Abstract
Members of the SoxB transcription factor family play critical roles in the regulation of neurogenesis. The SoxB1 proteins are required for the induction and maintenance of a proliferating neural progenitor population in numerous vertebrates, however the role of the SoxB2 protein, Sox21, is less clear due to conflicting studies. To clarify the role of Sox21 in neurogenesis, we examined its function in the Xenopus neural plate. Here we report that misexpression of Sox21 expands the neural progenitor domain, and represses neuron formation by binding to Neurogenin (Ngn2) and blocking its function. Conversely, we found that Sox21 is also required for neuron formation, as cells lacking Sox21 undergo cell death and thus are unable to differentiate. Together our data indicate that Sox21 plays more than one role in neurogenesis, where a threshold level is required for cell viability and normal differentiation of neurons, but a higher concentration of Sox21 inhibits neuron formation and instead promotes progenitor maintenance.
Keywords: Differentiation, Neural progenitor, Neurogenesis, Sox, Xenopus
Introduction
Neurogenesis, the progression of neural stem cells to committed neurons, is a tightly regulated process that is fundamental for the development of the central nervous system (CNS) and neuron regeneration. Although many of the proteins that are involved in this process have been identified (Moody et al., 2013; Rogers et al., 2009b; Suh et al., 2009, 2007), the coordination of the steps required to maintain a balance of proliferating neural stem cells and differentiating neurons is not well understood. To characterize the regulatory factors that drive neurogenesis, we looked to the SoxB family of transcription factors, which play critical roles in neural stem cell induction, maintenance, and neuronal differentiation (Bergsland et al., 2006; Bylund et al., 2003; Kiefer, 2007; Rogers et al., 2009b, 2008; Sandberg et al., 2005).
The 20 vertebrate Sox proteins contain a highly conserved high mobility group (HMG) DNA-binding domain which interacts with the minor groove of the DNA helix to cause it to bend (Kamachi and Kondoh, 2013; Kiefer, 2007). The SoxB proteins also share a conserved eight amino acid Group B homology domain located adjacent to the HMG domain (Kamachi et al., 1995; Uchikawa et al., 1999). The SoxB group is further divided into SoxB1 and SoxB2 subgroups based on both sequence homology and function (Uchikawa et al., 1999).
The SoxB1 proteins (Sox1-3), which act as transcriptional activators (Bylund et al., 2003; Kamachi et al., 1995; Rogers et al., 2009a), are required for induction of the CNS and maintenance of a proliferating neural progenitor population (Buescher et al., 2002; Bylund et al., 2003; Graham et al., 2003; Rogers et al., 2009a; Uchikawa et al., 1999). Overexpression of SoxB1 proteins represses differentiation thereby allowing for the maintenance of a proliferative progenitor population (Bylund et al., 2003; Graham et al., 2003; Holmberg et al., 2008; Mizuseki et al., 1998a; Rex et al., 1997; Rogers et al., 2009a), and the loss of SoxB1 protein expression promotes differentiation into neurons (Bylund et al., 2003). Whereas the SoxB1 proteins function primarily as activators, the closely related SoxB2 proteins (Sox14 and Sox21) function as repressors (Argenton et al., 2004; Sandberg et al., 2005; Uchikawa et al., 1999) and have been proposed to inhibit SoxB1 target genes to facilitate progression from progenitor cell to neuron (Sandberg et al., 2005; Uchikawa et al., 1999).
Although there has been intense study of the function of SoxB1 proteins, much less is known about how the SoxB2 proteins function during neurogenesis. Work in chick, mouse, and frog has demonstrated that the SoxB2 transcription factors are expressed in distinct regions of the CNS during different times in neural development, unlike the SoxB1 proteins which are pan-neural. Whereas sox14 is expressed later in development in distinct subsets of neurons (Cunningham et al., 2008; Hargrave et al., 2000; Uchikawa et al., 1999), sox21 is expressed in the neural plate during neurogenesis (Cunningham et al., 2008), and is later limited to select regions of the developing brain including the forebrain, midbrain-hindbrain boundary (MHB), the olfactory placodes (Cunningham et al., 2008; Ohba et al., 2004; Rimini et al., 1999; Uchikawa et al., 1999), and the spinal cord (Sandberg et al., 2005; Uchikawa et al., 1999). Like the soxB1 genes, sox21 is expressed in neural progenitors and cells that express the neurogenesis marker neurogenin (ngn2), but is not expressed in post-mitotic neurons (Ohba et al., 2004; Sandberg et al., 2005) as is its counterpart sox14 (Hargrave et al., 2000).
It is postulated that Sox21 regulates soxB1 expression in the developing CNS, however functional data are in conflict. Reports have shown that Sox21 counteracts SoxB1 activation of a reporter construct in chick lens cells (Uchikawa et al., 1999), promotes neuronal differentiation in the chick spinal cord (Sandberg et al., 2005) and the dentate gyrus of the adult hippocampus (Matsuda et al., 2012), and promotes differentiation of mouse embryonic stem cells (ESCs) into neural stems cells (Mallanna et al., 2010) when overexpressed. Yet, Sox21 has also been implicated in the maintenance of progenitor proliferation rather than in promoting neuronal differentiation. For example, Sox21 decreases neurite outgrowth induced by nerve growth factor (NGF) in mouse PC12 cells (Ohba et al., 2004), indicating that it inhibits maturation. Likewise, Sox21 levels are elevated in small cell lung cancers suggesting a role in stimulating uncontrolled proliferation (Titulaer et al., 2009). Furthermore, Sox21 is a direct target of pluripotency factors such as Nanog, Oct4 and Sox2 in mouse ESCs (Chakravarthy et al., 2011; Mallanna et al., 2010) and is essential for maintaining progenitors, and induced pluripotency and reprogramming by Sox2 (Kuzmichev et al., 2012). These studies suggest that Sox21 functions similarly to SoxB1 proteins to maintain progenitors and prohibit neuronal outgrowth, and that down-regulation of Sox21 may be required for differentiation to occur. Together these conflicting results suggest that Sox21 does not have one global function, but instead may have distinct, contextual functions in regulating neurogenesis.
In this study, we performed gain- and loss-of-function analyses of Sox21 in Xenopus laevis embryos and explants to determine its mechanism of action during primary neurogenesis. Our gain-of-function assays show that Sox21 expanded the SoxB1 neural progenitor domain and inhibited neuronal differentiation by binding to and counteracting the activity of Ngn2. Conversely, our loss-of-function studies demonstrate that the loss of Sox21 also reduced neuron formation and induced cell death. Together our data indicate that Sox21 plays more than one role in neurogenesis. A threshold level is required for differentiation of neurons, whereas a higher concentration of Sox21 inhibits neurogenesis and instead promotes expansion of SoxB1 expression and progenitor maintenance. Therefore our data suggest that, like other Sox proteins, Sox21 functions in a dose/context dependent manner. With this report we have clarified the relationship between Sox21 and SoxB1 proteins, suggesting that they have cooperative functions in progenitor maintenance and inhibition of differentiation. More importantly, we demonstrate that Sox21 enhances progenitor gene expression during early neurogenesis in vivo by offsetting Ngn2 function and regulates the decision to remain as a progenitor cell or become a neuron based on its level of expression.
Results
Sox21 expands neural progenitors and inhibits neuronal differentiation
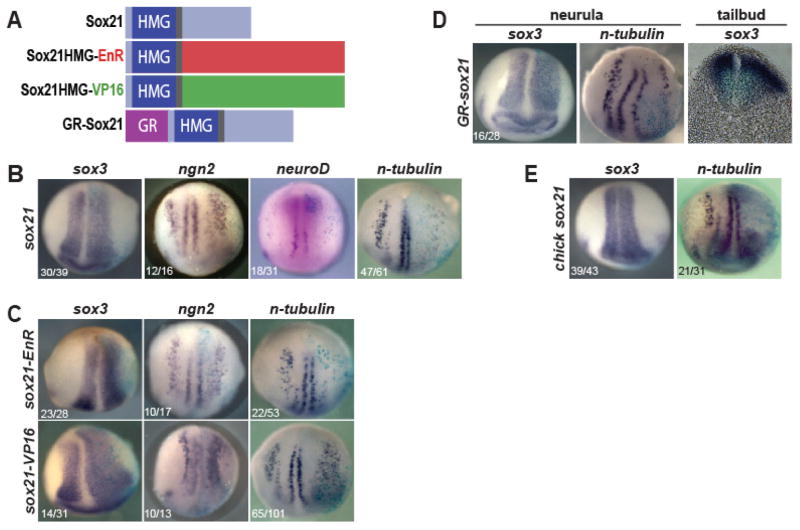
To elucidate the function of Sox21 during neurogenesis, we injected sox21 mRNA into one blastomere of a two-cell Xenopus embryo, collected neurula embryos, and analyzed the expression of sox3, ngn2, neuroD and n-tubulin, which are progenitor (Bylund et al., 2003), proneural (Lee et al., 1995; Ma et al., 1996) and neuronal (Chitnis et al., 1995) genes expressed in cells undergoing neurogenesis, respectively. Overexpression of sox21 expanded the neural plate, as evidenced by the expanded expression domains of sox3 and ngn2 (Fig. 1B). However, the expression of proneural gene neuroD, a direct target of Ngn2 (Ma et al., 1996; Seo et al., 2007), and that of primary neuron marker n-tubulin were both severely reduced (Fig. 1B). This suggests that Sox21 functions to expand neural progenitors and repress neuronal differentiation.
Fig. 1. Sox21 functions as a repressor to expand neural progenitors and inhibit neuronal differentiation.
(A) Diagram of Sox21 proteins used in overexpression studies. (B) In situ hybridization (ISH) of neurula stage embryos injected with sox21 mRNA on right side and analyzed for sox3, ngn2, neuroD or n-tubulin expression. (C) ISH of neurula stage embryos injected with sox21-EnR or sox21-VP16 mRNA on right side and analyzed for sox3, ngn2 or n-tubulin expression. (D) ISH of embryos injected with hormone-inducible GR-sox21 mRNA and analyzed for sox3 or n-tubulin expression. Right: Transverse section of GR-sox21 injected tailbud embryo following ISH for sox3. (E) ISH of embryos injected with chick sox21 mRNA and analyzed for sox3 or n-tubulin expression.
To determine if the X. laevis Sox21 acts as a repressor or activator, we replaced the C-terminal domain of Sox21 with the Engrailed (EnR) repression domain or the VP16 activation domain (Fig. 1A). We injected mRNA into one blastomere of two-cell embryos and analyzed the expression of sox3, ngn2, and n-tubulin. As in embryos injected with sox21, sox21 EnR-injected embryos expanded the neural plate as marked by sox3, and reduced neuron formation as indicated by a repression of n-tubulin expression. Sox21-VP16 grossly expanded both sox3 and n-tubulin (Fig. 1C). This indicates that Sox21 functions as a repressor in its role of expanding neural progenitors and inhibiting neuronal differentiation.
The onset of sox21 expression occurs during early gastrulation, however its expression peaks at mid-neurula stage (Cunningham et al., 2008). To examine the effect of overexpression later in development, we tested the function of the hormone-inducible form of Sox21, in which the Glucocorticoid receptor (GR) binding domain is fused to the N-terminus of full length Sox21 (Fig. 1A). The induction of GR-Sox21 by incubation of gastrulae in dexamethasone (DEX) phenocopied sox21 mRNA injections: expansion of sox3 and repression of n-tubulin in neurula and tailbud embryos (Fig. 1D). This indicates that the effects of Sox21 overexpression do not depend on its time of expression.
These results were unexpected, considering that in the chick embryonic spinal cord, increasing levels of Sox21 were proposed to promote neuronal differentiation (Sandberg et al., 2005). Even though chick and frog Sox21 proteins exhibit a high degree of conservation in their amino acid compositions (Cunningham et al., 2008), misexpression of Xenopus Sox21 inhibits differentiation. To determine if this function was specific to X. laevis Sox21, we injected chick sox21 mRNA (cSox21) into one blastomere of two-cell Xenopus embryos. cSox21 also expanded sox3 expression and repressed n-tubulin (Fig. 1E). Additionally, we analyzed the expression of mouse Sox21 mRNA (mSox21) in the cortex during neurogenesis (E10.5-P10) using RT-PCR, and found that its expression mirrors that of mSox3 and mSox2, and is reduced prior to the onset of neuronal differentiation marked by the expression of proneural gene Ngn2 and neuronal gene Tubb3 (unpublished data). These results support the hypothesis that the early role of Sox21 is to promote neural progenitor formation or maintenance and inhibit neuronal differentiation, and the phenotypic differences are not due to species differences in protein sequence or function.
Sox21 enhances neural progenitor induction by Noggin
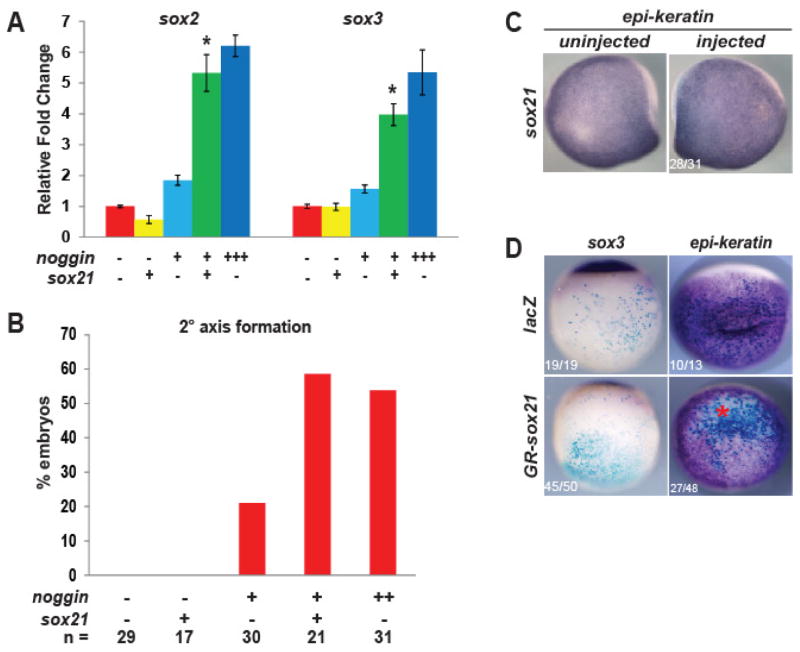
To determine if Sox21 alone is sufficient to induce the formation of neural progenitors or facilitate the induction of progenitors by Noggin, we employed an ectodermal explant assay (Slack and Forman, 1980), which allows for the assessment of molecular function in the absence of other fate-inducing signals provided by the embryo. We injected one-cell embryos with sox21, low or high levels of noggin, or the combination of noggin and sox21 mRNAs, and used quantitative RT-PCR to analyze the expression of neural progenitor genes in neurula staged explants. Noggin is sufficient to induce the expression of progenitor genes sox2 and sox3 (Rogers et al. 2008; Fig. 2A, dark blue bars) and neural competence genes foxd5a (Yan et al., 2009) and zicr1 (Kuo et al., 1998) with little neuron formation (Lamb et al, 1993; Rogers et al, 2011; Fig. S1, dark blue bars). Neither a low level of noggin (light blue bars), nor sox21 alone (yellow bars) induced expression of sox2, sox3 (Fig. 2A), foxd5a or zicr1 (Fig. S1). However, the combination of low noggin with sox21 (green bars) induced the expression of sox2, sox3, foxd5a, and zicr1 to levels comparable to that of high noggin alone (dark blue bars) (Fig. 2A, Fig. S1). This indicates that Sox21 and Noggin synergistically induce progenitor gene expression in naïve ectoderm. We verified this synergy in vivo by in situ hybridization (ISH) of sox3 in embryos injected with low noggin and/or sox21 mRNA in the ventral blastomeres of four-cell embryos. Low levels of noggin induced a secondary axis marked by sox3 in 20% of injected embryos (n= 4/19), however sox21 and noggin together induced a secondary axis in 60% of injected embryos (n= 17/29) (Fig. 2B. This capacity to convert ectoderm to a neural fate was strictly due to collaboration with noggin, as sox21 alone did not affect epidermis formation marked by epi-keratin expression (Fig. 2C), nor did it induce a secondary axis when overexpressed in ventral cells (Fig. 2B, n= 23/29 unchanged). To determine if a higher amount of sox21 expression has the capacity to convert ventral ectoderm to neural tissue, we injected 400 pg of GR-sox21 mRNA and induced expression during late gastrula stage. While there was a slight decrease in epi-keratin expression, sox3 was not expressed in these ventral cells (Fig. 2D). This suggests that at a high level Sox21 interferes with epidermis formation, but does not induce a neural fate in non-neural ectoderm.
Fig. 2. Sox21 facilitates neural progenitor induction by Noggin.
(A) Quantitative RT-PCR analysis of sox2 and sox3 expression in neurula stage ectodermal explants isolated from embryos injected with low (+) or high (++) levels of noggin mRNA with or without sox21 mRNA at the 1-cell stage. * indicates significance by Student’s t-test (p<0.05) compared to both sox21 and low noggin alone. (B) Graph of secondary axis phenotype for embryos injected with low (+) or high (++) nogginA3 mRNA with or without sox21 into one ventral blastomere, and analyzed by ISH for sox3. (C) ISH of a single neurula stage embryo injected with sox21 mRNA and analyzed for epi-keratin expression. (D) ISH of embryos injected into one ventral blastomere with hormone-inducible GR-sox21 mRNA, and analyzed for sox3 or epi-keratin expression. * indicates reduced expression.
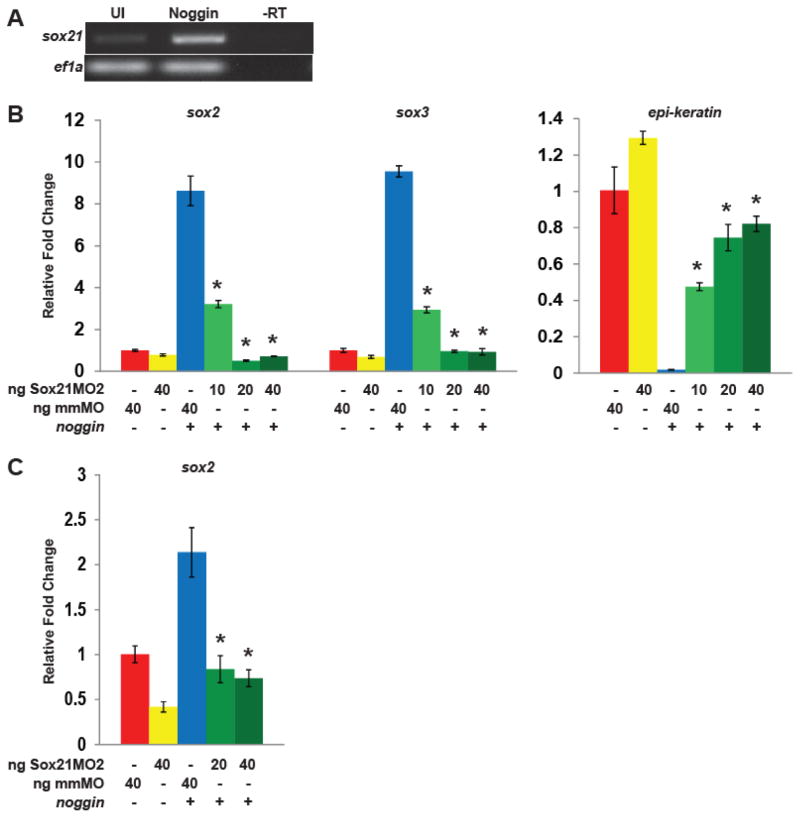
These data demonstrate that Sox21 facilitates neural progenitor induction by Noggin. In fact, Noggin induces sox21 expression in ectodermal explants (Fig. 3A). To determine if Noggin requires Sox21 to induce neural progenitor gene expression, we inhibited Sox21 protein translation in ectodermal explants with antisense morpholinos (MOs). We injected noggin mRNA with increasing amounts of Sox21 MO2, and used quantitative RT-PCR to analyze the expression of progenitor genes in mid-neurula staged explants. We found that with increasing amounts of MO2, sox2 and sox3 expression significantly decreased and epi-keratin expression significantly increased in a dose-dependent manner (Fig. 3B). Thus, Noggin requires Sox21 to maintain or induce the expression of neural progenitor markers in neurulae. We next asked if Sox21 was required for onset of Sox2 expression, which is induced in mid-gastrula stage explants (Rogers et al., 2008). When Sox21 levels were reduced, sox2 was not expressed in response to Noggin by the mid-gastrula stage (Fig. 3C), suggesting that Noggin requires Sox21 to induce a neural fate.
Fig. 3. Sox21 is required for the induction and maintenance of sox2 expression.
(A) RT-PCR analysis of sox21 expression in neurula stage ectodermal explants isolated from untreated embryos and those injected with noggin at the 1-cell stage. Reference gene ef1a was used as a loading control. (B) Quantitative RT-PCR analysis of sox2, sox3, and epi-keratin expression in mid-neurula stage ectodermal explants isolated from embryos injected with noggin, Sox21MO2, mismatch MO (mmMO) or in combination (as indicated) at the 1-cell stage. * indicates significance by Student’s t-test (p<0.05) compared to noggin alone. (C) Quantitative RT-PCR analysis of sox2 expression in early gastrula (stage 11) ectodermal explants isolated from embryos injected as in B.
Sox21 interferes with Ngn2 function
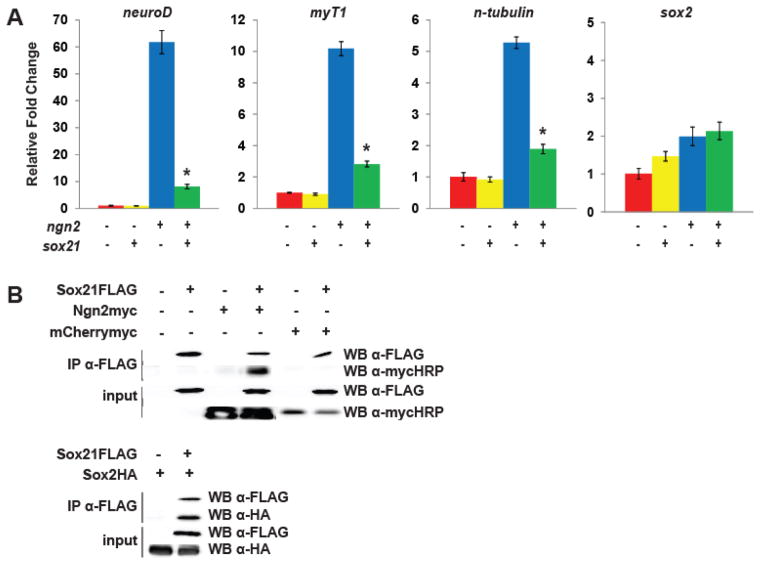
Our data show that Sox21 promotes progenitor maintenance and inhibits neuronal differentiation, as the expression of neuroD and n-tubulin (Fig. 1B) is repressed by Sox21 misexpression. However, overexpression of Sox21 does not decrease the level of ngn2 (Fig. 1B). One possibility is that Sox21 prevents neuronal differentiation by blocking Ngn2 function, but does not affect ngn2 transcription. To test this, we injected sox21, ngn2 or the combination of mRNAs into embryos and analyzed the expression of Ngn2 target genes in neurula stage explants using quantitative RT-PCR (Fig. 4A). While Ngn2 is sufficient to induce the expression of neuroD, myT1 and n-tubulin in explants (Bellefroid et al. 1996; Ma et al. 1996; Seo et al. 2007; Fig. 4A, blue bars), the addition of Sox21 significantly reduced the expression of these genes (Fig. 4A, green bars), indicating that Sox21 inhibits neurogenesis by counteracting Ngn2 function.
Fig. 4. Sox21 inhibits the expression of neuronal differentiation genes and binds to Ngn2 to counteract its function.
(A) Quantitative RT-PCR analysis of neuroD, myT1, n-tubulin and sox2 expression in neurula stage ectodermal explants isolated from embryos injected with ngn2, sox21, or ngn2+sox21. * indicates significance by Student’s t-test (p<0.05) compared to ngn2 alone. (B) Co-Immunoprecipitation (co-IP) of in vitro translated (IVT) Sox21-FLAG protein with Ngn2-myc or mCherry-myc proteins (top), or Sox2-HA (bottom) protein.
Ngn2 binds directly to enhancer regions to activate transcription of target genes such as neuroD and myT1 (Seo et al., 2007). To determine if Sox21 binds to Ngn2 to interfere with Ngn2 function, we performed a co-immunoprecipitation (co-IP) assay using in vitro translated Sox21-FLAG and Ngn2-myc proteins. We found that Ngn2-myc co-precipitates with Sox21-FLAG, and this interaction was specific to Ngn2 as mCherry-myc did not co-precipitate with Sox21-FLAG (Fig. 4B, top). We also examined the interaction of Sox21 with Sox2, which have been shown to be expressed in the same cells (Kuzmichev et al., 2012; Ohba et al., 2004; Sandberg et al., 2005) and interact in mouse embryonic stem cells (Mallanna et al., 2010), and confirmed that Xenopus Sox2 and Sox21 proteins also interact as reported (Fig. 4B, bottom). Hence, Sox21 interacts with both Ngn2 and Sox2, and these interactions may alter its function during neurogenesis.
Sox21 is required for neuron formation
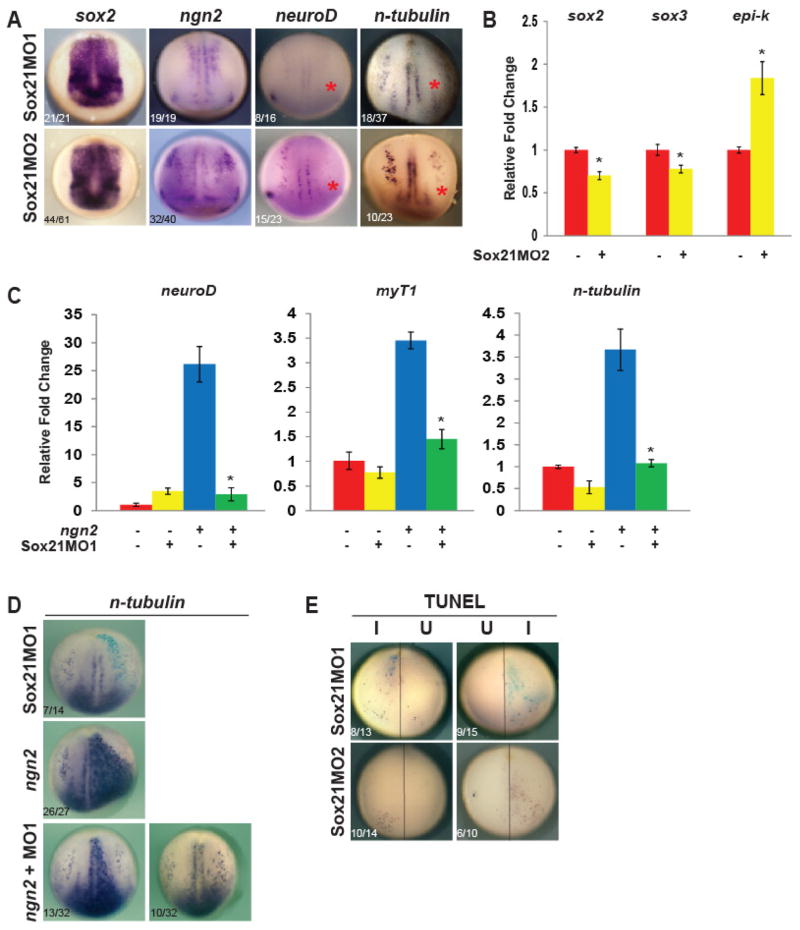
Our data indicate that Sox21 plays an important role in regulating the progression of neurogenesis; wherein it inhibits neuron formation in favor of progenitor cell expansion. To determine if Sox21 is required in the embryo for progenitor formation, we injected Sox21 MOs into one dorsal blastomere of four-cell embryos and analyzed the resulting expression of progenitor, proneural and neuronal genes by ISH. Knocking down Sox21 did not affect expression of progenitor marker sox2 (Fig. 5A), or the proneural gene ngn2 (Fig. 5A). To confirm this result, we injected Sox21MO2 into one-cell embryos and used quantitative RT-PCR to quantify the expression of sox2, ngn2, and epi-keratin. At stage 11 the expression of sox2 and ngn2 were reduced by 30% compared to untreated controls (Fig. 5B). Additionally, the expression of epi-keratin was slightly increased (Fig. 5B), suggesting that with a decrease in Sox21, more ectodermal cells undergo an epidermal fate. We also found that the expression levels of neuroD and the neuron marker n-tubulin were drastically reduced (Fig. 5A, red asterisks), suggesting that Sox21 is also required for subsequent neuron formation. To test this, we injected ngn2 mRNA and Sox21MO1 into embryos and analyzed changes in Ngn2 target gene expression in neurula explants by quantitative RT-PCR (Fig. 5C). The reduction of Sox21 severely decreased the expression of Ngn2 targets neuroD and myT1 and of n-tubulin, indicating that even though high levels of Sox21 inhibits Ngn2, some Sox21 is required for activation of Ngn2 targets and ultimately for neuron formation (Fig. 5C). We confirmed this in embryos by injecting ngn2 and Sox21MO1 into one blastomere of four-cell embryos and assaying for ectopic neurons. The lack of Sox21 reduced induction of ectopic n-tubulin by Ngn2 (Fig. 5D). Given that neuronal and progenitor gene expression was absent or reduced in Sox21-deficient cells, we asked if this was in part due increased cell death. We assayed for cell death using TUNEL in embryos injected with Sox21MO1 or MO2. We found that embryos injected with Sox21MO exhibited increased TUNEL staining on the injected side compared to the uninjected side (Fig. 5E).
Fig. 5. The loss of Sox21 reduces neuron formation and increases apoptosis in embryos.
(A) ISH of neurula stage embryos injected on right side with Sox21 morpholino MO1 (top) or fluorescent morpholino MO2 (bottom) and analzed for expression of neurogenesis genes. Asterisks indicate a loss of expression. (B) Quantitative RT-PCR analysis of sox2, ngn2 and epi-keratin expression in gastrula stage embryos injected with Sox21MO2. * indicates significance by Student’s t-test (p<0.05) compared to uninjected embryos. (C) Quantitative RT-PCR analysis of neuroD, myT1, and n-tubulin expression in neurula stage ectodermal explants isolated from embryos injected with ngn2 mRNA, Sox21MO1, or the combination as indicated. * indicates significance by Student’s t-test (p<0.05) compared to ngn2 alone. (D) ISH of neurula stage embryos injected on right side with ngn2 mRNA and MO1 (as indicated) and analyzed for n-tubulin expression. (E) TUNEL staining of embryos injected with MO1 or MO2.
Our data indicate that both reduction and overexpression of Sox21 block neuron formation. One possibility is that the level of Sox21 must be finely tuned in a developing cell in order for neuron formation, and that the level of Sox21 expression determines its function during neurogenesis. To examine this further, we injected increasing amounts of GR-sox21 mRNA into one blastomere of four-cell embryos, induced expression of Sox21 at late gastrula, and analyzed changes in the expression of sox3 and n-tubulin by ISH in neurula embryos (Fig. 6). We found that 25 pg of GR-sox21 expanded sox3 in approximately 18% of injected embryos (n= 3/17), 50–100 pg expanded sox3 in 43% of injected embryos (n= 13/30, 9/21 respectively), and 400 pg of GR-sox21 expanded sox3 in 87% of injected embryos (n= 14/16). In contrast, 25 pg of GR-sox21 was sufficient to reduce n-tubulin expression in 46% of injected embryos (n= 13/28), and 50 pg reduced n-tubulin in 69% (n= 22/32). Thus, a lower concentration of GR-sox21 (50 pg) is sufficient to inhibit differentiation, whereas eight-fold higher concentration is required for progenitor expansion. Considered with the MO knockdown data, this indicates that a low level of Sox21 is needed for n-tubulin expression and cell survival, whereas a high level of Sox21 expands sox3 inhibits n-tubulin expression. This suggests that the level of Sox21 expression dictates its targets, and therefore its functions during neurogenesis. Ultimately, the level of Sox21 expression directs if and how a neural progenitor cell will progress through neuronal differentiation.
Fig. 6. Sox21 affects neural gene expression in a dose-dependent manner.
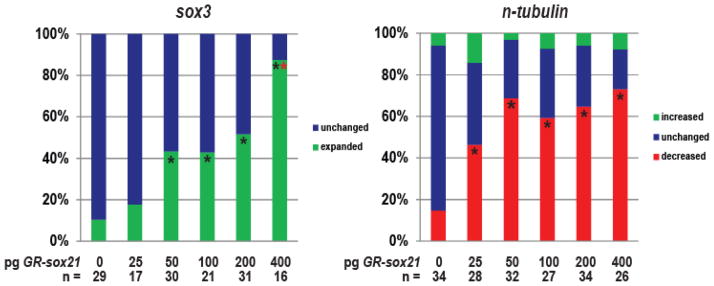
Embryos were injected with the indicated amounts of GR-sox21 mRNA and analyzed by ISH for changes in sox3 and n-tubulin expression. Graphs are data from a single representative experiment, which was performed three times and displayed the same trend. n = total number of injected embryos analyzed in one experiment. * Indicates significance by Chi-square test (p<0.05) compared to uninjected (0 pg) alone. * Indicates significance by Chi-square test (p<0.05) compared to other doses (25–200 pg).
Discussion
SoxB transcription factors play important roles in the regulation of neurogenesis. Prior studies have indicated that Sox21 drives neuronal differentiation (Sandberg et al, 2005; Uchikawa et al, 1999), while others indicate that it is involved in the maintenance of progenitor populations (Ohba et al, 2004; Kuzmichev et al, 2012; Titulaer et al, 2009). Due to these observed differences, we sought to clarify its role and to define its mechanism for action during neurogenesis in the neural plate.
In this study, we investigated the role of Sox21 during neurogenesis using gain- and loss-of-function. At a high level of expression, Sox21 expanded progenitor cells at the expense of differentiation. This is in line with other reports suggesting that Sox21 functions in the maintenance of a proliferating progenitor cell population (Kuzmichev et al., 2012; Ohba et al., 2004). We also discovered that Sox21 inhibited neurogenesis by specifically blocking Ngn2 from activating its downstream targets neuroD, myT1 and subsequently n-tubulin (Fig. 4A). We determined that Sox21 interacts with Ngn2 (Fig. 4B), indicating that together they bind and repress Ngn2 target genes, or that Sox21 sequesters Ngn2 and prevents it from activating target genes, thus allowing for enhanced progenitor gene expression.
Previous studies have shown that SoxB1 protein Sox2 activates Sox21 expression (Chakravarthy et al., 2011; Kuzmichev et al., 2012), and we show here that sox21 overexpression enhances sox2 expression (Fig. 1F). Likewise, we have found that the reduction of Sox21 reduces sox2 expression (Fig. 3B,C), and a decrease in Sox2 reduces sox21 expression (unpublished data). Thus, a Sox2-Sox21 partnership could present a feedback loop by which Sox2 and Sox21 maintain each other’s expression to generate and/or maintain a progenitor pool and inhibit differentiation. As Sox21 is a known target of reprogramming factors like Sox2 (Kuzmichev et al., 2012), our work suggests that it may be useful for future work investigating induced pluripotency.
Sox21 is not sufficient to directly induce SoxB1 expression in ectoderm tissue. Instead, Sox21 works synergistically with Noggin, a BMP antagonist, to induce ectoderm cells to undergo a neural fate (Fig. 2, green bars; Fig. 2B,D). Therefore it is likely that Sox21 facilitates Noggin activity, or is used as an additional aide to maintain cells in a progenitor state, rather than as a neural induction agent. Since Noggin both activates Sox21 as well as promotes SoxB1 expression, one possibility is that Noggin employs Sox21 to induce, maintain, or enhance sox2 and sox3 in ectodermal explants.
This role in progenitor maintenance is also supported by the expression pattern of sox21; it is highly expressed in the forebrain, mid-hindbrain boundary (MHB) and olfactory placodes (Cunningham et al., 2008), which are regions characterized by delayed neurogenesis (Baek et al., 2006; King-Robson, 2011; Stigloher et al., 2008). This suggests that Sox21 may be utilized during brain development to maintain a pool of progenitors as other cells differentiate, perhaps to form a properly sized brain. Sox21 is also expressed in the ventricular zone of the embryonic mouse cortex and crypts of the gut, where it overlaps with the expression of progenitor marker Nestin (Ohba et al., 2004) and SoxB1 proteins (Kuzmichev et al., 2012; Sandberg et al., 2005), as well as in neurospheres isolated from the mouse cortex (unpublished data). We have also seen that during corticogenesis, its expression mirrors that of progenitor markers sox2 and sox3, and its expression decreases as differentiation factors such as ngn2 and tubb3 increase to signal neuron development (unpublished data). These findings provide evidence that the behavior of Sox21 may be contextual and regulated by other environmental factors that differ between regions of the nervous system.
While our gain-of-function data show that Sox21 plays a role in progenitor maintenance and differentiation inhibition, our loss-of-function data suggest that Sox21 is required for cell viability and early neuron development. Therefore at some minimal threshold level, Sox21 must be expressed for neuron formation to occur in a normally developing embryo. We note that in ectodermal explants, Sox21 is required for the expression of SoxB1 progenitor genes. This was not immediately evident in whole embryos, however quantitative RT-PCR revealed a decrease in the expression of soxB1 genes (Fig. 5B). In the embryo, there are multiple pathways that converge to induce neural genes (Marchal et al., 2009; Rogers et al., 2011). In our ectodermal explants, Noggin is the only neuralizing factor present. Thus while Noggin may require Sox21 for its induction of SoxB1 proteins, other factors that are present in the embryo may compensate and allow for some SoxB1 expression in a Sox21-reduced environment. However despite this compensation, there are fewer neural progenitors as well as an increase in cell death in the absence of Sox21. One interpretation is that progenitors cannot exit the cell cycle, or that they exit the cell cycle but cannot differentiate, and as a result apoptose. In support of this, a study showed that the protein Cdh1 is required for both cell cycle exit and differentiation, and that its loss delayed cell cycle exit and led to increased apoptosis (Delgado-Esteban et al., 2013). There is no evident accumulation of progenitor cells due to the reduction of Sox21, therefore we conclude that it is not likely that the cells cannot exit the cell cycle, but instead that the progenitor cells that are present are unable to differentiate and therefore die as a result. This suggests that Sox21 plays some early role in cell survival. We have also observed that increasing the amount of sox21 mRNA injected by 1.5X is lethal during embryonic gastrulation (unpublished data). Therefore it is possible that Sox21 acts on a target involved in cell cycle regulation, and precise regulation of its expression is required for cell survival and differentiation.
Thus, we find that Sox21 has multiple roles based on its level and spatiotemporal expression. At a high level, Sox21 inhibits neuron formation by interacting with Ngn2 to promote progenitor maintenance (Fig. 7); however some Sox21 expression is required for cell survival and ultimately neuron formation. As such, our model proposes that the level of Sox21 expression within a neural cell regulates how it will progress during neurogenesis. When Sox21 is severely reduced, neural progenitor cells will undergo cell death as cells exit the cell cycle but are unable to differentiate, or as cells are stuck in the cell cycle. With a minimal level of Sox21 expression, these cells differentiate to become neurons. Yet when slightly increased, Sox21 inhibits neurogenesis, and as more Sox21 is expressed, it also expands the neural plate by enhancing SoxB1 expression, potentially through its interaction with Sox2.
Fig. 7. Model for function of Sox21 during neurogenesis.
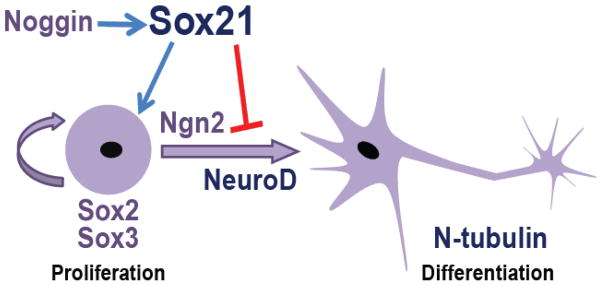
Sox21 is induced by Noggin and maintains cells in a progenitor state by binding to Ngn2 to inhibit neuronal differentiation and expand the expression of the soxB1 genes when expressed at a high level.
Other Sox proteins, including Sox2, have been shown to function in a dose-dependent manner (Hutton and Pevny, 2011; Taranova et al., 2006). Because all SOX proteins recognize the same DNA-binding sequence (A/T) (A/T) CAA(A/T)G (Harley et al., 1994), it is understood that Sox proteins cooperate with other region-specific transcription factor partners to regulate expression of their specific down-stream target genes (Kamachi et al., 2000). Therefore in different cells or at different times in development, the Sox proteins interact with different partners and as a result have different functions. However, despite their importance very few partner interactions have been confirmed in vivo. We have shown that interaction with a partner Oct4 allows for the three SoxB1 proteins to activate unique neural targets during neurogenesis (Archer et al., 2011). Another Sox protein, Sox10, performs multiple functions during glial development based on its interaction with different transcription factors and epigenetic modifiers (Weider et al., 2013). Sox21 function and its role in neurogenesis may also depend on the interaction of different partner proteins based on its level of expression. In addition to its roles in neurogenesis, Sox21 has also been shown to be required for the development of other organs, including sensory epithelium in developing cochlea (inner ear) of mice (Hosoya et al., 2011) and the cuticle layer and the progenitor cells of the hair shaft in both mouse and human (Kiso et al., 2009). Again this suggests that its multiple roles in development may be specified by cooperation with different partners in different environments at different times during development.
Materials and Methods
Embryo culturing and manipulations
Xenopus laevis embryos were obtained using standard methods (Sive, Grainger et al. 2000) and staged according to Nieuwkoop and Faber (1994). Ectodermal explants were obtained by excising naïve ectoderm from the animal pole of stage 8–9 embryos, and culturing them in 0.75X Normal Amphibian Medium (NAM) (Slack and Forman, 1980) with gentamycin. Explants were collected in sets of 20 per treatment when uninjected sibling embryos reached stages 11, 13, 14, or 16.
Plasmid construction
Sox21HMG-VP16 was constructed by amplifying the N-terminus and sox21 HMG box via PCR using the forward SP6 primer and reverse: 5′-CGAGATCTCCCTGTAAACCCATAGGGC-3′ primer, and ligating the digested PCR product into a p-Bluescript vector containing the VP16 activator domain (Brickman et al., 2000). Sox21HMG-EnR was constructed in a similar manner via PCR using the following primers: forward 5′-CCGCTCGAGGGGACCCCGGGCAACGAG-3′ and reverse 5′-CCATCGATGTCCCCTGTAAACCCATAGGG-3′. The digested PCR product was ligated into a p-Bluescript vector containing amino acids 2–298 of the Drosophila Engrailed (EnR) protein, then subcloned into the pCS2+ expression vector. Chick sox21 was obtained from J. Muhr, and subcloned into the pCS2+ expression vector. Sox21-FLAG was constructed by amplifying full length sox21 via PCR using the following primers: forward 5′-GCAGGATCCATGTCTAAACCGCTGGATCATG-3′ and reverse 5′-TGCCTCGAGTCACTTGTCATCGTCATCTTTATAATCTAATGCTGCCGCATAGGCTG-3′ (FLAG sequence underlined), and ligating into pCS2+ expression vector. Sox2-HA was constructed in a similar manner by PCR-amplifying sox2 using the following primers: forward 5′-GCAGAATTCATGTACAGCATGATGGAGACC-3′ and reverse 5′-TGCCTCGAGTCAAGCGTAATCTGGAACATCGTATGGGTACATGTGCGACAGAGGCA GC-3′ (HA sequence underlined), and ligating into pCS2+ expression vector.
mRNA and morpholino microinjections
Synthetic capped mRNA was made by in vitro transcription using mMessage mMachine kits (Ambion). Amounts injected for in situ hybridization (ISH) analyses in whole embryos were as follows: 150 pg sox21, 200 pg sox21HMG-EnR, 250 pg sox21HMG-VP16, 200 or 400 pg GR-sox21, 150 pg chick sox21, 25 pg or 50 pg noggin A3 (Smith and Harland, 1992), 40 ng Sox21MO1 (Gene Tools), and 40 ng Sox21MO2 (Gene Tools). For the dose dependence assay, 25, 50, 100, 200, and 400 pg GR-sox21 mRNA were injected. GR was induced with 10 μM dexamethasone + 0.2% BSA at stage 12 or 12.5. All embryos were injected at the two- or four-cell stage and include 300 pg lacZ mRNA as a tracer, except for Sox21MO2 which is tagged with lissamine as a fluorescent tracer. Embryos were cultured until stages 14–15 and analyzed by ISH. For ectodermal explant assays, the following amounts were injected: 100 pg sox21 mRNA, 8 pg (low) or 28 pg (high) noggin mRNA (Knecht et al., 1995), 290 pg ngn2 (Ma et al., 1996) mRNA, 10–40 ng Sox21MO2, 40 ng mismatch MO (Gene Tools), and 40 ng Sox21MO1. Embryos were injected at the one-cell stage, and the explants were analyzed by reverse transcription- polymerase chain reaction (RT-PCR) or quantitative RT-PCR.
RT-PCR and qRT-PCR
RT-PCR was performed as described (Wilson and Hemmati-Brivanlou 1995). Total RNA was extracted from 20 ectodermal explants using either RNAqueous (Ambion) or PureLink (Invitrogen) RNA extraction kits. cDNA was generated using random hexamers and either MMLV reverse transcriptase (Fisher) or Tetro cDNA synthesis kit (Bioline). Following cDNA synthesis, PCR was performed using primers for ef1α (XMMR) to confirm cDNA production, and muscle actin (Hemmati-Brivanlou and Melton, 1994) to detect mesoderm contamination. RT-minus samples were also assayed to detect genomic DNA contamination.
qPCR was performed with Express SYBR Green Supermix (Invitrogen) or SensiMix SYBR Green Mastermix (Bioline) in MX3000P system (Strategene), or with SensiFAST SYBR Green Mastermix (Bioline) in CFX96 system (BioRad). Relative quantification of gene expression was determined using the ΔΔCt method (Livak and Schmittgen, 2001). All samples were normalized to levels of reference gene ef1a, which was used as the loading control, and analyzed relative to untreated or control ectodermal explants. All samples were analyzed in triplicate, and experiments were repeated at least twice. Graphs in figures represent the mean fold change in expression of a representative experiment, relative to control explants ± standard deviation. See Table S1 for primer sequences and qPCR conditions.
ISH and β-galactosidase assay
Whole mount in situ hybridization (ISH) was performed as described (Harland, 1991; Hemmati-Brivanlou et al., 1990) except RNAse treatments were omitted. For lineage tracing, β-galactosidase activity was visualized with X-gal (Research Organics). Digoxygenin labeled mRNA probes were generated for sox2 (Mizuseki et al., 1998b), sox3 (Penzel et al., 1997), ngn2 (Ma et al., 1996), neuroD (Lee et al., 1995), epi-keratin (Jonas et al., 1985) and n-tubulin (Richter et al., 1988).
Western Blots and Co-IPs
In vitro translation (IVT) with 250 ng of mRNA was performed using the TnT SP6 High-Yield Protein Expression System (Promega) to synthesize tagged proteins. 66% of this reaction was subjected to co-IP while 33% was saved as input.
Co-immunoprecipitation (co-IP) was performed according to (Singh et al., 2013). For each co-IP reaction, 10 μl reaction mix (66%) was mixed with 650 μl cold TNSG lysis buffer (20 mM Tris pH 8, 137 mM NaCl, 10% glycerol, 1% NP-40) containing a protease inhibitor cocktail tablet (Roche) and 2 μg/ml of anti-FLAG (Sigma), and incubated for one hour on ice. Next, 25 μl Protein A/G agarose beads (Santa Cruz) were added and the reaction was incubated at 4°C overnight on an orbital mixer. Following this, beads were briefly pelleted at 4°C and washed three times in ice-cold TNSG buffer. Buffer was decanted and beads were resuspended in 50 μl 1X SDS sample buffer and boiled at 100°C for 5–10 minutes. 12 μl of IP sample and 2.5 μl of input (also boiled in 6X SDS sample buffer) were loaded onto 12.5% Tris-glycine SDS polyacrylamide gels, resolved by electrophoresis, transferred to nitrocellulose, and blocked in 5% non-fat dry milk in PBST (PBS+ 0.1% Tween-20) for one hour at room temperature. Following PBST rinse, blots were incubated with anti-FLAG, anti-HA (Applied Biological Materials), or anti-myc-HRP (Santa Cruz) primary antibodies for one hour at room temperature, washed multiple times with PBST, then incubated with anti-mouse lgG-HRP secondary antibody (Santa Cruz) for one hour at room temperature. After PBST washes, blots were incubated with Clarity chemiluminescent ECL substrate (Bio-Rad) and imaged using ImaqeQuant LAS-4000 mini digital imager (GE Healthcare).
Supplementary Material
Highlights.
Sox21 expands and is required for soxB1 gene expression in neural progenitor cells
Sox21 inhibits neuronal gene expression by binding to Ngn2 to repress its function
Sox21 is required for cell viability and differentiation
The level of Sox21 expression regulates the progression of neurogenesis
Acknowledgments
We thank Sally Moody, Richard Harland, Chris Kinter, Jonas Muhr, and Julie Devine for plasmids. We thank Richard Harland and the Casey lab for helpful discussion. We also thank Ira Daar for providing detailed co-IP protocols. This work was supported by NIH Grants NS048918 and NS078741 to ESC.
Footnotes
Author contributions
NCW, DDC, and EMS conceived the research, designed experiments and analyzed data. NW, DDC, KL, and DD performed experiments. EMS, DDC, and NCW wrote and edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Niteace Whittington, Email: Ncw7@georgetown.edu.
Doreen Cunningham, Email: ddc24@georgetown.edu.
Kim Le, Email: kimle1029@gmail.com.
David De Maria, Email: ddemaria@gmail.com.
Elena M. Silva, Email: elena.silva@georgetown.edu.
References
- Archer TC, Jin J, Casey ES. Interaction of Sox1, Sox2, Sox3 and Oct4 during primary neurogenesis. Dev Biol. 2011;350:429–40. doi: 10.1016/j.ydbio.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argenton F, Giudici S, Deflorian G, Cimbro S, Cotelli F, Beltrame M. Ectopic expression and knockdown of a zebrafish sox21 reveal its role as a transcriptional repressor in early development. Mech Dev. 2004;121:131–42. doi: 10.1016/j.mod.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Baek JH, Hatakeyama J, Sakamoto S, Ohtsuka T, Kageyama R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development. 2006;133:2467–76. doi: 10.1242/dev.02403. [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, Bourguignon C, Hollemann T, Ma Q, Anderson DJ, Kintner C, Pieler T. X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell. 1996;87:1191–202. doi: 10.1016/s0092-8674(00)81815-2. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–86. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman JM, Jones CM, Clements M, Smith JC, Beddington RS. Hex is a transcriptional repressor that contributes to anterior identity and suppresses Spemann organiser function. Development. 2000;127:2303–15. doi: 10.1242/dev.127.11.2303. [DOI] [PubMed] [Google Scholar]
- Buescher M, Hing FS, Chia W. Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development. 2002;129:4193–203. doi: 10.1242/dev.129.18.4193. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–8. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Chakravarthy H, Ormsbee BD, Mallanna SK, Rizzino A. Rapid activation of the bivalent gene Sox21 requires displacement of multiple layers of gene-silencing machinery. FASEB J. 2011;25:206–18. doi: 10.1096/fj.10-166926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Cunningham DD, Meng Z, Fritzsch B, Casey ES. Cloning and developmental expression of the soxB2 genes, sox14 and sox21, during Xenopus laevis embryogenesis. Int J Dev Biol. 2008;52:999–1004. doi: 10.1387/ijdb.082586dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Esteban M, García-Higuera I, Maestre C, Moreno S, Almeida A. APC/C-Cdh1 coordinates neurogenesis and cortical size during development. Nat Commun. 2013;4:2879. doi: 10.1038/ncomms3879. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–65. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Hargrave M, Karunaratne a, Cox L, Wood S, Koopman P, Yamada T. The HMG box transcription factor gene Sox14 marks a novel subset of ventral interneurons and is regulated by sonic hedgehog. Dev Biol. 2000;219:142–53. doi: 10.1006/dbio.1999.9581. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–95. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou a, Frank D, Bolce ME, Brown BD, Sive HL, Harland RM. Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development. 1990;110:325–30. doi: 10.1242/dev.110.2.325. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou a, Melton Da. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–81. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Holmberg J, Hansson E, Malewicz M, Sandberg M, Perlmann T, Lendahl U, Muhr J. SoxB1 transcription factors and Notch signaling use distinct mechanisms to regulate proneural gene function and neural progenitor differentiation. Development. 2008;135:1843–51. doi: 10.1242/dev.020180. [DOI] [PubMed] [Google Scholar]
- Hosoya M, Fujioka M, Matsuda S, Ohba H, Shibata S, Nakagawa F, Watabe T, Wakabayashi KI, Saga Y, Ogawa K, Okano HJ, Okano H. Expression and function of sox21 during mouse cochlea development. Neurochem Res. 2011;36:1261–9. doi: 10.1007/s11064-011-0416-3. [DOI] [PubMed] [Google Scholar]
- Hutton SR, Pevny LH. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev Biol. 2011;352:40–7. doi: 10.1016/j.ydbio.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Jonas E, Sargent TD, Dawid IB. Epidermal kerain gene expressed in embryos of Xenopus laevis. Proc Natl Acad Sci U S A. 1985;82:5413–5417. doi: 10.1073/pnas.82.16.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–44. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Sockanathan S, Liu Q, Breitman M, Lovell-Badge R, Kondoh H. Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 1995;14:3510–9. doi: 10.1002/j.1460-2075.1995.tb07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Kondoh H, Biology C. Pairing SOX off. 2000;9525:2–7. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- Kiefer JC. Back to basics: Sox genes. Dev Dyn. 2007;236:2356–66. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- King-Robson J. Encouraging regeneration in the central nervous system: is there a role for olfactory ensheathing cells? Neurosci Res. 2011;69:263–75. doi: 10.1016/j.neures.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Kiso M, Tanaka S, Saba R, Matsuda S, Shimizu A, Ohyama M, Okano HJ, Shiroishi T, Okano H, Saga Y. The disruption of Sox21-mediated hair shaft cuticle differentiation causes cyclic alopecia in mice. Proc Natl Acad Sci U S A. 2009;106:9292–7. doi: 10.1073/pnas.0808324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klisch TJ, Souopgui J, Juergens K, Rust B, Pieler T, Henningfeld Ka. Mxi1 is essential for neurogenesis in Xenopus and acts by bridging the pan-neural and proneural genes. Dev Biol. 2006;292:470–85. doi: 10.1016/j.ydbio.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Good PJ, Dawid IB, Harland RM. Dorsal-ventral patterning and differentiation of noggin-induced neural tissue in the absence of mesoderm. Development. 1995;121:1927–35. doi: 10.1242/dev.121.6.1927. [DOI] [PubMed] [Google Scholar]
- Kuo JS, Patel M, Gamse J, Merzdorf C, Liu X, Apekin V, Sive H. Opl: a zinc finger protein that regulates neural determination and patterning in Xenopus. Development. 1998;125:2867–82. doi: 10.1242/dev.125.15.2867. [DOI] [PubMed] [Google Scholar]
- Kuzmichev AN, Kim SK, D’Alessio AC, Chenoweth JG, Wittko IM, Campanati L, McKay RD. Sox2 acts through Sox21 to regulate transcription in pluripotent and differentiated cells. Curr Biol. 2012;22:1705–10. doi: 10.1016/j.cub.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Knecht aK, Smith WC, Stachel SE, Economides aN, Stahl N, Yancopolous GD, Harland RM. Neural induction by the secreted polypeptide noggin. Science. 1993;262:713–8. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- Lee J, Hollenberg S, Snider L, Turner D, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science (80-) 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Mallanna SK, Ormsbee BD, Iacovino M, Gilmore JM, Cox JL, Kyba M, Washburn MP, Rizzino A. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010;28:1715–27. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal L, Luxardi G, Thomé V, Kodjabachian L. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc Natl Acad Sci U S A. 2009;106:17437–42. doi: 10.1073/pnas.0906352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Kuwako K, Okano HJ, Tsutsumi S, Aburatani H, Saga Y, Matsuzaki Y, Akaike A, Sugimoto H, Okano H. Sox21 promotes hippocampal adult neurogenesis via the transcriptional repression of the Hes5 gene. J Neurosci. 2012;32:12543–57. doi: 10.1523/JNEUROSCI.5803-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998a;125:579–87. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998b;125:579–87. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Moody Sa, Klein SL, Karpinski Ba, Maynard TM, Lamantia AS. On becoming neural: what the embryo can tell us about differentiating neural stem cells. Am J Stem Cells. 2013;2:74–94. [PMC free article] [PubMed] [Google Scholar]
- Ohba H, Chiyoda T, Endo E, Yano M, Hayakawa Y, Sakaguchi M, Darnell RB, Okano HJ, Okano H. Sox21 is a repressor of neuronal differentiation and is antagonized by YB-1. Neurosci Lett. 2004;358:157–60. doi: 10.1016/j.neulet.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Penzel R, Oschwald R, Chen Y, Tacke L, Grunz H. Characterization and early embryonic expression of a neural specific transcription factor xSOX3 in Xenopus laevis. Int J Dev Biol. 1997;41:667–77. [PubMed] [Google Scholar]
- Rex M, Orme a, Uwanogho D, Tointon K, Wigmore PM, Sharpe PT, Scotting PJ. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev Dyn. 1997;209:323–32. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Richter K, Grunz H, Dawid IB. Gene expression in the embryonic nervous system of Xenopus laevis. Proc Natl Acad Sci U S A. 1988;85:8086–90. doi: 10.1073/pnas.85.21.8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimini R, Beltrame M, Argenton F, Szymczak D, Cotelli F, Bianchi ME. Expression patterns of zebrafish sox11A, sox11B and sox21. 1999;89:167–171. doi: 10.1016/s0925-4773(99)00199-9. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Archer TC, Cunningham DD, Grammer TC, Casey EMS. Sox3 expression is maintained by FGF signaling and restricted to the neural plate by Vent proteins in the Xenopus embryo. Dev Biol. 2008;313:307–319. doi: 10.1016/j.ydbio.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Ferzli GS, Casey ES. The response of early neural genes to FGF signaling or inhibition of BMP indicate the absence of a conserved neural induction module. BMC Dev Biol. 2011;11:74. doi: 10.1186/1471-213X-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Harafuji N, Archer T, Cunningham DD, Casey ES. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech Dev. 2009a;126:42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth Defects Res C Embryo Today. 2009b;87:249–62. doi: 10.1002/bdrc.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg M, Källström M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Winterbottom EF, Ji YJ, Hwang YS, Daar IO. Abelson interactor 1 (ABI1) and its interaction with Wiskott-Aldrich syndrome protein (wasp) are critical for proper eye formation in Xenopus embryos. J Biol Chem. 2013;288:14135–46. doi: 10.1074/jbc.M112.445643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM, Forman D. An interaction between dorsal and ventral regions of the marginal zone in early amphibian embryos. J Embryol Exp Morphol. 1980;56:283–99. [PubMed] [Google Scholar]
- Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–40. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- Stigloher C, Chapouton P, Adolf B, Bally-Cuif L. Identification of neural progenitor pools by E(Spl) factors in the embryonic and adult brain. Brain Res Bull. 2008;75:266–73. doi: 10.1016/j.brainresbull.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D’Amour Ka, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–28. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Deng W, Gage FH. Signaling in adult neurogenesis. Annu Rev Cell Dev Biol. 2009;25:253–75. doi: 10.1146/annurev.cellbio.042308.113256. [DOI] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titulaer MJ, Klooster R, Potman M, Sabater L, Graus F, Hegeman IM, Thijssen PE, Wirtz PW, Twijnstra A, Smitt PaES, van der Maarel SM, Verschuuren JJGM. SOX antibodies in small-cell lung cancer and Lambert-Eaton myasthenic syndrome: frequency and relation with survival. J Clin Oncol. 2009;27:4260–7. doi: 10.1200/JCO.2008.20.6169. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Kamachi Y, Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech Dev. 1999;84:103–20. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Weider M, Reiprich S, Wegner M. Sox appeal - Sox10 attracts epigenetic and transcriptional regulators in myelinating glia. Biol Chem. 2013;394:1583–93. doi: 10.1515/hsz-2013-0146. [DOI] [PubMed] [Google Scholar]
- Yan B, Neilson KM, Moody SA. foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev Biol. 2009;329:80–95. doi: 10.1016/j.ydbio.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.