Abstract
The numerous and diverse eukaryotic viruses with large double-stranded DNA genomes that at least partially reproduce in the cytoplasm of infected cells apparently evolved from a single virus ancestor. This major group of viruses is known as Nucleocytoplasmic Large DNA Viruses (NCLDV) or the proposed order Megavirales. Among the “Megavirales”, there are three groups of giant viruses with genomes exceeding 500 kb, namely Mimiviruses, Pithoviruses, and Pandoraviruses that hold the current record of viral genome size, about 2.5 Mb. Phylogenetic analysis of conserved, ancestral NLCDV genes clearly shows that these three groups of giant viruses have three distinct origins within the “Megavirales”. The Mimiviruses constitute a distinct family that is distantly related to Phycodnaviridae, Pandoraviruses originate from a common ancestor with Coccolithoviruses within the Phycodnaviridae family, and Pithoviruses are related to Iridoviridae and Marseilleviridae. Maximum likelihood reconstruction of gene gain and loss events during the evolution of the “Megavirales” indicates that each group of giant viruses evolved from viruses with substantially smaller and simpler gene repertoires. Initial phylogenetic analysis of universal genes, such as translation system components, encoded by some giant viruses, in particular Mimiviruses, has led to the hypothesis that giant viruses descend from a fourth, probably extinct domain of cellular life. The results of our comprehensive phylogenomic analysis of giant viruses refute the fourth domain hypothesis and instead indicate that the universal genes have been independently acquired by different giant viruses from their eukaryotic hosts.
Introduction
The discovery of giant viruses infecting protists, sometimes called giruses, pioneered by the isolation of Acanthamoeba polyphaga mimivirus (APMV), is one of the most unexpected and spectacular breakthroughs in virology in decades (Claverie, 2006; Claverie and Abergel, 2010; Claverie, Abergel, and Ogata, 2009; Claverie et al., 2006; Koonin, 2005; La Scola et al., 2003; Raoult et al., 2004; Van Etten, 2011; Van Etten, Lane, and Dunigan, 2010). The giant viruses shatter the textbook definition of viruses as “filterable” infectious agents because their virions do not pass bacterial filters and obliterate all boundaries between viruses and cellular life forms in terms of size. Indeed, not only are the particles of giant viruses larger than the cells of numerous bacteria and archaea but also the genomes of Pandoraviruses, the current record holders at approximately 2.5 Mb (Philippe et al., 2013), are larger and more diverse in gene content than many bacterial and archaeal genomes, from both parasites and free-living microbes (Koonin and Wolf, 2008). The recent identification of Pandoraviruses and Pithoviruses (Legendre et al., 2014) that are not only huge, by the standards of the virology, but also possess a previously unseen, asymmetrical virion structure, shows that the true diversity of giant viruses has been barely tapped into.
The unexpected, “cell-like” features of giant viruses led several researchers to propose fundamental concepts that go far beyond the study of these particular viruses and beyond virology in general. The foremost of these conceptual developments is the proposition that giant viruses represent a “fourth domain of life” that is distinct from but comparable to the three cellular domains, bacteria, archaea and eukaryotes (Claverie et al., 2006; Colson et al., 2012; Colson et al., 2011; Desnues, Boyer, and Raoult, 2012; Legendre et al., 2012; Raoult et al., 2004). It seems useful to distinguish the fourth domain concept as a general idea and as a specific hypothesis. As a general notion, the claim that giant viruses represent a fourth domain of life simply refers to the “cell-like” character of these viruses in terms of size of the virions and genomes and, in addition, to the observation that many genes of these viruses have no detectable homologs and so might come from some unknown source. With these general statements, the fourth domain concept does not make any falsifiable predictions. In contrast, the specific fourth domain hypothesis is steeped directly in the original definition of the three domains of cellular life. These three domains, bacteria, archaea and eukaryota, correspond to the three major trunks in the unrooted phylogenetic tree of 16S ribosomal RNA (Pace, 1997; Pace, 2006; Pace, Olsen, and Woese, 1986; Woese, 1987; Woese and Fox, 1977; Woese, Kandler, and Wheelis, 1990; Woese, Magrum, and Fox, 1978) that is topologically consistent with the phylogenies of most of the other (nearly) universal genes that encode primarily components of the translation and the core transcription machineries (Brown and Doolittle, 1997; Brown et al., 2001; Puigbo, Wolf, and Koonin, 2009; Puigbo, Wolf, and Koonin, 2013). Strikingly, and unlike other viruses, the giant viruses encode several proteins that are universal among cellular life forms, in particular translation system components, such as aminoacyl-tRNA synthetases and translation factors. The presence of these universal genes provides for the opportunity to formally incorporate the giant viruses into the tree of life (Raoult et al., 2004). The outcome of the phylogenetic analysis of the universal genes is (at least, in principle) readily interpretable: the placement of the viral genes outside the three traditional domains of cellular life is compatible with the fourth domain hypothesis whereas their placement within any of the three domains is not. Several studies, starting with the original analysis of the mimivirus genome, have reported phylogenetic trees that appeared compatible with giant viruses comprising a fourth domain (Colson et al., 2012; Colson et al., 2011; Nasir, Kim, and Caetano-Anolles, 2012; Raoult et al., 2004). However, such observations could be inherently problematic. Indeed, accelerated evolution of viral genes that is likely to have occurred, especially immediately following the acquisition of the respective genes from the host, has the potential to obscure their affinity with homologs from cellular organisms within one of the recognized domains (a common problem in the analysis of deep phylogenies (Felsenstein, 2004). A subsequent re-analysis of the phylogenies of several universal genes has failed to find support for the fourth domain hypothesis (Williams, Embley, and Heinz, 2011).
Notwithstanding their unusual size, genetic complexity and the presence of some universal cellular genes, all giant viruses contain a set of core genes that define an expansive group of eukaryotic double-stranded (ds) DNA viruses that is referred to as Nucleo-Cytoplasmic Large DNA viruses (NCLDV) (Iyer, Aravind, and Koonin, 2001; Iyer et al., 2006; Koonin and Yutin, 2010) or the proposed order Megavirales (Colson et al., 2012; Colson et al., 2013; Iyer, Aravind, and Koonin, 2001; Iyer et al., 2006; Koonin and Yutin, 2010). Hereinafter we refer to this major group of viruses as “Megavirales” to signal our support of this amendment to virus taxonomy while indicating that the order so far has not been officially adopted by the International Committee for the Taxonomy of Viruses. The “Megavirales” unite 7 families of viruses infecting diverse eukaryotes, namely Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Marseilleviridae, Phycodnaviridae, and Mimiviridae, as well as the recently discovered giant Pandoraviruses and Pithoviruses that could found new families. Evolutionary reconstructions have mapped about 50 genes encoding essential viral functions to the putative common ancestor of the “Megavirales” although some of these putative ancestral genes have been lost in certain groups of viruses (Koonin and Yutin, 2010; Yutin et al., 2013; Yutin et al., 2009). This ancestral gene set does not include genes for components of the translation system or any other genes that might be considered suggestive of a cellular nature of the common ancestor of the “Megavirales” implied by the fourth domain hypothesis.
Phylogenetic analysis of the universal “Megavirales” genes reveals apparent evolutionary relationships between giant and smaller viruses. Specifically, Mimiviruses cluster with the so-called Organic Lake phycodnaviruses and Phaeocystis globosa viruses (Santini et al., 2013; Yutin et al., 2013), Pandoraviruses with Phycodnaviruses, in particular Coccolithoviruses (Yutin and Koonin, 2013), and Pithoviruses with Marseilleviruses and Iridoviruses (Legendre et al., 2014). Combined with the results of evolutionary reconstructions based on the phyletic patterns of “Megavirales” genes (i.e. matrices of gene presence and absence), these relationships suggest that different groups of giant viruses could have independently evolved from smaller ancestral viruses (Yutin and Koonin, 2013).
There is an obvious tension between the fourth domain of life hypothesis and the monophyly of the “Megavirales”. The fact that giant viruses encode the large set of ancestral “Megavirales” genes, some of which are “virus hallmark genes” without close homologs encoded in cellular life forms (Koonin, Senkevich, and Dolja, 2006), constrains the fourth domain hypothesis to a specific version. Specifically, one would have to postulate that a viral ancestor of the giant viruses reproduced in a host that belonged to a fourth domain of cellular life and acquired numerous genes including some that are universal in cellular life forms. After the fourth cellular domain went extinct, the resulting giant viruses would remain the only “living fossils” of their original hosts.
We sought to formally test the fourth domain hypothesis as comprehensively as possible and additionally to address the origins of the gene repertoires of giant viruses, and their evolutionary relationships with other “Megavirales”. The results of this phylogenomic analysis effectively falsify the fourth domain hypothesis, reveal diverse origins of the genes of giant viruses, and reaffirm the origin of giant viruses from simpler ancestors.
RESULTS and DISCUSSION
Origins of universal cellular genes present in giant viruses: testing the fourth domain hypothesis
The three domains of (cellular) life were originally introduced from the topology of the phylogenetic tree of the 16S rRNA (Pace, 1997; Pace, Olsen, and Woese, 1986; Woese, 1987; Woese and Fox, 1977; Woese, Kandler, and Wheelis, 1990). Subsequently, these domains have been validated by phylogenetic analysis of multiple, (nearly) universal genes all of which encode components of the translation and transcription systems (Brown and Doolittle, 1997; Brown et al., 2001; Ciccarelli et al., 2006). The high topological congruence between the phylogenies of all these genes has been demonstrated indicating that each of them can be used to test the fourth domain hypothesis (Puigbo, Wolf, and Koonin, 2009; Puigbo, Wolf, and Koonin, 2013).
Table 1 lists the (nearly) universal genes of cellular life forms that are represented in each of the virus families that comprise the “Megavirales”. These genes fall into two distinct, uneven-sized groups with contrasting phyletic patterns across the “Megavirales”. The two large subunits of the RNA polymerase (RNAP) are present in all “Megavirales” except for most of the phycodnaviruses that apparently have lost these genes upon evolving a nuclear phase of the reproduction cycle (Koonin and Yutin, 2010; Yutin et al., 2009). In contrast, the translation system components, i.e. aminoacyl-tRNA synthetases and translation factors, typically occur in one or two groups of the “Megavirales”. The translation system components are represented primarily in giant viruses as opposed to the members of the “Megavirales” with smaller virions and genomes (for the purpose of this work, we define giant viruses strictly, as those with genomes in excess of 500 kb; this leaves only three groups in the giant category: mimiviruses, pandoraviruses and pithoviruses). Specifically, these genes are (nearly) missing in the families Poxviridae, Asfarviridae, Iridoviridae and Ascoviridae (Table 1). Even within the family Mimiviridae, translation-associated genes show a patchy distribution, the exception being tyrosyl-tRNA synthetase that is encoded in all mimiviruses sensu stricto(Table 1). CroV, although a smaller virus than the mimiviruses and pandoraviruses, encodes the largest number of translation-associated proteins. Conversely, the most common translation-associated protein in giant viruses is the cap-binding subunit of translation initiation factor 4E (Table 1). This, however, is not a universal but rather a eukaryote-specific protein.
Table 1.
Genes encoding translation system components in members of the “Megavirales”
| KOG/COG annotation | Acanthamoeba polyphaga mimivirus | Acanthamoeba castellanii mamavirus | Acanthamoeba polyphaga lentillevirus | Acanthamoeba polyphaga moumouvirus | Moumouvirus | Moumouvirus Monve | Moumouvirus goulette | Courdo11 virus | Terra1 virus | Megavirus chiliensis | Megavirus courdo7 | Megavirus courdo11 | Cafeteria roenbergensis virus BV-PW1 | OLPV | OLPV1 | OLPV2 | Pglob16 | Pglob14 | Pglob12 | Marseillevirus | Lausannevirus | Pandoravirus salinus | Pandoravirus dulsis | Acanthocystis turfacea Chlorella virus | Paramecium bursaria Chlorella virus | Ranaviruses* | Diachasmimorpha longicaudata entomopoxvirus | Canarypox virus | Fowlpox virus | Pithovirus sibericum |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNA polymerase, subunit alpha | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||
| RNA polymerase, subunit beta | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||
| Arginyl-tRNA synthetase | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||||||||||||||||||||
| Aspartyl/asparaginyl-tRNA synthetase | Y | Y | Y | Y | Y | |||||||||||||||||||||||||
| Cysteinyl-tRNA synthetase | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||||||||||||||||||||
| Isoleucyl-tRNA synthetase | Y | Y | Y | Y | Y | Y | ||||||||||||||||||||||||
| Methionyl-tRNA synthetase | Y | Y | Y | Y | Y | Y | Y | Y | Y | |||||||||||||||||||||
| Tryptophanyl-tRNA synthetase | Y | Y | Y | |||||||||||||||||||||||||||
| Tyrosyl-tRNA synthetase | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||||||||||||||||
| Pseudouridine synthase | Y | |||||||||||||||||||||||||||||
| Peptidyl-tRNA hydrolase | Y | Y | ||||||||||||||||||||||||||||
| Peptide chain release factor 1 (eRF1) | Y | Y | Y | Y | Y | Y | Y | |||||||||||||||||||||||
| Putative translation initiation inhibitor, yjgF family | Y | Y | Y | |||||||||||||||||||||||||||
| Putative translation factor (SUA5) | Y | |||||||||||||||||||||||||||||
| translation elongation factor eEF3 | Y | Y | Y | Y | Y | |||||||||||||||||||||||||
| Translation elongation factor EF-1alpha (GTPase) | Y | Y | ||||||||||||||||||||||||||||
| Translation initiation factor 1 (eIF-1/SUI1) | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||||||||||||||||||||
| Translation initiation factor 1A (eIF-1A) | Y | |||||||||||||||||||||||||||||
| Translation initiation factor 2 (IF-2; GTPase) | Y | |||||||||||||||||||||||||||||
| Translation initiation factor 2, alpha subunit (eIF-2alpha) | Y | Y | ||||||||||||||||||||||||||||
| Translation initiation factor 2, beta subunit (eIF-2beta)/eIF-5 N-terminal domain | Y | Y | ||||||||||||||||||||||||||||
| Translation initiation factor 2, gamma subunit (eIF-2gamma; GTPase) | Y | |||||||||||||||||||||||||||||
| Translation initiation factor 3, subunit g (eIF-3g) | Y | |||||||||||||||||||||||||||||
| Translation initiation factor 4F, cap binding subunit (eIF-4E) and related cap-binding proteins | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | |||||||||||||
| Translation initiation factor 4F, helicase subunit (eIF-4A) and related helicases | Y | Y | Y | Y | Y | Y | Y |
Bold type shows the giant viruses analyzed in detail in this work.
- Ambystoma tigrinum virus
- Andrias davidianus ranavirus
- Bohle iridovirus
- Chinese giant salamander iridovirus
- Common midwife toad ranavirus
- Epizootic haematopoietic necrosis virus
- European catfish virus
- Frog virus 3
- Ictalurus melas ranavirus
- Rana catesbeiana virus Z
- Rana esculenta iridovirus
- Silurus glanis ranavirus
- Tiger frog virus
We performed a comprehensive phylogenetic analysis of the genes of “Megavirales” that are homologous to genes widely represented in at least two domains of cellular life forms and encode proteins involved in transcription and translation, with the specific aim to test the fourth domain hypothesis. To this end, we adopted the following criterion: if a giant virus gene reliably placed inside a subtree corresponding to one of the three domains of cellular life, the respective tree was taken to be incompatible with the fourth domain hypothesis. Conversely, when a giant virus gene placed outside any of the three cellular domains, the outcome was considered to be compatible with the fourth domain hypothesis. In addition to the bootstrap values that reflect the reliability of each internal branch in the tree, we used the approximately unbiased (AU) test to compare the likelihoods of alternative tree topologies, namely those compatible and incompatible with the fourth domain hypothesis (Yutin and Koonin, 2012).
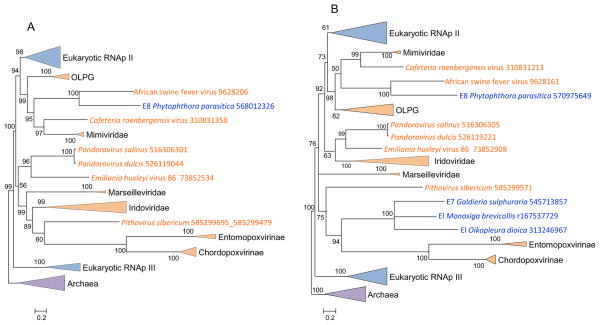
Figure 1 shows the phylogenies of the two large RNAP subunits. As noticed previously (Yutin and Koonin, 2012), the RNAPs of the NLCDV appear to be polyphyletic, and in particular, both large RNAP subunits of the mimiviruses and asfarviruses confidently clustered with eukaryotic RNAP II. The constrained tree, in which the mimivirus branch was joined with the rest of the “Megavirales”, had a significantly lower likelihood than the original tree according to the AU test (Supplementary Table S1). The rest of the “Megavirales” formed astrongly supported sister group to RNAP II in both trees (Figure 1ab). As proposed previously, the ancestral RNAP of the “Megavirales” probably was replaced by RNAP II of the eukaryotic host during the stem phase of the mimivirus evolution (Yutin and Koonin, 2012). Thus, the phylogenies of the two RNAP subunits, genes that are nearly universal and definitely ancestral among the “Megavirales”, do not conform to the fourth domain hypothesis.
Figure 1. Phylogenies of the large subunits of DNA-dependent RNA polymerase.
(a) Subunit a
(b) Subunit b
Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. OLPG: Organic Lake phycodnavirus – Phaeocystic globosa virus clade.
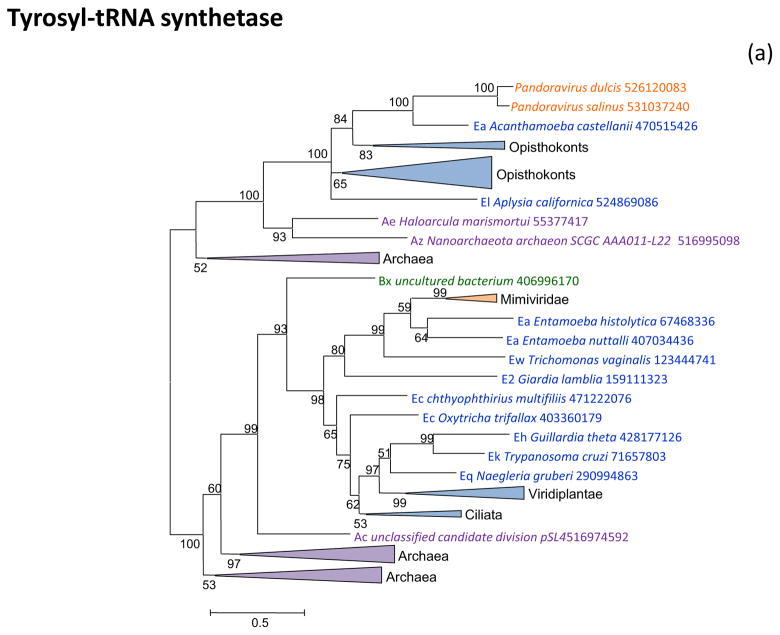
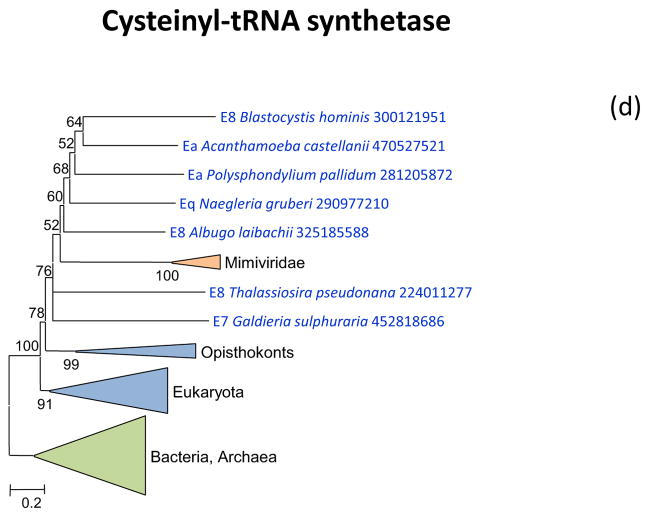
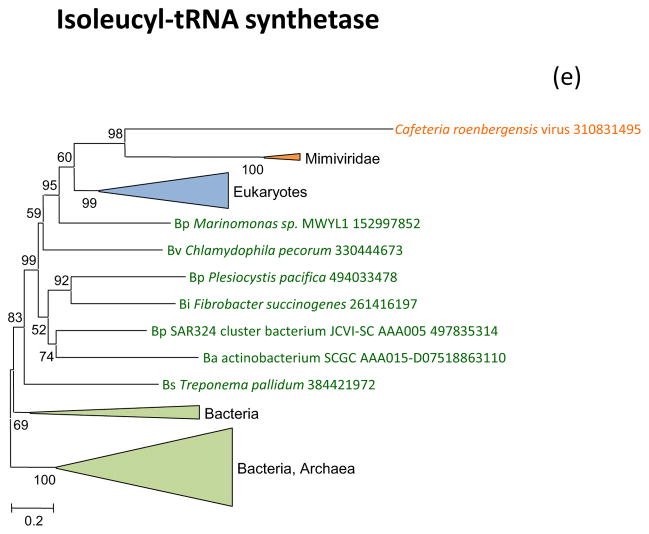
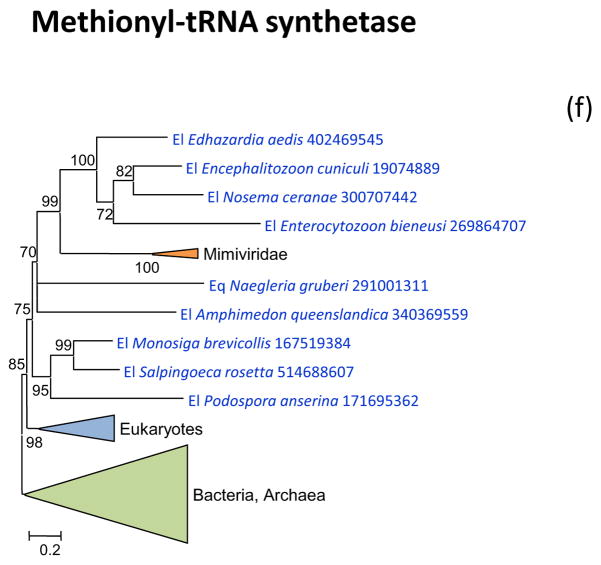
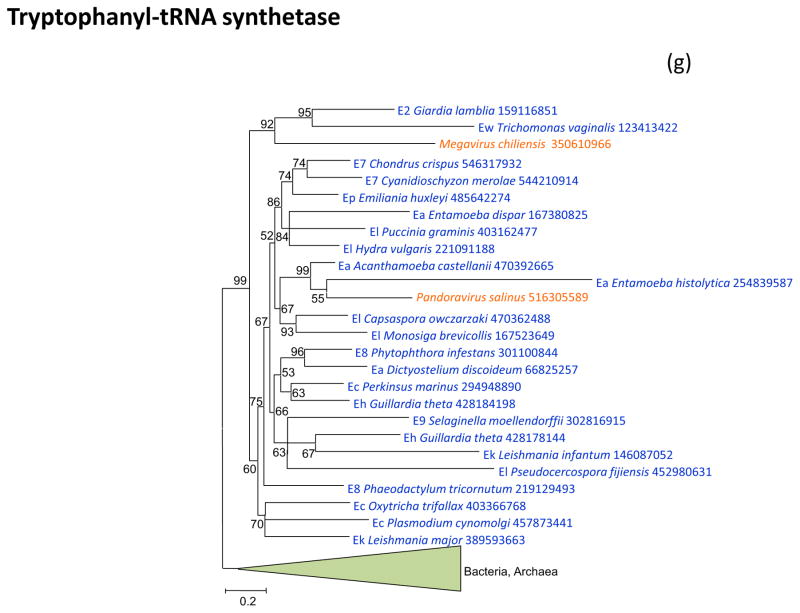
Figure 2 shows the phylogenetic trees for all aaRS encoded in “Megavirales” genomes. Tyrosyl-aaRS is encoded by all mimiviruses and pandoraviruses, and strikingly, the genes of the two “Megavirales” do not share a common origin. The mimivirus TyrRS clusters with homologs from Entamoeba whereas the Pandoravirus TyrRS is highly similar to the Acanthamoeba homolog with which it forms a tight cluster in the tree (Figure 2a). It should be noticed that the phylogeny of TyrRS is complex, with the two major eukaryotic branches apparently evolved from distinct archaeal ancestors. Further investigation of the evolution of this universal enzyme is beyond the scope of the present work, but it should be emphasized that the TyrRS from two families of “Megavirales” confidently place each within one of the two eukaryotic branches (Figure 2a). The conclusion that TyrRS was independently acquired by mimiviruses and pandoraviruses from distinct eukaryotic hosts appears inescapable.
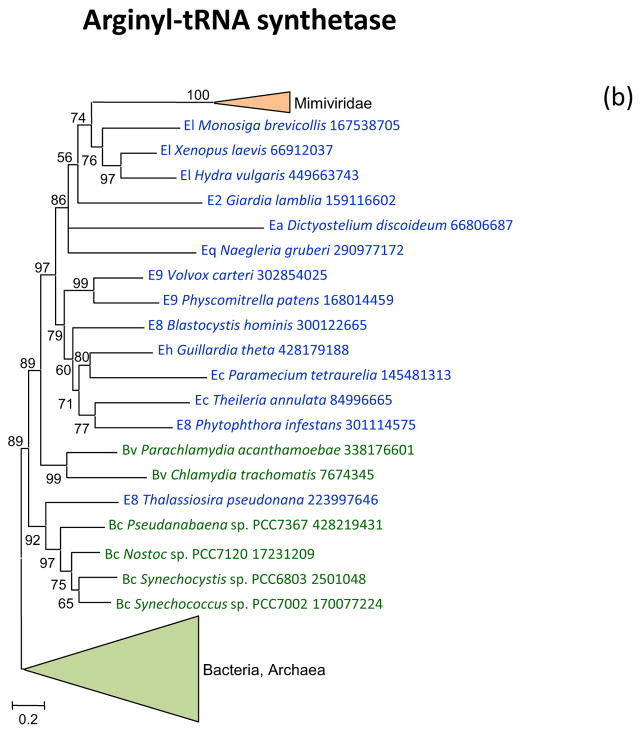
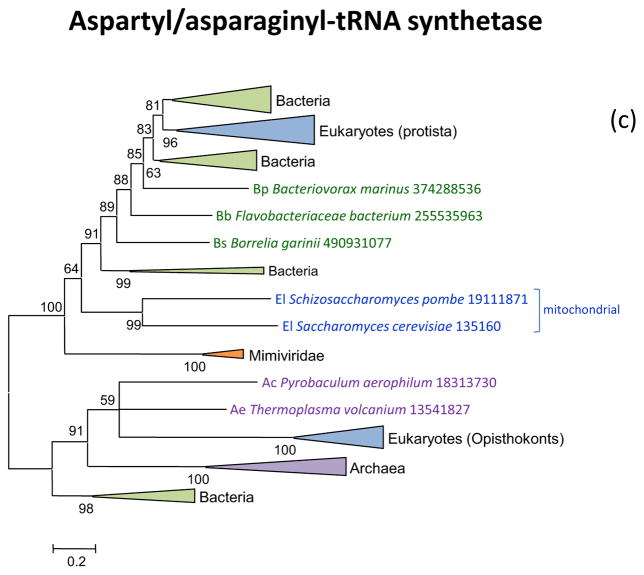
Figure 2. Phylogenies of aminoacyl-tRNA synthetases encoded by giant viruses.
(a) Tyrosyl-tRNA synthetase
(b) Arginyl-tRNA synthetase
(c) Aspartyl/asparaginyl-tRNA synthetases
(d) Cysteinyl-tRNA synthetase
(e) Isoleucyl-tRNA synthetase
(f) Methionyl-tRNA synthetase
(g) Tryptophanyl-tRNA synthetase
Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, bacterial sequences in green, archaeal sequences in purple. Taxa abbreviations: Ac, Crenarchaeota; Ae, Euryarchaeota; Az, unclassified Archaea; Ba, Actinobacteria; Bb, Bacteroidetes/Chlorobi group; Bc, Cyanobacteria; Bi, Acidobacteria; Bp, Proteobacteria; Bs, Spirochaetes; Bv, Chlamydiae/Verrucomicrobia group; E2, Fornicata; E7, Rhodophyta; E8, stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalidea.
Arginyl-tRNA synthetase, cysteinyl-tRNA and methionyl-tRNA synthetases are encoded by the majority of the mimiviruses and in the respective phylogenetic trees, are all deeply embedded within the eukaryotic subtree (Figure 2bcd). Again, these aaRS seem to have been acquired from the eukaryotic hosts, an evolutionary scenario that is incompatible with the fourth domain hypothesis.
Aspartyl- and asparaginyl-tRNA synthetases have evolved under a complex scenario whereby opisthokonts inherited the archaeal enzyme whereas the rest of the eukaryotes possess the bacterial version. The asparaginyl-tRNA synthetases present in a subset of the mimiviruses appears to be of the bacterial variety, suggestive of acquisition from a respective eukaryotic host (Figure 2e).
Tryptophanyl-tRNA synthetase is the second, after TyrRS, aaRS that is present both in mimiviruses (in this case, only one species, Megavirus chilensis) and pandoraviruses. As in the case of Tyr-RS, the two viral TrpRS appear to be of distinct origins, each originating from a different group of eukaryotes (Figure 2f). Notably, in both cases, Pandoraviruses clustered with Acanthamoeba, compatible with a relatively recent acquisition of the respective aaRS genes from this host. In constrast, the mimiviruses belonged to a composite protist branch, suggestive of a more ancient acquisition, possibly from a different host (Figures 1a and 1f).
The only giant virus aaRS that forms a sister group to the eukaryotes (albeit with a low bootstrap support), as opposed to placing within the eukaryotic subtree, is IleRS that is represented in several mimiviruses and in CroV (Figure 2g). Thus, among the 7 aaRS encoded by giant viruses (Table 1), there is only one case where the phylogenetic tree topology is formally compatible with the fourth domain hypothesis. Furthermore, the two aaRS that are encoded in two families of giant viruses showed a clear polyphyletic origin, most likely due to independent acquisition of the respective aaRS genes from distinct eukaryotic hosts.
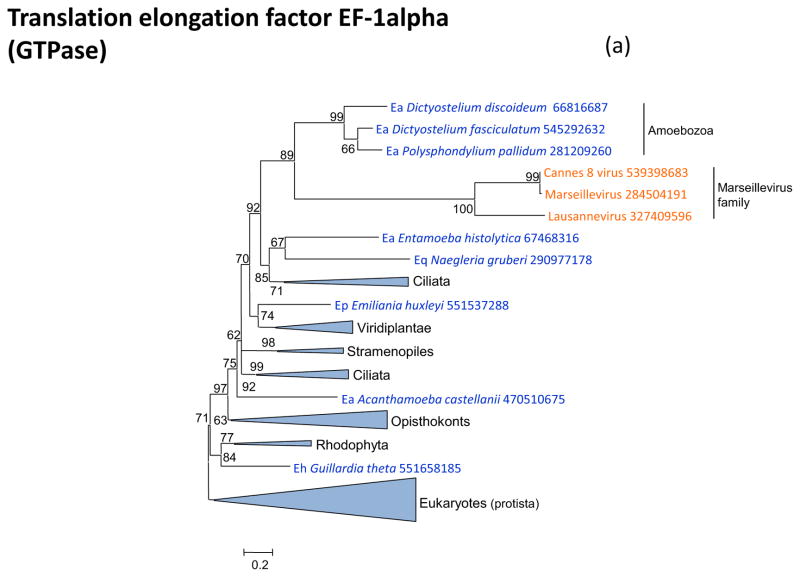
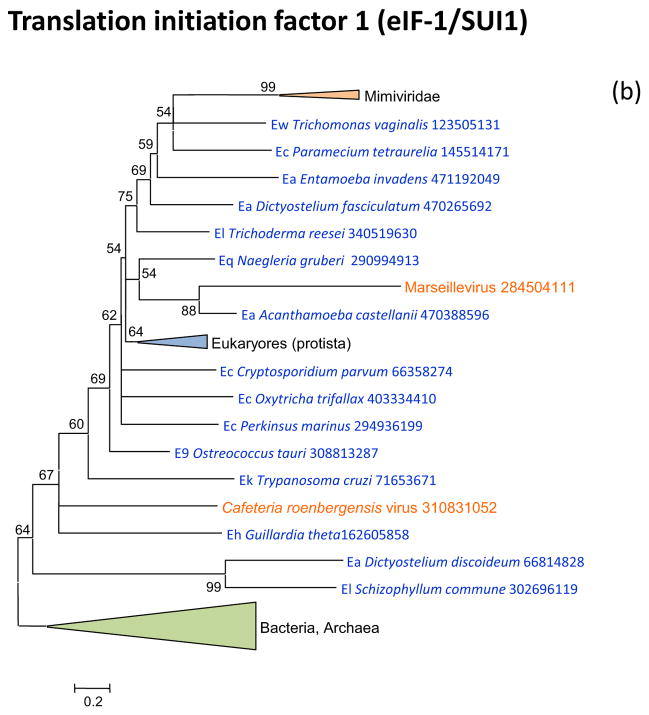
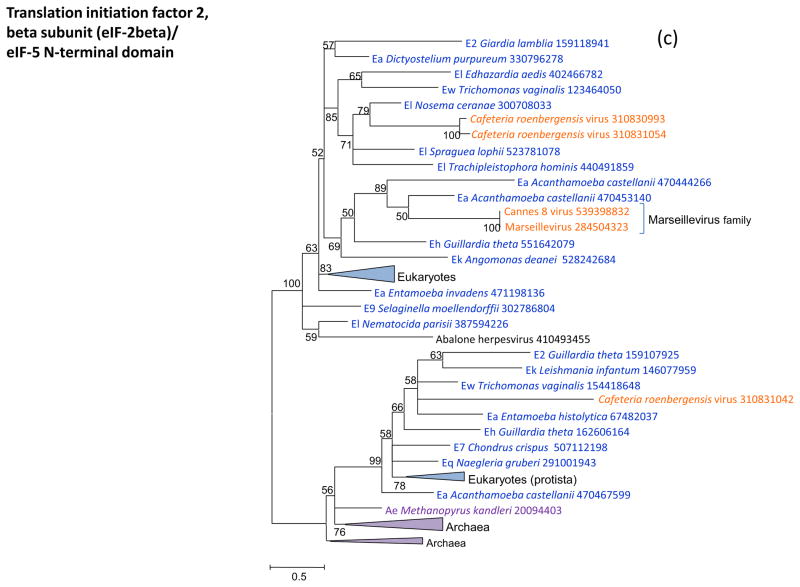
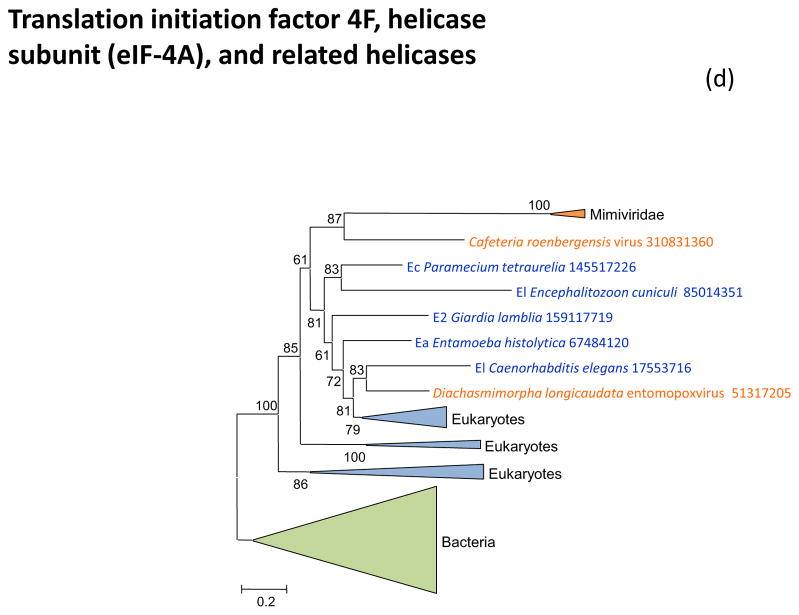
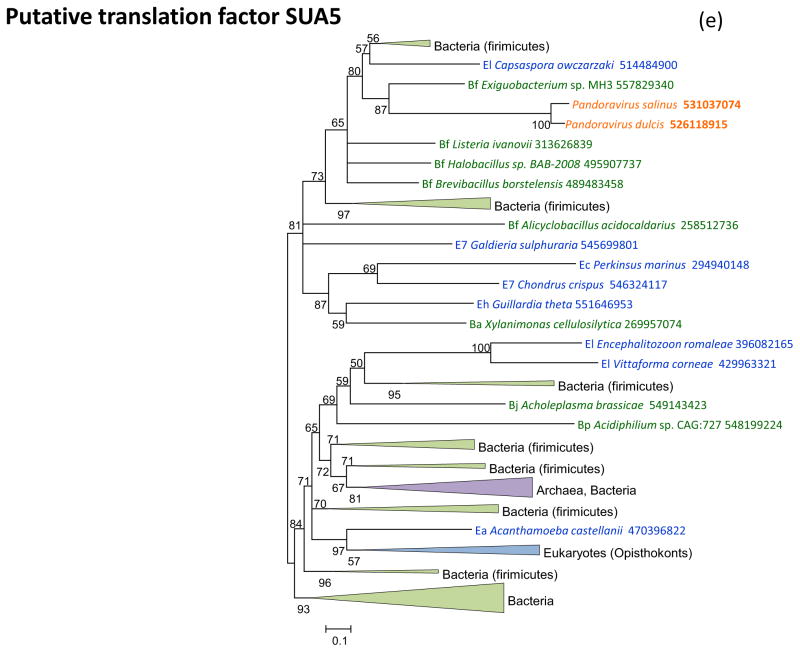
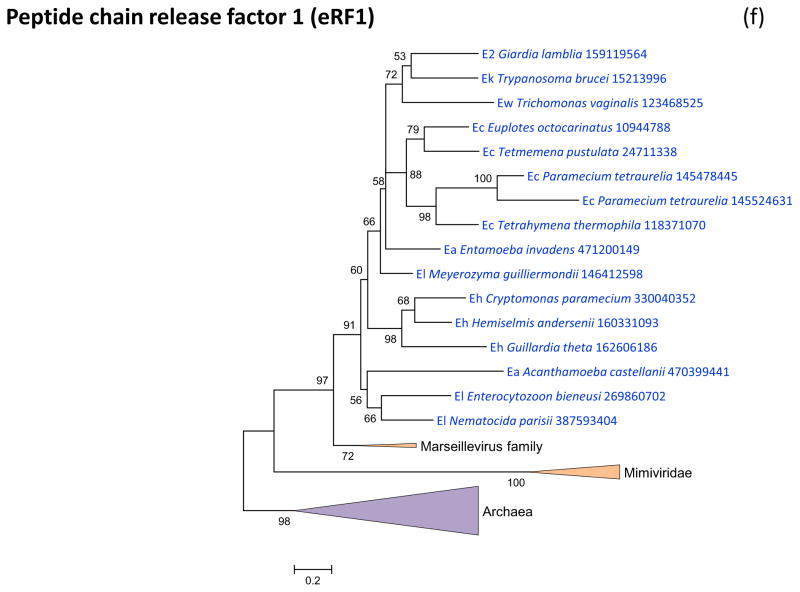
Figure 3 shows the phylogenies of translation factors encoded by giant viruses. Two of these translation factors, EF1-a and eIF1 (SUI1), are universal in cellular life forms. The first of these is only represented in the family Marseilleviridae (large but not giant viruses under the definitions adopted in this work), with the viral branch deeply embedded within the eukaryotic subtree (Figure 3a). The eIF1 tree generally is of poor quality due to the small size of the protein. Nevertheless, it is notable that the genes of mimiviruses, CroV and Marseillevirus appear to be polyphyletic and place in different parts of the eukaryotic subtree, suggestive of multiple acquisitions from eukaryotes (Figure 3b). A similar tree topology, with polyphyletic Megavirales, was observed for the archaeo-eukaryotic translation factor, the beta subunit of eIF5 (Figure 3c) and for a translation factor of apparent bacterial origin, eIF-4a (Figure 3d). The initiation factor SUA5 appears to have a complex history, with apparent multiple acquisitions by eukaryotes from bacteria; the SUA5 protein encoded by pandoraviruses belongs to one of the eukaryotic groups embedded within a bacterial branch (Figure 3e). Finally, the phylogeny of the archaeo-eukaryotic peptide chain release factor eRF1 showed the typical pattern of polyphyletic “Megavirales” inside the eukaryotic subtree (Figure 3f).
Figure 3. Phylogenies of translation encoded in “Megavirales” genomes.
(a) elongation factor EF-1alpha
(b) initiation factor eIF-1 (SUI1)
(c) initiation factor eIF-2beta
(d) initiation factor eIF-4A
(e) initiation factor SUA5
(f) peptide chain release factor eRF1
Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. Taxa abbreviations: Ae, Euryarchaeota; Ba, Actinobacteria; Bf, Firmicutes; Bj, Tenericutes; Bp, Proteobacteria; E2, Fornicata; E7, Rhodophyta; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Ep, Haptophyceae; Eq, Heterolobosea; Ew, Parabasalidea.
Altogether, we analyzed the phylogenies of 13 translation-associated genes of the “Megavirales” that are widely represented in at least two recognized domains of cellular life and thus provide for a meaningful test of the fourth domain hypothesis. Among these genes, only one showed a poorly supported tree with a topology that is formally compatible with the origin of the giant viruses from a fourth cellular domain. In most of the trees, the viral branches were separated from the eukaryotic root by multiple edges with high bootstrap support. Moreover, whenever a gene is present in more than one family of Megavirales, these viral genes appeared to be polyphyletic. Statistical testing using the constrained tree approach and the AU test rejected the fourth domain-compatible topology in only a subset of these cases as can be expected for trees that include highly diverged sequences. However, except in a single case, the likelihood of the constrained tree was lower than the likelihood of the original tree (Supplementary table S1). Collectively, the results of this phylogenetic analysis appear to be incompatible with the fourth domain hypothesis and instead strongly suggest that the (nearly) universal cellular genes were acquired by the giant viruses from their eukaryotic hosts at different stages of evolution.
Where do the genes of giant viruses come from: a phylogenomic analysis
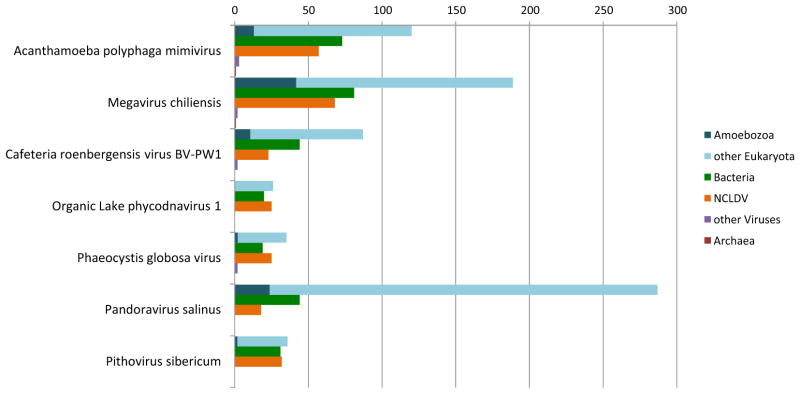
We developed a computational phylogenomic pipeline aimed at genome-wide inference of the origins of the genes of giant viruses (see Methods for details). The phylogenomic analysis was performed for 7 giant and large viruses that represent the four major branches of the extended family Mimiviridae (Yutin et al., 2013), pandoraviruses and pithoviruses. It is well known that numerous genes of large and especially giant viruses are ORFans, with no homologs detectable apart from closely related isolates. Many other viral genes have few homologs and/or show limited similarity to the detectable homologs, resulting in uninformative phylogenetic trees. Nevertheless, overall 1292 trees passed the criteria for origin inference. The results indicate that, apart from the small core of “Megavirales” genes, these phylogenetically tractable genes of giant and large dsDNA viruses appeared to be primarily of eukaryotic origin, with a sizable minority of genes of likely bacterial descent (Figure 4). The viruses differed dramatically in the phylogenetic breakdown of their genes. Pandoraviruses are particularly rich in eukaryotic genes, followed by some of the mimiviruses. In contrast, the Pithovirus and especially the smaller members of the extended family Mimiviridae (Organic Lake phycodnaviruses and Phaeocystis globosa viruses) had few phylogenetically tractable genes such that the eukaryotic and bacterial contributions are comparable with the core of “Megavirales” genes. Notably, the Pithovirus appeared to possess nearly as many bacterial as eukaryotic genes (Figure 4). These observations point to distinct evolutionary histories of the giant viruses that have shaped substantially different gene repertoires.
Figure 4.
Phylogenomic breakdown of giant virus genes
Evolutionary relationships between giant viruses and other “Megavirales”
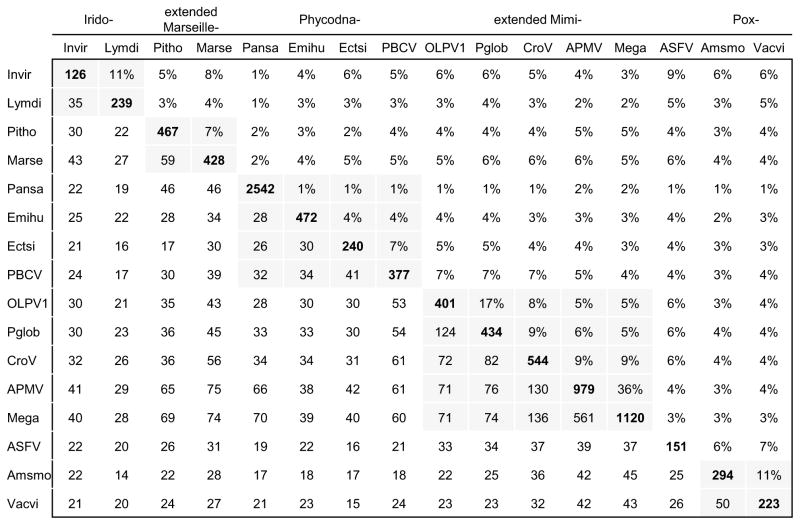
Previous studies on the evolution of the “Megavirales” have suggested evolutionary connections between giant viruses and other, smaller members of the Megavirales, based primarily on the phylogenies of the core genes. With the current updated collection of viral genomes, we revisited these relationships. The updated version of the NCVOGs was used to extract the patterns of gene presence-absence across all members of the “Megavirales”; the matrix of shared genes (Figure 5a) was then used to construct a tree of the relationships between the gene complements of the viruses (Figure 5b).
Figure 5. Relationships between the gene contents of giant viruses and their smaller relatives.
(a) matrix of shared genes
Lower left: number of shared gene families. Upper right: Jaccard similarity of gene complements. Diagonal: number of annotated genes in the genome.
Intra-family comparisons are shaded.
(b) tree of gene contents
Bold lines indicate branches with high (>70%) bootstrap support; thin lines indicate branches with low bootstrap support. Branches that disagree with the tree, reconstructed with the universal core genes, are highlighted in red (except the poorly resolved branches inside the Poxviridae), dashed lines indicate the relationships expected from the core phylogeny.
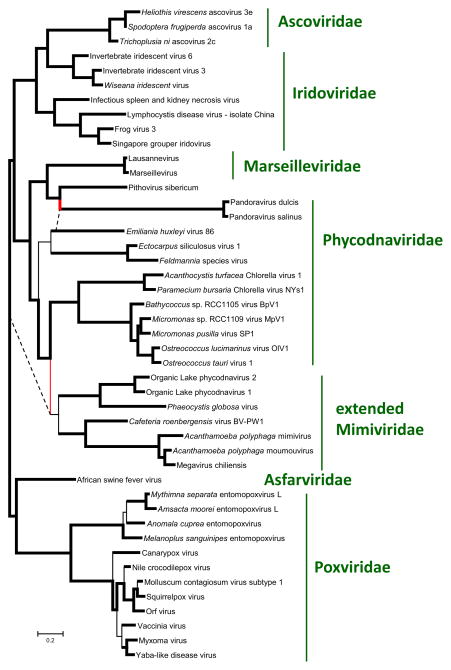
We then updated the phylogenetic tree of the “Megavirales” using concatenated alignments of 6 (nearly) universal core genes. The main features of the resulting phylogeny are compatible with the previous observations (Iyer et al., 2006; Yutin and Koonin, 2012; Yutin and Koonin, 2013; Yutin et al., 2009). The giant viruses fall within three distinct groups of “Megavirales”: i) Mimiviruses within the extended family Mimiviridae that is the sister group of Phycodnaviridae; ii) Pandoraviruses inside the family Phycodnaviridae, as the sister group of coccolithoviruses; iii) Pithovirus as the sister group of Marseilleviridae, within the branch that also includes the families Iridoviridae and Ascoviridae (Figure 6). In each of these cases, the sister group of the giant viruses includes viruses with substantially smaller genomes.
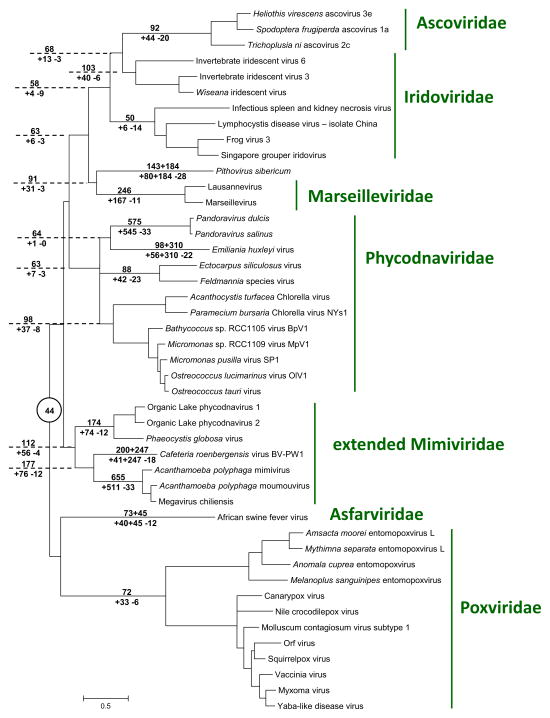
Figure 6. Phylogenetic tree of the (nearly) universal core genes of the “Megavirales” and reconstruction of gene gain and loss.
Numbers above the branch indicate the estimated number of NCVOG families (plus the number of singletons for extant genomes) at the end of the branch. Numbers below the branch indicate the estimated number of gained and lost NCVOG families (plus the number of acquired singletons for extant genomes). Dashed lines extend the branches The estimated number of “Megavirales” ancestral families is indicated in a circle at the tree root.
The topology of the tree constructed using the matrix of shared genes (Figure 5b) was similar to the topology of the phylogenetic tree (Figure 6), which is indicative of a general congruence of the evolution of the extended core gene sets of the “Megavirales” with the evolution of the universal genes that were used as phylogenetic markers. Two exceptions involved giant viruses: Pandoraviruses and Pithoviruses became a clade that was the sister group of Mareilleviruses, whereas the Mimiviridae place within the phycodnavirus clade. The similarities of gene complements that led to these changes in the tree topology might reflect a combination of ancestral gene conservation, intervirus gene transfer (particularly, in coinfected amoeba) and parallel acquisition of homologous genes from hosts. However, these affinities were based on small numbers of shared genes (Figure 5a). Therefore, on the whole, the results of gene composition analysis emphasizes distinct histories of genome evolution in the giant viruses.
Finally, we combined the phyletic patterns extracted from the NCVOGs and the phylogenetic tree of the core genes (Figure 6) to obtain a maximum likelihood reconstruction of the gene complements of the “Megavirales”. When superimposed over the phylogenetic tree, the results suggest evolution from moderate-sized ancestors, with massive gene gain inferred for all three groups of giant viruses (Figure 6).
Discussion
The results of the present phylogenomic analysis clarify the status of giant viruses by showing that their evolution is part and parcel of the evolutionary history of the “Megavirales”, in a general agreement with previous observations (Iyer et al., 2006; Yutin and Koonin, 2012; Yutin and Koonin, 2013; Yutin et al., 2009). Indeed, all three groups of giant viruses share the core genes of the “Megavirales” albeit with an unusual extent of loss in the Pandoraviruses ((Yutin and Koonin, 2013) and Figure 5a). Moreover, phylogenetic analysis of the core genes firmly places each of the three groups of giant viruses inside subtrees of the “Megavirales” that otherwise consist of viruses with moderate-sized genomes.
The evolutionary reconstruction for the gene complements of the “Megavirales” (Figure 6) suggests that the giant viruses evolved independently, through extensive gene gain. The paucity of shared genes between different groups of giant viruses (Figure 5a) effectively rules out their origin from a common giant ancestor, with extensive gene losses in the related smaller viruses. The only alternative, however non-parsimonious, to the massive gene gain scenario appears to be independent early emergence of multiple ancestral giant viruses followed by massive losses in the branches leading to the smaller extant viruses. Deletion of large portions (up to 20%) of the mimivirus genome during the cultivation of the virus in amoeba has been reported (Boyer et al., 2011). However, these deletions show a distinct pattern whereby genes are lost from the terminal regions of the genome that primarily encode proteins involved in virus-host interaction. A similar pattern of deletion involving non-essential genes in the terminal regions has been observed in other members of the “Megavirales” as well, in particular in poxviruses (Kotwal and Moss, 1988; Perkus et al., 1991). This limited evolutionary process is unlikely to produce the extent of gene loss that the evolution of moderate-size viruses from giant ones would have required.
In and by themselves, the presence of the core “Megavirales” genes in the giant virus genomes and the evolutionary connections between the giant viruses and other “Megavirales” do not invalidate some versions of the fourth domain hypothesis. In particular, one could imagine that a moderate-sized member of the “Megavirales” that reproduced in a host cell that belonged to an extinct fourth domain acquired numerous genes including those for translation system components, and thus remains the only extant relic of that fourth domain of cellular life. Actually, given the above argument on the implausibility of a common giant ancestor of the three giant virus groups, one would have to postulate not one but three unknown domains of cellular life at the respective roots of these viral lineages.
The fourth domain (more precisely, multiple domains as discussed above, but we will continue to use the more popular phrase “fourth domain”) hypothesis is falsifiable through phylogenetic analysis of genes that are universal in cellular life forms, conform to the three-domain tree topology and are also represented in giant viruses. The genes that meet these criteria are those encoding RNAP subunits and universal translation system components. As shown here and elsewhere (Williams, Embley, and Heinz, 2011; Yutin and Koonin, 2012), the phylogenies of almost all of these genes are incompatible with the fourth domain hypothesis. Instead, these phylogenies consistently derive the respective viral genes from within one of the three known domains of cellular life, namely eukaryotes. Moreover, in those few cases when different giant viruses encode the same component of the translation systems, these genes show affinities with different eukaryotic lineages. In particular, the translation-associated genes in Pandoraviruses might represent relatively recent acquisitions from the amoebal hosts whereas the functionally similar genes in Mimiviruses could be older acquisitions from different protist sources.
A more complete, automated phylogenomic analysis points to preferential capture of eukaryotic genes by giant viruses but also a substantial contribution of bacterial genes, in a general agreement with several previous analyses (Filee and Chandler, 2008; Filee and Chandler, 2010; Filee, Pouget, and Chandler, 2008; Filee, Siguier, and Chandler, 2007; Moreira and Brochier-Armanet, 2008). A large fraction of genes in giant viruses remain ORFans (Colson and Raoult, 2010; Saini and Fischer, 2007) but it is inconceivable that these genes are heritage of missing domains of cellular life. The implausibility of the latter hypothesis follows from the very fact that the ORFans lack any recognizable structural domains and thus hardly could have come from extinct cellular domains. Indeed, in archaea, bacteria and eukaryotes, proteins containing detectable domains and structural features, such as metabolic enzymes, transporters, transcriptional regulators, and signaling system components, represent a substantial majority (Koonin et al., 2004; Koonin and Wolf, 2008). Notably, ORFans are abundant also in the comparatively small genomes of bacteriophages and especially archaeal viruses, and appear to be fast evolving proteins, often small in size (Prangishvili, Garrett, and Koonin, 2006; Yin and Fischer, 2008). The high prevalence of ORFans reflects the vastness of viral gene pools but not the existence of unknown cellular domains (Kristensen et al., 2013).
Taken together, these findings are fully compatible with the scenario of evolution of giant viruses from smaller viruses by gene accretion (Filee, 2013; Filee and Chandler, 2010; Yutin and Koonin, 2013) which apparently occurred on at least three independent occasions. All the giant viruses so far discovered reproduce in protists, in particular in amoeba. These phagocytic unicellular eukaryotes routinely harbor diverse endosymbionts and parasites and hence apparently present an environment that is highly conducive to gene exchange, and in some lineages, extensive gene accumulation (Raoult and Boyer, 2010). Understanding the factors that led to genome explosion in some but not other lineages of protist viruses is of major interest. It has been proposed that giant and large viruses evolve under a “genomic accordion” model whereby phases of genome expansion alternate with contraction phases (Filee, 2013). So far the reconstruction of gene gain and loss in the evolution of the “Megavirales” failed to identify phases of major genome reduction (Figure 6). However, given the apparent dominance of genome reduction in the evolution of cellular life forms (Wolf and Koonin, 2013), the existence of such phases in the evolution of large viruses appears highly likely and can be expected to become apparent with further genome sequencing of diverse viruses.
Finally, shifting the discussion from the falsifiable fourth domain hypothesis to the fourth domain as a general concept, it should be noted that the refutation of the hypothesis by no account undermines the fundamental distinctness of large DNA viruses and viruses in general. On the contrary, these findings emphasize the primary divide of organisms into cellular life forms and selfish, virus-like agents (Koonin, Senkevich, and Dolja, 2006; Raoult and Forterre, 2008). In many respects, the differences between major classes of viruses and virus-like agents run deeper than the differences between the three cellular domains: to name a most obvious issue, some of the major groups of viruses share no homologous genes (Koonin and Dolja, 2013; Koonin and Wolf, 2012). Outside the applicability of straightforward phylogenetic approaches, classification of biological entities, especially those that cross traditional boundaries, such as giant viruses, can become complicated and inevitably, to some extent, arbitrary (Raoult, 2013). Whether or not different classes of viruses should be called domains, is a question of semantics. It might be advisable to keep the term for its original usage as a primary division of cellular organisms identifiable from consistent phylogenies of universal genes. Such terminological conservatism certainly should not and would not diminish the impact of the research on giant viruses which are among the most remarkable denizens of the vast virus world.
Methods
Update of the NCVOGs
For the updated version of NCVOGs, the following genomes were retrieved from GenBank: Pithovirus sibericum, Pandoraviruses, Megavirus chiliensis, Cafeteria roenbergensis virus BV-PW1, Acanthamoeba polyphaga moumouvirus, OLPG clade viruses, Prasinoviridae, Lausannevirus, Wiseana iridescent virus, two entomopoxviruses, and Squirrelpox virus. Three genomes, Marseillevirus, Acanthamoeba polyphaga mimivirus, and Acanthamoeba castellanii mamavirus, were updated (see Supplementary table S2 for the full list of species and their GenBank accession codes). Multiple alignments of viral protein sequences from the previous version of NCVOGs were used as seeds for the initial psi-COGnitor procedure. Remaining sequences were clustered using GOCtriangle and proceeded as previously described (Kristensen et al., 2010). Briefly, the procedure included the following steps: 1) Initial clusters based on previous NCVOG profiles and triangles of symmetrical best hits were constructed; 2) Multiple alignments of the initial cluster members were constructed using the MUSCLE program (Edgar, 2004). The alignments were used to generate position-specific scoring matrices (PSSM) for a PSI-BLAST search (Altschul et al., 1997) against the original protein dataset. Significantly similar proteins were added to the corresponding clusters; 3) Clusters with nearly complementary phyletic patterns and high inter-cluster sequence similarity were manually examined and merged whenever appropriate. The updated NCVOGs are available at ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/NCVOG/.
Phylogenetic analysis of (nearly) universal cellular genes present in giant viruses
Translation-related genes in giant viruses were identified using the RPS-BLAST search against the NCBI CDD database (Table 1). Homologs of giant virus sequences were identified in the NCBI NR database using BLAST search. Nearly identical sequences were eliminated using BLASTCLUST (http://www.ncbi.nlm.nih.gov/Web/Newsltr/Spring04/blastlab.html website). Protein sequences were aligned using the MUSCLE program with default parameters (Edgar, 2004); columns containing a large fraction of gaps (greater than 30%) and non-homogenous columns defined as described previously (Yutin et al., 2008) were removed from the alignment prior to phylogenetic analysis. A preliminary maximum-likelihood tree was constructed using the FastTree program with default parameters (JTT evolutionary model, discrete gamma model with 20 rate categories (Price, Dehal, and Arkin, 2010)). The preliminary tree and the alignment were then used to determine the best substitution matrix using Prottest (Darriba et al., 2011). The best matrices found by Prottest are shown in Supplementary table S3. The final maximum-likelihood trees were constructed using TreeFinder (1,000 replicates, Search Depth 2), with the substitution matrix that was found to be the best for a given alignment (Jobb, von Haeseler, and Strimmer, 2004). The Expected-Likelihood Weights (ELW) of 1,000 local rearrangements were used as confidence values of TreeFinder tree branches (Jobb, von Haeseler, and Strimmer, 2004). For tree topology testing, whenever applicable, alternative (constrained) topologies were constructed and compared to the initial trees using TreeFinder (Jobb, von Haeseler, and Strimmer, 2004). Approximately unbiased (AU) test P value cutoff 0.05 was used for rejecting tree topologies (Shimodaira, 2002).
Phylogenetic analysis of the “Megavirales” core genes
Multiple alignments of 6 core genes (DNApol, Packaging ATPase, D5 helicase, Superfamily II helicase, RNApol a and RNApol b) that are nearly universal in 45 “Megavirales” were constructed using the MUSCLE program and concatenated. Phylogeny was reconstructed using the TreeFinder program (Jobb, von Haeseler, and Strimmer, 2004) with the LG+G+F evolutionary model.
Phylogenomic analysis
For automated pipeline, seven genomes were retrieved from the non-redundant database at the National Center for Biotechnology Information (NIH, Bethesda): Pandoravirus salinus (2,542 proteins; KC977571), Acanthamoeba polyphaga mimivirus (979 proteins; NC_014649.1), Megavirus chiliensis (1,120 proteins; JN258408), Cafeteria roenbergensis virus BV-PW1 (544 proteins; NC_014637), Organic Lake phycodnavirus 1 (401 proteins; HQ704802), Phaeocystis globosa virus strain 16T (434 proteins; NC_021312), and Pithovirus sibericum isolate P1084-T (467 proteins; NC_023423).
For each protein, the following procedure was run. A protein was used as a query for BLAST searches against nr and Refseq databases (e-value cutoff 0.01, composition-based statistics); first 200 hits from nr database and first 2,000 hits from Refseq database were combined; a new BLAST search was run using the same query against the collected proteins, with composition-based statistics turned off. The latter run produced proper ranking of the hits. Further, the number of hits was reduced by BLASTClust (http://www.ncbi.nlm.nih.gov/Web/Newsltr/Spring04/blastlab.html): first 20 hits were clustered with 95% sequence identity, next 500 hits – with 75%, and the remaining sequences – with 65% sequence identity. The resulting set of sequences was aligned using the MUSCLE program with default parameters; poorly aligned sequences and columns containing a large fraction of gaps (greater than 30%) and non-homogenous columns defined as described previously (Yutin et al., 2008) were removed from the alignment. Alignments retaining 40 or more sequences and 100 or more positions, were subjected to phylogenetic analysis using the FastTree program (JTT evolutionary model, discrete gamma model with 20 rate categories (Price, Dehal, and Arkin, 2010)). Trees were rooted using the least-square modification of the mid-point method (Wolf et al., 1999). Interpretation of these trees used a slightly modified version of the NCBI taxonomy whereas all “Megavirales”s were collected into a separate group and further resolved to the finer clades. For each query genome listed above, a “native clade” was defined as indicated (Supplementary Figure S1). Trees were traversed starting from the leaf corresponding to the query sequence, toward the root. Nodes with low support (<0.8 except for the root) were ignored. At each supported node, the taxonomic affiliations of all descending leaves (including the query) were collected and classified into 5 categories (Archaea, Bacteria, Eukaryotes, non-”Megavirales” Viruses and “Megavirales”) plus the native group. Categories represented by only one species were ignored. If all non-native leaves belonged to the same category, the respective tree was considered phylogenomically resolved, with the query and its native group affiliated with that category; otherwise, the tree was considered unresolved. This procedure produced 1141 resolved trees.
Trees that are formally unresolved (according to the above criteria) still can contribute to the breakdown of the phylogenomic affiliations of genes of the giant viruses. If the set of tree nodes was not “mostly “Megavirales”“ (i.e. <90% belonged to “Megavirales”) and contained representatives of only one of the 3 cellular domains (criteria for domain exclusion were <0.5% of Archaea, <5% of Bacteria and <5% of Eukaryotes), the query was considered affiliated with the respective domain. These criteria allowed classification of 151 additional trees.
Neighbor-Joining tree based on the phyletic patterns
Presence-absence data of NCVOGs was collected for the 45 virus genomes. For each pair of genomes (i,j) the number of shared NCVOGs (Sij) was used to compute the distance between the genomes as Dij = −ln(Sij/sqrt(Ni*Nj)), where Ni and Nj is the number of NCVOGs in the two genomes (Yutin et al., 2009). A neighbor-joining tree was constructed from the distance matrix D using the NEIGHBOR program of PHYLIP package (Felsenstein, 1996). Support values were obtained using 1,000 bootstrap resamplings of the families.
Reconstruction of gene content evolution
The tree reconstructed from the concatenated alignment of Neighbor-Joining gene content tree of (nearly) universal core genes and the gene presence-absence matrix for the NCVOGs were used to reconstruct the gene loss and gain events in the evolution of the “Megavirales” using the COUNT program(Csuros, 2010), as previously described (Yutin et al., 2009).
Supplementary Material
Giant viruses include mimiviruses, pandoraviruses and pithoviruses
Giant viruses belong to the proposed order Megavirales
Giant viruses evolved from smaller viruses in the order “Megavirales”
Numerous genes of giant virus were acquired from eukaryotic hosts
Giant virus do not represent a fourth domain of cellular life
Acknowledgments
We thank Mart Krupovic for critical reading of the manuscript and helpful suggestions. The authors’ research is supported by intramural funds of the US Department of health and Human Services (to the National Library of Medicine).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Azza S, Barrassi L, Klose T, Campocasso A, Pagnier I, Fournous G, Borg A, Robert C, Zhang X, Desnues C, Henrissat B, Rossmann MG, La Scola B, Raoult D. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc Natl Acad Sci U S A. 2011;108(25):10296–301. doi: 10.1073/pnas.1101118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev. 1997;61(4):456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. Universal trees based on large combined protein sequence data sets. Nat Genet. 2001;28(3):281–5. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(5765):1283–7. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7(6):110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie JM, Abergel C. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genet. 2010;26(10):431–7. doi: 10.1016/j.tig.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Claverie JM, Abergel C, Ogata H. Mimivirus. Curr Top Microbiol Immunol. 2009;328:89–121. doi: 10.1007/978-3-540-68618-7_3. [DOI] [PubMed] [Google Scholar]
- Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117(1):133–44. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Colson P, de Lamballerie X, Fournous G, Raoult D. Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales. Intervirology. 2012;55(5):321–32. doi: 10.1159/000336562. [DOI] [PubMed] [Google Scholar]
- Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol. 2013 doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, Gimenez G, Boyer M, Fournous G, Raoult D. The giant Cafeteria roenbergensis virus that infects a widespread marine phagocytic protist is a new member of the fourth domain of Life. PLoS One. 2011;6(4):e18935. doi: 10.1371/journal.pone.0018935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, Raoult D. Gene repertoire of amoeba-associated giant viruses. Intervirology. 2010;53(5):330–43. doi: 10.1159/000312918. [DOI] [PubMed] [Google Scholar]
- Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26(15):1910–2. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnues C, Boyer M, Raoult D. Sputnik, a virophage infecting the viral domain of life. Adv Virus Res. 2012;82:63–89. doi: 10.1016/B978-0-12-394621-8.00013-3. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–27. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- Filee J. Route of NCLDV evolution: the genomic accordion. Curr Opin Virol. 2013;3(5):595–9. doi: 10.1016/j.coviro.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Filee J, Chandler M. Convergent mechanisms of genome evolution of large and giant DNA viruses. Res Microbiol. 2008;159(5):325–31. doi: 10.1016/j.resmic.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Filee J, Chandler M. Gene exchange and the origin of giant viruses. Intervirology. 2010;53(5):354–61. doi: 10.1159/000312920. [DOI] [PubMed] [Google Scholar]
- Filee J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses. BMC Evol Biol. 2008;8:320. doi: 10.1186/1471-2148-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filee J, Siguier P, Chandler M. I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 2007;23(1):10–5. doi: 10.1016/j.tig.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci U S A. 2010;107(45):19508–13. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75(23):11720–34. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo cytoplasmic large DNA viruses. Virus Res. 2006;117(1):156–84. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koonin EV. Virology: Gulliver among the Lilliputians. Curr Biol. 2005;15(5):R167–9. doi: 10.1016/j.cub.2005.02.042. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV. A virocentric perspective on the evolution of life. Curr Opin Virol. 2013 doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Rogozin IB, Smirnov S, Sorokin AV, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Dolja VV. The ancient virus world and evolution of cells. Biol Direct. 2006;1(1):29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36(21):6688–719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Front Cell Infect Microbiol. 2012:2. doi: 10.3389/fcimb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53(5):284–92. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwal GJ, Moss B. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology. 1988;167(2):524–37. [PubMed] [Google Scholar]
- Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, Mushegian A. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics. 2010;26(12):1481–7. doi: 10.1093/bioinformatics/btq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen DM, Waller AS, Yamada T, Bork P, Mushegian AR, Koonin EV. Orthologous gene clusters and taxon signature genes for viruses of prokaryotes. J Bacteriol. 2013;195(5):941–50. doi: 10.1128/JB.01801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. A giant virus in amoebae. Science. 2003;299(5615):2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- Legendre M, Arslan D, Abergel C, Claverie JM. Genomics of Megavirus and the elusive fourth domain of Life. Commun Integr Biol. 2012;5(1):102–6. doi: 10.4161/cib.18624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Coute Y, Rivkina E, Abergel C, Claverie JM. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci U S A. 2014;111(11):4274–9. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir A, Kim KM, Caetano-Anolles G. Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya. BMC Evol Biol. 2012;12(1):156. doi: 10.1186/1471-2148-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Pace NR. Time for a change. Nature. 2006;441(7091):289. doi: 10.1038/441289a. [DOI] [PubMed] [Google Scholar]
- Pace NR, Olsen GJ, Woese CR. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986;45(3):325–6. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- Perkus ME, Goebel SJ, Davis SW, Johnson GP, Norton EK, Paoletti E. Deletion of 55 open reading frames from the termini of vaccinia virus. Virology. 1991;180(1):406–10. doi: 10.1016/0042-6822(91)90047-f. [DOI] [PubMed] [Google Scholar]
- Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341(6143):281–6. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Garrett RA, Koonin EV. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 2006;117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Wolf YI, Koonin EV. Search for a Tree of Life in the thicket of the phylogenetic forest. J Biol. 2009;8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Wolf YI, Koonin EV. Seeing the Tree of Life behind the phylogenetic forest. BMC Biol. 2013;11:46. doi: 10.1186/1741-7007-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D. TRUC or the need for a new microbial classification. Intervirology. 2013;56(6):349–53. doi: 10.1159/000354269. [DOI] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306(5700):1344–50. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Raoult D, Boyer M. Amoebae as genitors and reservoirs of giant viruses. Intervirology. 2010;53(5):321–9. doi: 10.1159/000312917. [DOI] [PubMed] [Google Scholar]
- Raoult D, Forterre P. Redefining viruses: lessons from Mimivirus. Nat Rev Microbiol. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- Saini HK, Fischer D. Structural and functional insights into Mimivirus ORFans. BMC Genomics. 2007;8:115. doi: 10.1186/1471-2164-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini S, Jeudy S, Bartoli J, Poirot O, Lescot M, Abergel C, Barbe V, Wommack KE, Noordeloos AA, Brussaard CP, Claverie JM. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc Natl Acad Sci U S A. 2013;110(26):10800–5. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51(3):492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Van Etten JL. Another really, really big virus. Viruses. 2011;3(1):32–46. doi: 10.3390/v3010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Lane LC, Dunigan DD. DNA viruses: the really big ones (giruses) Annu Rev Microbiol. 2010;64:83–99. doi: 10.1146/annurev.micro.112408.134338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Embley TM, Heinz E. Informational gene phylogenies do not support a fourth domain of life for nucleocytoplasmic large DNA viruses. PLoS One. 2011;6(6):e21080. doi: 10.1371/journal.pone.0021080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51(2):221–71. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977;74(11):5088–90. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Magrum LJ, Fox GE. Archaebacteria. J Mol Evol. 1978;11(3):245–51. doi: 10.1007/BF01734485. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9(8):689–710. [PubMed] [Google Scholar]
- Wolf YI, Koonin EV. Genome reduction as the dominant mode of evolution. Bioessays. 2013;35(9):829–37. doi: 10.1002/bies.201300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Fischer D. Identification and investigation of ORFans in the viral world. BMC Genomics. 2008;9:24. doi: 10.1186/1471-2164-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Colson P, Raoult D, Koonin EV. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol J. 2013;10:106. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Hidden evolutionary complexity of Nucleo-Cytoplasmic Large DNA viruses of eukaryotes. Virol J. 2012;9(1):161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Pandoraviruses are highly derived phycodnaviruses. Biol Direct. 2013;8:25. doi: 10.1186/1745-6150-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol Biol Evol. 2008;25(8):1619–30. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.