Highlights
-
•
We present an overview of the evolution of the virus world and its multifaceted, complex interaction with cellular life forms.
-
•
Several viral hallmark genes that encode essential proteins are shared by extremely diverse groups of viruses.
-
•
The existence of hallmark genes implies that viruses and related selfish elements evolved from the primordial gene pool.
-
•
The emergence of selfish elements is theoretically inevitable in any ensemble of evolving replicators.
-
•
Virus–host arms races and cooperation were among the decisive factors in the evolution of all life forms.
Abstract
Viruses and/or virus-like selfish elements are associated with all cellular life forms and are the most abundant biological entities on Earth, with the number of virus particles in many environments exceeding the number of cells by one to two orders of magnitude. The genetic diversity of viruses is commensurately enormous and might substantially exceed the diversity of cellular organisms. Unlike cellular organisms with their uniform replication-expression scheme, viruses possess either RNA or DNA genomes and exploit all conceivable replication-expression strategies. Although viruses extensively exchange genes with their hosts, there exists a set of viral hallmark genes that are shared by extremely diverse groups of viruses to the exclusion of cellular life forms. Coevolution of viruses and host defense systems is a key aspect in the evolution of both viruses and cells, and viral genes are often recruited for cellular functions. Together with the fundamental inevitability of the emergence of genomic parasites in any evolving replicator system, these multiple lines of evidence reveal the central role of viruses in the entire evolution of life.
Current Opinion in Virology 2013, 3:546–557
This review comes from a themed issue on Virus evolution
Edited by Valerian V Dolja and Mart Krupovic
For a complete overview see the Issue and the Editorial
Available online 12th July 2013
1879-6257/$ – see front matter, Published by Elsevier Ltd.
Introduction
Viruses were discovered at the end of the 19th century as peculiar plant and animal pathogens that were small enough to pass bacterial filters. Since then, the developments in virology and genomics have completely changed the concept of viruses. We now realize that viruses are ubiquitous and enormously abundant, in terms of both infecting all known cellular life forms and being present in all explored environments. Actually, viruses appear to be the most abundant biological entities on the planet, substantially outnumbering cells in most well-studied habitats [1, 2•, 3•]. The genetic diversity of viruses is enormous as well: the majority of distinct genes in the biosphere seem to reside in viral genomes [4]. Beyond their quantitative dominance, virus genome sizes, gene repertoires and particle sizes and shapes are extremely variable. Moreover, viruses exploit all theoretically conceivable strategies of genome replication and expression, in contrast to the uniform replication-expression cycle of cellular life forms.
The simple, size-based distinction between viruses and cells is gone: the discovery of giant viruses has overthrown the original virus definition because these viruses are bigger than many bacteria [5]. Commensurately, the genome size and complexity of the giant viruses overlap the genomic ranges of cellular life forms and exceed those of numerous parasitic bacteria [6].
Virus–host interaction is a theme that permeates the entire course of the evolution of life. This interaction is commonly pictured as an arms race in which the hosts constantly evolve new means of defense which viruses perpetually evolve to evade [7]. Although this scenario captures important aspects of virus–host coevolution, there is more to the story. Not only do viruses hijack host genes for counter-defense and other functions, but cellular hosts as well routinely recruit viral genes for diverse roles. Viruses that evolve at high rates might comprise the principal source of novel genes in the biosphere [8]. In particular, genetic material coming from virus-like retro-transcribing elements apparently dominates vertebrate and plant genomes [9, 10].
Here we explore the consilience of diverse lines of evidence that reveal essential roles played by viruses and virus-like selfish elements in the evolution of life and propound the ‘virocentric’ view of life's evolution.
Viruses as nature's genomic laboratory
All cellular life forms employ a single, strictly defined strategy for genome replication and expression whereby double-stranded (ds)DNA is the replicating form of the genome that is transcribed into single-stranded (ss)RNA some of which is then translated to produce proteins. The only variation on this theme is occasional reverse transcription of RNA that is typically non-essential for genome replication, an important exception being the telomere synthesis in eukaryotes. In a sharp contrast, viruses exploit effectively all possible combinations of DNA and RNA interconversions: the replicating genome that is incorporated into virions can be represented by either RNA or DNA, and the replication-expression cycles of RNA viruses can include a DNA intermediate and vice versa (Figure 1 ) [11, 12]. Furthermore, the selfish elements with the smallest known genomes, the viroids, are autonomously replicating small non-coding RNAs [13].
Figure 1.
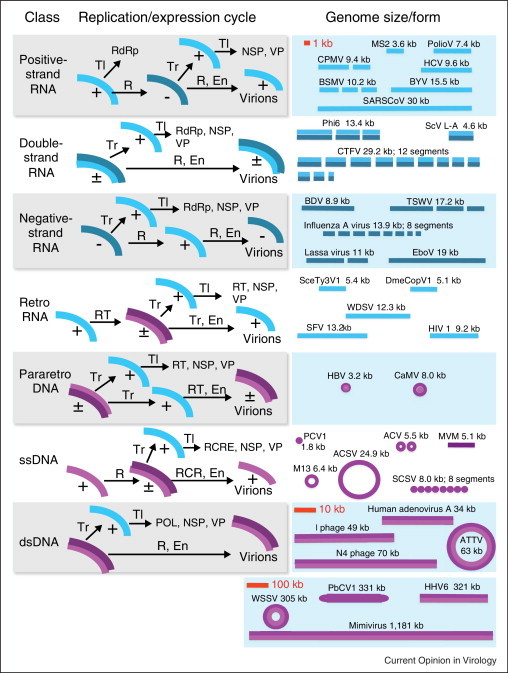
The diversity of the virus world. Seven major classes of viruses with distinct genome replication/expression cycles are shown at the left; representative examples of the corresponding genome sizes and architectures are shown at the right approximately to scale indicated in red. Abbreviations: Tl, translation; Tr, transcription; R, replication; RT, reverse transcription; RCR, rolling-circle replication; kb, kilobase. En, encapsidation; RdRp, RNA-dependent RNA polymerase; NSP, non-structural proteins; VP, virion proteins; RCRE, rolling-circle replication endonuclease. Designations: (+), (−), (±), positive-strand, negative-strand, and double-stranded nucleic acids, respectively. Virus acronyms: MS2, bacteriophage MS2; PolioV, Poliomyelitis virus; CPMV, Cowpea mosaic virus; HCV, Hepatitis C virus; BSMV, Barley stripe mosaic virus; BYV, Beet yellows virus; SARS CoV, SARS Coronavirus; Phi6, bacteriophage phi 5; ScV L-A, Saccharomyces cerevisiae virus L-A; CTFV, Colorado tick fever virus; BDV, Borna disease virus; TSWV, Tomato spotted wilt virus; EboV, Ebola virus; SceTy3V1, Saccharomyces cerevisiae Ty virus 1; DmeCopV1, Drosophilla melanogaster Copia virus 1; WDSV, Walleye dermal sarcoma virus; SFV, Simian foamy virus; HIV1, Human immunodeficiency virus 1; HBV, Hepatitis B virus; CaMV, Cauliflower mosaic virus; PCV1, Porcine circovirus 1; ACV, African cassava mosaic virus; MVM, Minute virus of mice; M13, bacteriophage M13; ACSV, Aeropyrum coil-shaped virus; SCSV, Subterranean clover stunt virus; ATV, Acidianus two-tailed virus; WSSV, White spot syndrome virus; PbCV1, Paramecium bursaria Chlorella virus 1; HHV6, Human herpesvirus 6.
In addition to the diversity of the replication-expression strategies, viruses display the complete repertoire of single-stranded and double-stranded, linear and circular RNA and DNA, and a distinct type of molecules, protein-linked DNA and RNA, as well as monopartite and multipartite genomes (Figure 1).
Viruses are generally perceived as miniscule entities with small genomes. Although on average, this is true compared to cellular life forms, the range of viral genome sizes spans three orders of magnitude (Figure 1), which is similar to the range of genome sizes of cellular life forms [14]. Moreover, the giant viruses clearly fall within the genome size range of prokaryotes. The virus genome sizes is tightly linked to the genome structure and replication-expression strategy: the genomes that are larger than ∼30 kb always consist of dsDNA, conceivably due to physical constraints on genome stability and topological malleability. Below this apparent limit, the genome ranges associated with all genomic strategies strongly overlap (Figure 1).
Thus, viruses exhaustively explore the space of genome structures and strategies. This exploration is feasible because viruses, being the ultimate parasites, possess small genomes that can be efficiently replicated as all forms of nucleic acids. Virus replication-expression cycles include the simplest scheme that is conceivable in the RNA-protein world (positive-strand RNA viruses use the same molecule for replication and translation), the transition from RNA to DNA genomes (reverse-transcribing viruses and elements), and the ‘invention’ of large dsDNA genomes. Therefore, it is tempting to think of the virus world as the primordial genomic ‘laboratory’, perhaps the direct descendant of a pre-cellular stage of evolution [12]. However, the emergence of different classes of viruses through the ‘escape’ of different forms of host nucleic acids is conceptually plausible as well. After discussing the global ecology of viruses, we harness argument against the escape hypothesis.
Global ecology of viruses: a virus-dominated biosphere
With the exception of some intracellular bacterial parasites, viruses and/or diverse mobile elements are associated with all cellular life forms. Even in organisms that for a long time have been considered virus-free, focused efforts typically result in virus isolation, recent cases in point being the identification of RNA viruses in nematodes [15•] and ssDNA viruses in copepods [16•]. In organisms that so far evaded such effort, multiple virus-related, genome-integrated mobile elements are detectable.
Over the last decade, studies of the distinct environmental viromes produced a completely unexpected conclusion: viruses are the most abundant biological entities on earth. This conclusion was substantiated by direct counting of virus particles and cells in marine samples (the environment harboring most of the Earth's biomass). These analyses consistently detect a 10–100 excess of virus particles over cells [3•, 17].
Even these striking numbers appear to be underestimates because the existing particle counting techniques only apply to large dsDNA viruses, primarily bacteriophages. An alternative method of nucleic acids quantification in the viral particle fraction has shown that small ssDNA and RNA viruses are at least as abundant as the dsDNA viruses [18••]. Thus, the water in the ocean is literally a virus solution with up to 109 virions/ml. Similar studies performed in soil or animal guts yielded comparable results establishing viruses as the most common biological objects on the planet [2•, 19, 20•]. Accordingly, viruses are a major ecological and geochemical force [21, 22]. Indeed, virus killing of marine bacteria and protists largely determines the composition of the biota, provides a major source of organic matter for consumption by heterotrophic organisms, and also defines the formation of marine sediments through the deposition of skeletons of killed plankton organisms such as foraminifera and diatoms.
The genetic diversity of viruses is no less startling than their physical abundance. In each sequenced virome, the great majority of the sequences represent ‘dark matter’, that is, have no detectable homologs in the current databases, and there is no sign of saturation as sequencing progresses [4, 23, 24]. Although complete viral genomes typically contain a greater fraction of genes with homologs than viromes, these genomes also encompass numerous ORFans, that is, genes that are restricted to a narrow virus group [25]. Analysis of the phyletic spread of bacteriophage genes using the Phage Orthologous Groups (POGs) has shown that an overwhelming majority of the POGs include a small number of phages and viruses of archaea, whereas none are represented in the majority of the analyzed genomes (Figure 2 ) [26•]. This seemingly unlimited diversity of virus genes contrasts the case of cellular organisms that include a substantial core of common genes [27]. Although precise estimates of the total gene pools of viruses and cellular organisms require better sampling and more sophisticated models of evolution than those currently available, it is almost certain that a majority of the distinct genes in the biosphere reside in viral genomes. Thus, viruses are likely to represent the principal reservoir of genetic diversity on earth.
Figure 2.
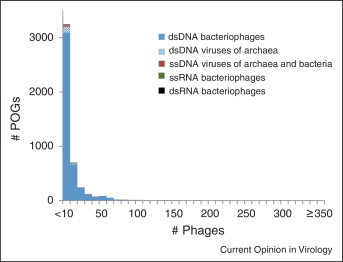
The size distribution of clusters of orthologous genes from prokaryotic viruses: the lack of a conserved core. POGs, Phage Orthologous Groups. Altogether, 487 genomes of bacteriophages and viruses of archaea were analyzed.
Modified, with permission, from Ref. [26•].
Viral hallmark genes and the spatial-temporal continuity of the virus world
There is no evidence of monophyly of all viruses in the traditional sense. Not a single gene is shared by all viruses, and as pointed out above, most viral genes are not wide spread and are poorly conserved. Given this background, it is all the more remarkable that a small set of genes are shared by a wide range of viruses that covers the entire diversity of genome strategies as well as the entire diversity of hosts (Figure 3 ) [12]. These ‘viral hallmark genes’ encode the essential viral functions including the capsid protein of icosahedral viruses, RNA and DNA polymerases, distinct helicases involved in genome replication, integrases that catalyze insertion of viral DNA into host genomes, and several others (Figure 3). Some of the smallest virus genomes (e.g. ssDNA parvoviruses or positive-strand RNA tombusviruses) consist mostly of the hallmark genes whereas in the largest viruses these genes comprise a minority. Moreover, some bacteriophages and archaeal viruses lack hallmark genes [12, 28], in some cases due to identifiable substitutions with functionally similar host-derived genes.
Figure 3.
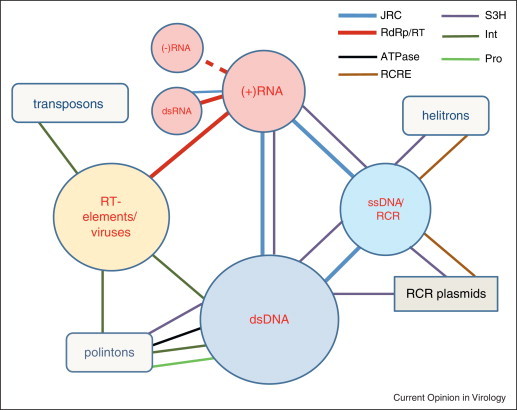
The network of hallmark genes connecting different classes of viruses and capsid-less selfish elements. The colored circles show classes of viruses and related capsid-less selfish elements (see Figure 1) and other shapes show distinct classes of non-viral selfish elements. The size of each shape roughly reflects the abundance and diversity of the respective class. The color-coded edges connecting the shapes denote shared hallmark genes; the thickness of each line roughly reflects the prevalence of the respective gene in the corresponding classes of viruses and selfish elements (in most cases, any given hallmark gene is present only in a subset of the class members). The dashed line reflects the tentative link between RNA-dependent RNA polymerases of positive-strand and negative-strand RNA viruses. JRC, Jelly Roll Capsid protein; RdRp, RNA-dependent RNA polymerase; RT, reverse transcriptase; S3H, superfamily 3 helicase; Int, integrase; ATPase, packaging ATPase of the FtsK family; Pro, C5-family thiol protease; RCRE, rolling circle replication (initiation) endonuclease. Compared with the original list of viral hallmark genes [12], integrase and thiol proteases were addionally included whereas several genes that are widespread among diverse dsDNA viruses but not found in other classes of selfish elements are not shown. The Int gene is present in numerous but not all prokaryotic and eukaryotic DNA transposons. Helitrons are eukaryotic transposons that replicate via the RCR mechanism. Polintons are large, self-replicating eukaryotic transposons.
The key observation on the viral hallmark genes is that they possess only distant homologs in cellular life forms (whenever close homologs of these genes are detected, they appear to be of viral origin) and yet form a network that connects almost the entire virus world (Figure 3). The parsimonious explanation of these findings appears to be that the hallmark genes became isolated from the cellular genomes at the earliest stages of evolution and ever since comprised the framework of the temporally and spatially continuous, expanding virus world. By contrast, the confinement of the hallmark genes to the virus world is hardly compatible with the escaped genes scenario. Furthermore, this ‘ancient virus world’ hypothesis [12] implies the primordial origin of diverse viral replication-expression strategies as opposed to the derivation of these strategies from elements of cellular information processing systems. Specifically, positive-strand RNA, retrotranscribing, ssDNA and dsDNA virus-like elements all can be inferred to have evolved within the primordial gene pool. The dsRNA and negative-strand RNA viruses that carry their replication machinery within the virions are likely to have evolved later. In particular, negative-strand RNA viruses probably originated in animals, with a subsequent horizontal transfer to plant hosts [29•].
The implications of this conclusion for the early evolution of life are far-reaching. Under this scenario, positive-strand RNA viruses are indeed direct descendants of the primordial RNA-protein world whereas the reverse-transcribing elements provide the means for the transition to the DNA world. The pre-cellular stage of evolution can be envisaged as a pool of small, virus-like genetic elements in which all the genomic strategies evolved and the separation between viruses and cellular life forms was precipitated by the gradual accretion of small dsDNA elements into large molecules (Figure 4 ) (the nature of the primordial compartmentalization is beyond the scope of this article; several models have been developed). The capsids might have evolved at this stage of evolution and would protect genomes of the primordial genetic elements and facilitate their dissemination [12]. This model goes beyond the notion that viruses coexisted with cells at all stages of evolution by suggesting that evolution of life actually started with a virus-like stage, with the advent of modern-type cells being a comparatively late event [30].
Figure 4.
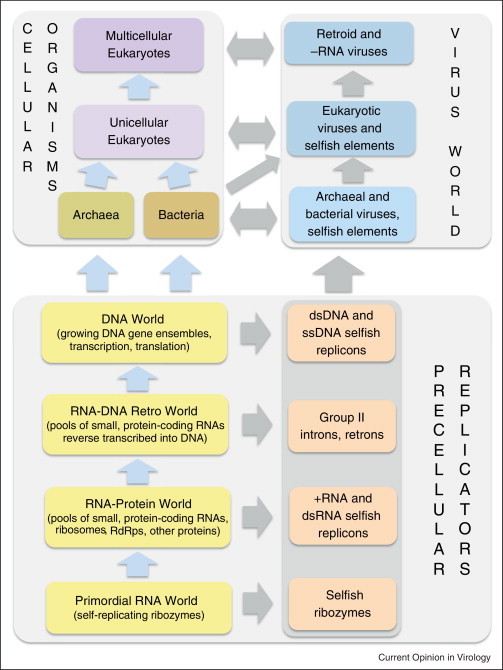
The model of the evolution of all extant life forms (top) from a virus-like primordial state (bottom).
Viruses and non-encapsidating selfish elements: not just capsid-encoding organisms
Viruses are traditionally conceived of as virions, particles with distinct, usually symmetrical architecture. The simplest virions are protein capsids that encase genomes. It has been argued that the fundamental divide among life forms lies between viruses, the capsid-encoding organisms, and cellular, ribosome-encoding organisms [31]. A related concept of the capsid as the viral ‘self’ is compatible with the extremely broad presence of the icosahedral virus capsid protein (the jelly roll fold) [32] in diverse viruses infecting bacteria, archaea, and eukaryotes although numerous distinct capsid proteins exist as well [33•]. Given the autonomy, vastness and diversity of the virus world, and its connectedness through the hallmark genes, the placement of the primary divide among life forms between viruses and cells appears unassailable. However, this division fails to give justice to the virus world that by no means is limited to typical capsid-encoding viruses. On the contrary, each of the major classes of viruses shows clear evolutionary connections to capsid-less genetic elements that are established through shared hallmark genes (and some additional genes) (Figure 3, Figure 4).
Apparently, evolution proceeded in both directions, from capsid-less elements to viruses and vice versa, on multiple occasions. The most prominent case of the former direction includes reverse-transcribing elements and viruses. In this class, bona fide infectious viruses (animal retroviruses and hepadnaviruses, and plant pararetroviruses) represent only one of the several branches that were identified by phylogenetic analysis of the reverse transcriptases, the signature gene for this class [34••]. Moreover, all prokaryotic reverse-transcribing elements are capsid-less, clearly indicating that retroviruses are derived forms. In addition, through the integrase, these elements are linked to the ubiquitous dsDNA transposons all of which are capsid-less. The rolling circle replicating ssDNA viruses show a clear-cut relationship with numerous plasmids that replicate via the same mechanism and with which they share two hallmark genes [35••]. In the case of plant geminiviruses, evidence has been presented in support of a particular direction of evolution: these viruses appear to have emerged via recombination between a bacterial plasmid and a plant RNA virus [36]. The opposite direction of evolution is exemplified by the positive-strand RNA viruses that gave rise to at least three distinct groups of capsid-less RNA elements, the narnaviruses, the endornaviruses and the hypoviruses [37].
Thus, the virus world is not limited to ‘capsid-encoding organisms’ but rather encompasses a panoply of diverse selfish genetic elements some of which do not possess a capsid. In full prudence, it would have been more appropriate to speak of the ‘world of diverse selfish genetic elements’ but for the sake of brevity and for historical reasons as well, we stick with the original ‘virus world’ designation [12]. Within the framework of the virus-like model of the primordial stage of life's evolution, capsid-less genetic elements most likely those capsid-encoding ones (Figure 4).
The incessant virus–host arms race and its role in cellular evolution
The history of life is a story of coevolution of selfish genetic elements and their cellular hosts. This coevolution is often described as arms race [7], and undoubtedly, this description reflects a key aspect of the interaction between the virus world and the cellular life forms. Indeed, all cellular organisms, with the exception of the most degraded intracellular parasitic bacteria, possess multilayered systems of antivirus defense, or more broadly defined, defense against invasion of foreign genetic material. Most if not all cellular organisms employ multiple principles of defense that include firstly, innate immunity, secondly, adaptive immunity, and thirdly, programmed cell death (induction of dormancy). Until recently, the two latter systems were considered eukaryotic innovations. However, the discovery of the prokaryotic system of heritable adaptive immunity, CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats [CRISPR] and CRISPR-Associated genes) [38•, 39] and the detailed characterization of the toxin–antitoxin (TA) systems that mediate cell death or induce dormancy [40, 41] have drastically transformed the entire concept of the evolution of antivirus defense. These findings show that all three major branches of antivirus defense are intrinsic to the survival of all cellular life forms and most likely emerged at the earliest stages of evolution (although innovation and diversification of these systems in multicellular eukaryotes are certainly spectacular). Moreover, comparative analysis of prokaryotic genomic loci encoding defense systems strongly suggests that the three branches of defense intimately interact, with cells ‘making decisions’ to proceed either via the route of active immune response or via the route of damage control by programmed cell death, depending on the genotoxic stress level [42, 43••]. Animal cells apparently face the same choices [44, 45••].
Genes encoding defense system components occupy up to 10% of prokaryotic genomes [43••], and even greater fractions of the eukaryotic protein-coding gene complements. Importantly, the defense genes themselves, especially in prokaryotes, show remarkable mobility and often possess properties of selfish genetic elements. The prokaryotic TA systems are typically encoded in small, compact operons that are transferred on plasmids and show addictive properties, that is, the toxin kills cells that lack the TA genes [46]. Restriction-modification systems, the principal innate immunity machinery in prokaryotes, similarly show mobile, selfish behavior [47]. Bacterial phage-defense islands, such as Staphylococcal pathogenicity islands (SaPIs), engage in a particularly striking form of selfish behavior. These defective prophage-like elements are excised from bacterial chromosmes, circularized and induced to replicate by helper phage infection with which they then interfere [48••, 49•].
The arms race certainly does not end with the host antivirus response: viruses have evolved a great variety of counter-defense measures that go far beyond simple evasion of the host defenses through fast mutation. Large viruses (e.g. dsDNA bacteriophages or animal poxviruses) encode multiple proteins that counteract immunity mechanisms or prevent programmed cell death [50, 51]. The repertoire of viral counter-defense proteins is constantly growing, and in large viruses, such genes appear to comprise the majority [52]. The study of the counter-defense, especially in viruses of prokaryotes, is only starting in earnest, and undoubtedly, numerous novel mechanisms that target specific defense systems of the host remain to be discovered [53•, 54••]. Remarkably, some bacteriophages incorporate host CRISPR-Cas systems and deploy them against other host phage-defense islands [55••]. Even small viruses often encode dedicated counter-defense genes, such as RNA interference suppressors that were identified in numerous plant RNA viruses [56, 57], whereas other small viruses encode dual function ‘security proteins’ that counteract programmed cell death [58]. Thus, the coevolution of multiple layers of defense and counter-defense is immanent to virus–host interaction and hence to life itself.
The arms race is not limited to virus-cell systems. Numerous viruses that parasitize other viruses have been discovered, and giant viruses host complex mobilomes that include small viruses (known as virophages), plasmid-like elements, transposons, and self-splicing introns [59, 60••, 61•].
A major consequence of the evolution of antivirus mechanisms is the subsequent recruitment of the respective genes for distinct cellular functions. In both prokaryotes and eukaryotes, such functions involve DNA repair that, similar to defense, employs various nucleases and helicases. Another striking example is the recruitment of CRISPR-Cas for control of bacterial gene expression [62••]. The extent of such conversion of defense systems in prokaryotes remains to be elucidated. In eukaryotes, conversion has taken truly dramatic forms. The quintessential eukaryotic regulation system, RNA interference, is thought to have evolved from an ancestral system of defense against RNA viruses [63]. Some of the key nucleases involved in RNA processing and degradation in eukaryotes evolved from prokaryotic toxin nucleases [64••, 65•]. Most of the proteins involved in DNA and protein (above all, histone) modification and chromatin remodeling in eukaryotes apparently evolved from prokaryotic ancestors that are involved in antivirus defense [66•]. Thus, the entire phenomenon of epigenetic inheritance that is central to the eukaryotic life appears to be a derivative of antivirus defense systems. Moreover, under a plausible evolutionary scenario, the nucleus itself evolved as a defense device against prokaryotic self-splicing introns that invaded the proto-eukaryote genome to later give rise to spliceosomal introns [67, 68]. In the same vein, the mechanism of programmed cell death that is essential for normal development in plants and animals [69] originally evolved as a distinct strategy of antivirus defense (see above) that already implies a primitive form of cell differentiation. As generalized by Aravind and colleagues [64••], “biological conflict systems served as evolutionary ‘nurseries’ for innovations in the protein world”, and such innovations were central to major evolutionary transitions, in particular eukaryogenesis.
Virus–host coevolution: extensive gene exchange and multifaceted cooperation
With all its intricacy and major biological consequences, the arms race is but one aspect of virus–host coevolution. The second side of the coevolution involves cooperation whereby viruses contribute to cellular functions whereas cellular genes are picked up by viruses and employed for counter-defense and other functions. Both processes are inherent to all virus–host systems. Although the virus world maintained its distinctness from the world of cellular organisms throughout the course of the evolution, the genetic exchange between the two parts of the biosphere always was extensive and highly consequential.
For temperate bacteriophages, genome integration into bacterial (and also archaeal) chromosomes is a regular phase of the life style and ‘domestication’ of phage genes accompanied by recruitment for various cellular functions appears to be a regular evolutionary process. One practically important phenomenon that critically depends on phage gene utilization is the evolution of bacterial pathogenicity [70, 71•].
Prophages routinely capture ‘normal’ bacterial genes and thus serve as vehicles for transduction, a major route of horizontal gene transfer among prokaryotes. A striking case in point is the phage-mediated transfer of photosystems among Cyanobacteria [72•]. Moreover, thanks to their fast evolution, phages provide the perfect media for functional innovation. Indeed, as pointed out above (see Figure 2), estimated sizes of prokaryotic and phage pan-genomes suggest that phages encompass the richest gene pool on earth and conceivably provide the principal reservoir of genetic novelty that is accessible to prokaryotes.
A striking case of prophage domestication is the evolution of gene transfer agents (GTAs), which are defective prophages that are present in diverse bacteria and archaea and encapsidate random fragments of prokaryotic chromosomes rather than the phage genome itself [73••]. The GTAs infect a broad range of prokaryotes and mediate gene transfer. The GTAs appear to have evolved into dedicated vehicles for HGT and could represent a major, still under-appreciated factor of evolution in prokaryotes.
The contributions of viruses and other selfish elements in the evolution of eukaryotes might be even more momentous than in prokaryotes. Notably, the introns, an intrinsic feature of eukaryotic gene architecture and an essential source of protein diversity in complex eukaryotes, are recognized to originate from self-splicing Group II introns of prokaryotes that are reverse-transcribing selfish elements [74, 75]. Moreover, the majority of the genomic DNA in many animals and plants (up to two-thirds in humans and 90% in maize) appears to derive from mobile elements, primarily retrotransposons [76, 77•]. Although most of these elements are non-functional, some are inevitably recruited for various roles, in particular regulatory ones, and given the extreme abundance of retroelements, the overall contribution of such recruitment appears to be quite substantial [9, 76]. Beyond these general trends, striking examples of key gene recruitment from selfish elements include the telomerase, an RT that is required from chromosome end replication in all eukaryotes and apparently was recruited from a bacterial Group II intron [34••, 78], and syncytins, membrane proteins that are essential for placental development in mammals and were derived from retrovirus envelope glycoprotein-encoding genes [79•].
The recruitment of host genes by viruses is as common as the recruitment of viral genes by cellular organisms. Far beyond the direct involvement of host derived genes in counter-defense (see above), the acquired genes also contribute to essential viral functions such as replication or protein processing, in many cases displacing ancestral viral hallmark genes with analogous activities [80•].
The theoretical inevitability of the virus–host differentiation, competition and cooperation
Viruses and other selfish elements are not simply ubiquitous companions of cellular life forms. Mathematical modeling of evolution shows that the emergence of genomic parasites is a fundamental property of any evolving replicator system that exceeds a certain threshold of minimal complexity [81]. In game theoretical terms, cheaters inevitably evolve as soon as there is a chance to cheat, that is, when functions evolve that can be utilized for replication in trans, such as a replicase [82, 83]. The extant analogies include the evolution of defective interfering particles that parasitize on many viruses after shedding the capsid protein genes and scavenging capsids from the host virus [84], and in vitro evolution experiments in which minimal viral genomes evolve that may not encode any proteins at all [85]. Thus, the evolution of any replicator system that exceeds minimal complexity is a story of host–parasite arms race and cooperation.
Such systems have been extensively studied in theoretical ecology and distinct evolutionary regimes and outcomes have been discovered. In a simple, unstructured host–parasite system, the inevitable outcome is stochastic extinction of both viruses and hosts. However, compartmentalization and, perhaps paradoxically, evolution of defense mechanisms stabilize the coevolving system as a whole [83]. Accordingly, cellular life forms evolved elaborate compartmentalization along with multiple defense strategies, embarking on the perennial arms race. Under this scenario, virus-like genomic parasites and the onset of the arms race far antedate the advent of modern-type cells and were key factors in the emergence of the cellular organization of life. Moreover, the first genomic parasites that consisted of RNA might have been the driving force behind the origin of DNA genomes because mathematical modeling suggests that dedicated information storage devices such as DNA can stabilize coevolution in a host–parasite system [86•].
The virocentric concept of the evolution of life
In the preceding sections we point out that viruses and virus-like selfish elements:
-
1.
Parasitize on all cellular life forms.
-
2.
Represent the most physically abundant and genetically diverse biological entities on Earth.
-
3.
Exploit all conceivable strategies of genome replication and expression in contrast with the single, universal strategy employed by cellular life forms.
-
4.
Form a coherent ‘virus world’ that is held together by a small set of virus hallmark genes that encode essential functions in a vast variety of viruses.
-
5.
Co-evolve with cellular hosts in an extremely complex process that combines an incessant arms race with various forms of cooperation.
Taken together, these features of the virus world translate into a ‘virocentric’ view of the history of life under which virus–host coevolution is the principal factor (or in the very least, one of the key factors) that defines the course of evolution of both cells and viruses. This coevolution in all likelihood has started within primitive replicator systems and was essential for the major evolutionary transitions such as the emergence of DNA genomes, cellular organization and later the eukaryotic cell.
Under the virocentric concept, the natural classification of life forms would necessarily include the primary divide between cells and selfish elements (viruses and capsid-less elements as well), the major classes of viruses being comparable in status with the three domains of cellular life (Figure 5 ) [87].
Figure 5.
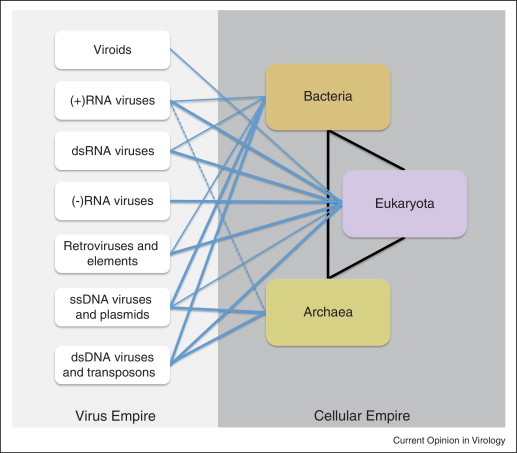
Classification of life forms: the viral and cellular empires. The lines denote parasite-host associations between selfish elements and cellular life forms as well as fluxes of genetic information. The thickness of the lines roughly reflects the prevalence of these associations and the intensity of the fluxes. The dashed line shows a putative RNA virus infecting a hyperthermophilic archaeon [88•].
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgement
EVK is supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine, NIH).
References
- 1.Bergh O., Borsheim K.Y., Bratbak G., Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 2•.Rosario K., Breitbart M. Exploring the viral world through metagenomics. Curr Opin Virol. 2011;1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]; An overview of the discovery of new viruses and virus–host associations through analysis of metagenomic data.
- 3•.Culley A.I. Virophages to viromes: a report from the frontier of viral oceanography. Curr Opin Virol. 2011;1:52–57. doi: 10.1016/j.coviro.2011.05.003. [DOI] [PubMed] [Google Scholar]; An overview of the discovery of novel marine viruses, including giant viruses and virophages, and their roles in the ecology of the oceans.
- 4.Rohwer F. Global phage diversity. Cell. 2003;113:141. doi: 10.1016/s0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 5.Colson P., de Lamballerie X., Fournous G., Raoult D. Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales. Intervirology. 2012;55:321–332. doi: 10.1159/000336562. [DOI] [PubMed] [Google Scholar]
- 6.Yutin N., Colson P., Raoult D., Koonin E.V. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol J. 2013;10:106. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forterre P., Prangishvili D. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann N Y Acad Sci. 2009;1178:65–77. doi: 10.1111/j.1749-6632.2009.04993.x. [DOI] [PubMed] [Google Scholar]
- 8.Daubin V., Ochman H. Start-up entities in the origin of new genes. Curr Opin Genet Dev. 2004;14:616–619. doi: 10.1016/j.gde.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Kazazian H.H., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 10.Cordaux R., Batzer M.A. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koonin E.V., Senkevich T.G., Dolja V.V. The ancient virus world and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores R., Ruiz-Ruiz S., Serra P. Viroids and hepatitis delta virus. Semin Liver Dis. 2012;32:201–210. doi: 10.1055/s-0032-1323624. [DOI] [PubMed] [Google Scholar]
- 14.Koonin E.V. Evolution of genome architecture. Int J Biochem Cell Biol. 2009;41:298–306. doi: 10.1016/j.biocel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Felix M.A., Ashe A., Piffaretti J. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first isolation of a virus from a classic model animal accompanied by the demonstration of the important role of RNAi in the antivirus defense of the nematode.
- 16•.Dunlap D.S., Ng T.F., Rosario K. Molecular and microscopic evidence of viruses in marine copepods. Proc Natl Acad Sci U S A. 2013;110:1375–1380. doi: 10.1073/pnas.1216595110. [DOI] [PMC free article] [PubMed] [Google Scholar]; The discovery of ssDNA viruses in copepods, the dominant members of the marine mesozooplankton and demonstration of the important contribution of virus infection to copepod mortality.
- 17.Zinger L., Gobet A., Pommier T. Two decades of describing the unseen majority of aquatic microbial diversity. Mol Ecol. 2012;21:1878–1896. doi: 10.1111/j.1365-294X.2011.05362.x. [DOI] [PubMed] [Google Scholar]
- 18••.Steward G.F., Culley A.I., Mueller J.A. Are we missing half of the viruses in the ocean? ISME J. 2013;7:672–679. doi: 10.1038/ismej.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]; Estimation of the virus content in marine samples by measuring the nucleic acid mass in the viral fraction shows that methods using fluorescence-based counting of virus particles yield substantial underestimates of virus abundance, presumably because they miss RNA viruses and ssDNA viruses that have small virions.
- 19.Srinivasiah S., Bhavsar J., Thapar K. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol. 2008;159:349–357. doi: 10.1016/j.resmic.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 20•.Minot S., Sinha R., Chen J. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of the diversity and variability of the human gut virome.
- 21.Suttle C.A. Marine viruses — major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 22.Rohwer F., Turber R.V. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 23.Angly F.E., Felts B., Breitbart M. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristensen D.M., Mushegian A.R., Dolja V.V., Koonin E.V. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin Y., Fischer D. Identification and investigation of ORFans in the viral world. BMC Genomics. 2008;9:24. doi: 10.1186/1471-2164-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Kristensen D.M., Waller A.S., Yamada T. Orthologous gene clusters and taxon signature genes for viruses of prokaryotes. J Bacteriol. 2013;195:941–950. doi: 10.1128/JB.01801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; Analysis of clusters of orthologous bacteriophage genes shows preponderance of rare genes, implying a vast phage pangenome.
- 27.Koonin E.V., Wolf Y.I. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prangishvili D., Garrett R.A., Koonin E.V. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 2006;117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 29•.Dolja V.V., Koonin E.V. Common origins and host-dependent diversity of plant and animal viromes. Curr Opin Virol. 2011;1:322–331. doi: 10.1016/j.coviro.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review article examines distinct evolutionary scenarios underlying the relationships between viruses that infect animals and plants including evolution from a common ancestral virus antedating the divergence of plants and animals; horizontal transfer of viruses, for example, by insect vectors; and, parallel origin of animal and plant viruses from related genetic elements. Comparative genomic evidence suggests that each of these scenarios was important in the evolution of different groups of viruses.
- 30.Koonin E.V. On the origin of cells and viruses: primordial virus world scenario. Ann N Y Acad Sci. 2009;1178:47–64. doi: 10.1111/j.1749-6632.2009.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raoult D., Forterre P. Redefining viruses: lessons from Mimivirus. Nat Rev Microbiol. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 32.Krupovic M., Bamford D.H. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat Rev Microbiol. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 33•.Krupovic M., Bamford D.H. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol. 2011;1:118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]; An overview of virion architectures with an emphasis on common structures among diverse viruses infecting hosts from all three domains of cellular life.
- 34••.Gladyshev E.A., Arkhipova I.R. A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci U S A. 2011;108:20311–20316. doi: 10.1073/pnas.1100266108. [DOI] [PMC free article] [PubMed] [Google Scholar]; A description of a novel group of reverse transcriptases, apparently recruited for unidentified cellular functions, and an updated phylogeny of retro-transcribing elements demonstrating that retroid viruses evolved from retrotransposons.
- 35••.Delwart E., Li L. Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 2012;164:114–121. doi: 10.1016/j.virusres.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; A review of the previously unsuspected, vast diversity of small ssDNA viruses that appear to infect all major groups of eukaryotes.
- 36.Krupovic M., Ravantti J.J., Bamford D.H. Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol Biol. 2009;9:112. doi: 10.1186/1471-2148-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolja V.V., Koonin E.V. Encyclopedia of Life Sciences. John Wiley & Sons Ltd.; Chichester: 2012. Capsid-less RNA viruses.http://www.els.net [Google Scholar]
- 38•.Makarova K.S., Haft D.H., Barrangou R. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]; A definitive description of the diversity of CRISPR-Cas system and their classification into three major types and 10 subtypes, with distinct signature genes and substantial differences in domain composition and molecular mechanisms.
- 39.Westra E.R., Swarts D.C., Staals R.H. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 40.Makarova K.S., Wolf Y.I., Koonin E.V. Comprehensive comparative-genomic analysis of type 2 toxin–antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blower T.R., Salmond G.P., Luisi B.F. Balancing at survival's edge: the structure and adaptive benefits of prokaryotic toxin–antitoxin partners. Curr Opin Struct Biol. 2011;21:109–118. doi: 10.1016/j.sbi.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Makarova K.S., Anantharaman V., Aravind L., Koonin E.V. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Makarova K.S., Wolf Y.I., Koonin E.V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review of the genomics of the prokaryotic defense systems that demonstrates the existence of a previously unsuspected diversity of defense mechanisms along with the integration of immunity and programmed cell death systems that implies intimate interactions between different defense strategies.
- 44.Guidotti L.G., Chisari F.V. To kill or to cure: options in host defense against viral infection. Curr Opin Immunol. 1996;8:478–483. doi: 10.1016/s0952-7915(96)80034-3. [DOI] [PubMed] [Google Scholar]
- 45••.Leonova K.I., Brodsky L., Lipchick B. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110:E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of the interplay between silencing and cell suicide in the response of mammalian cells to the expression of mobile elements.
- 46.Van Melderen L. Toxin–antitoxin systems: why so many, what for? Curr Opin Microbiol. 2010;13:781–785. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Ram G., Chen J., Kumar K. Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proc Natl Acad Sci U S A. 2012;109:16300–16305. doi: 10.1073/pnas.1204615109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents a dramatic demonstration of the selfish element-like behavior of bacterial antivirus defense systems, in particular Staphylococcal pathogenicity islands. These islands effectively are satellite phages that depend on virulent helper phages for their replication and inhibit the reproduction of the helper phages via several specific and non-specific mechanisms, thus rescuing the infected cell.
- 49•.Christie G.E., Dokland T. Pirates of the Caudovirales. Virology. 2012;434:210–221. doi: 10.1016/j.virol.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; An up to date review of satellite phages that exploit helper phages for their reproduction and in some cases defend the host cells from the helper phages. These phage ‘pirates’ also are vehicles for HGT.
- 50.Seet B.T., Johnston J.B., Brunetti C.R. Poxviruses and immune evasion. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 51.Martinez-Borra J., Gonzalez S., Lopez-Larrea C. The origin of the bacterial immune response. Adv Exp Med Biol. 2012;738:1–13. doi: 10.1007/978-1-4614-1680-7_1. [DOI] [PubMed] [Google Scholar]
- 52.Rus F., Morlock K., Silverman N. Characterization of poxvirus-encoded proteins that regulate innate immune signaling pathways. Methods Mol Biol. 2012;890:273–288. doi: 10.1007/978-1-61779-876-4_16. [DOI] [PubMed] [Google Scholar]
- 53•.Stern A., Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]; The coevolution of phages and bacterial host and in particular the Red Queen effect are considered to be the defining factors in the evolution of the prokaryotic world.
- 54••.Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes the discovery of bacteriophage genes that encode proteins specifically targeting and inactivating the host CRISPR-Cas systems.
- 55••.Seed K.D., Lazinski D.W., Calderwood S.B., Camilli A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]; A phage-encoded CRISPR/Cas system is shown to target a phage inhibitory chromosomal island of the bacterial host providing for a productive lytic infection.
- 56.Li F., Ding S.W. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burgyan J., Havelda Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011;16:265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Agol V.I., Gmyl A.P. Viral security proteins: counteracting host defences. Nat Rev Microbiol. 2010;8:867–878. doi: 10.1038/nrmicro2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desnues C., Boyer M., Raoult D. Sputnik, a virophage infecting the viral domain of life. Adv Virus Res. 2012;82:63–89. doi: 10.1016/B978-0-12-394621-8.00013-3. [DOI] [PubMed] [Google Scholar]
- 60••.Desnues C., La Scola B., Yutin N. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes the discovery of transpovirions, abundant, small linear plasmids associated with giant mimiviruses, and presents a comparative analysis of the diversified mobilome of giant viruses.
- 61•.Yutin N., Raoult D., Koonin E.V. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol J. 2013;10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive comparative genomic analysis reveals complex evolutionary relationships involving multiple gene exchanges between virophages that infect giant viruses, a class of eukaryotic self-replicating transposons (polintons/mavericks), transpovirons and viruses infecting eukaryotes and bacteria.
- 62••.Sampson T.R., Saroj S.D., Llewellyn A.C. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that, in addition to their defense function, at least some CRISPR-Cas systems can target and silence endogenous bacterial genes which in the case of the bacterium Francisella novicida studied in this work is required for pathogenicity.
- 63.Shabalina S.A., Koonin E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64••.Aravind L., Anantharaman V., Zhang D. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front Cell Infect Microbiol. 2012;2:89. doi: 10.3389/fcimb.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]; An up to date review of the numerous cases of recruitment of bacterial defense genes and viral counterdefense genes for diverse functions in eukaryotes. The authors convincingly argue that, due to their rapid evolution, the genes involved in biological conflicts comprise a unique reservoir of molecular innovations.
- 65•.Anantharaman V., Makarova K.S., Burroughs A.M. HEPN: a major nucleic-acid targeting domain involved in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive analysis of a vast, poorly characterized family of nculease domains that seem to play key roles in prokaryotic immunity and prgrammed cell death systems and have been recruited for non-defense functiosn, such as RNA processing, in eukaryotes.
- 66•.Iyer L.M., Abhiman S., Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]; This article shows that the great majority of eukaryotic chromatin-associated protein domains evolved from bacterial defense system components.
- 67.Martin W., Koonin E.V. Introns and the origin of nucleus-cytosol compartmentation. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 68.Lopez-Garcia P., Moreira D. Selective forces for the origin of the eukaryotic nucleus. Bioessays. 2006;28:525–533. doi: 10.1002/bies.20413. [DOI] [PubMed] [Google Scholar]
- 69.Baehrecke E.H. How death shapes life during development. Nat Rev Mol Cell Biol. 2002;3:779–787. doi: 10.1038/nrm931. [DOI] [PubMed] [Google Scholar]
- 70.Asadulghani M., Ogura Y., Ooka T. The defective prophage pool of Escherichia coli O157: prophage–prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 2009;5:e1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71•.Busby B., Kristensen D.M., Koonin E.V. Contribution of phage-derived genomic islands to the virulence of facultative bacterial pathogens. Environ Microbiol. 2013;15:307–312. doi: 10.1111/j.1462-2920.2012.02886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comparative genomic analysis shows that bacterial pathogens are substantially enriched in phage-derived DNA sequences compared to related non-pathogenic strains.
- 72•.Alperovitch-Lavy A., Sharon I., Rohwer F. Reconstructing a puzzle: existence of cyanophages containing both photosystem-I and photosystem-II gene suites inferred from oceanic metagenomic datasets. Environ Microbiol. 2011;13:24–32. doi: 10.1111/j.1462-2920.2010.02304.x. [DOI] [PubMed] [Google Scholar]; Extension of previous observations on cyanophage genomes showing that these phages can carry and transfer the complete repertoire of photosynthetic genes.
- 73••.Lang A.S., Zhaxybayeva O., Beatty J.T. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol. 2012;10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]; An up to date review of the gene transfer agents that are shown to be widespread in bacteria and archaea and appear to function as dedicated vehicles for HGT.
- 74.Rogozin I.B., Carmel L., Csuros M., Koonin E.V. Origin and evolution of spliceosomal introns. Biol Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koonin E.V., Csuros M., Rogozin I.B. Whence genes in pieces: reconstruction of the exon-intron gene structures of the last eukaryotic common ancestor and other ancestral eukaryotes. Wiley Interdiscip Rev RNA. 2013;4:93–105. doi: 10.1002/wrna.1143. [DOI] [PubMed] [Google Scholar]
- 76.Gogvadze E., Buzdin A. Retroelements and their impact on genome evolution and functioning. Cell Mol Life Sci. 2009;66:3727–3742. doi: 10.1007/s00018-009-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77•.Solyom S., Kazazian H.H., Jr. Mobile elements in the human genome: implications for disease. Genome Med. 2012;4:12. doi: 10.1186/gm311. [DOI] [PMC free article] [PubMed] [Google Scholar]; An up to date overview of the genomic content of mobile elements in animal genomes and their roles in evolution and disease.
- 78.Koonin E.V. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79•.Dupressoir A., Lavialle C., Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]; Discussion of the key role of retrovirus infection in the origin of placental mammals.
- 80•.Yutin N., Koonin E.V. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J. 2012;9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article demonstrates that evolution of large DNA viruses involved multiple displacements of ancestral viral genes with homologs from cellular hosts and other viruses.
- 81.Takeuchi N., Hogeweg P. Evolutionary dynamics of RNA-like replicator systems: a bioinformatic approach to the origin of life. Phys Life Rev. 2012;9:219–263. doi: 10.1016/j.plrev.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Szathmary E., Maynard Smith J. From replicators to reproducers: the first major transitions leading to life. J Theor Biol. 1997;187:555–571. doi: 10.1006/jtbi.1996.0389. [DOI] [PubMed] [Google Scholar]
- 83.Takeuchi N., Hogeweg P. Evolution of complexity in RNA-like replicator systems. Biol Direct. 2008;3:11. doi: 10.1186/1745-6150-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roux L., Simon A.E., Holland J.J. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oehlenschlager F., Eigen M. 30 years later — a new approach to Sol Spiegelman's and Leslie Orgel's in vitro evolutionary studies, Dedicated to Leslie Orgel on the occasion of his 70th birthday. Orig Life Evol Biosph. 1997;27:437–457. doi: 10.1023/a:1006501326129. [DOI] [PubMed] [Google Scholar]
- 86•.Takeuchi N., Hogeweg P., Koonin E.V. On the origin of DNA genomes: evolution of the division of labor between template and catalyst in model replicator systems. PLoS Comput Biol. 2011;7:e1002024. doi: 10.1371/journal.pcbi.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]; The model of evolution developed in this work implies that the evolution of DNA, as a dedicated information storage media, might have been driven by genomic parasites.
- 87.Koonin E.V. The two empires and three domains of life in the postgenomic age. Nat Educ. 2010;3:27. [Google Scholar]
- 88•.Bolduc B., Shaughnessy D.P., Wolf Y.I. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J Virol. 2012;86:5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first isolation of a putative RNA virus from an archaea-dominated thermal habitat.


