Abstract
We describe inhibition of Mycobacterium tuberculosis topoisomerase I (MttopoI), an essential mycobacterial enzyme, by two related compounds, imipramine and norclomipramine, of which imipramine is clinically used as an antidepressant. These molecules showed growth inhibition of both Mycobacterium smegmatis and M. tuberculosis cells. The mechanism of action of these two molecules was investigated by analyzing the individual steps of the topoisomerase I (topoI) reaction cycle. The compounds stimulated cleavage, thereby perturbing the cleavage-religation equilibrium. Consequently, these molecules inhibited the growth of the cells overexpressing topoI at a low MIC. Docking of the molecules on the MttopoI model suggested that they bind near the metal binding site of the enzyme. The DNA relaxation activity of the metal binding mutants harboring mutations in the DxDxE motif was differentially affected by the molecules, suggesting that the metal coordinating residues contribute to the interaction of the enzyme with the drug. Taken together, the results highlight the potential of these small molecules, which poison the M. tuberculosis and M. smegmatis topoisomerase I, as leads for the development of improved molecules to combat mycobacterial infections. Moreover, targeting metal coordination in topoisomerases might be a general strategy to develop new lead molecules.
INTRODUCTION
Tuberculosis (TB) is a major health concern with 9 million new cases being added annually (1). The disease claims approximately 1.4 million lives every year (2). The etiological agent, Mycobacterium tuberculosis, requires a treatment regimen of four front-line drugs (pyrazinamide, isoniazid, rifampin, and ethambutol) for an initial 2-month period followed by isoniazid and rifampin treatment for an additional minimum of 4 months (1, 3). Inadequate compliance to the treatment with the front-line drugs has led to the emergence of multidrug-resistant TB (MDR-TB) (1, 4). MDR-TB needs treatment by second-line drugs, which include fluoroquinolones, aminoglycosides, etc. (1). Insufficient treatment of MDR-TB due to a variety of reasons has contributed to the emergence of extensively drug-resistant TB (XDR-TB) (5).
The drugs in use presently to combat the disease, such as rifampin, target RNA polymerase (6), and quinolones and fluoroquinolones (FQs) target DNA gyrase (7, 8). Various other molecules, including aminoglycosides, which target different steps of protein synthesis, also constitute a portfolio of molecules active against the pathogen. Despite the fact that the recently developed FQs gatifloxacin and moxifloxacin are effective against TB (9), there is a concern for development of resistance against them (10, 11). Hence, urgent efforts to develop new antitubercular drugs are underway, and, as a consequence, several other compounds for TB treatment are undergoing clinical trials (12, 13). The prolonged time of treatment and the upsurge of strains resistant to the currently used drugs underscore the requirement for development of novel molecules which can inhibit the function of essential proteins (14) by interfering with their reaction, leading to the cytotoxicity.
DNA topoisomerases constitute one such essential class of enzymes which function to maintain topological homeostasis within the cell during a variety of DNA transaction processes such as replication, transcription, and chromosome segregation (15). On the basis of their structure and mechanism, they are broadly classified as type I and type II (16, 17). Bacterial type I DNA topoisomerases belong to the type IA subclass based on their reaction characteristics. They catalyze the relaxation reaction by forming a transient 5′-phosphotyrosine covalent adduct and change the linking number in steps of one by generating a single-stranded nick and passing the intact strand across the broken DNA strand held by the enzyme (15, 16). In contrast, type II topoisomerases change the linking number in steps of two by generating a double-strand break and passing the intact duplex DNA through the cleaved DNA gate (15, 18). Several molecules which hamper the resealing of the topoisomerase II (topoII)-mediated DNA breaks have been characterized and clinically validated as drugs (19–21). For example, DNA gyrase has been extensively exploited to develop antibacterial agents, and FQs stabilize the protein-DNA cleavage complex to induce severe cytotoxicity (22). However, there is a dearth of such inhibitors of bacterial topoisomerase I. Molecules that trap the topoI-DNA covalent complex, causing DNA lesions and cytotoxicity, are currently needed. In this article, we report two molecules, imipramine and norclomipramine, that target mycobacterial topoI. Imipramine is a clinically used tricyclic antidepressant (23), while norclomipramine (also known as N-desmethylclomipramine) is the active metabolite of another tricyclic antidepressant, clomipramine (24). The molecules were identified as likely inhibitors by in silico screening using a homology model of the enzyme. The molecules inhibit the DNA relaxation reactions catalyzed by topoisomerase I from M. tuberculosis and from M. smegmatis but not from Escherichia coli. Importantly, both the molecules perturb the cleavage-religation equilibrium of the relaxation reaction catalyzed by topoI and are active in a whole-cell assay against M. smegmatis and M. tuberculosis.
MATERIALS AND METHODS
Enzyme, DNA constructs, and compounds.
M. tuberculosis topoisomerase I (MttopoI) (25), M. smegmatis topoisomerase I (MstopoI) (26), and E. coli topoisomerase I (EctopoI) (27) were purified as described previously. Norclomipramine and imipramine were purchased from Sigma-Aldrich (St. Louis, MO, USA), and a 10 mM stock was prepared in ultrapure H2O. A negatively supercoiled pUC18 plasmid DNA substrate for the relaxation assay was purified by Qiagen midiprep kits. For overexpression of TopoI in mycobacterial cells, both M. tuberculosis and M. smegmatis topoI genes were excised from their respective constructs, pAVN1 (25) and pPVN123 (26), by digestion with NdeI and EcoRV and cloned into the pMIND vector (28) linearized with the same restriction enzymes. The constructs were electroporated into M. smegmatis mc2 155 or M. tuberculosis H37Ra cells, and positive colonies were selected on kanamycin (25 µg/ml) 7H9 agar plates.
Homology modeling and in silico docking of molecules.
Three bacterial topoI structures from the Protein Data Bank (PDB) were used to build a homology model of MttopoI. These were 1ECL (closed state, no DNA or Mg2+ bound), 1MW8 (closed state with noncovalent DNA bound, no Mg2+ bound), and 1MW9 (closed state, no DNA or Mg2+ bound). A homology model of MttopoI in a closed state, no DNA or Mg2+ bound (A2VM29 based on 1ECL/1MW9), was also available in ModBase (29). The bacterial topoII structure 2RGR and the topoIII structure 1I7D were also available. Therefore, a homology model for the EctopoI was initially created with the site open and with Mg2+ bound by aligning topoI subdomains with topoIII subdomains. The Mg2+ site was also generated from the topoIII residue coordinates. The MttopoI homology model with the gate open and Mg2+ bound was created by using the same sequence alignment as that used for 1ECL in ModBase and the EctopoI homology model as a scaffold. This was achieved after downloading the sequence P0A620 in FASTA format and using the “align sequence to template” protocol in Discovery Studio (Biovia, San Diego, CA) (sequence identity 38.3 and sequence similarity 54.8). The model was used to create a homology model with Mg2+ and a covalently bound DNA fragment. The DNA in this final topoI model is based on the DNA position in the EctopoII crystal structure 2RGR and was achieved using pyMOL (30).
The MttopoI homology model with Mg2+ and a DNA fragment bound in the open state was used for docking using LibDock (Discovery Studio) (31). The proposed binding site was centered on Mg2+ with an 8-Å diameter. The protocol included 10 hotspots and docking tolerance (0.25). The FAST conformation method was also used along with steepest descent minimization with CHARMm. Further parameters followed the default settings. A set of FDA-approved drugs was collected and exported from the Collaborative Drug Discovery database (Burlingame, CA). This and other previously described sets of drugs approved by the FDA (SCUT database [32, 33]) were used for docking in the homology model. The molecules that scored well were visualized, and their two-dimensional (2D) interaction plots were generated and selected for follow-up. The model and complete Discovery Studio protocol used in docking are available from the authors upon written request.
DNA relaxation assay.
The relaxation of supercoiled pUC18 DNA was carried out as described previously (34). Briefly, 500 ng DNA was incubated with 1 unit of various type I topoisomerases for 30 min at 37°C in buffer containing 40 mM Tris-HCl (pH 8.0), 20 mM NaCl, 5 mM MgCl2, and 1 mM EDTA. The enzyme inhibition assays were carried out with a preincubation of the enzyme and increasing concentrations of the compounds at 37°C for 15 min, followed by the addition of the substrate DNA. After incubation at 37°C for 30 min, the samples were electrophoresed in a 1.2% agarose gel for 12 h at 2.5 V/cm and stained with ethidium bromide (EtBr) (0.5 μg/ml), and the DNA bands were visualized using a gel documentation system (Bio-Rad, Hercules, CA, USA).
Oligonucleotide cleavage assay.
Cleavage assays were carried out with a 5′-end-labeled 32-mer harboring the strong topoisomerase site (STS) annealed to a complementary sequence. The double-stranded substrate was preincubated with MttopoI in a buffer containing 40 mM Tris-HCl (pH 8.0), 20 mM NaCl, 1 mM EDTA, and 5 mM MgCl2 on ice for 15 min. Following this, various concentrations of imipramine or norclomipramine were added, and the reaction mixtures were incubated at 37°C for 30 min. The reactions were stopped with 45% formamide and heating at 95°C for 2 min. The products were resolved by 12% denaturing PAGE and analyzed by phosphorimager. A similar assay was also carried out with the 5′-end-labeled single-strand specific 32-mer.
Growth inhibition.
Mycobacterial cells (expressing normal levels of or overexpressing wild-type [WT] mycobacterial topoI) were grown to an optical density at 595 nm (OD595) of 0.6 in Middlebrook 7H9 broth supplemented with 0.2% glycerol and 0.05% Tween 80. The culture was diluted to a final OD595 of 0.05 with fresh medium and aliquoted into a 100-well growth plate. Serial dilutions of the compounds were added to the culture. The untreated culture was taken as a control. The growth was monitored at 595 nm with continuous shaking at 200 rpm. The readings were analyzed by GraphPad Prism software (version 5.0). To determine the MIC values of the compounds, a resazurin reduction microplate assay (REMA) (35) was carried out. The cultures were grown for 2 days for M. smegmatis or 7 days for M. tuberculosis in the presence of the compounds. Following this, resazurin dye was added to the cultures at a final concentration of 0.02% with further incubation for 1 h for M. smegmatis or for 14 h for M. tuberculosis. To assess the cell lethality of imipramine and norclomipramine, M. smegmatis cells were grown to an OD595 of 0.6 to 0.8 and treated with 1×, 2.5×, and 5× MICs of the molecules for 12 h. Following this treatment, serial dilutions of the cells were plated on Middlebrook 7H9 agar, and the CFU/ml values were determined. The untreated culture was taken as a control.
Combinatorial effect of imipramine and moxifloxacin on mycobacterial growth.
In order to assess the effect of combining imipramine with a known type II topoisomerase poison, M. tuberculosis cells were grown in the presence of a sub-MIC of moxifloxacin and various concentrations of imipramine or vice versa. The cells were cultured at 37°C for 8 days following which resazurin dye was added to the cultures at a final concentration of 0.02% with further incubation for 14 h, and conversion of resazurin (blue) to resorufin (pink) was monitored to score the viability of the cells.
RESULTS
TopoI model and in silico screening.
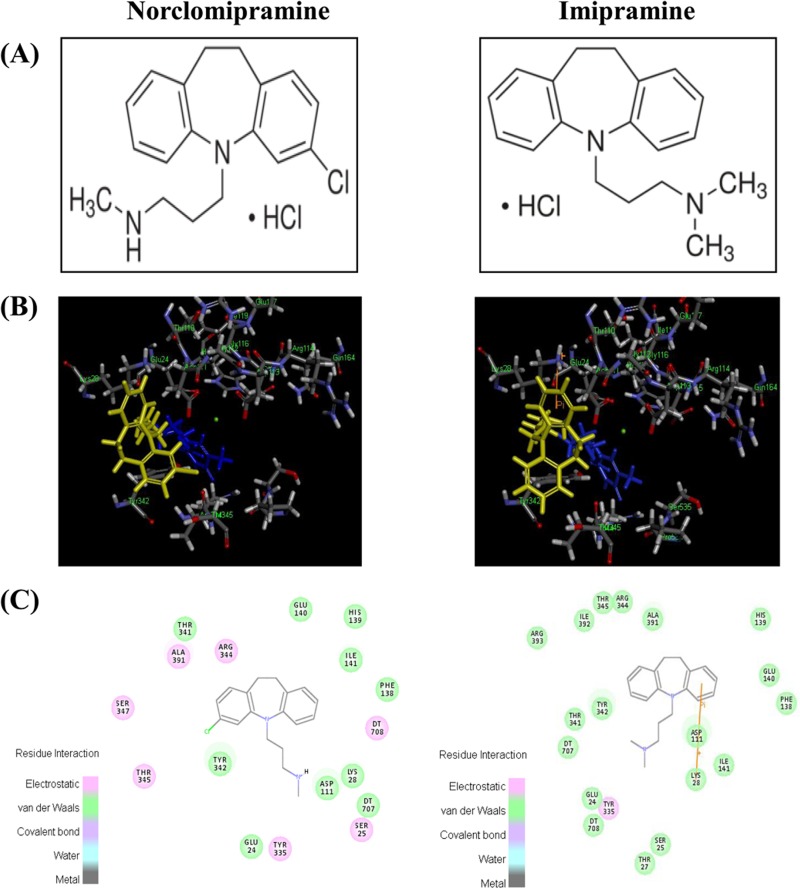
Various libraries of drugs approved by the FDA were docked into the homology model for MttopoI. Out of the 812 molecules tested, 88 were found to be docked and ranked by the LibDock score (range, ∼46.4 to 126.3). The interaction of several compounds with the modeled MttopoI structure suggested that molecules with a favorable docking score might affect the enzyme function. Among them, norclomipramine (LibDock score of 95.2) (Fig. 1) and imipramine (LibDock score of 103) (Fig. 1) were selected for further biochemical and whole-cell-based studies.
FIG 1.
Chemical structure and docking of norclomipramine and imipramine. (A) Chemical structure of norclomipramine and imipramine. (B) Docking of norclomipramine (LibDock score of 95.2) and imipramine (LibDock score of 103) shown in yellow in the topoI homology model with a DNA fragment (blue) and Mg2+ (green). (C) The region harbors the residues involved in Mg2+ coordination.
Imipramine and norclomipramine specifically inhibit the activity of mycobacterial topoI.
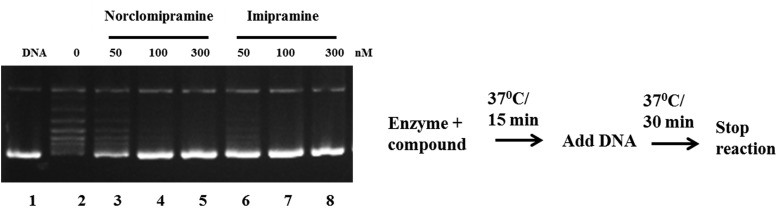
In order to determine the inhibitory potential of the molecules, DNA relaxation assays were carried out with MttopoI. Both of the compounds inhibited the activity of the enzyme. Assays with increasing concentrations of the inhibitors revealed a complete inhibition of the MttopoI activity at 0.1 μM norclomipramine and imipramine (Fig. 2). However, in assays carried out with EctopoI, no significant inhibition of DNA relaxation activity is seen, indicating that the molecules are likely to be specific inhibitors of the mycobacterial enzyme (see Fig. S1 in the supplemental material).
FIG 2.
Inhibition of DNA relaxation activity of MttopoI by norclomipramine and imipramine. One unit of MttopoI was incubated with various concentrations of the molecules at 37°C for 15 min following which 500 ng of supercoiled pUC18 was added. The incubation was further continued at 37°C for 30 min, and the reactions were terminated by addition of 0.6% SDS-agarose dye. The reaction products were resolved on a 1.2% agarose gel followed by staining with EtBr. Lane 1, supercoiled pUC18; lane 2, relaxation reaction in the absence of the compounds; lanes 3 to 5, various concentrations (50 to 300 nM) of norclomipramine; lanes 6 to 8, various concentrations (50 to 300 nM) of imipramine.
Antibacterial activity of norclomipramine and imipramine.
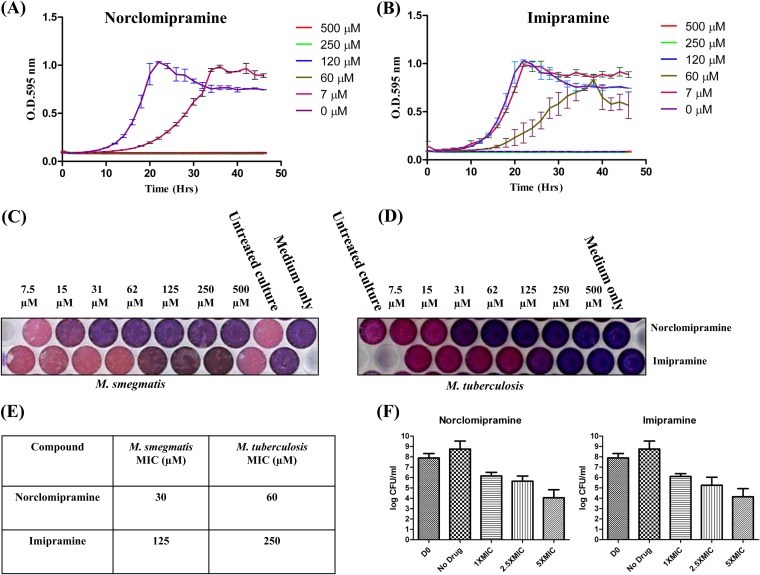
Topoisomerases carry out the essential function of maintaining genome topology during DNA replication and transcription and are hence indispensable for cellular functions. To assess the cell growth inhibitory potential and specificity of inhibition of the compounds, M. smegmatis and M. tuberculosis cells were treated with various concentrations of norclomipramine and imipramine. M. smegmatis cells showed delayed growth at a 7 μM concentration of norclomipramine, and the growth was completely inhibited at 30 μM (Fig. 3A). Similarly, cell growth was delayed at 60 μM and completely inhibited at a 125 μM concentration of imipramine (Fig. 3B). The MIC values of norclomipramine and imipramine for M. smegmatis and M. tuberculosis were determined by the resazurin assay (35) (Fig. 3C, D, and E). The MIC values of norclomipramine were 30 μM and 60 μM for M. smegmatis and M. tuberculosis, respectively. Similarly, the MICs of imipramine for the two species were 125 μM and 250 μM, respectively. Thus, as with the growth studies, the M. smegmatis cells showed high susceptibility to the molecules. The observed differences in the MIC levels between the two species might be attributed to various factors (see Discussion). Further, the cell lethality of the molecules was assessed by exposing exponential-phase M. smegmatis cells to various concentrations of the compounds. Compared to the untreated control, the CFU/ml value at 1× MIC for both compounds showed a 2-log reduction, and at 5× MIC, the CFU/ml value was reduced by 4 log values (Fig. 3F). The reductions in the CFU/ml values at increasing concentrations thus indicated the cell-killing effect.
FIG 3.
Inhibition of mycobacterial growth by norclomipramine and imipramine and determination of MIC values. M. smegmatis cells were grown in the presence of various concentrations of norclomipramine (A) or imipramine (B). The growth was followed over a period of 48 h with the OD being measured every 2 h. The growth curve was plotted. The sterile medium and untreated culture were used as controls. M. smegmatis (C) or M. tuberculosis (D) cells were grown in the presence of various concentrations of the compounds. Resazurin dye was added to a final concentration of 0.02% to each well. (E) The plate was incubated at 37°C to determine the MIC values. The sterile medium and untreated culture were used as controls. (F) M. smegmatis cells in the log phase were exposed to 1×, 2.5×, and 5× MIC of imipramine and norclomipramine. Untreated cells were taken as controls. Serial dilutions of the treated and untreated cells were plated on Middlebrook 7H9 agar plates, and the CFU/ml values were determined. The values were plotted as log (CFU/ml) versus concentration. Error bars indicate the standard deviations obtained in three independent experiments.
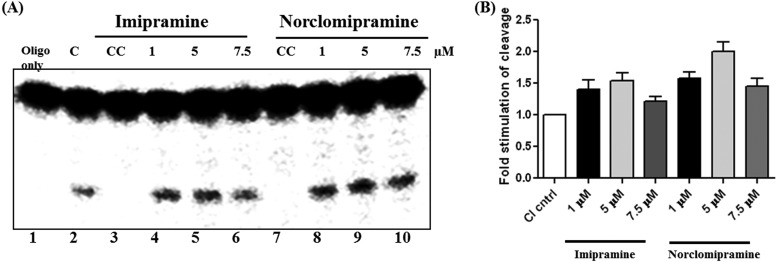
Imipramine and norclomipramine stimulate DNA cleavage activity of topoI.
Next, we addressed the mechanism of action of these molecules by carrying out assays on individual steps of the DNA relaxation cycle of the enzyme. The reaction catalyzed by topoI involves DNA binding, cleavage, and strand passage followed by religation. Binding of the molecules did not affect the binding of the enzyme to the DNA (data not shown). Next, cleavage assays were carried out using a double-stranded 32-mer harboring the strong topoisomerase site (STS) with MttopoI and different concentrations of the molecules. Imipramine and norclomipramine stimulated the topoI-mediated cleavage of the DNA by ∼1.7- and 2-fold, respectively (Fig. 4). When the stimulation of cleavage by the inhibitors was evaluated in the plasmid context, nicking and linearization of the plasmid molecule were observed at similar concentrations of the compounds (see Fig. S2 in the supplemental material). The compounds may act by primarily binding to the enzyme, the DNA, or the enzyme-DNA complex. In order to evaluate the preference for binding, addition reactions were carried out. The compounds affected stimulation of DNA cleavage more with the preformed protein-DNA complex than with preincubation with only the enzyme or DNA (see Fig. S3 in the supplemental material). According to the currently accepted model of action of FQs on DNA gyrase, the FQs primarily target the enzyme-DNA complex (36). Although the imipramine- and norclomipramine-induced cleavage is not as pronounced as that of FQs, the pattern is similar to the action of FQs on DNA gyrase.
FIG 4.
Imipramine and norclomipramine stimulate the DNA cleavage activity. (A) MttopoI was incubated with a 5′-end-labeled specific 32-mer annealed to a complementary sequence on ice for 15 min following which various concentrations of imipramine or norclomipramine were added, and the reaction was allowed to proceed at 37°C for 30 min. The reactions were terminated by the addition of 45% formamide and heating at 95°C for 2 min. The products were resolved by 12% denaturing PAGE and analyzed by phosphorimager. Lane 2 indicates the cleavage reaction by the enzyme in the absence of compound (C), and lanes 3 and 7 indicate compound control (CC) for imipramine and norclomipramine, respectively. (B) Quantification of cleavage products. Error bars represent the standard deviations obtained in three experiments.
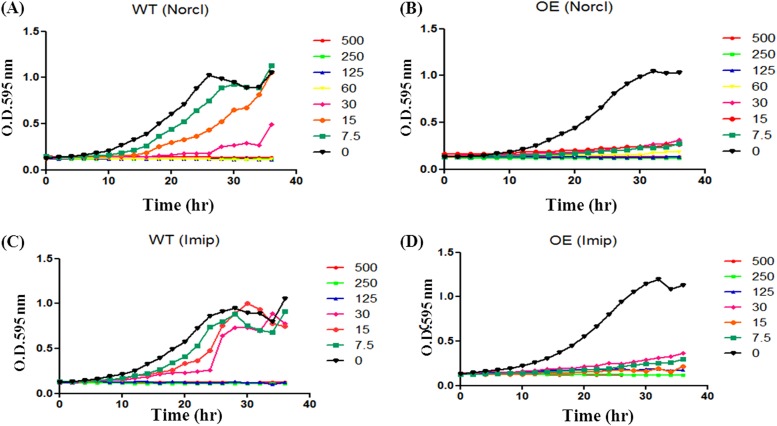
Norclomipramine and imipramine affect the growth of cells overexpressing the enzyme.
Stimulation of topoI-mediated DNA cleavage by imipramine and norclomipramine (Fig. 4; see also Fig. S2 in the supplemental material) indicated that the molecules might act as cytotoxic agents. To verify their cytotoxic potential, mycobacterial cells overexpressing the enzyme were used. Norclomipramine and imipramine inhibited the growth of M. smegmatis cells expressing normal levels of topoI at 30 μM and 125 μM, respectively (Fig. 5A and C). However, the cells overexpressing the enzyme were more susceptible to both compounds, and the growth was inhibited at lower concentrations of the molecules (Fig. 5B and D). Similarly, M. tuberculosis cells expressing normal levels of topoI exhibited MIC values of 125 μM for norclomipramine and 250 μM for imipramine while cells overexpressing the enzyme exhibited MIC values of 60 μM for norclomipramine and 125 μM for imipramine (see Fig. S4A and B in the supplemental material). The decreased MIC of the molecules in topoI-overexpressing cells is likely due to the stimulation of topoI-mediated DNA cleavage in vivo leading to cell death, indicating that the enzyme is the cellular target for the compounds.
FIG 5.
Increased cytotoxicity upon topoI overexpression in M. smegmatis. M. smegmatis cells overexpressing (OE) MstopoI or normal level (N) of MstopoI were grown in the presence of various concentrations (0 to 500 μM) of norclomipramine (A and B) or imipramine (C and D). The growth was followed over a period of 40 h with OD being measured every 2 h. The growth curve was plotted using GraphPad Prism (version 5.0). The sterile medium and untreated cultures were used as controls.
Activity of the metal binding mutants is affected differently by the compounds.
All type IA topoisomerases contain a TOPRIM domain in the amino-terminal region. The domain harbors the metal coordination motif DxDxE (37). Mg2+ coordination with these residues is essential for enzyme function. The mutations in these acidic residues lead to impairment in the catalytic activity and cytotoxicity (38, 39), indicating that targeting the topoI through these residues may impair the enzyme function. In our earlier studies, we showed that the mutations of the individual amino acids, D108 and E112, of the motif in MstopoI to alanine resulted in 20- and 5-fold losses of activity, respectively (38), while mutations of the acidic residues in MttopoI led to complete loss of activity (25). In silico docking studies using the model of MttopoI indicated that imipramine and norclomipramine bind near the metal binding site of the enzyme (Fig. 1). Binding of the molecules to the topoisomerase may be abrogated in the enzymes, harboring mutations in the metal binding site. In such a case, the metal binding mutants may show resistance to the inhibition by these molecules. Imipramine and norclomipramine were able to inhibit the activity of D108A at concentrations comparable to those for the inhibition of the WT enzyme (Fig. 6A). In contrast, the relaxation activity of E112A was unaffected by either of the compounds (Fig. 6B). From these results, it is apparent that the molecules interact with the enzyme in the TOPRIM region in the vicinity of the metal coordination motif, proximal to the E112 residue of the enzyme.
FIG 6.
Metal binding mutants of MstopoI are affected differently. One unit of D108A (lane 5) or E112A (lane 9) was incubated with various concentrations of imipramine (A) or norclomipramine (B) (lanes 6 and 7 and 10 and 11, respectively) at 37°C for 15 min. Following this, 500 ng of supercoiled pUC18 was added to each reaction tube and incubated for an additional 30 min. The reactions were terminated by the addition of 0.6% SDS-agarose dye. The reaction products were resolved on a 1.2% agarose gel followed by staining with EtBr. Monoclonal antibody (mAb) 2F3G4, which inhibits the DNA relaxation reaction of mycobacterial topoisomerase I, was used as a positive control.
Combination of imipramine and moxifloxacin lowers the MIC value.
Moxifloxacin is a well-known type II topoisomerase poison which targets mycobacterial DNA gyrase (40, 41). The experiments described above indicated that imipramine poisons the topoI reaction. In order to assess the effect of targeting both gyrase and topoI, mycobacterial cells were exposed to combinations of moxifloxacin and imipramine. First, imipramine was used in a range of concentrations and moxifloxacin was used at a sub-MIC (i.e., 0.075 μM). In such a combination, the MIC was lowered compared to that for the cells which were exposed to imipramine alone (see Fig. S5A in the supplemental material). In a reciprocal experiment, moxifloxacin was used at various concentrations, and imipramine was kept at a sub-MIC (i.e., 125 μM). Again, the MIC was seen to be lower than the MIC when the cells were exposed to moxifloxacin alone (see Fig. S5B in the supplemental material). The reduction in the MIC might be a consequence of the enhancement of DNA breaks due to a combination of topoisomerase poisons targeting both enzymes.
DISCUSSION
M. tuberculosis is a formidable pathogen which evades the host defense system and continues to persist in the host (42, 43). The organism efficiently manages its central metabolic pathways to successfully survive in the host environmental conditions. Several antimycobacterial agents target the essential metabolic pathways of the organism, viz. transcription, translation, and cell wall synthesis (44). However, the emergence of MDR-TB and XDR-TB demands the discovery of novel drug targets and improved chemotherapeutic agents, which can reduce the time of treatment. MttopoI (Rv3646c), an essential enzyme for the bacterial growth (45), offers one such potential candidate for a target. Currently, a sustained worldwide effort is underway to find small molecule inhibitors against eubacterial topoI. Here, we describe the specific inhibition of mycobacterial topoI by two small molecules identified by computational screening. Norclomipramine and imipramine inhibited mycobacterial topoI activity at concentrations not described so far for any bacterial type I topoisomerases. Studies with the metal binding mutants confirmed the in silico prediction that these molecules bind in the proximity of the TOPRIM domain. Moreover, these molecules stimulate the DNA cleavage activity and consequently show increased lethality in cells overexpressing topoI.
The stimulation of DNA cleavage observed when the molecules were incubated with a preformed enzyme-DNA complex suggests that they act on this reaction intermediate. Importantly, the stimulation of DNA cleavage might be visualized only in the context of double-stranded DNA. No stimulation of cleavage was observed with the single-stranded substrate (data not shown). However, the molecules do not seem to bind DNA on their own, but the DNA-enzyme complex appears to be the preferred target. In this respect, the mode of their interaction is analogous to that of FQs. Although initially FQs were thought to bind DNA and inhibit DNA gyrase, now it has been established that they bind the DNA gyrase-DNA complex to arrest the reaction (46–49). Thus, the observed effect of the molecules when the cells overexpress topoI may be a consequence of increased accumulation of DNA breaks as seen with experiments described in Fig. 4 (see also Fig. S2 in the supplemental material). In the topoI overexpression strain, more topoI-mediated breaks might be generated, leading to enhanced cell death. The inhibitors of topoisomerases can be broadly classified into two categories: those which inhibit the enzyme reaction by binding to the enzyme or those which arrest the reaction, leading to double-strand breaks. The latter class, which includes fluoroquinolones, has attained success therapeutically as they not only inhibit the enzyme but also induce cell poisoning. Imipramine and norclomipramine are the first molecules which appear to function as eubacterial topoI poisons, albeit at a lower potency.
The difference seen in the MICs of the two molecules for M. smegmatis and M. tuberculosis is not very surprising. These species are known to exhibit different degrees of susceptibility to rifampin and other anti-TB drugs (50). We have considered the following possibilities to account for the differential susceptibility to imipramine and norclomipramine. First, the differences in the cell wall and cell membrane composition between the pathogenic and the saprophytic mycobacterial species may contribute to the differences in the permeabilities of the molecules into the cell. Alternately, the difference in the topoI level between the two species might account for the observed difference in the MICs. Our recent estimates indicate that M. smegmatis has a higher level of topoI than M. tuberculosis (51). Given that the molecules tested impair the cleavage-religation equilibrium and overexpression of topoI leads to lowering of the MIC (Fig. 6; see also Fig. S4 in the supplemental material), the species with more enzyme should be more susceptible to these drugs. That was indeed the case.
Unlike E. coli and many other bacteria, which encode two type I and two type II topoisomerases, M. tuberculosis has a single type I and a single type II topoisomerase to carry out all the topological functions inside the cell. Thus, targeting both the essential enzymes simultaneously would hasten the cell death. Accordingly, in experiments where a combination of imipramine and moxifloxacin was used, the susceptibility of the cells was enhanced. Accumulation of DNA breaks brought about by poisoning both type I and type II topoisomerases simultaneously by molecules that are more efficient than the molecules used here to target topoI might be a step forward in countering mycobacterial and other eubacterial infections.
Earlier studies from our laboratory on the metal binding mutants of mycobacterial topoisomerase I revealed the role played by the triad of acidic amino acids within the TOPRIM domain in coordinating the Mg2+ ion (25, 38). The Mg2+ binding mutants have been shown to be compromised in maintaining the cleavage-religation equilibrium, indicating that a disturbance in the metal coordination might abrogate the breakage and resealing mechanism of the enzyme. From these studies, we predicted that small molecules which target the metal binding motif might affect the enzyme activity (38). Indeed, in the oligonucleotide cleavage assay with the wild-type enzyme in the presence of the molecules, stimulation of the cleavage reaction was observed (Fig. 4). The mutation of the first aspartate residue, D108, to alanine did not affect the inhibition brought about by imipramine and norclomipramine, suggesting that D108 may not contribute significantly to the interaction of these molecules with the enzyme. In contrast, the E112A mutant was resistant to inhibition, indicating that the glutamate residue might be involved in the interaction between the molecules and the enzyme in the TOPRIM domain. Mg2+ is an integral component in all topoisomerase reactions and is necessary for enzyme activity. The metal ion is not needed for the DNA cleavage but is important for the second transesterification reaction (38, 52). The catalytic Mg2+ is considered to be loosely bound with an ability to move within the enzyme (53). If indeed the catalytic Mg2+ is a “moving metal ion” (38, 53), the inhibitor interacting at the E112 residue would likely hinder such movement, arresting the reaction. High-resolution structures of the enzyme-inhibitor complex would validate such a mode of action.
The reaction cycle of all topoisomerases involves a vulnerable step of formation of a protein-DNA covalent adduct, the accumulation of which leads to DNA strand breakage and inevitable cell lethality. A number of molecules target this most vulnerable step of the reaction of various topoisomerases. Hence, topoisomerase poisons, viz. fluoroquinolones, camptothecin, amsacrine, etc., have been successfully utilized in antibacterial and anticancer therapy. Imipramine and norclomipramine, the tricyclic antidepressants used in this study, also seem to act as topoisomerase poisons since they induce protein-mediated DNA breaks and affect the cell growth. They form the first set of molecules which poison mycobacterial topoI. They are also the first molecules described that target the metal binding pocket of a topoisomerase. Their activity against M. tuberculosis cells may potentiate further development of novel anti-TB agents. To sum up, we demonstrate the use of a computational docking approach to filter libraries of FDA-approved drugs to identify molecules as inhibitors of an essential target in M. tuberculosis. We illustrate the possibility of repurposing known drugs that are not antibacterials as potential agents against M. tuberculosis (54). The use of combinations of agents may be even more desirable. Future lead optimization studies with imipramine and norclomipramine are necessary to increase the whole-cell potency while retaining the activity at the target.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the members of the laboratory for critical reading of the manuscript and useful suggestions. Shruti Menon is acknowledged for technical assistance.
V.N. is a J.C. Bose fellow of the Department of Science and Technology, Government of India, and partner 14 in MM4TB, an EU consortium project. S.E. is partner 17 in MM4TB. This work was supported by a grant from the European Community's Seventh Framework Program (MM4TB). Biovia is kindly acknowledged for providing Discovery Studio to S.E.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04516-14.
REFERENCES
- 1.Zumla A, Raviglione M, Hafner R, von Reyn CF. 2013. Tuberculosis. N Engl J Med 368:745–755, 2013. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2013. Global tuberculosis report 2013. Report WHO/HTM/TB/2013.11. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC, Friedman LN, Fujiwara P, Grzemska M, Hopewell PC, Iseman MD, Jasmer RM, Koppaka V, Menzies RI, O'Brien RJ, Reves RR, Reichman LB, Simone PM, Starke JR, Vernon AA. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am J Respir Crit Care Med 167:603–662. doi: 10.1164/rccm.167.4.603. [DOI] [PubMed] [Google Scholar]
- 4.Zignol M, van Gemert W, Falzon D, Sismanidis C, Glaziou P, Floyd K, Raviglione M. 2012. Surveillance of anti-tuberculosis drug resistance in the world: an updated analysis, 2007-2010. Bull World Health Organ 90:111D–119D. doi: 10.2471/BLT.11.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi NR, Nunn P, Dheda K, Schaaf HS, Zignol M, van Soolingen D, Jensen P, Bayona J. 2010. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 6.McClure WR, Cech CL. 1978. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem 253:8949–8956. [PubMed] [Google Scholar]
- 7.Gellert M, Mizuuchi K, O'Dea MH, Itoh T, Tomizawa JI. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A 74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CR, Malik M, Snyder M, Drlica K. 1996. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol 258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 9.Ginsburg AS, Grosset JH, Bishai WR. 2003. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect Dis 3:432–442. doi: 10.1016/S1473-3099(03)00671-6. [DOI] [PubMed] [Google Scholar]
- 10.van der Heijden YF, Maruri F, Blackman A, Mitchel E, Bian A, Shintani AK, Eden S, Warkentin JV, Sterling TR. 2013. Fluoroquinolone susceptibility in Mycobacterium tuberculosis after prediagnosis exposure to older- versus newer-generation fluoroquinolones. Int J Antimicrob Agents 42:232–237. doi: 10.1016/j.ijantimicag.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlman DC, El Sadr WM, Heifets LB, Nelson ET, Matts JP, Chirgwin K, Salomon N, Telzak EE, Klein O, Kreiswirth BN, Musser JM, Hafner R. 1997. Susceptibility to levofloxacin of Mycobacterium tuberculosis isolates from patients with HIV-related tuberculosis and characterization of a strain with levofloxacin monoresistance. Community Programs for Clinical Research on AIDS 019 and the AIDS Clinical Trials Group 222 Protocol Team. AIDS 11:1473–1478. [DOI] [PubMed] [Google Scholar]
- 12.Wong EB, Cohen KA, Bishai WR. 2013. Rising to the challenge: new therapies for tuberculosis. Trends Microbiol 21:493–501. doi: 10.1016/j.tim.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zumla A, Nahid P, Cole ST. 2013. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov 12:388–404. doi: 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
- 14.Lamichhane G, Freundlich JS, Ekins S, Wickramaratne N, Nolan ST, Bishai WR. 2011. Essential metabolites of Mycobacterium tuberculosis and their mimics. mBio 2:e00301–00310. doi: 10.1128/mBio.00301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champoux JJ. 2001. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 16.Forterre P, Gribaldo S, Gadelle D, Serre MC. 2007. Origin and evolution of DNA topoisomerases. Biochimie 89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Corbett KD, Berger JM. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct 33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 18.Schoeffler AJ, Berger JM. 2005. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem Soc Trans 33:1465–1470. doi: 10.1042/BST20051465. [DOI] [PubMed] [Google Scholar]
- 19.Pommier Y. 2013. Drugging topoisomerases: lessons and challenges. ACS Chem Biol 8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pommier Y, Leo E, Zhang H, Marchand C. 2010. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collin F, Karkare S, Maxwell A. 2011. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl Microbiol Biotechnol 92:479–497. doi: 10.1007/s00253-011-3557-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxwell A. 1997. DNA gyrase as a drug target. Trends Microbiol 5:102–109. doi: 10.1016/S0966-842X(96)10085-8. [DOI] [PubMed] [Google Scholar]
- 23.Glowinski J, Axelrod J. 1964. Inhibition of uptake of tritiated-noradrenaline in the intact rat brain by imipramine and structurally related compounds. Nature 204:1318–1319. doi: 10.1038/2041318a0. [DOI] [PubMed] [Google Scholar]
- 24.Kelly MW, Myers CW. 1990. Clomipramine: a tricyclic antidepressant effective in obsessive compulsive disorder. DICP 24:739–744. [DOI] [PubMed] [Google Scholar]
- 25.Godbole AA, Leelaram MN, Bhat AG, Jain P, Nagaraja V. 2012. Characterization of DNA topoisomerase I from Mycobacterium tuberculosis: DNA cleavage and religation properties and inhibition of its activity. Arch Biochem Biophys 528:197–203. doi: 10.1016/j.abb.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Jain P, Nagaraja V. 2006. Indispensable, functionally complementing N and C-terminal domains constitute site-specific topoisomerase I. J Mol Biol 357:1409–1421. doi: 10.1016/j.jmb.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 27.Tse-Dinh YC, Beran-Steed RK. 1988. Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repetitive zinc-binding domains. J Biol Chem 263:15857–15859. [PubMed] [Google Scholar]
- 28.Blokpoel MC, Murphy HN, O'Toole R, Wiles S, Runn ES, Stewart GR, Young DB, Robertson BD. 2005. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res 33:e22. doi: 10.1093/nar/gni023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, Shen MY, Kelly L, Melo F, Sali A. 2006. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA: http://www.pymol.org. [Google Scholar]
- 31.Rao SN, Head MS, Kulkarni A, LaLonde JM. 2007. Validation studies of the site-directed docking program LibDock. J Chem Inf Model 47:2159–2171. doi: 10.1021/ci6004299. [DOI] [PubMed] [Google Scholar]
- 32.Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. 2006. Rapid identification of P-glycoprotein substrates and inhibitors. Drug Metab Dispos 34:1976–1984. doi: 10.1124/dmd.106.012351. [DOI] [PubMed] [Google Scholar]
- 33.Ekins S, Johnston JS, Bahadduri P, D'Souzza VM, Ray A, Chang C, Swaan PW. 2005. In vitro and pharmacophore based discovery of novel hPEPT1 inhibitors. Pharm Res 22:512–517. doi: 10.1007/s11095-005-2505-y. [DOI] [PubMed] [Google Scholar]
- 34.Bhaduri T, Bagui TK, Sikder D, Nagaraja V. 1998. DNA topoisomerase I from Mycobacterium smegmatis. An enzyme with distinct features. J Biol Chem 273:13925–13932. [DOI] [PubMed] [Google Scholar]
- 35.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drlica K. 1999. Mechanism of fluoroquinolone action. Curr Opin Microbiol 2:504–508. doi: 10.1016/S1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 37.Berger JM. 1998. Structure of DNA topoisomerases. Biochim Biophys Acta 1400:3–18. doi: 10.1016/S0167-4781(98)00124-9. [DOI] [PubMed] [Google Scholar]
- 38.Bhat AG, Leelaram MN, Hegde SM, Nagaraja V. 2009. Deciphering the distinct role for the metal coordination motif in the catalytic activity of Mycobacterium smegmatis topoisomerase I. J Mol Biol 393:788–802. doi: 10.1016/j.jmb.2009.08.064. [DOI] [PubMed] [Google Scholar]
- 39.Zhu CX, Tse-Dinh YC. 2000. The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J Biol Chem 275:5318–5322. doi: 10.1074/jbc.275.8.5318. [DOI] [PubMed] [Google Scholar]
- 40.Manjunatha UH, Dalal M, Chatterji M, Radha DR, Visweswariah SS, Nagaraja V. 2002. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res 30:2144–2153. doi: 10.1093/nar/30.10.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar R, Madhumathi BS, Nagaraja V. 2014. Molecular basis for the differential quinolone susceptibility of mycobacterial DNA gyrase. Antimicrob Agents Chemother 58:2013–2020. doi: 10.1128/AAC.01958-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flynn JL, Chan J. 2003. Immune evasion by Mycobacterium tuberculosis: living with the enemy. Curr Opin Immunol 15:450–455. doi: 10.1016/S0952-7915(03)00075-X. [DOI] [PubMed] [Google Scholar]
- 43.Behar SM, Divangahi M, Remold HG. 2010. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol 8:668–674. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole ST, Riccardi G. 2011. New tuberculosis drugs on the horizon. Curr Opin Microbiol 14:570–576. doi: 10.1016/j.mib.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 45.Ahmed W, Menon S, Godbole AA, Karthik PV, Nagaraja V. 2014. Conditional silencing of topoisomerase I gene of Mycobacterium tuberculosis validates its essentiality for cell survival. FEMS Microbiol Lett 353:116–123. doi: 10.1111/1574-6968.12412. [DOI] [PubMed] [Google Scholar]
- 46.Heddle JG, Barnard FM, Wentzell LM, Maxwell A. 2000. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids 19:1249–1264. doi: 10.1080/15257770008033048. [DOI] [PubMed] [Google Scholar]
- 47.Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob Agents Chemother 52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kreuzer KN, Cozzarelli NR. 1979. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol 140:424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snyder M, Drlica K. 1979. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol 131:287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- 50.Taneja NK, Tyagi JS. 2007. Resazurin reduction assays for screening of anti-tubercular compounds against dormant and actively growing Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. J Antimicrob Chemother 60:288–293. doi: 10.1093/jac/dkm207. [DOI] [PubMed] [Google Scholar]
- 51.Ahmed W, Menon S, Karthik PV, Nagaraja V. 16 December 2014. Reduction in DNA topoisomerase I level affects growth, phenotype and nucleoid architecture of Mycobacterium smegmatis. Microbiology. doi: 10.1099/mic.0.000014. [DOI] [PubMed] [Google Scholar]
- 52.Tse-Dinh YC. 1986. Uncoupling of the DNA breaking and rejoining steps of Escherichia coli type I DNA topoisomerase. Demonstration of an active covalent protein-DNA complex. J Biol Chem 261:10931–10935. [PubMed] [Google Scholar]
- 53.Sissi C, Palumbo M. 2009. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res 37:702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. 2011. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov Today 16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.