterial resistance to the drug. This is the first report on the relationship between the mutation in codon 548 of RpoB and rifampin resistance in tuberculosis. The novel mutational profile of the rpoB gene described here will contribute to the comprehensive understanding of rifampin resistance patterns and to the development of a useful tool for simple and rapid drug susceptibility tests.
INTRODUCTION
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis. Globally, an estimated 3.6% of new TB cases and 20.2% of previously treated TB cases are considered multidrug-resistant TB (MDR-TB) (1). In Taiwan in 2011, MDR-TB comprised 0.9% of new cases and 6.7% of previously treated cases of TB (2). Rifampin is a first-line anti-TB drug that is often associated with treatment failure (3, 4). The bactericidal mechanism of rifampin functions by binding to the β-subunit of RNA polymerase, the product of the rpoB gene (5), thus suppressing transcription in bacterial cells. More than 96% of rifampin-resistant strains have mutations within the 81-bp rifampin resistance-determining region (RRDR) of the rpoB gene (codons 507 to 533) (4). Additionally, the frequency of codon mutations in rpoB of rifampin-resistant M. tuberculosis isolates varies across different geographical regions. The most common mutations in the RRDR are found in codons 526 and 531 and account for 62.5% to ∼81.1% of rifampin-resistant strains (6–8). However, not all mutations within the RRDR lead to the same level of rifampin resistance. Amino acid alterations in codon 526 or 531 cause greater resistance in bacteria than do mutations in codon 511, 516, 518, or 522 (9). Outside the RRDR, rare rifampin-resistant mutations have been reported in codons 176 (Val176Phe) (10), 381 (Ala381Val) (11), 490 (Gln490His) (12), 500 (Ala500Val), 502 (Ile502Val), 505 (Phe505Ser), 538 (Leu538Phe) (13), 146 (Val146Phe), and 572 (Ile572Phe) (4, 14). Thus, reference laboratories using molecular methods to examine the 81-bp RRDR only may miss strains in which rifampin resistance is suspected but no mutation is found.
The molecular surveillance of rifampin-resistant M. tuberculosis isolates in western, northern, and southern Taiwan has been reported in the past decade (6, 15–17). However, similar studies in eastern Taiwan, which accounts for two-thirds of the country and is characterized by rugged mountains, have not been carried out to a sufficient degree. The ethnic groups and lifestyles of the people in eastern Taiwan, who account for approximately 4.4% of the total population, are very different from those in other regions of the country. Here, we studied the prevalence of rpoB mutations associated with rifampin-resistant M. tuberculosis isolates in eastern Taiwan. We found one novel rpoB allele and constructed recombinant M. tuberculosis and Mycobacterium smegmatis strains carrying this mutated rpoB gene to demonstrate that it plays a role in bacterial resistance to rifampin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The clinical M. tuberculosis isolates were collected from Tzu Chi Hospital in Hualien, located in eastern Taiwan, from 2011 to 2012. The isolation rate of M. tuberculosis is 7.67%. Among these isolates, 51 were resistant to rifampin. The other laboratory strains and plasmids used in this study are listed in Table 1. M. smegmatis strain mc2155 and M. tuberculosis strain H37Rv were used as mycobacterial hosts to carry recombinant plasmids for drug susceptibility testing. M. smegmatis mc2155 and M. tuberculosis H37Rv and their transformants were cultured in Middlebrook 7H9 broth (Difco Laboratories, Detroit, MI, USA) supplemented with 10% Tween 80 or on 7H11 agar supplemented with oleic acid-albumin-dextrose-catalase (OADC) (Difco Laboratories, Detroit, MI, USA). Escherichia coli strain DH5α was used for DNA cloning and was incubated at 37°C in Luria-Bertani (LB) medium. Bacteria containing the pGEM-T Easy vector (Promega, WI, USA) were grown in LB medium supplemented with ampicillin (50 μg/ml). The primers designed in this study are listed in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| M. tuberculosis | ||
| MTBR548H | Clinical isolates resistant to rifampin but not to isoniazid | This study |
| E. coli DH5α | supE44ΔlacU169 (f80 lacZΔM15)hsdR1 recA1 endA1 gyrA96 thi-1 relA1 | 32 |
| M. smegmatis mc2155 | Wild type | ATCC 700084 |
| CH003 | M. smegmatis mc2155/pMV261 vector only, Kmr | This study |
| CH004 | M. smegmatis mc2155/pMV261::rpoB_WTc, Kmr | This study |
| CH005 | M. smegmatis mc2155/pMV261::rpoB_R548Hc, Kmr | This study |
| CH006 | M. smegmatis mc2155/pMV261::rpoB_S531Lc, Kmr | This study |
| M. tuberculosis H37Rv | Wild type | ATCC 27294 |
| YY001 | M. tuberculosis H37Rv/ pMV261 vector only, Kmr | This study |
| YY002 | M. tuberculosis H37Rv/pMV261::rpoB_R548Hc, Kmr | This study |
| YY003 | M. tuberculosis H37Rv/ pMV261::rpoB_WTc, Kmr | This study |
| YY004 | M. tuberculosis H37Rv/pMV261::rpoB_S531Lc, Kmr | This study |
| Plasmids | ||
| pGEM-T easy | Cloning vector, Ampr | Promega, WI, USA |
| pMV261 | Mycobacterium species/E. coli shuttle vector, Kmr | 33 |
| pMV261::rpoB_WTc | pMV261 containing wild-type rpoB from M. tuberculosis H37Rv, Kmr | This study |
| pMV261::rpoB_R548Hc | pMV261 containing mutated rpoB from rifampin-resistant M. tuberculosis, codon 548 of RpoB was changed to histidine, Kmr | This study |
| pMV261::rpoB_S531Lc | pMV261 containing mutated rpoB from rifampin-resistant M. tuberculosis, codon 531 of RpoB was changed to lysine, Kmr | This study |
Kmr, kanamycin resistant; Ampr, ampicillin resistant.
TABLE 2.
Primers designed in this study
| Primer | Sequence (5′–3′) | Amplicon size |
|---|---|---|
| rpoB-FP | CCCGCCAGAGCAAAACAGC | 1,681 bp |
| rpoB-RP | TACTCCACCTCGCCCGCC | |
| rpoB-seq-F | AATATCTGGTCCGCTTGCAC | |
| rpoB-seq-R | ACGAGGGCACGTACTCCA | |
| rpoB-full-cF | GCGAATTCGGAAGGAAAAATGGCAGATTCCCGCCAG | 3,545 bp |
| rpoB-full-cR | GCAAGCTTTTACGCAAGATCCTCGACAC |
Specimen collection and processing.
Sputum specimens were liquefied and decontaminated by the standard N-acetyl-l-cysteine–sodium hydroxide (NALC-NaOH) method. The sediment from each specimen was inoculated onto the culture medium and used for acid-fast staining. The bacterial isolates were identified by conventional methods that included routine microscopy, culture, and positive nitrate and niacin tests (18).
Mycobacterial DNA extraction, PCR amplification, and DNA sequencing.
The mycobacterial chromosomal DNA grown on Middlebrook 7H11 agar plates was extracted according to procedure described in the previous study (19). In brief, an aliquot of the bacterial pellet was lysed in lysis buffer (KOH [pH 13.1]) at 95°C for 15 min before being neutralized by neutralization buffer (HCl and acetic acid [pH 1.2]). An aliquot of crude extract suspension was used in the PCRs. The rpoB fragments were amplified by PCR in a Biometra Uno-Thermoblock (Biometra, Göttingen, Germany) using the primer pair rpoB-FP/rpoB-RP (Table 2). The PCRs began with 5 min of denaturation at 95°C, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 57°C for 1 min, extension at 72°C for 2 min, and a final extension at 72°C for 2 min. Next, DNA sequencing of the PCR fragments was performed by Tri-I Biotech, Inc. (New Taipei City, Taiwan), using the rpoB-FP, rpoB-RP, rpoB-seq-F, or rpoB-seq-R primer. The rpoB-seq-F and rpoB-seq-R primers were designed for confirming the sequence of a 693-bp DNA region, including the RRDR. The sequences obtained from the 51 clinical isolates were compared with the known sequence of M. tuberculosis H37Rv rpoB (GenBank accession no. NC_000962.3).
Construction of a recombinant mycobacterial strain carrying exogenous rpoB.
Wild-type and mutated rpoB DNA fragments were amplified by PCR using the primer pair rpoB-full-cF/rpoB-full-cR (Table 2), and chromosomal DNA from M. tuberculosis H37Rv, a clinical M. tuberculosis isolate with the Ser531Leu mutation in rpoB, and a clinical M. tuberculosis isolate with the Arg548His mutation in rpoB (MTBR548H) was used as the template. The PCRs began with a 5-min denaturation at 95°C, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 58°C for 1 min, extension at 72°C for 2 min, and a final step at 72°C for 2 min. The PCR fragments were cloned into the pGEM-T Easy plasmid (Promega, WI, USA), followed by excision with EcoRI/HindIII and ligation into pMV261 (Table 1) (20). The constructs were then confirmed by DNA sequencing. To prepare competent M. tuberculosis and M. smegmatis cells, the bacteria were incubated in 7H9 broth containing 0.2 M glycine at 37°C, with shaking at 220 rpm, until the optical density at 600 nm (OD600) reached 0.8 to ∼1.0. Subsequently, the bacterial cells were washed three times with ice-cold 10% (wt/vol) glycerol. Finally, the competent cells were suspended in ice-cold 10% (wt/vol) glycerol, and the recombinant plasmid pMV261::rpoB_WTc, pMV261::rpoB_S531Lc, or pMV261::rpoB_R548Hc was transformed into competent M. tuberculosis H37Rv and M. smegmatis mc2155 cells by electroporation (BTX ECM 630 system; Harvard Apparatus, MA, USA) at 2,500 V, 1,000 Ω, and 25 μF.
Drug susceptibility testing.
Drug susceptibility testing for mycobacterial clinical isolates was determined by the standard agar proportion method, using 1 μg/ml rifampin as a breakpoint (21). The MIC was determined using the standard agar proportion method on 7H11 agar for recombinant M. tuberculosis and the broth microdilution method for recombinant M. smegmatis (22). In brief, aliquots of recombinant M. smegmatis (5 × 103 cells/ml) were inoculated into a 96-well microtiter plate containing 2-fold serial dilutions of rifampin (from 0.125 to 64 μg/ml) in Mueller-Hinton broth with OADC. Next, the bacteria were incubated at 37°C and 200 rpm for 3 to 5 days. Aliquots of recombinant M. tuberculosis and a 1:100 dilution control were inoculated on 7H11 agar containing OADC and 2-fold serial dilutions of rifampin (from 0.125 to 64 μg/ml). The MIC tests were performed three times. The MIC for each strain was defined as the lowest concentration of rifampin needed to inhibit bacterial growth.
Structural modeling.
Structural models of M. tuberculosis RpoB bound to rifampin or DNA were developed using SWISS-MODEL, a fully automated protein structure homology-modeling server, using Protein Data Bank (PDB) accession code 1I6V (Thermus aquaticus RNA polymerase [RNAP] complexed with rifampin) or 4G7Z (Thermus thermophilus RNAP complexed with DNA) as a template. The M. tuberculosis RpoB Arg548His (using E. coli residue numbering) mutant was generated by Coot (23), using the RpoB-DNA modeling structure. All structural models were optimized by energy minimization using REFMAC5 (24) in the CCP4 program suite (25, 26). Prior to generation of structural figures, all models were superimposed by the secondary-structure matching (SSM) algorithm of the PDBeFold server (27). The structural figures were produced using PyMOL (DeLano Scientific [http://www.pymol.org]).
RESULTS
Genetic analysis of rpoB genes in rifampin-resistant M. tuberculosis isolated from eastern Taiwan.
Fifty-one rifampin-resistant isolates were collected from eastern Taiwan. To determine whether the DNA sequence outside the RRDR of rpoB that codes for codons 507 to 533 of RpoB (using E. coli numbering) was associated with rifampin resistance in M. tuberculosis, a 1,681-bp DNA fragment containing the RRDR was amplified from each rifampin-resistant isolate by PCR and analyzed by DNA sequencing. After comparison with the rpoB sequence of M. tuberculosis H37Rv, the most commonly mutated sites in RpoB were found to be codons 531 (74.5%) and 526 (21.6%), which are both located in the RRDR (Table 3). Of the 51 rifampin-resistant isolates, 50 were mutated within the RRDR, while only one clinical isolate, named MTBR548H, had a single mutation outside this region. Although three isolates had a mutation at codons 500, 552 and 576, respectively, outside the RRDR, they had a second or third mutation in the RRDR (Table 3). Most rifampin-resistant isolates (72.5%) had one mutated codon in RpoB; however, 23.5% and 3.9% of the rifampin-resistant isolates had two and three mutated RpoB codons, respectively (Table 4).
TABLE 3.
Mutations in rpoB gene in 51 rifampin-resistant M. tuberculosis isolates
| Codon mutation | Nucleic acid change | No. of strains | Frequency (%) |
|---|---|---|---|
| Ala500Gly | GCC→GGC | 1a | 2.0 |
| Ser512Asnb | AGC→AAC | 1 | 2.0 |
| Met515Leub | ATG→TTG | 1 | 2.0 |
| Asp516Valb | GAC→GTC | 3 | 5.9 |
| His526Tyrb | CAC→TAC | 1 | 2.0 |
| His526Asnb | CAC→AAC | 10 | 19.6 |
| Ser531Leub | TCG→TTG | 38 | 74.5 |
| Leu533Prob | CTG→CCG | 10 | 19.6 |
| Arg548His | CGC→CACc | 1 | 2.0 |
| Pro552Ser | CCG→TCG | 1d | 2.0 |
| Pro567Ser | CCT→TCT | 1e | 2.0 |
Isolate had three mutated codons in RpoB: Ala500Gly, Met515Leu and Asp516Val.
Mutations located within the 81-bp RRDR of rpoB gene. There were 11 isolates having two mutated codons, both in the RRDR.
The novel allele was identified in this study.
Isolate had two mutated codons in RpoB: Ser531Leu and Pro552Ser.
Isolate had three mutated codons in RpoB: Ser531Leu, Leu533Pro, and Pro567Ser.
TABLE 4.
Frequencies of single or multiple mutations in RpoB in 51 rifampin-resistant M. tuberculosis isolates
| No. of codons mutated in RpoB | No. of strains | Frequency (%) |
|---|---|---|
| 1 | 37 | 72.5 |
| 2 | 12 | 23.5 |
| 3 | 2 | 3.9 |
| 4 | 0 | 0 |
Mutation of codon 548 in RpoB results in resistance to rifampin in recombinant M. tuberculosis and M. smegmatis.
In the rifampin-resistant isolates, one novel rpoB allele that contained a single mutation at codon 548, outside the 81-bp RRDR, was identified in the clinical mutant MTBR548H (Table 3). To determine whether the mutation of codon 548 (Arg548His) in RpoB was involved in bacterial resistance to rifampin, we constructed recombinant M. tuberculosis H37Rv and M. smegmatis mc2155 strains that carried mutated or wild-type rpoB (Table 1). A bacterium carrying rpoB with the mutated codon Ser531Leu was constructed as a positive control, due to an association between this common missense mutation of RpoB and high levels of mycobacterial rifampin resistance (Table 1). Next, the susceptibilities of the recombinant strains to rifampin were tested. The rifampin MIC for the recombinant M. smegmatis strain containing the wild-type M. tuberculosis rpoB gene (named CH004) was 2 μg/ml. Meanwhile, the recombinant M. smegmatis strains containing RpoB mutated at codon 531 and 548 (named CH006 and CH005, respectively) showed MICs of 16 μg/ml and 4 μg/ml, respectively, which were elevated 8-fold and 2-fold compared with those of the strains containing either the vector control or wild-type rpoB (Table 5). Additionally, an M. tuberculosis H37Rv strain harboring RpoB (Arg548His), named YY002, had an MIC of 2 to ∼4 μg/ml, which was elevated >16-fold compared with that of an M. tuberculosis strain harboring the wild-type rpoB (named YY003) (Table 6). The M. tuberculosis H37Rv strain harboring RpoB with mutated codon 531 (named YY004) was used as a positive control, which showed rifampin resistance (Table 6). Thus, our results indicated that a missense mutation in RpoB at codon 548 (Arg548His), which is located outside the RRDR, slightly increased the resistance of mycobacteria to rifampin.
TABLE 5.
Rifampin susceptibilities of the recombinant M. smegmatis strains with wild-type RpoB or mutated RpoB
| M. smegmatis strain | Mutation information | Rifampin MIC (μg/ml)a |
|---|---|---|
| CH003 | pMV261b | 2 |
| CH004 | Wild-type rpoB | 2 |
| CH005 | Arg548His | 4 |
| CH006 | Ser531Leu | 16 |
Each MIC test was performed three times.
Vector control.
TABLE 6.
Rifampin susceptibilities of the recombinant M. tuberculosis strains with wild-type RpoB or mutated RpoB using agar proportion method
| M. tuberculosis strain | Mutation information | Results at a dilution ofa: |
|||
|---|---|---|---|---|---|
| 10−2 |
10−4 |
||||
| Rifampin MIC (μg/ml)b | Susceptibilityc | Rifampin MIC (μg/ml)b | Susceptibilityc | ||
| YY001 | pMV261d | <0.125 | S | <0.125 | S |
| YY002 | Arg548His | 2 to ∼4 | R | 2 | R |
| YY003 | Wild-type rpoB | <0.125 | S | <0.125 | S |
| YY004 | Ser531Leu | >64 | R | >64 | R |
The 10−2 or 10−4 dilutions of standardized bacterial suspension in sterile water were prepared, followed by inoculation of a 0.1-ml suspension on 7H11 agar.
Each MIC test was performed three times.
Resistance is defined as the growth of >1% of an inoculum of bacterial cells in 1 μg/ml rifampin. S, susceptible; R, resistant.
Vector control.
Structural models of M. tuberculosis RpoB bound to rifampin or promoter DNA.
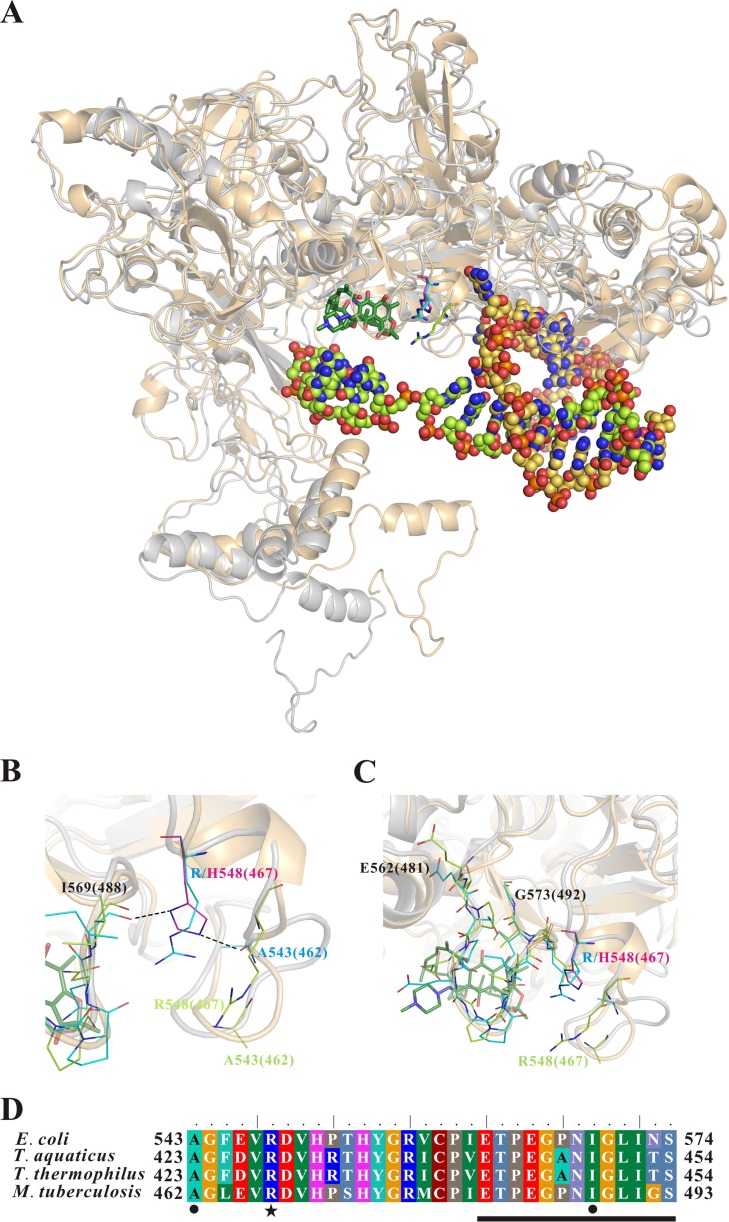
To address the structural effect of the Arg548His RpoB mutation on M. tuberculosis rifampin resistance, structural models of RpoB bound to rifampin or DNA were generated by a homology-modeling server. In the RpoB-rifampin complex model, it appeared that residue Arg548 was shifted away from the rifampin-binding pocket of RpoB and nonessential residues for RpoB binding to rifampin (Fig. 1). However, in the RpoB-DNA complex model, it appeared that residue Arg548 was close to the rifampin-resistant cluster II region of RpoB (Fig. 1) (5). In the Arg548His mutant of the RpoB-DNA complex model, the imidazole ring of His548 seemed to provide two hydrogen bonds, one between His548 and Ile569 and one between His548 and Ala543 (Fig. 1B), leading to increased rigidity of the rifampin-resistant cluster II region of RpoB (Fig. 1C). We also found dramatic variations in the side-chain orientation of the rifampin-resistant cluster II region of RpoB in the rifampin- and DNA-binding models (Fig. 1C). We speculated that the side chain of His548 could provide a more stable hydrogen bond-linking structure than that of Arg548, thus reducing the flexibility of the rifampin-resistant cluster II region of RpoB. Thus, the rifampin-resistant cluster II region of the Arg548His RpoB mutant was unable to interact effectively with rifampin, resulting in bacterial rifampin resistance.
FIG 1.
Structural models of M. tuberculosis RpoB that bound with rifampin or promoter DNA. (A) Superimposition of the models of M. tuberculosis RpoB that bound with rifampin (protein in gray) or promoter DNA (protein in light orange). The rifampin is shown in stick model (carbon atoms in dark green). The DNA is shown in sphere model (carbon atoms in light green and yellow-orange). The carbon atoms of the key residues for the RpoB-rifampin complex, RpoB-DNA complex, and RpoB mutant-DNA complex are shown in the light green, cyan, and magenta line models, respectively. Oxygen atoms are shown in red, nitrogen atoms are in blue, and phosphorus atoms are in orange. (B and C) Close-up view of the Arg548-located loop (B) and the rifampin-resistant cluster II region (C) of RpoB. The potential hydrogen bonds are indicated by black dashed lines. The color coding of the residue numbers is the same as that in the color painting of the line models. The residue numbers are labeled according to the E. coli RpoB protein sequence. The values in the parentheses are for the M. tuberculosis RpoB protein sequence. (D) Protein sequence alignment of the Arg548-located loop and the rifampin-resistant cluster II loop of four prokaryotic RpoB. The rifampin-resistant cluster II region of RpoB is marked with a black bar. The Arg548His mutant site and the residues interacting with His548 by a potential hydrogen bond are indicated by a black asterisk and dots, respectively.
DISCUSSION
Although mycobacterial resistance to rifampin is mediated primarily by mutations within the RRDR, many studies have reported novel mutations outside this region, such as those at codons 176 (Val176Phe) (10), 381 (Ala381Val) (11), 490 (Gln490His) (12), 500 (Ala500Val), 502 (Ile502Val), 505 (Phe505Ser), 538 (Leu538Pro) (13), 146 (Val146Phe), and 572 (Ile572Phe) (14). These mutations were discovered using DNA sequencing of clinical rifampin-resistant M. tuberculosis isolates; however, most of these findings lacked in vivo confirmation of the relationship between the mutated codon and drug resistance. An exception to this is the work by Siu et al. (14), who constructed recombinant M. smegmatis and M. tuberculosis strains to show that mutations in codons 146 (Val146Phe) and 572 (Val572Phe) lead to rifampin resistance (14). The topography and population composition in eastern Taiwan are different from those in western, northern, and southern Taiwan. Therefore, we screened the rpoB gene in 51 clinical rifampin-resistant M. tuberculosis isolates in eastern Taiwan and identified one isolate, MTBR548H, with a mutation in codon 548, which is located outside the RRDR and had not been reported, as a cause of bacterial rifampin resistance. To validate the association of this mutated genotype with the rifampin-resistant phenotype in MTBR548H, the mutated rpoB of MTBR548H was cloned in a multicopy-number plasmid and transformed into wild-type M. tuberculosis and M. smegmatis to produce resistant mutant strains, YY002 and CH005, respectively. The rifampin susceptibilities of the recombinant strains indicated that the Arg548His mutation in RpoB contributed to rifampin resistance in M. tuberculosis. This finding was supported by data obtained from RpoB-DNA structural modeling, which showed that the RpoB residue His548 reduced the interaction between RpoB and rifampin in the RpoB-DNA complex. Our findings might help improve the detection of mycobacterial rifampin resistance, and we suggest that position 548 be included when the RRDR is analyzed by DNA sequencing due its close proximity to the RRDR. This can be readily performed without increasing the turnaround time or reaction cost of single-sequencing reactions.
Here, the most common mutations involved in rifampin resistance were missense mutations in codon 531 of rpoB (Table 3). These results were comparable to those of previous studies (6), but the frequency of this mutation (74.5%) was much higher than that in earlier reports from other regions of Taiwan, including southern Taiwan (41.5% during 1996 to 1998), Taiwan overall (49.4% during 1998 to 2003), and central and northern Taiwan (68.2% during 1999 to 2011) (6, 15, 16). This difference in frequency might be due to the increasing prevalence of rifampin-resistant M. tuberculosis clones spreading through eastern Taiwan in recent years (2011 to 2012).
In a recent study in Canada, only 2.9% of the clinical rifampin-resistant isolates (1/35) had double point mutations in rpoB (28). However, we found high rates of clinical rifampin-resistant isolates having two or three point mutations in rpoB (23.5% and 3.9%, respectively; Table 4). In our study, most of the double or triple point mutations in rpoB were still located in the RRDR (Table 3). The relationship between the multiple mutations in rpoB and MICs is not clear and needs further study.
The population of aboriginal people in eastern Taiwan is greater than that in other regions in Taiwan. In a recent study, 5.43% of the drug-resistant TB cases were rifampin monoresistant in aboriginal people in eastern Taiwan, while 2.27% were rifampin monoresistant in aboriginal people in southern Taiwan (29). In nonaboriginal people in southern Taiwan from 2010 to 2011 and eastern Taiwan from 2005 to 2008, as well as in people in Canada during 2012, no rifampin-monoresistant clinical isolates were found (30, 31). The novel rpoB (R548H) mutant found in our study might occur due to this special population and geographical environment. Such a mutant strain may spread to other regions in the future if the efforts to control tuberculosis are not effective.
The mutation at codon 531 was associated with high-level resistance to rifampin in most clinical and recombinant strains (Tables 3, 5, and 6) (15, 20). Extrapolation from the crystal structure of the rifampin-RNAP (RNA polymerase) complex showed that Ser531 in the RpoB rifampin-binding pocket formed a direct hydrogen bond with the critical hydroxyl of rifampin, O-2. Thus, the mutation in codon 531 reduced the interaction between rifampin and RpoB, resulting in bacterial rifampin resistance (5). Siu et al. (14) reported that the mutations in residues Val146 and Ile572, which are outside the RRDR, resulted in M. tuberculosis resistance to rifampin. In a model of rifapentine-RRDR (rather than rifampin-RRDR), residue 572 was localized to the wall of the rifampin-binding pocket of RpoB, while residue 146 was located beneath the rifampin-binding pocket and did not directly interact with the drug. The mutation in residue 572 (Ile572Phe) likely reduced the affinity between that residue and rifampin, while the mutation in residue 146 (Val146Phe) likely affected the folding and packing of the rifampin-binding pocket (14). In our study, RpoB residue Arg548, which is outside the RRDR, was not located in the rifampin-binding pocket in the RpoB-rifampin model. Therefore, residue Arg548 does not directly interact with rifampin. However, a point mutation (CGC to CAC) in codon 548, which changes the residue from Arg to His, led to reduced flexibility in the RpoB structure (Fig. 1B and C). These results indicate that rifampin did not block the transcriptional activity of RNAP containing a mutated form of RpoB when the RNAP was bound to DNA. Together, our results provide a potential mechanism for the differences in rifampin resistance between the RpoB-DNA and RpoB-rifampin models.
ACKNOWLEDGMENTS
This work was supported partly by grants (contract NSC 101-2320-B-320-002 and MOST 103-2320-B-320-008-MY3) from the Ministry of Science and Technology and partly by grant TCIRP99002-04 from Tzu Chi University.
We thank the Core Facilities for Protein Structural Analysis in Academia Sinica (Taipei, Taiwan) for assistance in structural modeling and expanding. We also thank Chi-Hsien Fu and Ying-Huei Chen in the Department of Laboratory Medicine, Buddhist Tzu Chi General Hospital, for their kind help in collecting the clinical mycobacterial isolates.
REFERENCES
- 1.World Health Organization. 2013. Global tuberculosis report 2013. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf. [Google Scholar]
- 2.Centers for Disease Control, R.O.C. (Taiwan). 2012. Taiwan tuberculosis control report 2012. Centers for Disease Control, R.O.C (Taiwan), Taipei, Taiwan: http://www.cdc.gov.tw/infectionreportinfo.aspx?treeid=075874dc882a5bfd&nowtreeid=54c73190d38c0d49&tid=2E3ABF0F44B82D24. [Google Scholar]
- 3.Mitchison DA, Nunn AJ. 1986. Influence of initial drug resistance on the response to short-course chemotherapy of pulmonary tuberculosis. Am Rev Respir Dis 133:423–430. [DOI] [PubMed] [Google Scholar]
- 4.Somoskovi A, Parsons LM, Salfinger M. 2001. The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir Res 2:164–168. doi: 10.1186/rr54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 6.Jou R, Chen HY, Chiang CY, Yu MC, Su IJ. 2005. Genetic diversity of multidrug-resistant Mycobacterium tuberculosis isolates and identification of 11 novel rpoB alleles in Taiwan. J Clin Microbiol 43:1390–1394. doi: 10.1128/JCM.43.3.1390-1394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramirez MV, Cowart KC, Campbell PJ, Morlock GP, Sikes D, Winchell JM, Posey JE. 2010. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol 48:4003–4009. doi: 10.1128/JCM.00812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar P, Balooni V, Sharma BK, Kapil V, Sachdeva KS, Singh S. 2014. High degree of multi-drug resistance and hetero-resistance in pulmonary TB patients from Punjab state of India. Tuberculosis (Edinb) 94:73–80. doi: 10.1016/j.tube.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Ohno H, Koga H, Kohno S, Tashiro T, Hara K. 1996. Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother 40:1053–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heep M, Rieger U, Beck D, Lehn N. 2000. Mutations in the beginning of the rpoB gene can induce resistance to rifamycins in both Helicobacter pylori and Mycobacterium tuberculosis. Antimicrob Agents Chemother 44:1075–1077. doi: 10.1128/AAC.44.4.1075-1077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taniguchi H, Aramaki H, Nikaido Y, Mizuguchi Y, Nakamura M, Koga T, Yoshida S. 1996. Rifampicin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol Lett 144:103–108. doi: 10.1111/j.1574-6968.1996.tb08515.x. [DOI] [PubMed] [Google Scholar]
- 12.Minh NN, Van Bac N, Son NT, Lien VT, Ha CH, Cuong NH, Mai CT, Le TH. 2012. Molecular characteristics of rifampin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Vietnam. J Clin Microbiol 50:598–601. doi: 10.1128/JCM.05171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue J, Shi W, Xie J, Li Y, Zeng E, Wang H. 2003. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from China. J Clin Microbiol 41:2209–2212. doi: 10.1128/JCM.41.5.2209-2212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siu GK, Zhang Y, Lau TC, Lau RW, Ho PL, Yew WW, Tsui SK, Cheng VC, Yuen KY, Yam WC. 2011. Mutations outside the rifampicin resistance-determining region associated with rifampicin resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 66:730–733. doi: 10.1093/jac/dkq519. [DOI] [PubMed] [Google Scholar]
- 15.Hwang HY, Chang CY, Chang LL, Chang SF, Chang YH, Chen YJ. 2003. Characterization of rifampicin-resistant Mycobacterium tuberculosis in Taiwan. J Med Microbiol 52:239–245. doi: 10.1099/jmm.0.05045-0. [DOI] [PubMed] [Google Scholar]
- 16.Lin YH, Tai CH, Li CR, Lin CF, Shi ZY. 2013. Resistance profiles and rpoB gene mutations of Mycobacterium tuberculosis isolates in Taiwan. J Microbiol Immunol Infect 46:266–270. doi: 10.1016/j.jmii.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Tseng ST, Tai CH, Li CR, Lin CF, Shi ZY. The mutations of katG and inhA genes of isoniazid-resistant Mycobacterium tuberculosis isolates in Taiwan. J Microbiol Immunol Infect, in press. doi: 10.1016/j.jmii.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Nolte FS, Metchock B, Williams T, Diem L, Bressler A, Tenover FC. 1995. Detection of penicillin-resistant Streptococcus pneumoniae with commercially available broth microdilution panels. J Clin Microbiol 33:1804–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soo PC, Horng YT, Chang KC, Wang JY, Hsueh PR, Chuang CY, Lu CC, Lai HC. 2009. A simple gold nanoparticle probes assay for identification of Mycobacterium tuberculosis and Mycobacterium tuberculosis complex from clinical specimens. Mol Cell Probes 23:240–246. doi: 10.1016/j.mcp.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Nakata N, Kai M, Makino M. 2012. Mutation analysis of mycobacterial rpoB genes and rifampin resistance using recombinant Mycobacterium smegmatis. Antimicrob Agents Chemother 56:2008–2013. doi: 10.1128/AAC.05831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2011. Susceptibility testing of mycobacteria, Nocardia and other aerobic actinomycetes; approved standard, 2nd ed CLSI M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 22.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project No. 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 28.Jamieson FB, Guthrie JL, Neemuchwala A, Lastovetska O, Melano RG, Mehaffy C. 2014. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J Clin Microbiol 52:2157–2162. doi: 10.1128/JCM.00691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YY, Chang JR, Huang WF, Kuo SC, Yeh JJ, Lee JJ, Jang CS, Sun JR, Chiueh TS, Su IJ, Dou HY. 2014. Molecular epidemiology of Mycobacterium tuberculosis in aboriginal peoples of Taiwan, 2006–2011. J Infect 68:332–337. doi: 10.1016/j.jinf.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Public Health Agency of Canada. 2012. Tuberculosis: drug resistance in Canada–2012. Public Health Agency of Canada, Ottawa, Ontario, Canada: http://www.phac-aspc.gc.ca/tbpc-latb/pubs/tb-dr2012/index-eng.php. [Google Scholar]
- 31.Chen YY, Tseng FC, Chang JR, Kuo SC, Lee JJ, Yeh JJ, Chiueh TS, Sun JR, Su IJ, Dou HY. 2014. Distinct modes of transmission of tuberculosis in aboriginal and non-aboriginal populations in Taiwan. PLoS One 9:e112633. doi: 10.1371/journal.pone.0112633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 33.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR Jr, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]