Summary
The intestine is a key organ for lipid uptake and distribution, and abnormal intestinal lipid metabolism is associated with obesity and hyperlipidemia. Although multiple regulatory gut hormones secreted from enteroendocrine cells (EEs) regulate systemic lipid homeostasis, such as appetite control and energy balance in adipose tissue, their respective roles regarding lipid metabolism in the intestine are not well understood. We demonstrate that Tachykinins (TKs), one of the most abundant secreted peptides expressed in midgut EEs, regulate intestinal lipid production and subsequently control systemic lipid homeostasis in Drosophila, and that TKs repress lipogenesis in enterocytes (ECs) associated with the TKR99D receptor and PKA signaling. Interestingly, nutrient deprivation enhances the production of TKs in the midgut. Finally, unlike the physiological roles of TKs produced from the brain, gut-derived TKs do not affect behavior, thus demonstrating that gut TK hormones specifically regulate intestinal lipid metabolism without affecting neuronal functions.
Introduction
Under normal feeding conditions, lipids that are digested from dietary food are absorbed by enterocytes (ECs) and resynthesized into triglyceride (TG) and packaged into lipoprotein particles that are transported to peripheral tissues for energy supply (Warnakula et al., 2011). Defects in enteric lipid homeostasis have been implicated in obesity, type 2 diabetes, and cardiovascular diseases (Anzai et al., 2009; Warnakula et al., 2011). Thus, characterization of the molecular mechanisms that coordinate lipid uptake, synthesis and mobilization with lipid homeostasis in the intestine is critical for understanding the basis of lipid metabolic disorders.
Gut hormones secreted from enteroendocrine cells (EEs) play crucial roles in systemic lipid homeostasis, such as the control of appetite and lipid metabolism in peripheral tissues. For example, Cholecystokinin (CCK) from I cells, EEs in the mucosal epithelium of the small intestine, reduces food intake through CCK1 receptors on the vagal nerve (Sullivan et al., 2007). Ghrelin from B/D1 cells that are mainly located in stomach and duodenum reduces lipid mobilization in adipose tissues (Tschop et al., 2000). Interestingly, glucagon-like peptide-1 (GLP1) secreted from L cells in ileum and colon suppresses intestinal chylomicron release and postprandial plasma TG level (Qin et al., 2005), whereas, glucagon-like peptide-2 (GLP2), co-secreted with GLP1, stimulates free fatty acid uptake in the intestine and increases plasma TG level (Hsieh et al., 2009), suggesting that modulation of intestinal lipid metabolism is another important physiological role of gut hormones in maintaining systemic lipid homeostasis. However, probably due to gene redundancy and overlapping functions, loss-of-function studies in the mouse for gut hormones and their receptors have failed to associate them with severe metabolic changes (Mellitzer and Gradwohl, 2011). Thus, how these hormones coordinate lipid metabolism in the intestine is still not clear.
In recent years, Drosophila has emerged as a powerful genetic system to study intestine homeostasis. Although it is simpler than the mammalian gastrointestinal tract, the Drosophila gut is similar at both the cellular and molecular levels (Apidianakis and Rahme, 2011). In particular, the adult midgut contains EEs, marked by the transcription factor Prospero (Pros), that express nine major gut prohormones, AstA, AstB, AstC, NPF, sNPF, TK, DH31, and CCHamides 1 and 2, which are processed into over 24 mature peptides (Reiher et al., 2011). Previous studies have documented their expression patterns. For example, Tachykinin (TK), the most abundant one, is expressed in anterior, middle, and posterior midgut and encodes six mature peptides (TK1–6) (Poels et al., 2009; Siviter et al., 2000; Veenstra, 2009; Veenstra et al., 2008). Only in a few cases the gut-specific functions of these hormones have been reported. In vitro treatments have shown that TK1–5 can stimulate gut contraction (Siviter et al., 2000) and loss of DH31 EEs or gut hormones in the larval midgut result in impaired peristalsis (LaJeunesse et al., 2010). Besides these examples, the physiological roles of EE hormones in gut lipid metabolism are completely unknown.
In order to analyze the physiological functions of gut hormones, we first characterized a specific Gal4 driver that is expressed in TK EEs. Using this Gal4 driver, we demonstrate that TKs produced by midgut EEs regulate intestinal lipid metabolism via controlling lipid synthesis in ECs.
Results
Identification of a Gal4 driver specifically targeting TK EEs
Since TKs are expressed both in the CNS and midgut (Asahina et al., 2014; Birse et al., 2011; Reiher et al., 2011; Winther et al., 2006), we first characterized a Gal4 driver that would allow us to perform genetic manipulation in gut TK EEs only. We screened several TK-Gal4 transgenic lines (see Experimental Procedures) and identified one of them, referred to as “TK-gut-Gal4 (TKg-Gal4)”, driving gene expression solely in TK EEs, but not TK neurons (Fig. 1A–B).
Figure 1. Characterization of TKg-Gal4 as a specific driver for TK EEs.
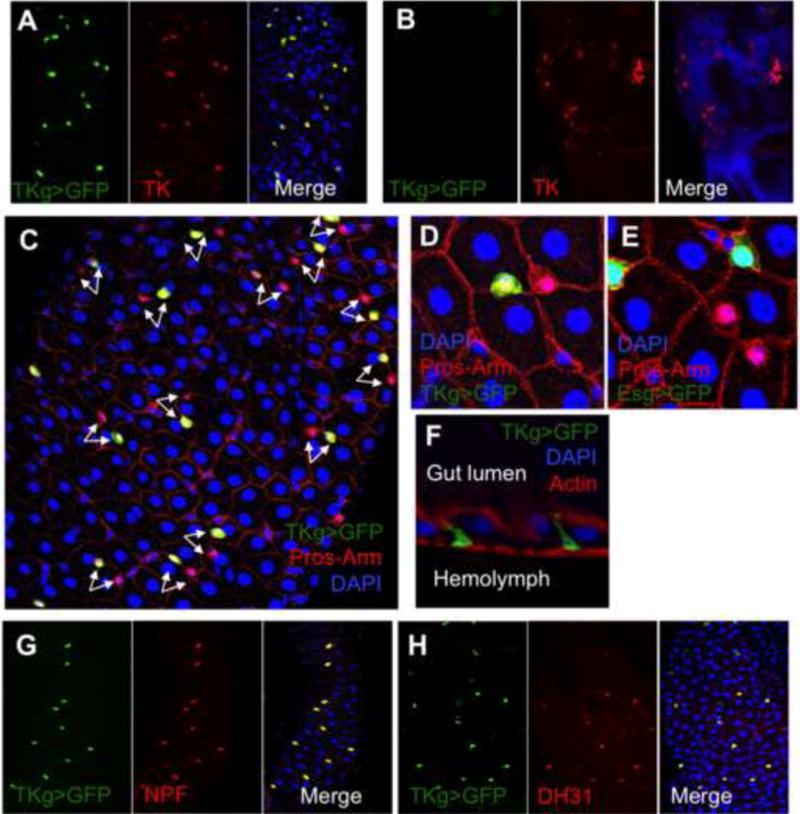
A, B, TKg-Gal4 specifically targets TK EEs but not TK brain cells. GFP expression driven by TKg-Gal4 perfectly co-localizes with TK positive cells in the gut (A) but is not detectable in TK brain cells (B) (TKg>GFP is UAS-srcGFP/+; TKg-Gal4/+, green; anti-TK, 1:500, red; DAPI, blue). C, D, TK positive cells (TKg>GFP, green) are one of heterologous pair of EEs (anti-Pros, Red nuclei). E, EEs and ISCs are intermingled among the large ECs. EEs are polygon shaped, arranged in heterologous pairs, and juxtaposed to two large-nuclear ECs, whereas ISCs are triangularly shaped and next to three ECs. ISCs are labeled with GFP (esg>GFP is UAS-GFP, esg-Gal4, green) and EEs are labeled with Prospero (anti-Pros, red nuclei). Outline of the cells are labeled with membrane-enriched Armadillo (anti-Arm, red). Nuclei are labeled with DAPI (blue). F, Confocal projection image showing that TK positive cells (TKg>GFP, green) simultaneously contact both gut lumen and hemolymph sides. Actin is labeled with phalloidin (red). G, H, TK EEs (TKg>GFP, green; DAPI, blue) also produce NPF (anti-NPF, red) in the middle midgut (G) and DH31 (anti-DH31, red) in the middle-posterior midgut (H).
EEs differ from intestine stem cells (ISCs) and are present in pairs between two ECs (Fig. 1C–E). TK EEs, which exist as one of the heterologous pair of EEs (Fig. 1C–D), are numerous in the anterior, mid, and posterior midgut with a characteristic shape, simultaneously contacting both hemolymph and gut lumen sides (Fig. 1F). Additionally, TK EEs also produce NPF in the middle midgut and DH31 in the middle-posterior midgut (Fig. 1G–H) (Veenstra et al., 2008).
TKs derived from TK EEs control intestinal lipid metabolism
To study the physiological role of TK gut hormones, we selectively ablated TK EEs by expressing the apoptotic gene reaper (RPR) under the control of TKg-Gal4. Compared to control that showed strong TK expression in both the gut and brain (TKg>Con), TK expression was lost only in the gut of TKg>RPR1 flies (Fig. 2A–B). TK EEs ablation also significantly decreased Pros-positive EEs number and impaired EEs paired appearance in midgut (Fig. S1A). Consistent with TK EEs depletion, NPF and DH31 mRNAs and proteins of TKg>RPR1 flies were dramatically reduced in the gut but remained at normal levels in the CNS (Fig. S1B–C). However, ablation of TK EEs did not result in significant gut contraction/emptiness defects as analyzed using the blue-dye food assay, but was associated with a slight increase in body weight and a slight decrease in food intake (Fig. S1D–F).
Figure 2. Gut TKs affect intestinal lipid metabolism.
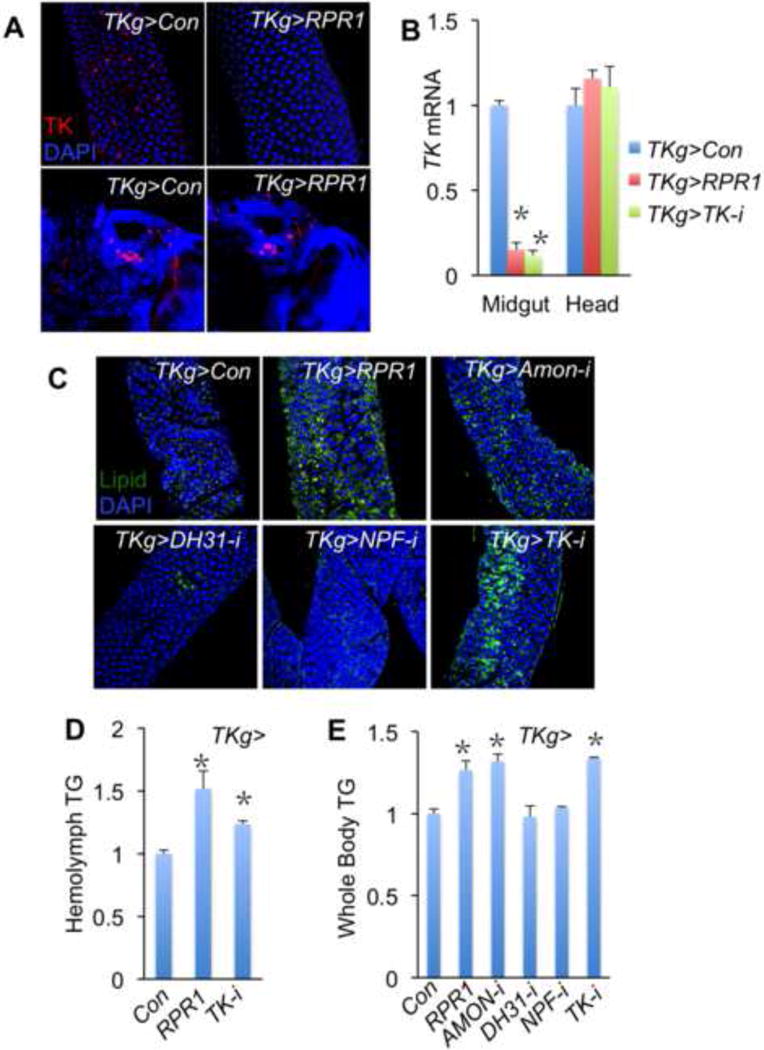
A, TKg-Gal4 allows specific ablation of TK EEs in the gut (upper) but not TK neurons (lower). Control is TKg>Con (TKg-Gal4/+) and cell ablation is achieved by expressing reaper (RPR1) in TKg>RPR1 (UAS-rpr1/+; TKg-Gal4/+) animals (anti-TK, 1:500, red; DAPI, blue). B, qPCR analysis showing the dramatic decrease of TK mRNA in TKg>RPR1 and TKg>TK-i (UAS-TK-RNAi/TKg-Gal4) guts (n=3, 30 guts or 60 heads per group). C, Lipid droplets accumulation marked with fluorescent dye in the gut (TKg>Amon-i is UAS-Amon-RNAi/+; TKg-Gal4/+. TKg>DH31-i is UAS-DH31-RNAi/+; TKg-Gal4/+. TKg>NPFi is TKg-Gal4/UAS-NPF-RNAi) (n=3, 30 guts per group). D, E, TK EEs ablation or TK knockdown in TK EEs increases both circulating TG in hemolymph (D) (n=3, 60 flies per group) and systemic TG storage (E) (n=3, 18 flies per group).
Specific ablation of TK EEs allowed us to examine whether gut peptides affects intestinal lipid metabolism. In wild type animals, intestinal TG level, the major form of neutral lipid, accounts for only about 1% of the total body TG content (Fig. S1G), reflecting the role of intestine in lipid transport. Moreover, neutral lipid droplets, detected with the neutral lipid Bodipy dye, are most abundant in the ECs located in the anterior and posterior regions of the adult midgut (Fig. S1H–I and S3D–E). Strikingly, in the absence of TK EEs, we observed a dramatic increase of neutral lipid level in midgut ECs (Fig. 2C and Fig. S3D, compare to TKg>Con control). As midgut lipids are transported throughout the body as energy supplies (Palm et al., 2012; Sieber and Thummel, 2009, 2012), elevation of intestinal lipid level may be due to an increase in lipid production in the midgut, a decrease in lipid transport, or both. To address this question, we measured the levels of circulating TG in the hemolymph. TKg>RPR1 flies showed 50% increase of TG level in hemolymph (Fig. 2D). Furthermore, whole body TG (Fig. 2E) and neutral lipid levels in the fat body (Fig. S3D), a major lipid storage organ, in TKg>RPR1 were dramatically increased, suggesting that TK EEs ablation increases intestinal lipids production and promotes systemic lipid distribution.
To confirm that the elevated lipid production in midgut was due to a deficiency in gut hormones from TK EEs, we suppressed gut hormone process and maturation by knocking down the expression of the proprotein convertase Amontillada (AMON) (Reiher et al., 2011). Similar to TK EEs ablation, AMON knockdown, which has been shown to diminish mature TKs production in TK EEs (Reiher et al., 2011), led to increased lipid levels in gut and whole body (Fig. 2C and 2E). Further, to identify the hormone(s) involved in lipid metabolic regulation, we knocked down the expression of each of the three gut hormones, TK, NPF, and DH31, in TK EEs (Fig. 2B and Fig. S1J). Surprisingly, only TK knockdown (TKg>TK-i) resulted in an increase of intestinal lipid production, hemolymph TG level and TG storage (Fig. 2C–E and Fig. S3D). Taken together, our observations indicate that TK hormones, but not NPF or DH31, secreted from TK EEs regulate intestinal lipid metabolism and subsequently affect systemic lipid storage.
Brain- and gut-derived TKs play distinct physiological roles
We wondered whether TKs produced from the gut or CNS regulates similar processes. Thus, we compared the phenotypes associated with TK knockdown only in the gut versus both in the brain and gut. RNAi against TK driven by ELAV-Gal4 (ELAV>TK-i) diminished TK expression in both the gut and CNS compared to control flies (ELAV>white-i) (Fig. S2A–B). These flies showed increased locomotor activity and reduced olfactory responses to certain chemicals (Fig. S2C–D) as previously reported (Ignell et al., 2009; Winther et al., 2006). However, TK knockdown only in the gut failed to affect locomotor activity or the olfactory response (Fig. S2C–D). Additionally, unlike the effect observed when brain TKs were depleted (Birse et al., 2011), we did not detect a change in dILP2 content in IPCs or body weight (Fig. S2E–G) when TKs were knocked down only in the gut. These data demonstrate that gut TKs specifically regulate intestinal lipid metabolism, and that they are not involved in behavior regulation or dILPs secretion, functions attributable to TK neuropeptides.
TKR99D/PKA regulates lipid metabolism in ECs in response to gut TKs
To determine how TK regulates lipid production in ECs, we tested whether removal of the TK receptor affects intestinal lipid metabolism. Two different G protein-coupled TK receptors, TKR99D and TKR86C, expressed in gut have been identified (Birse et al., 2006; Poels et al., 2009). Interestingly, while TKR86C is mainly expressed in gut muscles and a few EEs (Poels et al., 2009), TKR99D, as determined using a TKR99D-Gal4 line, appears highly expressed in lipid absorptive ECs (Fig. 3A), as well as in a few TK/NPF EEs (Fig. S3A–B). Strikingly, knockdown of TKR99D in ECs by about 60% (Fig. 3B and S3C) was sufficient to result in an increase in midgut lipid production and whole body TG storage (compare Myo1A>TKR99D-i to Myo1A>Con, Fig. 3D–E and S3E).
Figure 3. TKR99D/PKA signaling is essential for EC lipid metabolism.
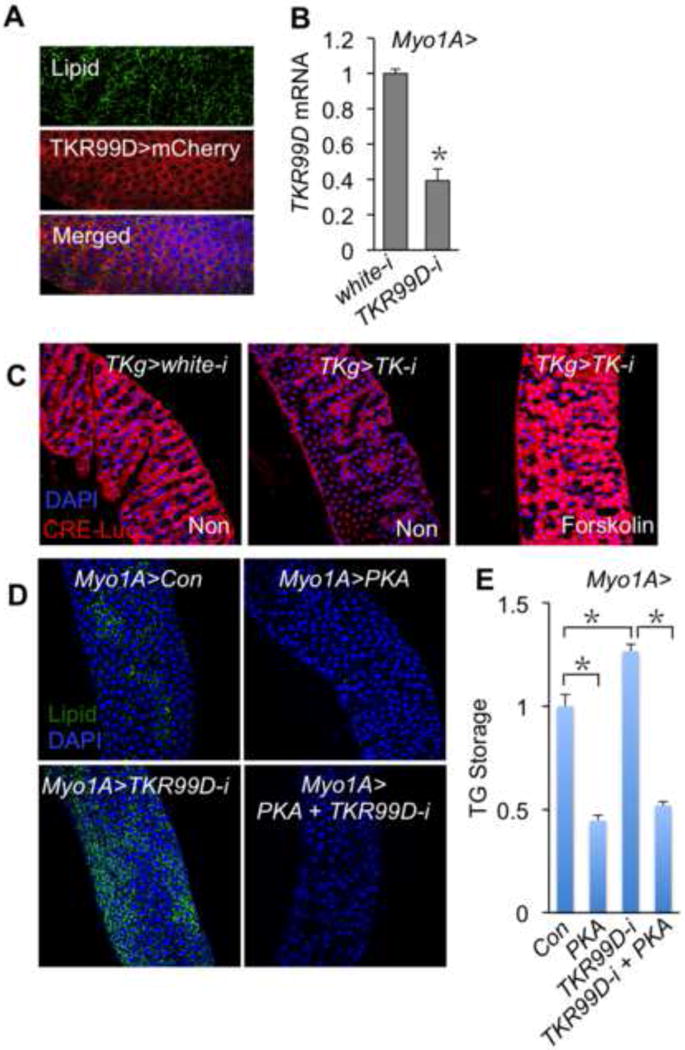
A, TKR99D is expressed in lipid absorptive ECs (lipid, green; TKR99D>mCherry is TKR99D-Gal4/UAS-mCherry, red; DAPI, blue). B, qPCR results of TKR99D expression in Myo1A>white-i (Myo1A-Gal4/+; UAS-white-RNAi/+) and Myo1A>TKR99D-i (Myo1A-Gal4/+; UAS-TKR99D-RNAi/+) guts (n=3, 30 guts per group). C, CREB transcriptional activity, detected using CRE-Luci, is decreased in TKg>TK-i guts under normal diet but restored to normal level when flies are fed Forskolin (10mM in normal fly food). D, E, Lipid level in guts (D) and TG storage (E) (n=3, 18 flies per group) of Myo1A>Con (Myo1A-Gal4/+; UAS-white-RNAi/+), Myo1A>PKA (Myo1A-Gal4/+; UAS-PKA-C1/+), Myo1A>TKR99D-i (Myo1A-Gal4/+; UAS-TKR99D-RNAi/+), and Myo1A>PKA + TKR99D-i (Myo1A-Gal4/+; UAS-TKR99D-RNAi/UAS-PKA-C1) animals.
To test whether gut TKs or TKR99D regulates enteric lipid metabolism through GPCR/cAMP/PKA signaling as previously suggested (Birse et al., 2006; Lundquist and Nassel, 1997), we tested the activity of CREB, a direct substrate of PKA, using a CRE-Luciferase reporter (Belvin et al., 1999). TK knockdown (TKg>TKi) dramatically decreased CREB transcriptional activity in ECs compared to control (TKg>white-i), whereas activation of PKA by feeding flies with Forskolin, a PKA agonist, restored CREB transcriptional activity to a normal level (Fig. 3C). Furthermore, overexpression of a catalytic form of PKA in ECs (Myo1A>PKA, TKR99D-i) was sufficient to reverse the increased intestinal lipid production and systemic TG storage (Fig. 3D–E) associated with TKR99D knockdown. Collectively, our results suggested that gut TKs regulate EC lipid metabolism through TKR99D/PKA signaling.
TKs suppress lipogenesis in the midgut
To identify the lipid processing pathways regulated by TKs in ECs, we analyzed the mRNA expression profile of genes involved in intestinal lipid metabolism. Interestingly, the intestinal lipases Magro (Sieber and Thummel, 2009) and CG2772 that regulate dietary lipid digestion in the gut lumen, and the two key enzymes of lipogenesis fatty acid synthase (FAS) and acetyl-CoA carboxylase (ACC) that control enteric lipid synthesis (Lim et al., 2011), were all up-regulated when TKs production was reduced (TKg>TK-i) (Fig. S3F), suggesting that TKs regulate intestinal lipid metabolism via multiple lipid processing pathways. On the other hand, expression of the lipid transporter NinaD involved in lipid absorption (Kiefer et al., 2002), the ER UPR sensor IRE1 and Microsomal Triglyceride Transfer Protein (MTP) that regulate lipoprotein particle package (Iqbal et al., 2008), and the intestinal lipase CG31089 that controls dietary lipid digestion, remained unaffected (Fig. S3F). Notably, mRNA expression of the FoxO target genes, 4EBP and insulin receptor (InR), in the midgut were not affected by removal of TKs (Fig. S3F), suggesting that gut TKs do not affect insulin signaling in the midgut.
The up-regulation of FAS induced by TK deficiency (Fig. 4A and Fig. S3F) suggested that TKs regulate midgut lipid metabolism, at least, via modulation of intestinal lipogenesis. To test this hypothesis, flies were fed with 14C-labelled glucose and the lipids derived from 14C-carbon backbones in the gut were measured. TK>TK-i flies contained more lipids derived from glucose carbon backbones in the midgut (Fig. 4C), suggesting that TK deficiency promotes midgut lipogenesis. Sterol Regulatory Element-Binding Protein (SREBP) is a conserved transcription factor of lipogenic genes, like FAS (Fig. 4B) (Kunte et al., 2006), and is negatively modulated by GPCR/cAMP/PKA signaling (Lu and Shyy, 2006). Consistent with this idea, TKR99D/PKA signaling in ECs suppressed FAS expression and lipogenesis in the midgut (Fig. 4B–C).
Figure 4. Gut TKs suppress intestinal lipogenesis.
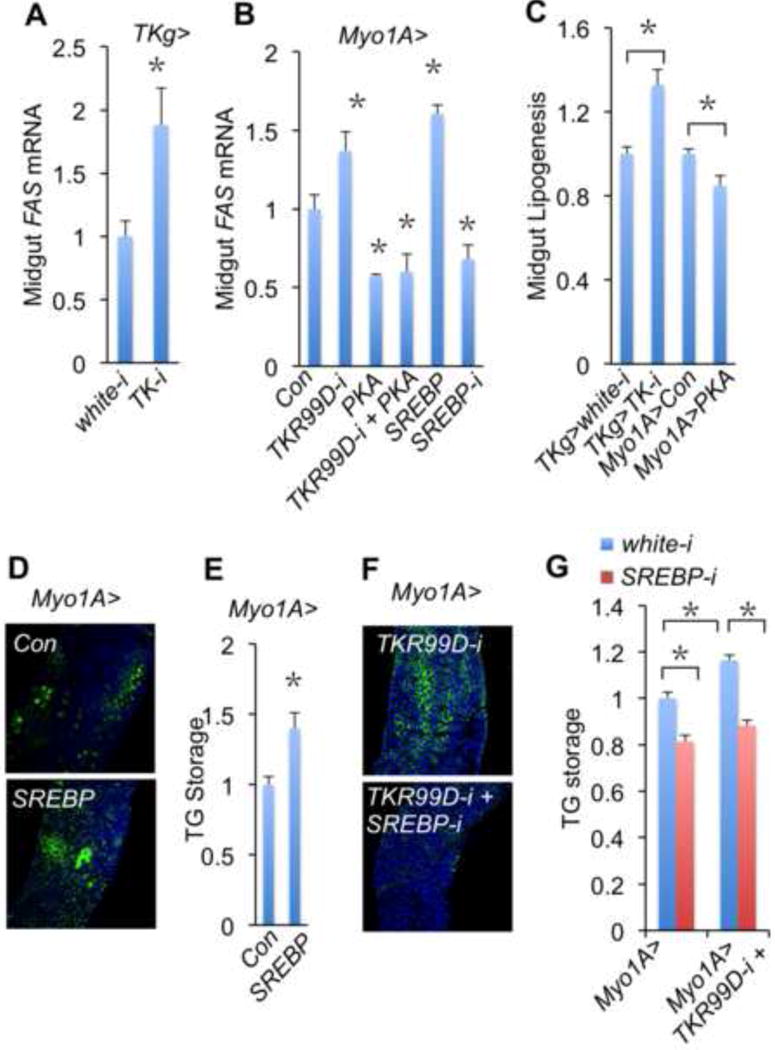
A, B, Expression levels of FAS in gut analyzed by qPCR (n=3, 30 guts per group). (A) FAS mRNA expression in guts of TKg>white-i (TKg-Gal4/UAS-white-RNAi) and TKg>TK-i (TKg-Gal4/UAS-TK-RNAi) flies. (B) FAS mRNA expression in the gut of Myo1A>Con, Myo1A>TKR99D-i, Myo1A>PKA, Myo1A>TKR99D-i + PKA, Myo1A>SREBP (Myo1A-Gal4/UAS-SREBP) and Myo1A>SREBP-i (Myo1A-Gal4/+; UAS-SREBP-RNAi/+) flies (n=3, 30 gut per group). C, Midgut lipogenesis was analyzed by 14C-labeled lipids derived from 14C-glucose in TKg>white-i, TKg>TK-i, Myo1A>Con, and Myo1A>PKA guts (n=3, 90 guts per group). D–G, Intestinal lipid level (D, F) and whole body TG storage (E, G) (n=3, 18 flies per group) of adult flies. (D, E) Myo1A>Con and Myo1A>SREBP adult flies. (F, G) Myo1A>white-i, Myo1A>TKR99D-i, Myo1A>SREBP-i, and Myo1A>TKR99D-i + SREBP-i (Myo1A-Gal4/+; UAS-TKR99D-RNAi/UAS-SREBP-RNAi) adult flies.
Intestinal lipogenesis contributes to TKs deficiency-induced midgut lipid production and systemic lipid storage
To test whether intestinal lipogenesis is sufficient to contribute to changes in midgut lipid production, we expressed an active form of SREBP in ECs (Myo1A>SREBP). As predicted, increases of midgut FAS mRNA expression, intestinal lipid production, and even whole body TG storage were observed in Myo1A>SREBP flies (Fig. 4B, 4D, and 4E). Conversely, specific SREBP knockdown in ECs (Myo1A>SREBP-i) decreased midgut FAS expression and body TG storage (Fig. 4B and 4G). Thus, intestinal lipogenesis is essential for midgut lipid production and systemic lipid storage.
We further tested whether SREBP-induced lipogenesis is required for TKs regulation of intestinal lipid metabolism. Surprisingly, SREBP knockdown in ECs potently blocked the increase of midgut lipid level and systemic TG storage associated with TKR99D knockdown (Fig. 4F–G). Collectively, our results indicate that gut TKs regulate intestinal lipid metabolism through, at least, repression of SREBP-induced lipogenesis.
The midgut produces TKs in response to nutrient availability
Release or production of gut hormones is regulated by diverse physiological conditions in different species. An increase of TKs released from gut into the hemolymph has been observed in the starved Locust (Winther and Nassel, 2001). Thus, we tested whether TK levels in the gut are affected by starvation. Flies, deprived from food resource for 24h, showed a significant increase of TK level in their midgut (Fig. S4A). To test whether increased intracellular TK levels was due to less TK secretion or more TK production, the expression of downstream targets of TK signaling in the midgut were measured. Strikingly, TK/PKA dependent CREB activity was potently increased (Fig. S4B), whereas, FAS mRNA suppressed by TK signaling was dramatically decreased (Fig. S4C), suggesting that starvation enhances TKs production in the midgut. Consistent with this idea, diminishing intestinal TK expression partially restored midgut FAS expression and lipid production under starvation (Fig. S4C–D). To determine which nutrient regulates TK production, flies were refed with different ingredients, such as sucrose, coconut oil, or yeast, after starvation. Interestingly, only yeast refeeding potently suppressed TK production in midgut (Fig. S4E). As amino acid is the major nutrient existing in yeast, our results suggest that the midgut produces TKs in response to amino acid availability.
To examine whether nutrient status or TKR99D signaling changes in ECs regulate TKs production in a feedback manner, we specifically modulated TKR99D signaling in ECs. Knockdown of TKR99D or overexpression of PKA in ECs showed altered lipid levels (Fig. 3D–E), but did not affect TKs levels in midgut EEs (Fig. S4F), suggesting that TK signaling or lipid levels in ECs do not regulate intestinal TKs production.
Interestingly, different from mammalian regulation of TKs production by gut pathogen infection, flies fed with the human pathogen Pseudomonas Aeruginosa 14 (PA14), previously shown to cause severe gut defects and gut stem cell proliferation in Drosophila (Apidianakis et al., 2009), failed to show any change of intracellular TK levels (Fig. S4G). These results suggest that the presence of pathogen does not affect production of gut TKs in Drosophila.
Discussion
Previous studies in mammals have indicated that a few gut secretory hormones, like GLP1/2, are involved in intestinal lipid metabolism (Qin et al., 2005). However, due to gene and functional redundancy, mammalian genetic models for gut hormones and/or their receptors with severe metabolic defects are not available. Here, we establish that Drosophila TKs produced from EEs coordinate midgut lipid metabolic processes. Our studies clarify the roles of TK hormones in intestinal lipogenesis and establish Drosophila as a genetic model to study the regulation of lipid metabolism by gut hormones.
Six mature TKs, TK1–6, are processed and secreted from TK EEs in both the brain and midgut (Reiher et al., 2011). Using a specific Gal4 driver line, we were able to specifically manipulate gene expression in TK EEs, leading us to demonstrate that loss of gut TKs results in an increase in midgut lipid production. Further, we showed that TKs regulate intestinal lipid metabolism associated with TKR99D, but not TKR86C, which is consistent with the expression of these receptors. Consistent with previous reports that TK/TKR99D signaling regulates cAMP level and PKA activation (Birse et al., 2006; Lundquist and Nassel, 1997), loss of gut TKs is associated with a reduction in PKA activity in ECs, and overexpression of a PKA catalytic subunit was able to reverse the increased intestinal lipid production associated with loss of TKR99D. In addition, the transcription factor SREBP that triggers lipogenesis was controlled by TK/TKR99D/PKA signaling. Taken together, our results suggest that TKs produced from EEs regulate midgut lipid metabolism via TKR99D/PKA signaling and regulation of, at least, SREBP-induced lipogenesis in ECs.
Interestingly, our study reveals that TKs derived from either the brain or gut exhibit distinct functions: TKs derived from gut control intestinal lipid metabolism, whereas TKs derived from brain control behavior. This is reminiscent of the distinct functions of mammalian secreted regulatory peptides, where different spatial expressions or deliveries of peptides can result in distinct physiological functions, like Ghrelin (Nakazato et al., 2001; Tschop et al., 2000). In addition, some prohormones encode multiple mature peptides that can have multiple functions. For example, processing of proglucagon in the pancreas α-cells preferentially give rise to glucagon that antagonizes the effect of Insulin. In intestine L cells, however, proglucagon is mostly processed into GLP1 to promote Insulin release (Brubaker and Drucker, 2004). Our studies of TKs exemplify how secreted regulatory peptides derived from different tissues can be associated with fundamentally diverse physiological functions. Clearly, additional studies examining the function of secreted peptides in a cell type and tissue specific manner are needed to fully appreciate and unravel their complex roles both in flies and mammals.
There is a growing body of studies emphasizing that intestinal lipid metabolism is key to the control of systemic lipid homeostasis. For example, chemicals, such as Orlistat (Heck et al., 2000), designed to inhibit dietary lipid digestion/absorption in the intestine efficiently reduce obesity. In addition, mammalian IRE1Jβ deficiency-induced abnormal chylomicron assembly in the small intestine results in hyperlipidemia (Iqbal et al., 2008). Similarly, in Drosophila, dysfunction of intestinal lipid digestion/absorption caused by Magro/LipA deficiency eventually decreases whole body lipid storage and starvation resistance (Karpac et al., 2013; Sieber and Thummel, 2009, 2012). Further, intestinal lipid transport, controlled by lipoproteins, is essential for systemic lipid distribution and energy supply in other tissues (Palm et al., 2012; Panakova et al., 2005). Consistent with these observations, we demonstrate that increased midgut lipid synthesis associated with gut TKs deficiency is sufficient to elevate systemic lipid storage. Although TK ligands and TK receptors show high homologies between mammals and fruit flies (Birse et al., 2006), whether mammalian TK signaling plays a similar role in intestinal lipid metabolism is largely unknown. Future studies will reveal whether mammalian TK signaling affects intestinal lipid metabolism as in Drosophila. If this is the case, it may provide a therapeutic opportunity for the treatment of intestinal lipid metabolic disorder and obesity.
Production and secretion of gut hormones are precisely regulated under various physiological conditions. Similar to previous observations that starvation induces gut TKs secretion in other insects (Winther and Nassel, 2001), we found that nutrient deprivation promotes TK production in EEs. Interestingly, feeding of amino acid-enriched yeast, but not coconut oil or sucrose, potently suppressed gut TK level, indicating that amino acid may act directly on TK production in EEs. It has been reported that dietary nutrients regulate gut hormones production through certain receptors located on EEs membrane in mammals (Reimann et al., 2012). Future studies will be necessary to elucidate the detailed mechanism by which nutrients regulate TKs production from EEs.
Experimental Procedures
Drosophila Strains
Expression patterns of different TK-Gal4 P-element lines that contain the 0.5–2.5kb fragment upstream of the TK gene were examined by crossing to UAS-srcGFP flies. One line that showed expression in TK EEs, but not in TK neurons, was referred to as TK-gut-Gal4 (TKg-Gal4) and used for this study. Other lines were obtained from Bloomington Drosophila Stock Center, TRiP at Harvard Medical School, and Vienna Drosophila Resource Center. See Supplemental Experimental Procedures for detailed information.
Immunostaining and Western Blot
Immunostaining of adult midgut and brain and Western blot were described previously (Karpowicz et al., 2010; Song et al., 2010). See Supplemental Experimental Procedures for detailed information.
TG measurement
TG measurement was performed as previously described (Song et al., 2010). See Supplemental Experimental Procedures for detailed information.
RT-qPCR
RT-qPCR was performed as previously described (Song et al., 2010). See Supplemental Experimental Procedures for detailed primer information.
Midgut lipogenesis measurement
Adult flies were fed with 0.2 mCi/mL 14C-glucose (PerkinElmer) for 3 days. 30 guts were dissected and homogenized in 200 μL of chloroform/methanol/H2O (2:1:1) mixture. The lysate was incubated at 37°C for 1 hour before 75 μL chloroform and 75 μL 1 M KCl were added. After centrifugation at 3,000rpm for 2 min, 14C-labeled lipids that are contained in chloroform phase were measured by liquid scintillation counting.
Behavior Assays
Behavior assays were performed as previously described (Winther et al., 2006).
Statistical Analyses
The data are presented as the mean ± SEM. Student’s t tests were used for comparisons between two groups. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Richard Binari for technical support, and Phillip Karpowicz, Young Kwon, Akhila Rajan, and Edward Owusu-Ansah, for comments on the manuscript. This work was supported by 5P01CA120964 and 5R01DK088718 from the NIH. N.P is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions. Wei Song conceived, designed, and performed all experiments. Jan A. Veenstra generated TKg-Gal4 lines and antibodies against TK and DH31 and made constructive comments. Wei Song and Norbert Perrimon discussed results and wrote the manuscript.
References
- Anzai K, Fukagawa K, Iwakiri R, Fujimoto K, Akashi K, Tso P. Increased lipid absorption and transport in the small intestine of zucker obese rats. J Clin Biochem Nutr. 2009;45:82–85. doi: 10.3164/jcbn.09-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apidianakis Y, Pitsouli C, Perrimon N, Rahme L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Natl Acad Sci U S A. 2009;106:20883–20888. doi: 10.1073/pnas.0911797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apidianakis Y, Rahme LG. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech. 2011;4:21–30. doi: 10.1242/dmm.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina K, Watanabe K, Duistermars BJ, Hoopfer E, Gonzalez CR, Eyjolfsdottir EA, Perona P, Anderson DJ. Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell. 2014;156:221–235. doi: 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PubMed] [Google Scholar]
- Birse RT, Johnson EC, Taghert PH, Nassel DR. Widely distributed Drosophila G-protein-coupled receptor (CG7887) is activated by endogenous tachykinin-related peptides. J Neurobiol. 2006;66:33–46. doi: 10.1002/neu.20189. [DOI] [PubMed] [Google Scholar]
- Birse RT, Soderberg JA, Luo J, Winther AM, Nassel DR. Regulation of insulin-producing cells in the adult Drosophila brain via the tachykinin peptide receptor DTKR. J Exp Biol. 2011;214:4201–4208. doi: 10.1242/jeb.062091. [DOI] [PubMed] [Google Scholar]
- Brubaker PL, Drucker DJ. Minireview: Glucagon-like peptides regulate cell proliferation and apoptosis in the pancreas, gut, and central nervous system. Endocrinology. 2004;145:2653–2659. doi: 10.1210/en.2004-0015. [DOI] [PubMed] [Google Scholar]
- Heck AM, Yanovski JA, Calis KA. Orlistat, a new lipase inhibitor for the management of obesity. Pharmacotherapy. 2000;20:270–279. doi: 10.1592/phco.20.4.270.34882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Longuet C, Maida A, Bahrami J, Xu E, Baker CL, Brubaker PL, Drucker DJ, Adeli K. Glucagon-like peptide-2 increases intestinal lipid absorption and chylomicron production via CD36. Gastroenterology. 2009;137:997–1005. doi: 10.1053/j.gastro.2009.05.051. 1005 e1001–1004. [DOI] [PubMed] [Google Scholar]
- Ignell R, Root CM, Birse RT, Wang JW, Nassel DR, Winther AM. Presynaptic peptidergic modulation of olfactory receptor neurons in Drosophila. Proc Natl Acad Sci U S A. 2009;106:13070–13075. doi: 10.1073/pnas.0813004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, Ron D, Tabas I, Hussain MM. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7:445–455. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpac J, Biteau B, Jasper H. Misregulation of an adaptive metabolic response contributes to the age-related disruption of lipid homeostasis in Drosophila. Cell Rep. 2013;4:1250–1261. doi: 10.1016/j.celrep.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, Sumser E, Wernet MF, Von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc Natl Acad Sci U S A. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 2006;3:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- LaJeunesse DR, Johnson B, Presnell JS, Catignas KK, Zapotoczny G. Peristalsis in the junction region of the Drosophila larval midgut is modulated by DH31 expressing enteroendocrine cells. BMC Physiol. 2010;10:14. doi: 10.1186/1472-6793-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HY, Wang W, Wessells RJ, Ocorr K, Bodmer R. Phospholipid homeostasis regulates lipid metabolism and cardiac function through SREBP signaling in Drosophila. Genes Dev. 2011;25:189–200. doi: 10.1101/gad.1992411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Shyy JY. Sterol regulatory element-binding protein 1 is negatively modulated by PKA phosphorylation. Am J Physiol Cell Physiol. 2006;290:C1477–1486. doi: 10.1152/ajpcell.00374.2005. [DOI] [PubMed] [Google Scholar]
- Lundquist CT, Nassel DR. Peptidergic activation of locust dorsal unpaired median neurons: depolarization induced by locustatachykinins may be mediated by cyclic AMP. J Neurobiol. 1997;33:297–315. doi: 10.1002/(sici)1097-4695(199709)33:3<297::aid-neu8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Mellitzer G, Gradwohl G. Enteroendocrine cells and lipid absorption. Curr Opin Lipidol. 2011;22:171–175. doi: 10.1097/MOL.0b013e32834622a2. [DOI] [PubMed] [Google Scholar]
- Nakazato M, Murakami N, Date Y, Kojima M, Matsuo H, Kangawa K, Matsukura S. A role for ghrelin in the central regulation of feeding. Nature. 2001;409:194–198. doi: 10.1038/35051587. [DOI] [PubMed] [Google Scholar]
- Palm W, Sampaio JL, Brankatschk M, Carvalho M, Mahmoud A, Shevchenko A, Eaton S. Lipoproteins in Drosophila melanogaster–assembly, function, and influence on tissue lipid composition. PLoS Genet. 2012;8:e1002828. doi: 10.1371/journal.pgen.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- Poels J, Birse RT, Nachman RJ, Fichna J, Janecka A, Vanden Broeck J, Nassel DR. Characterization and distribution of NKD, a receptor for Drosophila tachykinin-related peptide 6. Peptides. 2009;30:545–556. doi: 10.1016/j.peptides.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Qin X, Shen H, Liu M, Yang Q, Zheng S, Sabo M, D’Alessio DA, Tso P. GLP-1 reduces intestinal lymph flow, triglyceride absorption, and apolipoprotein production in rats. Am J Physiol Gastrointest Liver Physiol. 2005;288:G943–949. doi: 10.1152/ajpgi.00303.2004. [DOI] [PubMed] [Google Scholar]
- Reiher W, Shirras C, Kahnt J, Baumeister S, Isaac RE, Wegener C. Peptidomics and peptide hormone processing in the Drosophila midgut. J Proteome Res. 2011;10:1881–1892. doi: 10.1021/pr101116g. [DOI] [PubMed] [Google Scholar]
- Reimann F, Tolhurst G, Gribble FM. G-protein-coupled receptors in intestinal chemosensation. Cell Metab. 2012;15:421–431. doi: 10.1016/j.cmet.2011.12.019. [DOI] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009;10:481–490. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. Coordination of triacylglycerol and cholesterol homeostasis by DHR96 and the Drosophila LipA homolog magro. Cell Metab. 2012;15:122–127. doi: 10.1016/j.cmet.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siviter RJ, Coast GM, Winther AM, Nachman RJ, Taylor CA, Shirras AD, Coates D, Isaac RE, Nassel DR. Expression and functional characterization of a Drosophila neuropeptide precursor with homology to mammalian preprotachykinin A. J Biol Chem. 2000;275:23273–23280. doi: 10.1074/jbc.M002875200. [DOI] [PubMed] [Google Scholar]
- Song W, Ren D, Li W, Jiang L, Cho KW, Huang P, Fan C, Song Y, Liu Y, Rui L. SH2B regulation of growth, metabolism, and longevity in both insects and mammals. Cell Metab. 2010;11:427–437. doi: 10.1016/j.cmet.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CN, Raboin SJ, Gulley S, Sinzobahamvya NT, Green GM, Reeve JR, Jr, Sayegh AI. Endogenous cholecystokinin reduces food intake and increases Fos-like immunoreactivity in the dorsal vagal complex but not in the myenteric plexus by CCK1 receptor in the adult rat. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1071–1080. doi: 10.1152/ajpregu.00490.2006. [DOI] [PubMed] [Google Scholar]
- Tschop M, Smiley DL, Heiman ML. Ghrelin induces adiposity in rodents. Nature. 2000;407:908–913. doi: 10.1038/35038090. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Peptidergic paracrine and endocrine cells in the midgut of the fruit fly maggot. Cell Tissue Res. 2009;336:309–323. doi: 10.1007/s00441-009-0769-y. [DOI] [PubMed] [Google Scholar]
- Veenstra JA, Agricola HJ, Sellami A. Regulatory peptides in fruit fly midgut. Cell Tissue Res. 2008;334:499–516. doi: 10.1007/s00441-008-0708-3. [DOI] [PubMed] [Google Scholar]
- Warnakula S, Hsieh J, Adeli K, Hussain MM, Tso P, Proctor SD. New insights into how the intestine can regulate lipid homeostasis and impact vascular disease: frontiers for new pharmaceutical therapies to lower cardiovascular disease risk. Can J Cardiol. 2011;27:183–191. doi: 10.1016/j.cjca.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Winther AM, Acebes A, Ferrus A. Tachykinin-related peptides modulate odor perception and locomotor activity in Drosophila. Mol Cell Neurosci. 2006;31:399–406. doi: 10.1016/j.mcn.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Winther AM, Nassel DR. Intestinal peptides as circulating hormones: release of tachykinin-related peptide from the locust and cockroach midgut. J Exp Biol. 2001;204:1269–1280. doi: 10.1242/jeb.204.7.1269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


