Abstract
Aflatoxin B1 (AFB1) is one of the major risk factors for liver cancer globally. A recent study showed that sulforaphane (SF), a potent inducer of phase II enzymes that occurs naturally in widely consumed vegetables, effectively induces hepatic glutathione S-transferases (GSTs) and reduces levels of hepatic AFB1-DNA adducts in AFB1-exposed Sprague Dawley rats. The present study characterized the effects of SF pre-treatment on global gene expression in the livers of similarly treated male rats. Combined treatment with AFB1 and SF caused reprogramming of a network of genes involved in signal transduction and transcription. Changes in gene regulation were observable 4 h after AFB1 administration in SF-pretreated animals and may reflect regeneration of cells in the wake of AFB1-induced hepatotoxicity. At 24 h after AFB1 administration, significant induction of genes that play roles in cellular lipid metabolism and acetyl-CoA biosynthesis was detected in SF-pretreated AFB1-dosed rats. Induction of this group of genes may indicate a metabolic shift toward glycolysis and fatty acid synthesis to generate and maintain pools of intermediate molecules required for tissue repair, cell growth and compensatory hepatic cell proliferation. Collectively, gene expression data from this study provide insights into molecular mechanisms underlying the protective effects of SF against AFB1 hepatotoxicity and hepatocarcinogenicity, in addition to the chemopreventive activity of this compound as a GST inducer.
Keywords: Aflatoxin B1, sulforaphane, chemoprevention, membrane biosynthesis, gene expression profiling
Introduction
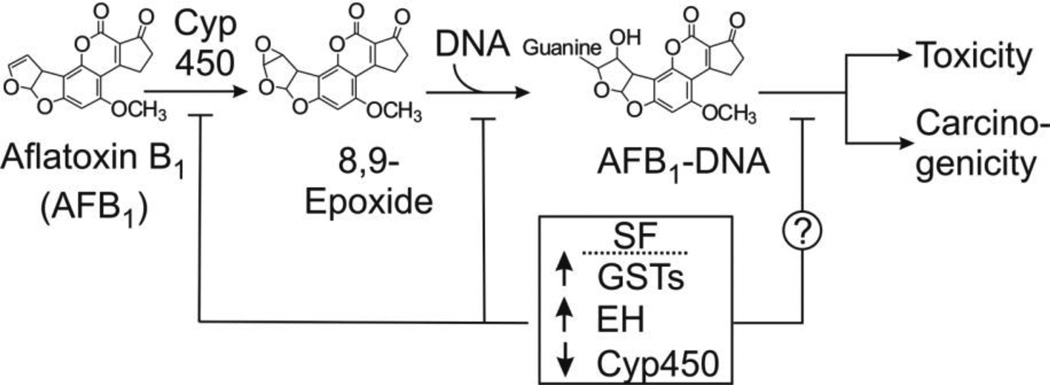
Aflatoxin B1 (AFB1) is a known human carcinogen that significantly contributes to the burden of hepatocellular carcinoma (HCC) in many parts of the world, especially in areas with a warm and moist climate such as Asia and sub-Saharan Africa (Kensler et al., 2011). A critical mechanism responsible for the hepatotoxic and carcinogenic potential of AFB1 is based on the balance of its bioactivation and detoxification (Figure 1). Several lines of evidence indicate that variation in the extent of glutathione (GSH) conjugation of the ultimate carcinogen, AFB1-8,9-epoxide, by glutathione S-transferases (GSTs) is an important detoxification pathway. Treatment of rats with oltipraz and 3H-1,2-dithiole-3-thione (D3T) leads to marked increases in the activity of liver GSTs, which resulted in reductions in both the extent of aflatoxin-DNA adduction and tumorigenesis (Kensler et al., 1987). The inducible A5 subunit of alpha-class GSTs in the rat has been identified as the GST isozyme that is primarily responsible for the enhanced detoxification of the AFB1-8,9-epoxide by chemopreventive agents (Hayes et al., 1998). Modulation of GST activity is only one mechanism by which exogenous agents can influence aflatoxin carcinogenesis. This paper probes additional pathways.
Figure 1.
Pathway of metabolic activation of aflatoxin B1 and disruption of the pathway by sulforaphane (SF). Aflatoxin is metabolized by cytochrome P450 (Cyp450) oxidases to the 8,9-epoxide, which reacts with DNA to form DNA adducts with guanine residues. The guanine adducts are presumably the functional precursors to carcinogenesis and significant contributors to toxicity. SF induces glutathione transferases (GSTs) and epoxide hydrolases (EH) and downregulates certain Cyp450s. Each of these alterations in enzyme activities would affect the intracellular concentration of the epoxide, hence modulating the levels of DNA adducts.
Sulforaphane (SF), a potent isothiocyanate derivative found in broccoli and other cruciferous vegetables (Fahey et al., 1997) has received attention as a chemopreventive agent due to its ability to activate the transcription factor Nrf2 and induce phase II detoxification enzymes, including the GSTs (Fimognari and Hrelia, 2007; Myzak and Dashwood, 2006; Thimmulappa et al., 2002). Recent evidence illustrates protective effects of SF against AFB1-induced hepatotoxicity in rats attributable to increased GST expression. Treatment of rats with SF hepatic total GST activity and a proportional reduction in the amounts of AFB1-N7-guanine (the principal DNA adduct of AFB1) formed in liver DNA (Fiala et al., 2011). Previous studies, however, have also identified additional chemopreventive mechanisms for SF that are independent of phase II enzyme induction. SF induces apoptosis in both in vitro (Carmela Fimognari et al., 2004; Karmakar et al., 2006) and in vivo models (Singh et al., 2004). SF-mediated cell cycle arrest has been reported in many previous studies, including induction of a dose-dependent growth arrest in prostate cancer cells by inhibiting the expression of cyclin D1 and DNA synthesis, along with a G1 cell cycle block (Chiao et al., 2002) and induction of G2/M accumulation and pre-metaphase arrest in bovine aortic endothelial (BAE) cells (Jackson et al., 2007). SF also exerts anti-inflammatory properties by inhibiting pro-inflammatory and pro-carcinogenic signaling factors such as IL-1β (Lin et al., 2008), COX-2 and TNF-α (Heiss et al., 2001). Understanding these phase II-independent pathways provides the rationale for the current project.
Cell regeneration and survival responses signal metabolic reprogramming that supports anabolic pathways required for tissue repair and growth (Ward and Thompson, 2012). The Keap1-Nrf2 complex, which is activated by SF, has been demonstrated to influence intermediary metabolism (Hayes and Dinkova-Kostova, 2014). This study aimed to assess the extent to which anabolic pathways modulated by SF provide protective mechanisms against AFB1 toxicity in vivo. The results revealed prominent reprogramming of gene sets involved in lipid synthesis in SF-pretreated rats, suggesting that SF facilitates regeneration of hepatic cells damaged by AFB1.
Materials and methods
Chemicals
R,S-Sulforaphane (SF) was purchased from LKT Laboratories (St. Paul, MN). Aflatoxin B1 (AFB1) was purchased from Sigma Chemical (St. Louis, MO). RNAlater, RNeasy Mini Kit, and One-Step QuantiTech SYBR Green RT-PCR were obtained from QIAGEN (Valencia, CA). One-Cycle Target Labeling and Control Reagents complete kit (P/N 900493) and GeneChip® Rat Genome 230 2.0 arrays (Rat 230 2.0) were obtained from Affymetrix, Inc. (Santa Clara, CA). R-Phycoerythrinstreptavidin (SAPE) was purchased from Molecular Probes (Eugene, OR) and biotinylated, anti-streptavidin goat antibody was purchased from Vector Laboratories (Burlingame, CA). Reagent grade goat IgG was purchased from Sigma-Aldrich (St. Louis, MO). Nuclease-free water was obtained from Ambion (Austin, TX). DNA primers for real time RT-PCR were ordered from Integrated DNA Technologies (Coralville, IA). Unless otherwise noted, all other chemicals and reagents were of ACS grade or better.
Animals
Male Sprague Dawley rats (21 days old; Charles River, Wilmington, MA) were fed the AIN76A diet (TestDiet, Richmond, IN) for one week prior to the start of the experiment. They were housed individually in facilities maintained at standard relative humidity and temperature, and 12 h: 12 h light: dark conditions, with food and water available ad libitum. All procedures involving animals followed NIH guidelines and protocols approved by the Massachusetts Institute of Technology Committee on Animal Care.
Treatment Protocol
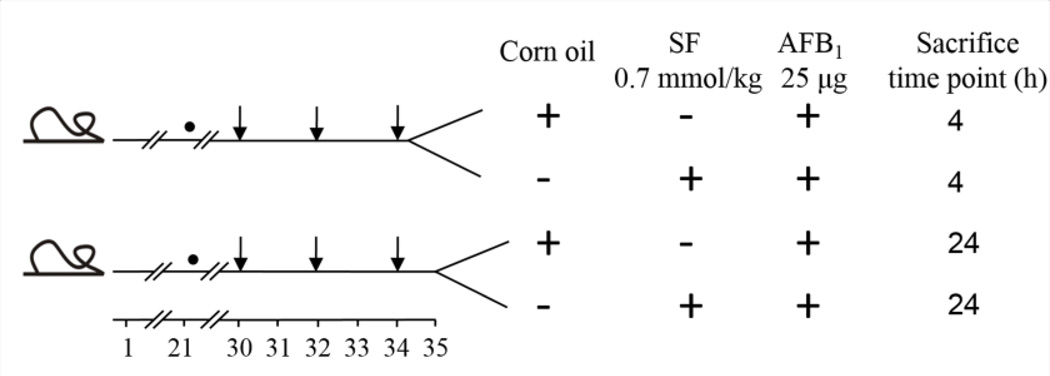
Treatment groups consisted of 12 male Sprague Dawley rats randomly assigned into four groups of three. Rats were gavaged with corn oil or with 0.7 mmol/kg SF dissolved in corn oil at 30, 32, and 34 days of age (Figure 2). A previously performed dose-response study in male rats revealed that 0.7 mmol/kg SF was optimal for GST induction (Fiala et al., 2011). Twenty-four h after the third dose, animals in each group were injected intraperitoneally with 25 µg AFB1 in DMSO. All animals were sacrificed at 4 h or 24 h after AFB1 administration. This experimental protocol is schematically illustrated in Figure 2. Livers were collected and submerged in RNAlater for further gene expression profiling by microarray and for validating the microarray results by RT-PCR.
Figure 2.
Experimental protocol. Effects of sulforaphane on transcriptional responses were evaluated in the livers of male Sprague-Dawley rats. ● Indicates start of AIN76A diet; ↓ Indicates gavage with sulforaphane or corn oil. Each group consisted of 3 animals.
RNA isolation procedure
Total RNA was isolated from RNAlater-stabilized rat livers using QIAshredder homogenization and RNeasy® Mini Kit from QIAGEN (Valencia, CA). Total RNA was isolated and purified by a procedure performed as described in QIAGEN’s RNeasy® Animal Tissues protocol. The isolation method included a DNase digestion step according to the manufacturer’s instructions. Concentration and purity of isolated total RNA were preliminarily verified by checking the absorbance at 260 nm and 280 nm using a DU 730 UV/Vis spectrophotometer (Beckman Coulter, CA, USA). The integrity of total RNA was assessed qualitatively by gel analysis using RNA 6000 Nano chips on an Agilent 2001 (Agilent technologies, Berlin, Germany). RNA samples were used for gene expression analysis by microarray and RT-PCR.
Microarray preparation procedure
All procedures were performed as described in detail in the Affymetrix GeneChip® Expression Analysis Technical Manual. Biotin-labeled cRNA samples for hybridization on GeneChip® Rat 230 2.0 arrays were prepared using the “One-Cycle Target Labeling and Control Reagents complete kit.” A set of poly-A controls supplied in the kit was used as a positive control to monitor the entire target labeling process. Total RNA (15 µg) was reverse transcribed using a T7-oligo(dT) Promoter Primer in the first-strand cDNA synthesis reaction. Following this step, the process of RNase H-mediated second-strand cDNA synthesis was performed. The double-stranded cDNA was purified according to the Sample Cleanup Module protocol supplied with the kit and served as a template in the subsequent in vitro transcription (IVT) reaction. The IVT reaction was performed in the presence of T7 RNA Polymerase and a biotinylated nucleotide analog/ribonucleotide mix for complementary RNA (cRNA) amplification and biotin labeling. The biotinylated cRNA was spectrophotometrically quantified prior to purifification and fragmentation in buffer supplied with the Sample Cleanup Module. Subsequently, the purified fragmented cRNA samples were hybridized to GeneChip® Rat 230 2.0 arrays (Rat 230 2.0) for 16 h at 45 °C in an Affymetrix Hybridization Oven 640. The microarrays were washed and stained with streptavidin-phycoerythrin (SAPE) on an Affymetrix Fluidics Station 450. A signal amplification step was included according to the manufacturer’s instructions. Fluorescent images were read using an Affymetrix®GeneChip®Scanner 3000.
Microarray data analysis
Raw data image files (DAT) were converted into CEL files using the Affymetrix Microarray Suite (MAS) 5.0. Probe-level intensity measurements were normalized using the Robust Multi-Array Average (RMA) method (Irizarry et al. 2003). The normalized signal intensities for each gene were averaged across three independent biological replicates (three rats for each experimental time point group). Differential expression was calculated using Spotfire DecisionSite® (TIBCO Software Inc., Palo Alto, CA). A significant difference in the expression of genes in the livers between SF-pretreated, AFB1-exposed rats and the corresponding non-SF pretreated, AFB1-exposed rats time-matched control groups was defined as the point at which the average fold change was greater than ±1.5 and the P-value was less than 0.05 (t-test). Spotfire was also used for visualization purposes, hierarchical clustering, and for searching the genes among categories. The WEB-based GEne SeT AnaLysis Toolkit (WebGestalt), which is available at http://bioinfo.vanderbilt.edu/webgestalt/, was used for statistical enrichment analyses of differentially expressed genes (Zhang et al., 2004, 2005). WebGestalt was used to analyze probe sets with ±1.5 fold or greater differential expression (P< 0.05) in the livers between SF-pretreated, AFB1-exposed rats and the corresponding non-SF pretreated, AFB1-exposed rats time-matched control groups against all probe sets available on the GeneChip® Rat 230 2.0 array. Gene ontology categories were considered statistically enriched when the P-value was 0.01 or less. Cytoscape, an open source bioinformatics software platform, was used for analyzing the gene-encoded protein interactions that were statistically deregulated in this study (Cline et al., 2007). GenMapp (Gene Map Annotator and Pathway Profiler), the open source bioinformatics software that is available on www.GenMapp.org (Gladstone Institutes, University of California at San Francisco), was used to visualize gene expression data on maps representing biological pathways.
Quantitative real-time RT-PCR for microarray results validation
Six to ten genes from each experimental group under study were selected as targets for real-time RT-PCR to validate the microarray results. All primer sets were designed using Primer3 (Rosen and Skaletsky, 2000), which is available at http://www-genome.wi.mit.edu/cgibin/primer/primer3.cgi/. The specificity of each primer was checked by BLAST searches. The oligonucleotide sequences of forward and reverse primers used for microarray results validation by RT-PCR in experimental groups of animals that were sacrificed at 4 h and at 24 h time points are shown in Tables 1 and 2, respectively. Reverse transcription and PCR were performed following the manufacturer’s instructions using One-Step QuantiTech SYBR Green RT-PCR. Reverse transcription and PCR amplification were performed on a DNA Engine Opticon Thermal Cycler (MJ research, Waltham, MA). Data were analyzed using Opticon Monitor Software Cycler (MJ research, Waltham, MA). The background fluorescence was subtracted using the global minimum function available in the software. A standard dilution of a control sample was used to generate the standard curve. Melting curve analysis was used for quality control and data from samples with outlier melting temperatures were not used. As an endogenous control, the mRNA level of the housekeeping enzyme GAPDH was used to normalize the mRNA level reported for each gene.
Table 1.
Oligonucleotide sequences of the forward and reverse RT-PCR primers used to validate microarray results. Rats were sacrificed 4 h after AFB1 administration.
| Gene | RefSeq ID | Oligonucleotide sequence | |
|---|---|---|---|
| Kng1 | NM_001009628 | Forward Primer Reverse Primer |
5' actgcagagtccccacaaac 3' 5' cgtgtgaagcagcatcctta 3' |
| Abcb1a | NM_133401 | Forward Primer Reverse Primer |
5' tgtaatgccagggctagagg 3' 5' tgaacctggagctccctatg 3' |
| Nmnat3 | NM_001013224 | Forward Primer Reverse Primer |
5' atgcatatgggagggacatc 3' 5' agagcatttgctgctcttctg 3' |
| Gpt2 | NM_001012057 | Forward Primer Reverse Primer |
5' aggcagctcagtcccataaa 3' 5' gtaggtgccttctcgctgtc 3' |
| Foxa3 | NM_017077 | Forward Primer Reverse Primer |
5'gcctagggttggggtattgt 3' 5' attgccttcacggtatgtcc 3' |
| Crat | NM_001004085 | Forward Primer Reverse Primer |
5' gatgtgaatcatggggaagg 3' 5' acagattctggcttggctgt 3' |
| Gapdh | NM_017008 | Forward Primer Reverse Primer |
5' gtgccaaaagggtcatcat 3' 5' ccacagtcttctgagtggca 3' |
Table 2.
Oligonucleotide sequences of the forward and reverse RT-PCR primers used to validate microarray results. Rats were sacrificed 24 h after AFB1 administration.
| Gene | RefSeq ID | Oligonucleotide sequence | |
|---|---|---|---|
| Srebf1 | XM_213329 | Forward Primer Reverse Primer |
5' gccaccctcttgctctgtag 3' 5' gggtgagagccttgagacag 3' |
| Fasn | NM_017332 | Forward Primer Reverse Primer |
5' agtcaccctgaaggcatcac 3' 5' caccaactgccaggtagtca 3' |
| Acly | NM_016987 | Forward Primer Reverse Primer |
5' acccttagccatcgtcacac 3' 5' gctttgcggtttccatttta 3' |
| Elovl6 | NM_134383 | Forward Primer Reverse Primer |
5' cccgtttgacctcacacata 3' 5' cagaggctgcagaaacacag 3' |
| Me3 | XM_341880 | Forward Primer Reverse Primer |
5' tggccttgtcttctccactt 3' 5' tcaagggaagtctggtcctg 3' |
| Pklr | NM_012624 | Forward Primer Reverse Primer |
5' actgctctgctgggtcagat 3' 5' ttctgggattctttgcctgt 3' |
| Mdh1 | NM_033235 | Forward Primer Reverse Primer |
5' tcatgaagccctgtgttcaa 3' 5' ggattggccatgaactcact 3' |
| Crat | NM_001004085 | Forward Primer Reverse Primer |
5' gatgtgaatcatggggaagg 3' 5' acagattctggcttggctgt 3' |
| Cpt1a | NM_031559 | Forward Primer Reverse Primer |
5' ctgactctcgctgctgtgac 3' 5' tgcattggtaactgctcagg 3' |
| Acf | NM_133400 | Forward Primer Reverse Primer |
5' ataactggggacagccagtg 3' 5' tagctttgggggtgtgaaag 3' |
| Gapdh | NM_017008 | Forward Primer Reverse Primer |
5' gtgccaaaagggtcatcat 3' 5' ccacagtcttctgagtggca 3' |
Statistical analysis
Gene expression changes are presented as the average fold change between SF-pretreated, AFB1-exposed rats and the corresponding non-SF pretreated, AFB1-exposed rats time-matched control groups. Differences between various groups were determined using a two sample t-test, with statistical significance set at P<0.05.
Results
Gene expression analysis
The goal of this study was to compare the response of rat liver to aflatoxin alone, or aflatoxin in concert with SF. The transcriptional responses in the livers of AFB1-treated male rats induced by SF were investigated using a DNA microarray, an established tool to assess global gene expression changes. Groups of rats were pretreated with 0.7 mmol/kg SF by gavage at ages 30, 32, and 34 days, a dose previously shown to induce GSTs significantly and result in a substantial reduction of AFB1-N7-guanine adduct levels in the livers of male rats (Fiala et al., 2011). Control animals received corn oil at a comparable volume. On day 35, 25 µg of AFB1 was given to all animals by intraperitoneal injection. This dose has previously been utilized in a multi-dose regimen to induce liver tumor formation in male Fischer rats (Egner et al., 1995). Livers from three animals per time point (4 h and 24 h) after AFB1 dosing were collected to investigate the response to SF treatment at the transcriptional level. Liver RNA was isolated as described, and subjected to microarray hybridization with arrays that contained 31,100 probe sets representing over 28,000 well-substantiated rat genes. Data were normalized using the Robust-Multi-Chip Average method. The deregulated genes were selected with a requirement of at least 1.5-fold change with respect to the mean intensity of the control groups. The numbers of genes are given both as probe sets and as non-redundant genes to account for the fact that one gene can be represented by several probe sets on the arrays. The number of probe sets and the corresponding numbers of up- and down-regulated genes obtained by these analyses are given in Table 3. At 4 h, 81 probe sets representing 78 non-redundant genes were significantly deregulated, with 53 genes up-regulated and 25 genes down-regulated. At 24 h, 110 probe sets representing 97 non-redundant genes were shown to be significantly deregulated; 51 genes upregulated and 46 genes downregulated.
Table 3.
Genes deregulated by SF in liver of AFB1-treated rats compared to the time-matched AFB1-treated controls.
| Parameter | 4 h group | 24 h group |
|---|---|---|
| Probe sets | 81 | 110 |
| Non-redundant genes | 78 | 97 |
| Up-regulated | 53 | 51 |
| Down-regulated | 25 | 46 |
Microarray validation
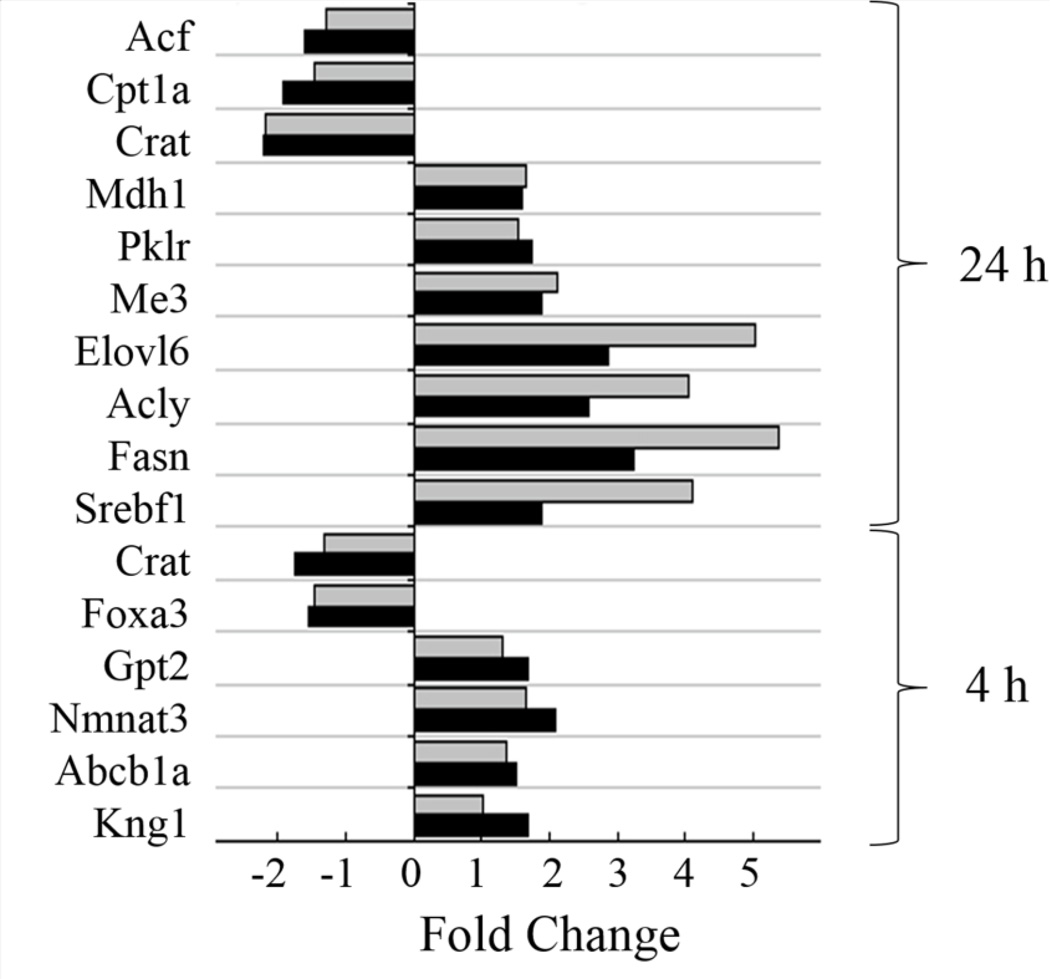
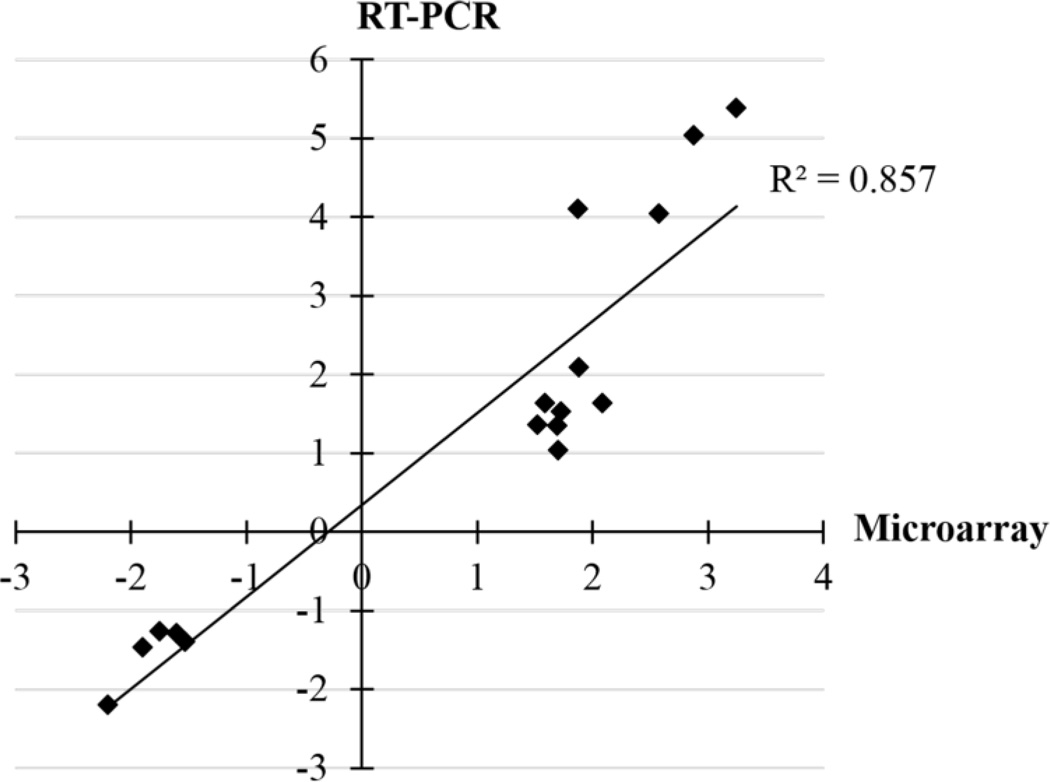
Six to ten genes from each experimental group were selected as targets for real-time RT-PCR to validate the microarray results. The mean fold-change of all samples from identically SF- and AFB1-treated animals versus time-matched AFB1-treated controls were compared between microarray and real-time RT-PCR results. As shown in Figure 3, the RT-PCR analysis corroborated the microarray results. A regression coefficient (Rs) of 0.926 and an R2 of 0.857 between the fold changes obtained from the two techniques confirmed that the gene expression changes detected by microarray represented actual deregulations (Figure 4).
Figure 3.
Validation of gene expression results from microarray by using RT-PCR. Each bar is the fold induction of the selected genes obtained by microarray (black bars) or RTPCR (gray bars).
Figure 4.
Correlation of fold changes obtained from microarray and RT-PCR analyses. The mean of fold change of all samples from identically SF and AFB1-treated animals versus time-matched AFB1-treated controls analyzed by microarray and real-time RT-PCR were plotted.
Functional analysis
To identify the pathways affected by SF in livers of AFB1-treated rats at 4 h and 24 h, each significantly deregulated gene was categorized according to its best known biological function. All recent gene annotations and information were obtained from NetAffx (Affymetrix, Inc.), the National Center for Biotechnology Information (NCBI) and Spotfire. It should be noted that gene categorization is imperfect as some genes are representative of more than one category. Numbers of significantly deregulated non-redundant genes assigned to their biological function categories at 4 h and 24 h are shown in Figures 5 and 6, respectively. Data of Expressed Sequence Tags (ESTs) and genes whose biological functions are not known or could not be specified with reasonable certainty were excluded.
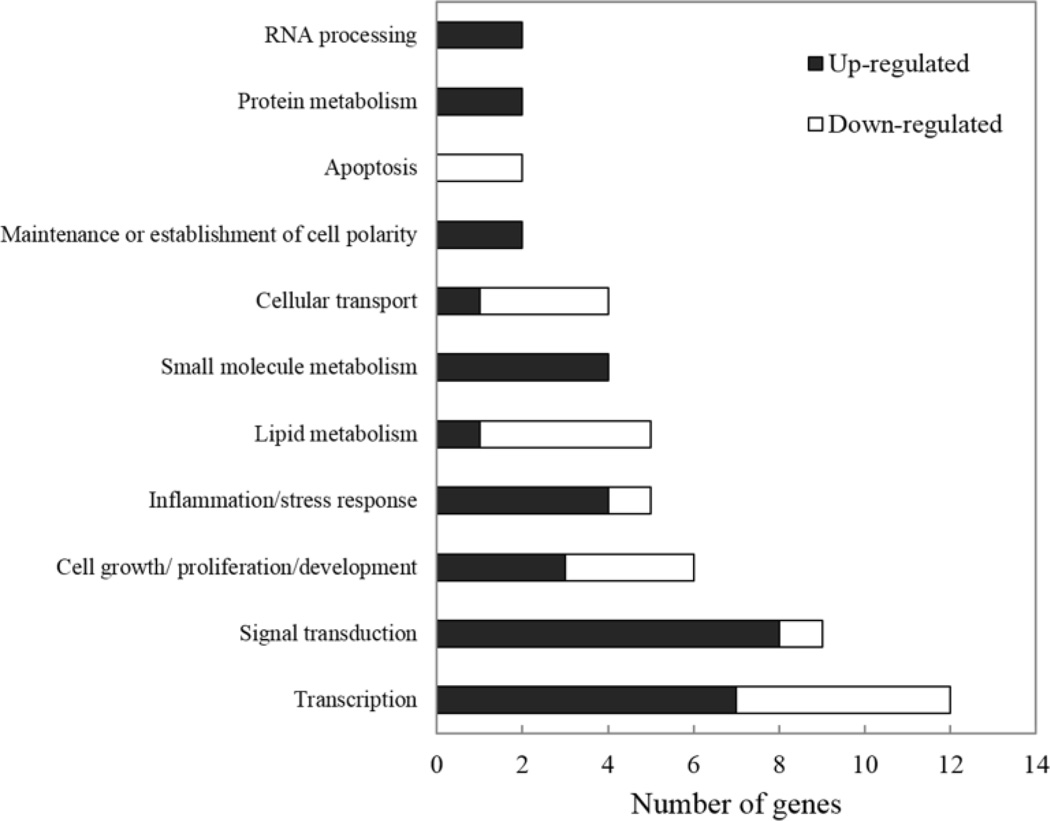
Figure 5.
Numbers of significant deregulated genes observed at 4 h in SF and AFB1-treated animals compared to the time-matched AFB1-treated controls which were assigned to their biological function categories.
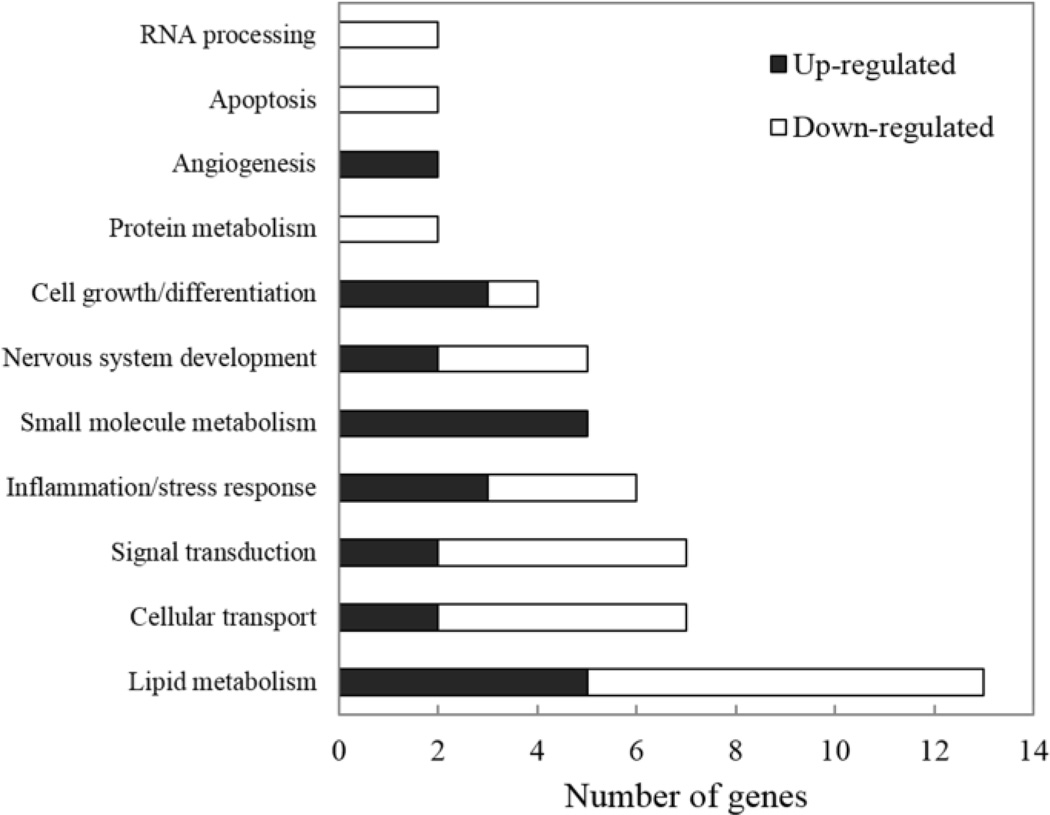
Figure 6.
Numbers of significant deregulated genes observed at 24 h in SF and AFB1-treated animals compared to the time-matched AFB1-treated controls which were assigned to their biological function categories.
The results showed that most of the genes deregulated at 4 h are involved in “Transcription”, whereas most of those deregulated at 24 h are involved in “Lipid metabolism”. Among the several functional groups classified, however, only three gene ontology categories related to biological processes were relatively enriched in the 4 h group. Table 4 shows gene ontology enrichment observed in the 4 h animals. The most highly enriched category in this experimental group was “Establishment and/or maintenance of cell polarity” (R=20; P=0.0045).
Table 4.
Gene ontology showing SF enhanced gene classes in rats sacrificed 4 h after treatment with AFB1.
| Gene ontology Category |
Gene ontology number |
O | E | R | P |
|---|---|---|---|---|---|
| Establishment or maintenance of cell polarity | GO:0007163 | 2 | 0.1 | 20.0 | 0.0045 |
| Positive regulation of development | GO:0010720 | 3 | 0.3 | 9.7 | 0.0034 |
| Transcription, DNA-dependent | GO:0006351 | 11 | 4.5 | 2.5 | 0.0031 |
O:Observed gene number in the GO category; E:Expected gene number in the GO category; R:Ratio of enrichment for the GO category; P:Significance of enrichment for the GO category
Cell polarity is an essential feature of many animal cells; during cell division, the mitotic spindle aligns to the axis of cell polarity to correct the positioning of daughter cells and ensure that the localized molecules are properly distributed to the daughter cells. Within this category, two genes were significantly up-regulated, namely Discs large homolog-1 (Dlgh1) and Protein kinase C, iota (PKCζ). These two genes also play a role in controlling cell growth and survival. Dlgh1 is a membrane-associated guanylate kinase that functions as a molecular scaffold for the organization of signal tranducers within cells. It plays a key role in the regulation of T cell function including activation of the mitogen-activated protein (MAP) kinase p38 (Round et al., 2007; Zanin-Zhorov et al., 2012). Cytokines and chemokines expressed by T cells have crucial roles in liver regeneration and tumor promotion (Grivennikov et al., 2010). The Dlgh1 protein can also function as a growth regulator through interaction with the carboxyl-end of the tumor suppressor adenomatous polyposis coli (APC) protein (Ishidate et al., 2000). PKCζ is a member of the atypical PKC group that is an important regulator of inflammatory and survival pathways. It acts as a positive regulator of NF-κB (Diaz-Meco and Moscat, 2012) which plays an important role in the innate immune response, inflammation and cell survival (Sun and Karin, 2008; Karin et al., 2002). The second highest enriched category was “Positive regulation of development” (R=9.7; P=0.0034) with the other enriched category being “Transcription, DNA-dependent” (R=2.5; P=0.0031). The list of deregulated genes in each enriched category in the 4 h experimental group is shown in Table 5.
Table 5.
List of SF-modulated genes in each enriched category in rats sacrificed 4 h after treatment with AFB1
| RefSeq ID | Gene | Description | Fold Change |
|---|---|---|---|
| Establishment and/or maintenance of cell polarity | |||
| XM_342223 | Prkci | Protein kinase C, iota | 1.58 |
| NM_012788 | Dlgh1 | Discs, large homolog 1 (Drosophila) | 1.52 |
| Positive regulation of development | |||
| NM_012788 | Dlgh1 | Discs, large homolog 1 (Drosophila) | 1.52 |
| XM_226988 | Fndc3b | Fibronectin type III domain containing 3B | 1.77 |
| NM_181086 | Tnfrsf12a | Tumor necrosis factor receptor superfamily, member 12a | −2.06 |
| Transcription, DNA-dependent | |||
| NM_022184 | Cask | Calcium/calmodulin-dependent serine protein kinase (MAGUK family) | 1.78 |
| NM_001014172 | Rsrc1 | Arginine/serine-rich coiled-coil 1 | 1.74 |
| NM_012988 | Nfia | Nuclear factor I/A | 1.62 |
| NM_153472 | Mnat1 | Menage a trois 1 | 1.57 |
| NM_021742 | Nr5a2 | Nuclear receptor subfamily 5, group A, member 2 | 1.54 |
| XM_215922 | Mybl2 | Myeloblastosis oncogene-like 2 | −1.52 |
| NM_017077 | Foxa3 | Forkhead box A3 | −1.53 |
| NM_057211 | Klf9 | Kruppel-like factor 9 | −1.61 |
| NM_001011930 | Trip13 | Thyroid hormone receptor interactor 13 | −1.63 |
| XM_218336 | Erf | Ets2 repressor factor | −1.76 |
| NM_031800 | Dedd | Death effector domain-containing | −1.61 |
Among several functional groups of genes classified in the 24 h group, there are eight gene ontology categories in biological processes that were shown to be relatively enriched (Table 6).
Table 6.
Gene ontology showing SF enhanced gene classes in rats sacrificed 24 h after treatment with AFB1.
| Gene ontology category |
Gene ontology number |
O | E | R | P |
|---|---|---|---|---|---|
| Acetyl-CoA biosynthesis | GO:0006085 | 2 | 0.04 | 50.00 | 0.0007 |
| Iron ion homeostasis | GO:0055072 | 2 | 0.12 | 16.67 | 0.0059 |
| Fatty acid metabolism | GO:0006631 | 5 | 1.05 | 4.76 | 0.0037 |
| Lipid biosynthesis | GO:0008610 | 5 | 1.21 | 4.13 | 0.0068 |
| Cellular lipid metabolism | GO:0044255 | 9 | 2.64 | 3.41 | 0.0010 |
| Organic acid metabolism | GO:0006082 | 9 | 2.77 | 3.25 | 0.0015 |
| Carboxylic acid metabolism | GO:0019752 | 9 | 2.75 | 3.27 | 0.0014 |
| Lipid metabolism | GO:0006629 | 10 | 3.21 | 3.12 | 0.0011 |
O:Observed gene number in the GO category; E:Expected gene number in the GO category; R:Ratio of enrichment for the GO category; P:Significance of enrichment for the GO category
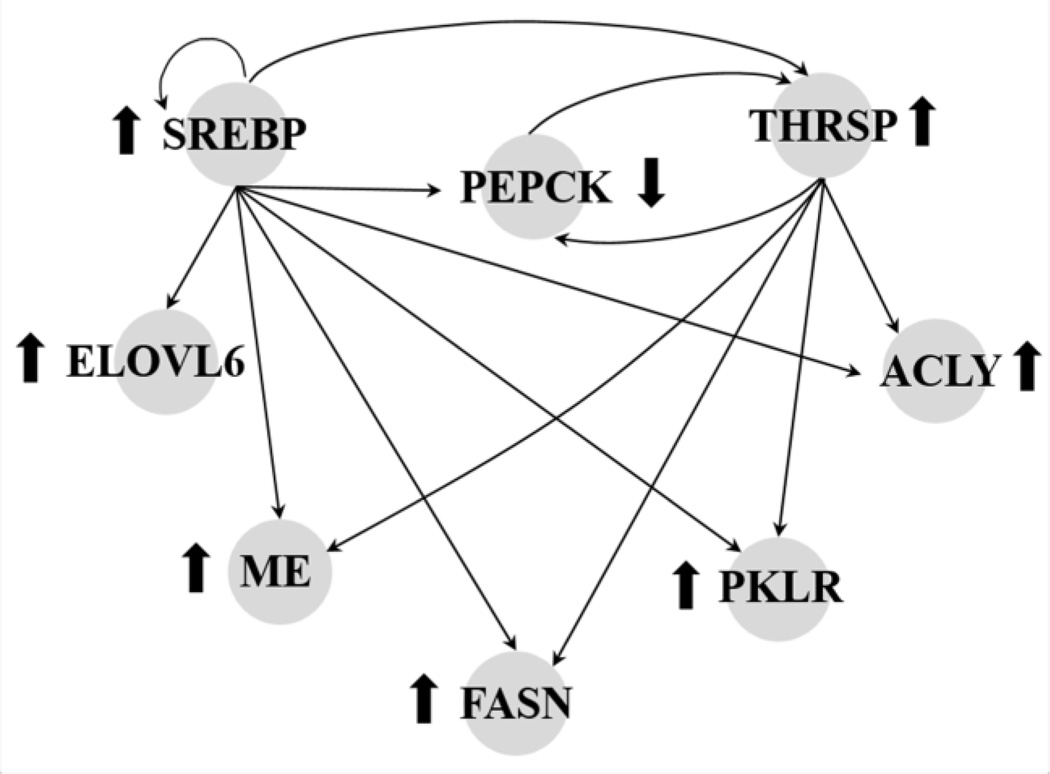
The most enriched of these was “Acetyl-CoA biosynthesis” (R= 50; P=0.0007), which contained two up-regulated genes, ATP citrate lyase (Acly) and dihydrolipoamide S-acetyltransferase (Dlat). The second most enriched category was “Iron ion homeostasis” (R=16.67; P=0.0059), which contained one highly up-regulated gene, Transferrin receptor (Tfrc), and one markedly down-regulated gene, Hepcidin antimicrobial peptide (Hamp). Both proteins play roles in cellular iron uptake and transport. The rest of the enriched categories in this experimental group were identified as “Cellular lipid metabolism”, “Lipid metabolism”, “Fatty acid metabolism”, “Organic acid metabolism”, “Carboxylic acid metabolism”, and “Lipid biosynthesis” (P=0.0068). As suggested by their names, these categories shared a similar list of genes (see Table 7). Eight molecules that play roles in glycolysis and lipid metabolism could be integrated into the network as shown in Figure 7.
Table 7.
List of SF-modulated genes in each enriched category in rats sacrificed 24 h after treatment with AFB1
| RefSeq ID | Gene | Description | Fold Change |
|---|---|---|---|
| Acetyl-CoA biosynthesis | |||
| NM_016987 | Acly | ATP citrate lyase | 2.57 |
| NM_031025 | Dlat | Dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 1.78 |
| Iron ion homeostasis | |||
| XM_340999 | Trfc | Transferrin receptor | 2.92 |
| NM_053469 | Hamp | Hepcidin antimicrobial peptide | −7.47 |
| Fatty acid / cellular lipid / organic acid / carboxylic and lipid metabolism Lipid biosynthesis | |||
| NM_017332 | Fasn | Fatty acid synthase | 3.25 |
| NM_012703 | Thrsp | Thyroid hormone responsive protein | 2.90 |
| NM_134383 | Elovl6 | ELOVL family member 6, elongation of long chain fatty acids | 2.87 |
| NM_016987 | Acly | ATP citrate lyase | 2.57 |
| XM_341880 | Me | Malic enzyme 3, NADP(+)-dependent, mitochondrial | 1.88 |
| XM_213329 | Srebf1 | Sterol regulatory element binding factor 1 | 1.87 |
| NM_031025 | Dlat | Dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 1.78 |
| NM_012624 | Pklr | Pyruvate kinase, liver and red blood cell | 1.73 |
| NM_001007620 | Pdhb | Pyruvate dehydrogenase (lipoamide) beta | 1.66 |
| NM_033235 | Mdh | Malate dehydrogenase 1, NAD | 1.59 |
| NM_031315 | Cte1 | Cytosolic acyl-CoA thioesterase 1 | −1.77 |
| NM_022594 | Ech1 | Enoyl coenzyme A hydratase 1, peroxisomal | −1.89 |
| NM_031559 | Cpt1a | Carnitinepalmitoyltransferase 1a, liver | −1.90 |
| NM_138907 | Mte1 | Mitochondrial acyl-CoA thioesterase 1 | −1.90 |
| NM_198780 | Pck1 | Phosphoenolpyruvatecarboxykinase 1 | −1.93 |
| NM_001004085 | Crat | Carnitineacetyltransferase | −2.20 |
Figure 7.
The molecular interactions among proteins encoded by the deregulated genes observed in the 24 h group.↑ Indicates upregulation of gene expression. ↓ Indicates downregulation of gene expression.
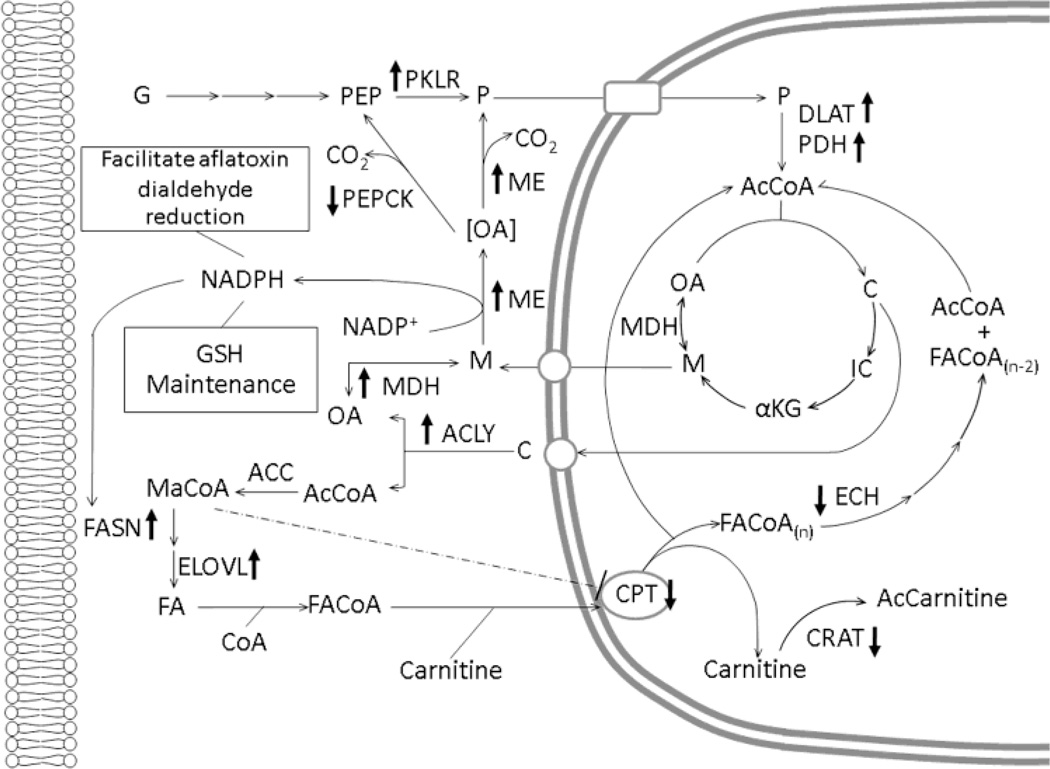
Molecules that were involved in metabolism were mapped into the common metabolic pathways of glycolysis, the TCA cycle, fatty acid synthesis and fatty acid β-oxidation. The pathways were combined and formatted to illustrate graphically how SF affects biochemical networks in AFB1-treated rat liver cells (see Figure 8).
Figure 8.
SF-induced alteration of common metabolic pathway in liver of 24 h rats. ↑ Indicates up-regulation of gene expression. ↓ Indicates down-regulation of gene expression. G, glucose; PEP, phosphoenolpyruvate; PKLR, pyruvate kinase; P, pyruvate; DLAT, E2 component of the pyruvate dehydrogenase complex; PDH, pyruvate dehydrogenase; AcCoA, acetyl-CoA; C, citrate; IC, isocitrate; αKG, α-ketoglutarate; M, malate; MDH, malate dehydrogenase; OA, oxaloacetate; ME, malic enzyme, PEPCK, phosphoenolpyruvate carboxykinase; ACLY, ATP citrate lyase; FASN, fatty acid synthase; ELOVL, elongation of long chain fatty acid protein; MaCoA, Malonyl-CoA; FA, fatty acid; FACoA, fatty acyl-CoA, CPT, carnitinepalmitoyltransferase; AcCarnitine; acylcarnitine; ECH, enoyl CoA hydratase; GSH, glutathione.
Discussion
AFB1 is one of the major risk factors for development of liver cancer, especially when it is present in concert with hepatitis B infection (Groopman and Kensler, 2005). Epidemiological investigations in human populations reveal an association of increased incidence of HCC with increasing dietary contamination by AFB1 (Wild and Gong, 2010). As the fungal species that produces AFB1 is ubiquitous, reduction of AFB1 exposure from ingestion may be problematic and impractical in certain areas of the world. Chemoprevention, that is the use of natural or synthetic agents to retard, block, or reverse the carcinogenic process, is an attractive strategy to mitigate the impact of AFB1 toxicity in high risk populations.
We recently showed that SF is a potent inducer of hepatic GST activity and reduces the amount of AFB1-N7-guanine formed in liver DNA in both sexes of Sprague Dawley and Fischer rats (Fiala et al., 2011). Our current study was carried out in an effort to understand further the in vivo mechanisms underlying the chemo-preventive action of SF against AFB1-induced liver cancer. Male animals were used because males are more sensitive to AFB1 than females. Effects of SF on biological networks of rat livers following AFB1-DNA adduction were explored by using microarray and RT-PCR techniques at two different time points, 4 h and 24 h, after AFB1 administration.
In the group of rats sacrificed 4 h after AFB1 administration, deregulation of a set of genes involved in signal transduction, transcription, and positive regulation of development was observed. While only three rats were used to keep the experiment at a manageable level, good statistical results were obtained. The results showed that SF enhanced the expression of genes involved in cytokine responses in a manner that would be expected to “favor” cell survival and regeneration. The expression changes observed in these gene sets may reflect the activation of signal transduction pathways required for the reprogramming of metabolic pathways to support anabolic growth in hepatocytes in the wake of cellular damage caused by AFB1 exposure.
At 24 h after AFB1 administration, up-regulation of transferrin receptor (Tfrc) expression is interpreted as a sign of hepatocyte regeneration. Transferrin receptors are expressed ubiquitously on proliferating cells (Besançon et al., 1987; Galbraith and Galbraith, 1981) and the interaction between transferrin and its receptor plays a crucial role in cell growth (Besançon et al., 1987). In addition, a shift in the dynamics of metabolic pathway regulation appears to have occurred upon SF pretreatment of AFB1-dosed male rats; for example, an induction of genes that play roles in cellular lipid biosynthesis and acetyl-CoA biosynthesis was evident 24 h after the AFB1 was administered. The enhanced gene expression profiles in these rats could be a response aimed at generating and maintaining pools of intermediate molecules needed for anabolism (DeBerardinis et al., 2007; Molenaar et al., 2009).
Some of the gene expression changes observed in livers of SF-pretreated, AFB1-dosed rats are reminiscent of changes induced during aerobic glycolysis (i.e., in tissues displaying the Warburg effect). The Warburg effect is postulated as a metabolic shift in energy production from oxidative phosphorylation to glycolysis, even in the presence of oxygen (Garber, 2004; Kim and Dang, 2006). In cancer cells, this metabolic switch allows the cell to maintain bioenergetics and a rigorous biosynthetic program during growth and proliferation (Jones and Thompson, 2009). Moreover, this metabolic switch toward glycolysis for energy production may favor cell growth in the tumor microenvironment (Gatenby and Gillies, 2004). Normal proliferating cells, such as primary lymphocytes, reveal similar patterns to those seen in cancer cells in their reliance on aerobic glycolysis to meet their bioenergetic needs (DeBerardinis et al., 2008; Jones and Thompson, 2007). In the present study, we see similar adaptations that effect pathways of glycolysis, the TCA cycle, fatty acid synthesis and fatty acid β-oxidation as shown in Figure 8.
Increases of pyruvate kinase (Pklr), dihydrolipoamide S-acetyltransferase (Dlat; the E2 component of the pyruvate dehydrogenase complex), and pyruvate dehydrogenase (Pdh) gene expression were observed in livers of male rats that received SF and were sacrificed 24 h after AFB1 administration. Increased expression of these enzymes, taken together, favor uptake of pyruvate into mitochondria, where it is converted to acetyl-CoA and enters the tricarboxylic acid (TCA) cycle. The acetyl-CoA entering the TCA cycle condenses with oxaloacetate (OA) to form citrate and then, rather than continuing in the TCA cycle, the citrate thereby formed is shunted to the cytoplasm of growing cells, where ATP citrate lyase (ACLY) converts it to cytoplasmic acetyl-CoA, which is required for lipogenesis (Ballard and Hanson, 1967). Lipid synthesis is very important in proliferating cells as phospholipids are a major structural component of biological membranes; examples include glycerolphospholipids, sphingolipids, and galactolipids (Nelson and Cox, 2009). Thus, this pathway’s upregulation by SF may be an important metabolic strategy to supply extra-mitochondrial acetyl-CoA for lipogenesis (Ballard and Hanson, 1967). The results of this study also show that malic enzyme (Me), the cytosolic version of malate dehydrogenase (Mdh), and ATP citrate lyase (Acly) were induced. In order to sustain lipogenesis, cells must have abundant citrate synthesis in mitochondria. Mitochondrial OA is consumed during citrate production, and its level must be restored after transport of citrate to the cytoplasm in advance of lipid biosynthesis. Cytosolic Mdh works in series with Me in a pathway shunt that helps maintain mitochondrial OA levels. In addition to providing cells with biosynthetic precursors, another important feature of this shunt is the trans-hydrogenation of NADH to NADPH by Me working in concert with Mdh. This series of reactions augments the available extra-mitochondrial NADPH needed to support lipogenesis (Ballard and Hanson, 1967). It is also possible that the generated NADPH would facilitate aflatoxin dialdehyde reduction as part of a detoxification strategy (Kelly et al., 2002). Moreover, aflatoxin treatment may deregulate glutathione metabolism, and the NADPH produced by this shunt may be used to maintain thiol pool homeostasis (Pashkov et al., 2005). We also note that glucose-6-phosphate dehydrogenase (G6pd) was increased in expression, although this enhancement did not reach the threshold for statistical significance. This enzyme is the portal to the pentose phosphate pathway, another major source of NADPH to support reductive biosynthesis. These changes are consistent with Nfr2-regulated genes that have been identified in Nrf2-deficient mouse models (Kwak et al., 2003; Wu et al., 2011).
Up-regulation of sterol regulatory element binding protein-1 (Srebp-1) in this group of SF-treated animals may also reflect active cellular membrane synthesis. SREBPs are a family of basic helix-loop-helix-leucine zipper transcription factors (Horton et al., 2002; Rawson, 2003) that play crucial roles in regulating lipid homeostasis in vertebrate cells (Horton et al., 2002). At present, three isoforms of Srebps have been identified; Srebp-1a, Srebp-1c, and Srebp-2 (Hua et al., 1993; Miserez et al., 1997). Srebp-1a and Srebp-1c are produced from a single gene with the use of alternate promoter start sites, while Srebp-2 is produced from a separate gene (Yokoyama et al., 1993). All are believed to form a homodimer to bind to the sterol regulatory elements (SREs) (Yokoyama et al., 1993). The three isoforms have different functions in the regulation of lipid synthesis. Evidence from many previous in vivo studies suggests that Srebp-1c is more active in enhancing lipogenic enzyme gene expression, whereas Srebp-1a and Srebp-2 are more specific to cholesterogenic genes. As shown in Figure 7, in this study, key lipogenic enzymes that are Srebp-1 target genes were up-regulated; Fatty acid synthase (Fasn), Elongation of long chain fatty acids protein 6 (Elovl6), Acly and Me (Horton et al., 2002; Kumadaki et al., 2008). FASN is a multi-enzyme complex that plays a central role in fatty acid synthesis (Nelson and Cox, 2009). ELOVL6 exhibits fatty acyl-CoA elongase activity specific for long chain fatty acids and is important for tissue fatty acid composition (Kumadaki et al., 2008). ACLY, as mentioned above, plays an early biochemical role in de novo fatty acid biosynthesis. Thus, the increase of Srebp-1 expression observed in this study is consistent with a role for SF as an inducer of lipogenic enzymes in AFB1-teated animals.
In addition to lipid-generating enzyme genes, the results showed the induction of the thyroid hormone responsive protein (Thrsp) mRNA transcript. Thrsp expression is also driven by SREBP-1 (Azzout-Marniche et al., 2000; Martel et al., 2006). Lipogenesis in tissues that synthesize fatty acids for use as fuel, such as liver, adipose and lactating mammary gland, is often regulated by thyroid hormone (Brown et al., 1997). Induction of the THRSP protein is likely to have occurred because its protein level usually parallels that of its mRNA (Kinlaw et al., 1989). The study of Brown and coworkers (1997) revealed that THRSP protein participates in induction of lipogenic enzyme genes, Fasn, Acly and Me, and glycolytic enzyme genes, such as Pklr and the gluconeogenic gene for phosphoenolpyruvate carboxykinase (Pepck). The mRNA transcripts for all of these enzymes were elevated in this study, except Pepck. Although THRSP can up-regulate Pepck gene expression, SREBP-1 has a negative role in Pepck transcription. It has been proposed that SREBP-1c is an intermediate in the action of insulin on Pepck gene transcription (Chakravarty et al., 2001). SREBP-1c markedly decreases the basal level of cytosolic Pepck mRNA in isolated hepatocytes by blocking the interaction of specific transcription factors involved in cAMP regulation of Pepck gene transcription with cAMPbinding protein (Chakravarty et al., 2001). This result is consistent with an in vivo study, where hepatic Pepck mRNA levels were suppressed by overexpression of Srebp-1a and -1c in the SREBP transgenic mice (Yamamoto et al., 2004). The down-regulation of Pepck observed in this study seems logical. Pepck is a gluconeogenic enzyme and its expression would be counter to expectations given our other observations indicating strong activation of glycolysis. Gluconeogenesis and glycolysis thus appear to be reciprocally regulated in this study, as expected.
In summary, the gene expression data presented herein provide a snapshot of the genetic network perturbed by SF treatment in AFB1-treated rats. The data suggest a SF-mediated adaptive response of cells following AFB1-induced cell damage. The gene expression data provide new insights into the molecular environment that exists in damaged cells pre-treated with the cancer chemopreventive agent, SF. Studies of this nature may prove useful in continuing efforts to identify novel pathways that could be induced or suppressed in an effort to ameliorate the risk for liver cancer created by exposure to aflatoxin.
Highlights.
This study revealed sulforaphane (SF)-deregulated gene sets in aflatoxin B1 (AFB1)-treated rat livers.
SF redirects biochemical networks toward lipid biosynthesis in AFB1-dosed rats.
SF enhanced gene sets that would be expected to favor cell repair and regeneration.
Acknowledgments
This work was supported by National Institutes of Health grants ES016313, P30-ES002109; P01 ES006052; P30 ES003819; P30 CA006973. The authors thank Drs. Stephen Slocum and Daam Settachan for critically reading the manuscript.
Abbreviations
- SF
Sulforaphane
- AFB1
Aflatoxin B1
- GST
Glutathione S-Transferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest. The authors declare no conflicts of interest in the support, conception, execution or publication of the work described herein.
References
- Azzout-Marniche D, Bécard D, Guichard C, Foretz M, Ferré P, Foufelle F. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem J. 2000;350:389–393. [PMC free article] [PubMed] [Google Scholar]
- Ballard FJ, Hanson RW. The citrate cleavage pathway and lipogenesis in rat adipose tissue: replenishment of oxaloacetate. J Lipid Res. 1967;8:73–79. [PubMed] [Google Scholar]
- Besançon F, Silbermann F, Dron M, Tovey MG, Thang MN, Bourgeade MF. Relationship between inhibition of cell growth and of transferrin receptor expression by interferon (IFN) alpha: studies in IFN-sensitive and IFN-resistant Daudi cells. J Gen Virol. 1987;68:2647–2654. doi: 10.1099/0022-1317-68-10-2647. [DOI] [PubMed] [Google Scholar]
- Brown SB, Maloney M, Kinlaw B. “Spot 14” protein functions at the pretranslational level in the regulation of hepatic metabolism by thyroid hormone and glucose. J Biol Chem. 1997;272:2163–2166. [PubMed] [Google Scholar]
- Busby WF, Wogan GN. Aflatoxins. In: Searle CE, editor. Chemical carcinogens, ACS Monograph 182. 1984. pp. 945–1136. [Google Scholar]
- Chakravarty K, Leahy P, Becard D, Hakimi P, Foretz M, Ferre P, Foufelle F, Hanson RW. Sterol regulatory element-binding protein-1c mimics the negative effect of insulin on phosphoenolpyruvate carboxykinase (GTP) gene transcription. J Biol Chem. 2001;276:34816–34823. doi: 10.1074/jbc.M103310200. [DOI] [PubMed] [Google Scholar]
- Chiao JW, Chung FL, Kancherla R, Ahmed T, Mittelman A, Conaway CC. Sulforaphane and its metabolite mediate growth arrest and apoptosis in human prostate cancer cells. Int J Oncol. 2002;20:631–636. doi: 10.3892/ijo.20.3.631. [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Julian JL, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Meco MT, Moscat J. The atypical PKCs in inflammation: NF-κB and beyond. Immunol Rev. 2012;246:154–167. doi: 10.1111/j.1600-065X.2012.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner PA, Gange SJ, Dolan PM, Groopman JD, Muñoz A, Kensler TW. Levels of aflatoxin-albumin biomarkers in rat plasma are modulated by both long-term and transient interventions with oltipraz. Carcinogenesis. 1995;16:1769–1773. doi: 10.1093/carcin/16.8.1769. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zhang Y, Talalay P. Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci USA. 1997;94:10367–10372. doi: 10.1073/pnas.94.19.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JL, Egner PA, Wiriyachan N, Ruchirawat M, Kensler KH, Wogan GN, Groopman JD, Croy RG, Essigmann JM. Sulforaphane-mediated reduction of aflatoxin B1-N7-guanine in rat liver DNA: impacts of strain and sex. Toxicol Sci. 2011;121:57–62. doi: 10.1093/toxsci/kfr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimognari C, Hrelia P. Sulforaphane as a promising molecule for fighting cancer. Mutat Res. 2007;635:90–104. doi: 10.1016/j.mrrev.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Fimognari C, Nüsse M, Berti F, Iori R, Cantelli-Forti G, Hrelia P. Isothiocyanates as novel cytotoxic and cytostatic agents: molecular pathway on human transformed and non-transformed cells. Biochem Pharmacol. 2004;68:1133–1138. doi: 10.1016/j.bcp.2004.03.044. [DOI] [PubMed] [Google Scholar]
- Galbraith RM, Galbraith GM. Expression of transferrin receptors on mitogen-stimulated human peripheral blood lymphocytes: relation to cellular activation and related metabolic events. Immunology. 1981;44:703–710. [PMC free article] [PubMed] [Google Scholar]
- Garber K. Energy boost: the Warburg effect returns in a new theory of cancer. J Natl Cancer Inst. 2004;96:1805–1806. doi: 10.1093/jnci/96.24.1805. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Robert JG. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groopman JD, Kensler TW. Role of metabolism and viruses in aflatoxin-induced liver cancer. Toxicol Appl Pharmacol. 2005;206:131–137. doi: 10.1016/j.taap.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Judah DJ, McLellan LI, Kerr LA, Peacock SD, Neal GE. Ethoxyquin-induced resistance to aflatoxin B1 in the rat is associated with the expression of a novel alpha-class glutathione S-transferase subunit, Yc2, which possesses high catalytic activity for aflatoxin B1-8,9-epoxide. Biochem J. 1991;279:385–398. doi: 10.1042/bj2790385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Pulford DJ, Ellis EM, McLeod R, James RF, Seidegard J, Mosialou E, Jernström B, Neal GE. Regulation of rat glutathione S-transferase A5 by cancer chemopreventive agents: mechanisms of inducible resistance to aflatoxin B1. Chem Biol Interact. 1998;111–112:51–67. doi: 10.1016/s0009-2797(97)00151-8. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Heiss E, Herhaus C, Klimo K, Bartsch H, Gerhäuser C. Nuclear factor-{kappa}B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J. Biol. Chem. 2001;276:32008–32015. doi: 10.1074/jbc.M104794200. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, Wang X. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ishidate T, Matsumine A, Toyoshima K, Akiyama T. The APC-hDLG complex negatively regulates cell cycle progression from the G0/G1 to S phase. Oncogene. 2000;19:365–372. doi: 10.1038/sj.onc.1203309. 2000. [DOI] [PubMed] [Google Scholar]
- Jackson SJ, Singletary KW, Venema RC. Sulforaphane suppresses angiogenesis and disrupts endothelial mitotic progression and microtubule polymerization. Vascul Pharmacol. 2007;46:77–84. doi: 10.1016/j.vph.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Revving the Engine: Signal Transduction Fuels T Cell Activation. immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. 2002. [DOI] [PubMed] [Google Scholar]
- Karmakar S, Weinberg MS, Banik NL, Patel SJ, Ray SK. Activation of multiple molecular mechanisms for apoptosis in human malignant glioblastoma T98G and U87MG cells treated with sulforaphane. Neuroscience. 2006;141:1265–1280. doi: 10.1016/j.neuroscience.2006.04.075. [DOI] [PubMed] [Google Scholar]
- Kelly VP, Sherratt PJ, Crouch DH, Hayes JD. Novel homodimeric and heterodimeric rat gamma-hydroxybutyrate synthases that associate with the Golgi apparatus define a distinct subclass of aldo-keto reductase 7 family proteins. Biochem J. 2002;366(Pt 3):847–861. doi: 10.1042/BJ20020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Roebuck BD, Wogan GN, Groopman JD. Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci. 2011;120(Suppl 1):S28–S48. doi: 10.1093/toxsci/kfq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Egner PA, Dolan PM, Groopman JD, Roebuck BD. Mechanism of protection against aflatoxin tumorigenicity in rats fed 5-(2-pyrazinyl)-4-methyl-1,2-dithiol-3-thione (oltipraz) and related 1,2-dithiol-3-thiones and 1,2-dithiol-3-ones. Cancer Res. 1987;47:4271–4277. [PubMed] [Google Scholar]
- Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- Kinlaw WB, Ling NC, Oppenheimer JH. Identification of rat S14 protein and comparison of its regulation with that of mRNA S14 employing synthetic peptide antisera. J Biol Chem. 1989;264:19779–19783. [PubMed] [Google Scholar]
- Kumadaki S, Matsuzaka T, Kato T, Yahagi N, Yamamoto T, Okada S, Kobayashi K, Takahashi A, Yatoh S, Suzuki H, Yamada N, Shimano H. Mouse Elovl-6 promoter is an SREBP target. Biochem Biophys Res Commun. 2008;368:261–266. doi: 10.1016/j.bbrc.2008.01.075. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- Lin W, Wu RT, Wu T, Khor TO, Wang H, Kong AN. Sulforaphane suppressed LPS-induced inflammation in mouse peritoneal macrophages through Nrf2 dependent pathway. Biochem Pharmacol. 2008;76:967–973. doi: 10.1016/j.bcp.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Roebuck BD, Yager JD, Groopman JD, Kensler TW. Protection by 5-(2-pyrazinyl)-4-methyl-1,2-dithiol-3-thione (oltipraz) against the hepatotoxicity of aflatoxin B1 in the rat. Toxicol Appl Pharmacol. 1988;93:442–451. doi: 10.1016/0041-008x(88)90047-6. [DOI] [PubMed] [Google Scholar]
- Martel PM, Bingham CM, McGraw CJ, Baker CL, Morganelli PM, Meng ML, Armstrong JM, Moncur JT, Kinlaw WB. S14 protein in breast cancer cells: direct evidence of regulation by SREBP-1c, super induction with progestin, and effects on cell growth. Exp Cell Res. 2006;312:278–288. doi: 10.1016/j.yexcr.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Miserez AR, Cao G, Probst LC, Hobbs HH. Structure of the Human Gene Encoding Sterol Regulatory Element Binding Protein 2 (SREBF2) Genomics. 1997;40:31–40. doi: 10.1006/geno.1996.4525. [DOI] [PubMed] [Google Scholar]
- Molenaar D, van Berlo R, de Ridder D, Teusink B. Shifts in growth strategies reflect tradeoffs in cellular economics. MolSyst Biol. 2009;5:323. doi: 10.1038/msb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myzak, Melinda C, Roderick H, Dashwood Chemoprotection by sulforaphane: keep one eye beyond Keap1. Cancer Lett. 2006;233:208–218. doi: 10.1016/j.canlet.2005.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger Principles of Biochemistry. fifth ed. New York: W.H. Freeman Publishers; 2009. [Google Scholar]
- Pashkov AN, Popov SS, Semenikhina AV, Rakhmanova TI. Glutathione system and activity of nadph-generating enzymes in the liver of intact rats and animals with toxic hepatitis receiving melatonin. Bull Exp Biol Med. 2005;139:565–568. doi: 10.1007/s10517-005-0346-7. [DOI] [PubMed] [Google Scholar]
- Rawson RB. The SREBP pathway - insights from insigs and insects. Nat Rev Mol Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- Rosen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Round JL, Humphries LA, Tomassian T, Mittelstadt P, Zhang M, Miceli MC. Scaffold protein Dlgh1 coordinates alternative p38 kinase activation, directing T cell receptor signals toward NFAT but not NF-kappaB transcription factors. Nat Immunol. 2007;8:154–161. doi: 10.1038/ni1422. [DOI] [PubMed] [Google Scholar]
- Singh AV, Xiao D, Lew KL, Dhir R, Singh SV. Sulforaphane induces caspasemediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis. 2004;25:83–90. doi: 10.1093/carcin/bgg178. [DOI] [PubMed] [Google Scholar]
- Sun B, Karin M. NF-kappaB signaling, liver disease and hepatoprotective agents. Oncogene. 2008;27:6228–6244. doi: 10.1038/onc.2008.300. [DOI] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- Ward PS1, Thompson CB. Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol. 2012;4:a006783. doi: 10.1101/cshperspect.a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP, Gong YY. Mycotoxins and human disease: a largely ignored global health issue. Carcinogenesis. 2010;31:71–82. doi: 10.1093/carcin/bgp264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Cui JY, Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Shimano H, Nakagawa Y, Ide T, Yahagi N, Matsuzaka T, Nakakuki M, Takahashi A, Suzuki H, Sone H, Toyoshima H, Sato R, Yamada N. SREBP-1 interacts with hepatocyte nuclear factor-4 alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J Biol Chem. 2004;279:12027–12035. doi: 10.1074/jbc.M310333200. [DOI] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- Zanin-Zhorov A, Lin J, Scher J, Kumari S, Blair D, Hippen KL, Blazar BR, Abramson SB, Lafaille JJ, Dustin ML. Scaffold protein Disc large homolog 1 is required for T-cell receptor-induced activation of regulatory T-cell function. Proc Natl Acad Sci USA. 2012;109:1625–1630. doi: 10.1073/pnas.1110120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a webbased platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16–16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov SA, Snoddy JR. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]