Abstract
Genome engineering with designer nucleases is a rapidly progressing field, and the ability to correct human gene mutations in situ is highly desirable. We employed fibroblasts derived from a patient with Fanconi anemia as a model to test the ability of the clustered regularly interspaced short palindromic repeats/Cas9 nuclease system to mediate gene correction. We show that the Cas9 nuclease and nickase each resulted in gene correction, but the nickase, because of its ability to preferentially mediate homology-directed repair, resulted in a higher frequency of corrected clonal isolates. To assess the off-target effects, we used both a predictive software platform to identify intragenic sequences of homology as well as a genome-wide screen utilizing linear amplification-mediated PCR. We observed no off-target activity and show RNA-guided endonuclease candidate sites that do not possess low sequence complexity function in a highly specific manner. Collectively, we provide proof of principle for precision genome editing in Fanconi anemia, a DNA repair-deficient human disorder.
Introduction
The FANCC gene on chromosome 9 encodes a protein that is a constituent of an eight-protein Fanconi anemia (FA) core complex that functions as part of the FA pathway responsible for genome surveillance and repair of DNA damage.1 A predominant cause of FA complementation group C (FA-C) is the c.456+4A>T (previously c.711+4A>T; IVS4+4A>T) point mutation that results in a cryptic splice site that causes aberrant splicing and the in-frame deletion of FANCC exon 4.2,3 The loss of exon 4 prevents FANCC participation in the formation of the core complex and results in a decrease in DNA repair ability. Typically, FA-C patients exhibit congenital skeletal abnormalities and progressive cytopenias culminating in bone marrow failure.4 Furthermore, FA-C patients exhibit a high incidence of hematological and solid tumors.5 People with FA who experience bone marrow failure, and for whom a suitable donor exists, are currently treated with allogeneic hematopoietic cell transplantation (HCT).6 However, the risks associated with HCT provide an incentive to gene-correct autologous cells by gene addition or genome editing.7,8 Previous studies have shown that FANCC gene replacement with functional copies for the FANCC cDNA can rescue the FA phenotype.9,10 Because of the premalignant phenotype FA patients possess, a key consideration for any gene therapy is safety. Because of the magnified risk of insertional mutagenesis associated with the delivery of functional copies of the FANCC gene borne on integrating viral or nonviral vectors,11,12 we sought to determine whether precision gene targeting could be achieved using genome-modifying proteins.
Efficient genome editing relies on engineered proteins that can be rapidly synthesized and targeted to a specific genomic locus. Currently, the major candidates able to mediate genome modification are the zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced palindromic repeat (CRISPR/Cas9) nucleases. ZFNs and TALENs are comprised of DNA-binding elements that provide specificity and are tethered to the nonspecific FokI nuclease domain. Dimerization of the complex at the target site results in the generation of a double-stranded DNA break (DSB). The generation of ZFNs can be challenging and requires the acquisition of specialized starting materials and methodologies that somewhat limits their broader application.13 In contrast, the starting materials to generate the multirepeat TALEN complexes are publicly available, and assembly of reagents by this method is much simpler than those required for ZFNs.14,15 The newly described Streptococcus pyogenes CRISPR/Cas9 platform is also highly user-friendly and contains two components: the Cas9 nuclease and a guide RNA (gRNA).16,17 The gRNA is a short transcript that can be designed for a unique genomic locus possessing a GN20GG sequence motif and that serves to recruit the Cas9 protein to the target site where it induces a DSB.16–18 gRNAs direct Cas9 using complementarity between the 5′-most 20 nts and the target site, which must have a protospacer adjacent motif (PAM) sequence of the form NGG.17
Possible uses for gene-editing reagents to achieve gene correction include (1) the introduction of a full-length cDNA at a so-called genomic safe harbor, or (2) true in situ mutation-specific targeting. The AAVS1 locus on chromosome 19 was identified as an integration hotspot for wild-type AAV and as encoding the PPP1R12C gene that functions as a subunit of myosin phosphatase.19–21 This locus has been targeted for integration of genetic material that then regulated by the PPP1R12C promoter or an exogenous promoter contained in the targeting construct.22,23 Because these scenarios result in continuous gene expression that is not subject to locus-specific promoter control, and because it has been shown that constitutive FANCC expression can result in cellular apoptotic resistance,24,25 we considered this approach suboptimal for FA. Instead, we explored the possibility of FANCC c.456+4A>T-specific gene targeting.
At the onset of our studies it was unknown if gene-editing reagents that cleave single- or double-stranded DNA targets are able to be repaired in the context of the FA phenotype that is characterized by an inability to repair DNA lesions. As such, we carefully considered the optimal cell type to determine the usefulness of precision gene targeting in FA. Hematopoietic progenitors are the ideal population for correction for therapeutic use; however, the paucity of these cells in patients does not support their use for proof-of-principle studies. Instead, we employed transformed fibroblasts from an FA-C patient because of their relative ease of culture and their ability to be transfected at more efficient rates than patient-derived lymphoblastoid cells.10 Moreover, despite FA-C patient cells being highly refractory to reprogramming to pluripotency, establishing proof of concept would support gene editing in other FA subtypes for subsequent reprogramming. Therefore, in order to unambiguously assess whether FA cells can be corrected by gene editing, we used homozygous FA-C patient fibroblasts and generated a mutation-proximal gRNA for recognition and cleavage by Cas9. The nuclease and nickase versions of Cas9 were employed and the highest rates of gene correction were obtained with the Cas9 nickase. These data are the first use of CRISPR/Cas9 for gene editing in FA and provide a platform for translation to therapeutically viable cell populations.
Materials and Methods
Research subject cell line generation and culture
Informed consent was obtained from the parents of a child possessing the c.456+4A>T mutation, and a skin punch biopsy was performed in accordance with the University of Minnesota Institutional Review Board and Declaration of Helsinki requirements for research on human subjects. A fibroblast cell line was derived by dicing the skin tissue, covering it with a microscope slide, and adding complete DMEM (20% FBS, 100 U/ml nonessential amino acids, 0.1 mg/ml each of penicillin and streptomycin, and EGF and FGF at a concentration of 10 ng/ml, and 1× antioxidant supplement [Sigma, St. Louis, MO]) with culture under hypoxic conditions. A TERT–green fluorescent protein (GFP) lentiviral construct was then added to the cells at an MOI of 10, and then cells were FACs sorted to purity.
CRISPR and donor construction
The Cas9 and Cas9 D10A plasmids were obtained from Addgene (plasmids 41815 and 41816), the U6 promoter and FANCC-specific gRNA were synthesized as a G-block (IDT DNA technologies, Coralville, IA), and TA cloned into the pCR4 TOPO vector (Invitrogen, Carlsbad, CA) as previously described.17 The right donor arm was cloned from the human genome and consisted of an 849 bp sequence. The left arm was synthesized from overlapping G-block fragments in order to introduce the corrective base and silent mutations at the CRISPR cut sites. The donor arms flanked a floxed PGK-puromycin-T2A-FANCC cDNA selection cassette; the full donor sequence is provided as Supplementary Fig. S1 (Supplementary Material are available online at www.liebertpub.com/hum).
Gene transfer
For 293T transfections, CRISPR/Cas9 nuclease and nickase with gRNA were delivered with Lipofectamine 2000 (Invitrogen) at a concentration of 1 μg each. Fibroblast gene transfer was performed using the Neon Transfection System (Invitrogen) using 1500 V, 20 ms pulse width, and a single pulse. Concentrations of DNA for gene correction were as follows: Cas9 nuclease/nickase=1 μg, gRNA=200 ng, and donor=5 μg. For 48 hr after gene transfer, all cells were incubated at 31°C.26
Surveyor nuclease
Genomic DNA was isolated from 293 cells at 72 hr post-CRISPR gene transfer and was amplified for 35 cycles with FANCC forward (5′-AGACCACCCCCATGTACAAA-3′) and FANCC reverse (5′-GGAAAACCCTTCCTGGTTTC-3′). It was then subjected to Surveyor nuclease (Transgenomic, Omaha, NE) treatment, and products were resolved on a 10% TBE PAGE gel (Invitrogen).27 Gel images were utilized to determine rates of cleavage using the following equation: % gene modification=100×(1 − [1 − fraction cleaved] 1/2).27 The fraction cleaved is determined using ImageJ analysis software and is the densitometric value of the cleavage products divided by the total densitometric value for all of the peaks.27 The exposure times for the gels in Fig. 1E and F and Supplementary Fig. S2 were 750 ms, 1.5 s, and 3 s, respectively.
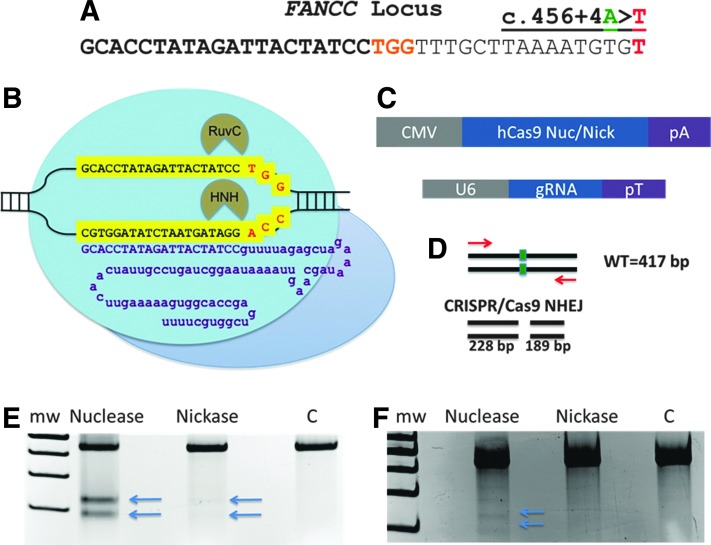
FIG. 1.
FANCC c.456+4A>T gene targeting. (A) The FANCC locus with the c.456+4A>T mutation shown at far right; base highlighted in red. The CRISPR gRNA recognition site is in bold and the PAM sequence in orange. (B) CRISPR architecture and FANCC gene target recognition. A gRNA chimeric RNA species has a gene-specific component (purple upper-case letters) that recognizes a 23 bp sequence in the FANCC gene (highlighted yellow sequence) with the 3′ terminal NGG protospacer; adjacent motif shown in red letters. The remainder of the gRNA (purple lower-case letters) are constant regions that contain secondary structure that interacts with the Streptococcus pyogenes Cas9 nuclease protein (blue circles). The Cas9 RuvC- and HNH-like domains mediate noncomplementary and complementary DNA strand cleavage. A D10A mutation in the RuvC domain converts the complex to a nickase.17 (C) DNA expression platforms. Cas9 nuclease or RuvC D10A nickase were expressed from a plasmid containing the CMV promoter and bovine growth hormone polyadenylation signal (pA). gRNA gene expression was mediated by the U6 polymerase III promoter and a transcriptional terminator (pT). (D) Nuclease activity assessment by the Surveyor assay. The FANCC locus in cells that received CRISPR/Cas9 nuclease or nickase with corresponding gRNA (target site shown as a green box), or a GFP-treated control group (labeled “C”), were amplified with primers (red arrows) yielding a 417 bp product. Nuclease- or nickase-generated insertions or deletions from NHEJ result in heteroduplex formation with unmodified amplicons that are cleaved by the mismatch-dependent Surveyor nuclease.27 For CRISPR/Cas9, these cleavage products are 228 and 189 bp. Surveyor analysis of 293T cells (E) or FA-C fibroblasts (F). Equivalent amounts of DNA were amplified using the primers in 1D and showed post-Surveyor fragmentation patterns consistent with CRISPR/Cas9 activity, with arrows indicating the cleavage bands. Data shown are representative gels of four experiments each. Gel exposure time for 293T cell Surveyor group was 750 msec and for FA-C cells was 1.5 sec. C, GFP-treated cells serving as the control; Mw, molecular weight standards; FA-C, Fanconi anemia complementation group C; GFP, green fluorescent protein; gRNA, guide RNA; NHEJ, nonhomologous end-joining; PAM, protospacer adjacent motif.
Traffic light reporter cell line generation and testing
The PGE-200 pRRL TLR2.1 sEF1a Puro WPRE parental plasmid was digested with SbfI and SpeI for ligation of the following oligonucleotides that were annealed and inserted at the FANCC CRISPR target site into the interrupted GFP portion of the plasmid: 5-GGCACCTATAGATTACTATC CTGGA-3 and 5- CTAGTCCAGGATAGTAATCTATAG GTGCCTGCA-3. Lentiviral particles were prepared by packaging with Addgene plasmids: 12259 (pMD2.G), 12251 (pMDLg/pRRE), and 12253 (pRSV-Rev) in 293T cells transfected with Lipofectamine 2000 (Invitrogen). The cell culture volume for viral production was 20 ml, viral particles were collected for 48 hr, and 20 μl of the supernatant was added to 293T cells followed by puromycin selection with 0.3 μg/ml. This reporter line was transfected with 1 μg each of the Cas9 nuclease or nickase and 1 μg of the gRNA with the indicated concentrations of the pCVL SFFV d14GFP Donor (Addgene 31475). Green or red fluorescence was analyzed 72 hr posttransfection using the BD LSRFortessa Cell Analyzer (BD Biosciences, San Jose, CA).
Selection and transgene excision
Seven days after gene transfer, cells were selected in bulk in 0.2 mg/ml puromycin. Resistant cells were then plated at low density (∼500 cells in a 10 cm2 dish) for 3 days, followed by silicone grease-coated cloning disk placement (Corning, Corning, NY). Isolated colonies were progressively passed to larger culture vessels so that cell culture confluency was maintained between 50% and 70% under hypoxic culture conditions. Cells confirmed to have undergone HDR were seeded into 24-well plates and serially transfected with a CAGGs promoter-driven Cre-recombinase (Addgene: 13775) or transduced with an adenoviral cre (Vector Biolabs, Philadelphia, PA) at an MOI of 10. Excision was confirmed using the following primers:
FC CRP 491 F: GAAACCAGGAAGGGTTTTCC
FC CRP 1163R: CAACCCCCATCTTCTCATGT
Cell correction molecular screening and H2AX staining
Primer pairs were designed to amplify a junction between the donor right arm and endogenous locus using donor-specific forward (5′-GCCACTCCCACTGTCCTTTCCT-3′) and FANCC reverse (5′-ccaagtccctcagtcccaga-3′), and to confirm HDR on the left portion of the correction template, the FANCC genomic forward (5′-CAGACACACCCCTGGAAGTC-3′) and donor reverse (5′-CTTTTGAAGCGTGCAGAATGCC-3′). For RT-PCR, total cellular RNA was isolated and reverse transcribed using SuperScript Vilo (Invitrogen) followed by amplification with FANCC allele-specific RT forward (5′-GGTGTATTAAGCCATATTCTGAGC-3′) and reverse (5′-ACAACCCGGAATATGGCAGG-3′). PCR products were cloned into the pCR 4 TOPO vector (Invitrogen) for Sanger sequencing confirmation of the entire amplicon using the M13 forward and reverse primers. H2AX staining was performed on cells seeded at a concentration of 120,000 total cells in a T25 flask in the presence of 2 mM hydroxyurea (Sigma) for 48 hr using the H2AX phosphorylation assay kit according to the manufacturer's instructions (EMD Millipore, Billerica, MA). Flow cytometry was performed using the BD LSRFortessa Cell Analyzer (BD Biosciences).
Off-target analysis
CRISPR/Cas9 nuclease/nickase and gRNA plasmids (1 μg each) were delivered to 293T cells by lipofection. At 24 hr post-gene transfer, an integrase-deficient GFP virus at an MOI of 5 was added. The viral cargo was prepared by packaging the pllCMV-GFP expression vector with the pCMV-ΔR8.2 plasmid harboring the D64V integrase mutation28,29 and the pMD2.VSV-G (Addgene 12259) envelope-encoding plasmids that were delivered to the 293T viral-1producing line with Lipofectamine 2000 (Invitrogen). Seven days after integrase-deficient lentiviral (IDLV) addition, the cells were sorted for GFP and then expanded. In silico-predicted OT sites were amplified with OT1 (F: 5′-TGGGTGGAGGTAGTTTCCTG-3′ and R: 3′-AGTGGGAAGAGGGCTGATTT-3′), OT2 (F: 5′-TCTGGGCATAAAGAAGGTGTG-3′ and R: 5′-ATTGACTCATCTCGGGCATT-3′), OT3 (F: 5′-GACCTGGGCTTGAATGTGTT-3′ and R: 5′-GCAGTTGCTGTAGAATAGGCTGT-3′), OT4 (F: 5′-CCCAGAGCAAAACCATTCAT-3′ and R: 5′-CACCTGTTGCAGACTCCTCA-3′), and OT5 (F: 5′-AGGAGCTGGGACACTGCTAA-3′ and R: 5′-ACACATGCCTGTCCTTCTCC-3′). These amplicons were then subjected to Surveyor analysis as above. IDLV:FANCC or OT detection PCR was performed with the long terminal repeat (LTR) forward primer (5′-GTGTGACTCTGGTAACTAGAG-3′) and the corresponding FANCC or OT reverse primers from above. IDLV:FANCC junction amplicons were cloned and Sanger sequenced.
Genome-wide screening
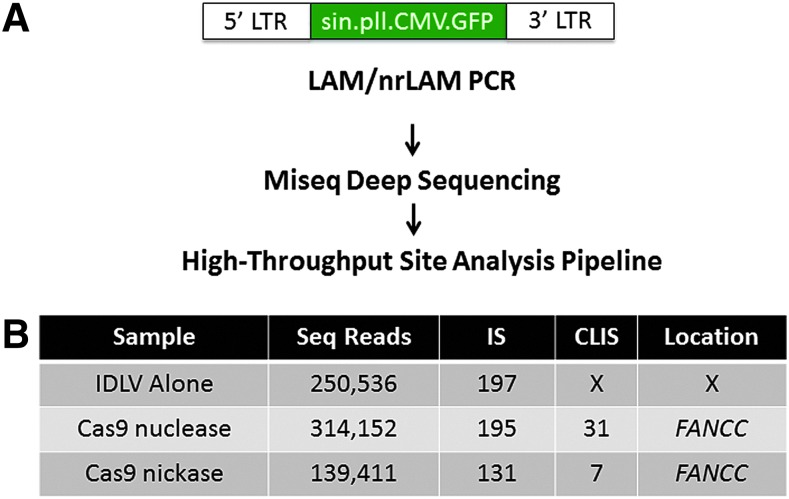
Duplicate samples underwent nrLAM PCR or LAM PCR with MseI or MluCI as previously described30–32 except that these deep-sequencing data were generated with the Illumina MiSeq platform (San Diego, CA). Dataset analysis, vector trimming, genome alignment, and integration sites (IS)/clusters of integrations (CLIS) identification was determined using the high-throughput insertion site analysis pipeline.33 The dataset from this screen is available by request to the corresponding author.
Human CD34 culture, isolation, and gene transfer
Umbilical cord blood (UCB) was collected in accordance with the University of Minnesota Institutional Review Board requirements for research on human subjects. Total UCB was placed in IMDM expansion media comprising 100 ng/ml of IL-3, Il-6, GM-SCF, Flt-3l, and stem cell factor with 1× penicillin/streptomycin, 10% human plasma, and 1 μM SR1 aryl hydrocarbon receptor antagonist.34 CD34 cells were isolated using the EasySep Human CD34 Positive Selection Kit according to the manufacturer's (Stem Cell Technologies, Vancouver, BC) recommendations and placed back in expansion media overnight. Gene transfer was performed using the Neon Electroporator (Invitrogen) with the following settings: 1400 V, 10 ms pulse, with 3 pulses. Dose of DNA was 1 μg GFP and 1 μg each of Cas9 (nuclease and nickase) and gRNA. Seventy-two hours after transfection, the genomic DNA was harvested for FANCC locus Surveyor analysis as above.
Results
Gene-editing platform architecture, activity, and preferred DNA repair pathway
Using the CRISPR Design Tool,35 we assessed the FANCC gene sequence on chromosome 9 proximal to the c.456+4A>T locus for available target sites and identified one within 15 bp of the mutation (Fig. 1A). A CRISPR gRNA contacts the target locus and is recognized by the Cas9 protein that contains RuvC and HNH domains, each responsible for generating single-strand DNA breaks (“nicks”) on opposite strands of the DNA helix (Fig. 1B).16,17,36 Inactivation of one of these domains converts Cas9 into a DNA nickase capable of cutting only one strand.16 We encoded a FANCC c.456+4A>T-specific gRNA with the CRISPR/Cas9 nuclease and nickase on DNA vectors (Fig. 1C) and delivered them to 293T and FA-C cells in order to assess rates of DNA cutting using the Surveyor assay. To perform this assay, the target locus in CRISPR/Cas9-treated cells was PCR amplified, resulting in the generation of unmodified and modified alleles that were repaired by nonhomologous end-joining (NHEJ)-mediated repair because of nuclease-generated DNA lesions.27 The hybridization of these two populations results in imperfect pairing and cleavage by the mismatch-specific Surveyor enzyme, allowing for the generation of a predictable fragmentation pattern for a locus-specific nuclease (Fig. 1D).
The densitometric analyses showed ∼15% rate of activity for CRISPR/Cas9 nuclease in 293s and ∼5% in FA-C fibroblasts (Fig. 1E and F and Supplementary Fig. S2). Because of the higher activation rate of the CRISPR/Cas9 system, we focused on it for determining activity rates in FA-C fibroblasts. For both the 293Ts and FA-C fibroblasts, the nuclease version of Cas9 resulted in higher rates of activity, compared with the nickase, using the Surveyor assay that is reliant on NHEJ-induced mutations (Fig. 1E and F and Supplementary Fig. S2). To insure that this differential activity profile was not because of unequal rates of gene transfer, an mCherry reporter was included at the time of transfection. The mean fluorescence intensity for each treatment group was nearly identical (Supplementary Fig. S2). Our results support those of others suggesting that the use of nickases promotes higher levels of error-free HDR compared with NHEJ-mediated insertions/deletions.16,37,38
To definitively and quantitatively determine if DNA nicking induced by the Cas9 D10A nickase resulted in preferential employment of the HDR arm of DNA repair, we utilized the traffic light reporter (TLR) system that allows for simultaneous quantification of NHEJ and HDR.37 This platform allows for a user-defined nuclease target sequence to be inserted into a portion of an inactive GFP gene that is upstream of an out-of-frame mCherry cDNA (Fig. 2A). At its basal state, the TLR construct does not express a functional fluorescent protein; however, following cleavage of the target sequence in the context of an exogenous GFP donor repair template, GFP expression can be restored by HDR repair (Fig. 2A). Conversely, target site cleavage and repair by the error-prone NHEJ results in an in-frame mCherry (Fig. 2A). Therefore, we generated a 293T cell line with an integrated copy of the TLR containing the CRISPR/Cas9 FANCC target site, and assessed rates of HDR and NHEJ for the nuclease and nickase versions of Cas9 using three different donor concentrations. Importantly, the basal rates of green or red fluorescence were minute for either untransfected cells or cells receiving the donor template only (Fig. 2B and C). Nuclease delivery resulted in substantial rates of both mCherry and GFP fluorescence, showing that both mutagenic NHEJ and error-free HDR can occur in response to a DSB (Fig. 2D). In contrast, a single-stranded nick mediated by the D10A Cas9 nickase resulted in minimal levels of NHEJ-induced red fluorescence, with a preference toward HDR (Fig. 2E).
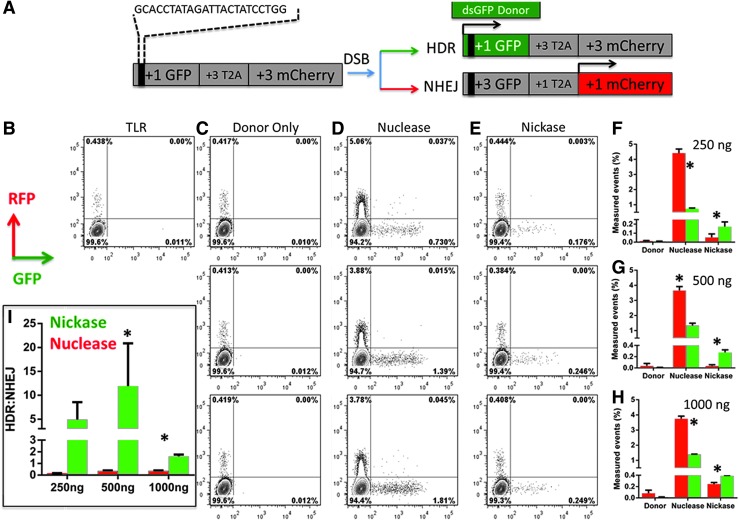
FIG. 2.
Traffic light reporter assessment of DNA repair fates. (A) Schematic of the TLR reporter. The FANCC CRISPR/Cas9 target sequence is contained within the dashed lines and was inserted into the GFP portion of the construct, resulting in an out-of-frame GFP. The +3 picornaviral 2A sequence allows the downstream nonfunctional +3 mCherry to escape degradation of the nonfunctional GFP.37 Following target site cleavage in the presence of an exogenous GFP donor (box labeled “dsGFP donor”) the GFP gene is repaired by HDR and expresses GFP (expression indicated by green box; inactive mCherry shown as gray box). DNA repair by NHEJ can result in a frameshift that restores the mCherry ORF, resulting in red fluorescence (indicated by red-filled mCherry box; gray indicates inactive GFP). (B–E) Representative FACs plot of 293T TLR cell line treatment with CRISPR/Cas9 nuclease and nickase. A stable 293T cell line with an integrated copy of the FANCC TLR construct was generated that, at its basal level, was GFP negative and expressed <0.5% mCherry (B). This low level of expression is because of integration errors.37 (C) Donor-only-treated cells showing no endogenous HDR. (D and E) FANCC-TLR-293T cell line transfected with the target gRNA, GFP donor, and the Cas9 nuclease (D) or nickase (E) with GFP (x-axis) and mCherry (y-axis) measured at 72 hr posttransfection. (F–H) Quantification and graphical representation of four different experiments and three different donor concentrations observed from FACs plots similar to those shown in (B–E). GFP data are plotted as green bars, and mCherry is plotted as red. The three different GFP donor concentrations (250, 500, and 1000 ng) are detailed. The basal level of mCherry from 2B was subtracted from all treatment groups. For the nuclease, p<0.05 for NHEJ (mCherry) vs. GFP (HDR). For the nickase, p<0.05 for GFP (HDR) vs. mCherry (NHEJ). (I) HDR ratio for nuclease and nickase. To determine the HDR ratio, the percentage of cells expressing GFP was divided by those expressing mCherry at each donor concentration. Mean±SD are graphed, and for nickase vs. nuclease at 250 ng of donor, p= 0.7, at 500 ng of donor p= 0.04, and at 1000 ng of donor p<0.001. TLR, traffic light reporter.
In aggregate, over three doses of donor concentration, the nuclease mediated the highest levels of GFP by HDR; however, there was a concomitant increase in NHEJ-induced mCherry (Fig. 2F–H). The nickase showed lower overall rates of HDR compared with the nuclease, but there was minimal NHEJ (Fig. 2F–H). When these data are expressed as a ratio of HDR to NHEJ, the repair of DNA nicks shows a clear preference for HDR (Fig. 2I). These data show that the nickase version of Cas9 promotes HDR and minimizes NHEJ similarly to previously categorized nuclease and nickase DNA-targeting enzymes.37
CRISPR on-target and off-target analysis
An important factor in gene-editing-based correction strategies is the potential for off-target (OT) effects because of sequence homology between the target site and other genomic loci. Therefore, we sought to assess the safety profile of the CRISPR/Cas9 reagent because of concerns of high levels of OT effects with this system.39
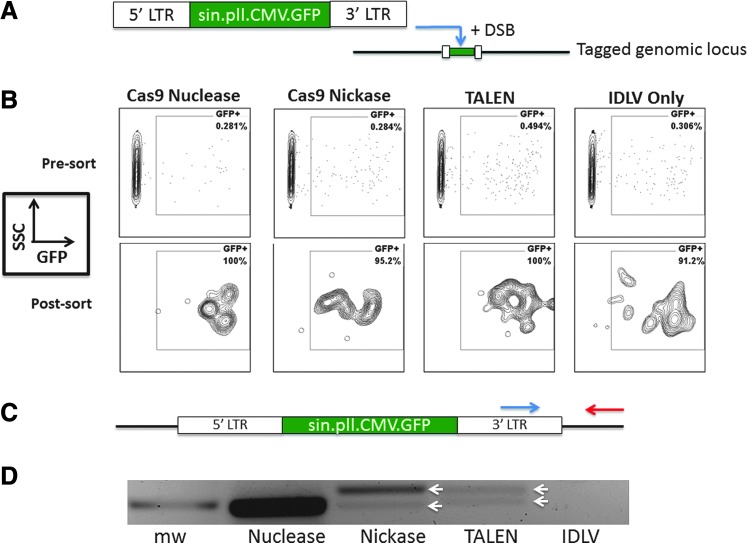
The CRISPR Design Tool analysis software from the Zhang laboratory has shown accuracy in predicting OT sites35 and revealed five such sites for our FANCC CRISPR that were within genes (Fig. 3A). To rigorously assess whether the FANCC CRISPR/Cas9 nuclease and nickase exhibited intragenic OT activity, we employed the dual methodologies of the Surveyor assay and an IDLV reporter gene-trapping technique.30 Surveyor analysis showed no demonstrable activity for the nuclease or nickase at the predicted intragenic OT sites (Fig. 3B). Because the limit of detection with the Surveyor methodology has been reported to be ∼1%,27 we also assessed OT effects by the tandem delivery of the CRISPR/Cas9 nuclease or nickase with a GFP IDLV, and used a PCR-based gene-trapping approach in order to maximize sensitivity (Fig. 4A). IDLV transduction of 293Ts resulted in ∼80% GFP expression at 48 hr that rapidly diminished because of loss of episomal vector genomes during cell division, which resulted in a low level of stable GFP cells that were then sorted to purity and expanded (Fig. 4B). PCR analysis using a 3′ LTR forward primer and a FANCC locus reverse primer (Fig. 4C) yielded a PCR product for the Cas9 nuclease- and nickase-treated cells but not the IDLV-only control cells (Fig. 4D) Sequencing of these products showed an LTR:FANCC genomic junction immediately upstream of the CRISPR PAM (Supplementary Fig. S3). These results are consistent with previous studies showing that the delivery of a GFP IDLV into cells results in trapping the viral cargo at the site of a DSB.30,31 The results also validate the methodology as a means to detect loci at which the nuclease is active.
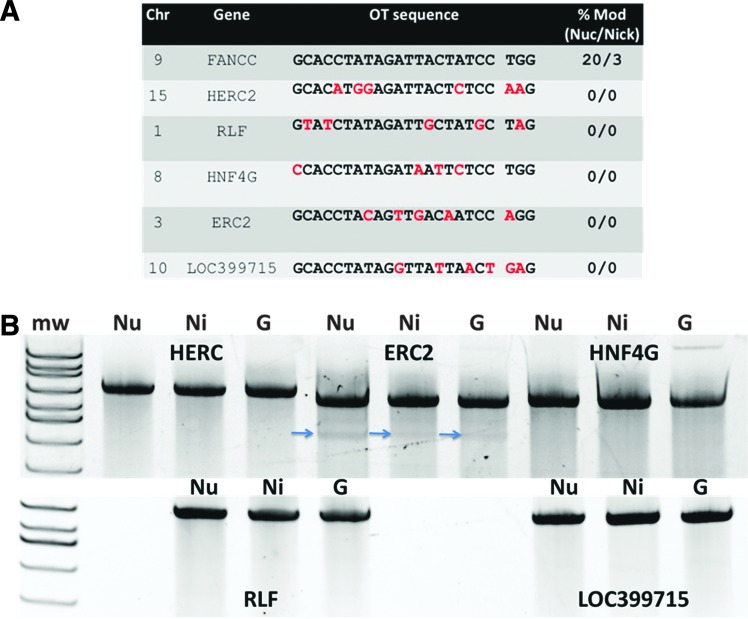
FIG. 3.
Off-target sequence analysis. (A) In silico off-target site acquisition. The CRISPR Design Tool identified five intragenic OT sites. Chromosomal location and gene name are shown with the FANCC target locus at top. Mismatches between FANCC target and OT sites are in red. (B) Surveyor nuclease assessment of OT sites. OT alleles for 293T cells treated with nuclease (“Nu”), nickase (“Ni”), or GFP (“G”) were amplified and assayed by the Surveyor procedure. Blue arrow indicates a cleavage product present in all three treatment groups, which indicates the presence of a natural polymorphism. At right in (A) is the % modification (“% Mod”) using the CRISPR nuclease (“nuc”) or nickase (“nick”) at each target site determined by Surveyor.
FIG. 4.
Integrase-deficient lentiviral gene tagging. (A) Diagram of self-inactivating integrase-deficient GFP lentiviral cassette with expression regulated by the CMV promoter (sin.pll.CMV.GFP). In the presence of the CRISPR/Cas9 reagent that generates DNA DSBs or nicks, a full copy of the viral cassette can be trapped at the on- or off-target break site where it remains permanently. (B) FACS analysis of IDLV treatment groups. Seven days post-IDLV treatment±concomitant nuclease and nickase delivery, the cells were assessed for GFP (labeled “7 days”). The sorted cell (“Post sort”) populations were analyzed 5 days after the initial sort. (C) PCR screen for IDLV at FANCC and OT sites. PCR assay using a 3′ LTR primer (blue arrow) and a FANCC or OT locus-specific primer (red arrow) was performed. (D) FANCC locus-specific IDLV integration was observed and white arrows show amplicons that were sequenced. DSB, double-stranded DNA break; IDLV, integrase-deficient lentiviral; LTR, long terminal repeat. Color images available online at www.liebertpub.com/hum
These analyses were biased toward specific loci predicted in silico. In order to fully evaluate the safety profile of these reagents, we performed an unbiased, genome-wide screen. To identify the sites of integration of the IDLV, the samples were tested using LAM PCR and by nonrestrictive (nr)LAM PCR that is not reliant on a nearby restriction endonuclease site.32,40 The workflow and analysis is summarized in Fig. 5A. Deep sequencing resulted in approximately 3.9 million individual paired sequencing reads, with nearly 900,000 that could be mapped to the human genome using the high-throughput insertion site analysis pipeline.33 Within this, IS were identified with an IS being classified as a junction between viral LTR and a genomic sequence. Each treatment group contained between 130 and 200 IS (Fig. 5B) that could be further analyzed for the formation of CLIS. CLIS are defined as a minimum of two integration events within a ∼500 bp range of genomic DNA.30 Such limited-range clusters in this context are considered to occur because of the recognition of a target DNA sequence (on- or off-target) by the gene-editing reagent with subsequent DNA cutting and IDLV trapping.30,32,33,40 Further, we also screened the human genome for CLIS at putative OT-binding sites that contained up to five mismatches between the OT locus and the FANCC CRISPR/Cas9 target sequence.
FIG. 5.
Unbiased genome-wide screen for OT loci. (A) Experimental workflow. Duplicate samples of 293T cells with integrated IDLV were subjected to nrLAM PCR and LAM PCR using MseI or MluCI enzymes and next-generation sequencing with Illumina MiSeq deep sequencing. The dataset was then refined using the High-Throughput Site Analysis Pipeline (HISAP).33 HISAP trims the sequence reads to remove vector and linker nucleotides in order to retain only the host genomic fragment amplicons. Redundant/identical sequences are consolidated and then mapped and annotated using the BLAT UCSC Genome Informatics database. The prevalence of CLIS in proximity to a locus is then assessed. (B) CLIS identification of IDLV integrants. The sample identifiers and number of sequence reads analyzed for each are indicated at left. The total number of IS for each sample is shown and the number of CLIS (X=no CLIS identified for IDLV-only treatment group) observed. For all of the reagents, the CLIS were localized only to the FANCC locus and were located within a 80 bp window. CLIS, clusters of integrations. Color images available online at www.liebertpub.com/hum
Our results documented CLIS frequencies of 5–31 at the intended target site, while no CLIS were recovered at loci containing partial target site homology (Fig. 5B). Cumulatively, our data show highly specific CRISPR/Cas9 reagents and support their application for precision gene-editing approaches and support new studies showing a favorable safety profile for this system.41
Homology-directed repair
To test the ability of our custom-designed reagents to mediate FANCC gene HDR, we derived a transformed skin fibroblast culture from an FA-C patient homozygous for the c.456+4A>T mutation. The cells were treated with a donor plasmid and either the CRISPR/Cas9 nuclease or nickase genome-editing reagents. The donor plasmid functions as the repair template following the generation of a DSB and spans a region of the FANCC gene from the third exon to the fifth intron (Fig. 6A and B). It also contains a puromycin selectable marker and the FANCC cDNA that are flanked by loxp sites, as well as silent DNA polymorphisms designed to allow tracking of HDR events and to prevent FANCC-specific nuclease cutting of the FANCC donor sequences (Fig. 6A and B and Supplementary Fig. S1).
FIG. 6.
FANCC donor design and homology-directed repair. (A) The FANCC locus with the c.456+4A>T intronic mutation is indicated with a red arrow with asterisk. Blue arrows indicate the endogenous genomic primers used for HDR screening. (B) Gene correction donor. The donor is shown in alignment relative to the endogenous locus. The plasmid donor contains a 1.3 kb left arm of homology comprised of FANCC genomic sequences, silent mutations to prevent nuclease cutting of the donor, and the normalized base for the c.456+4A>T mutation (green line). Following this was a loxP-flanked PGK promoter-regulated puromycin-T2A-FANCC expression cassette and a right donor arm that is 0.8 kb in length. Blue arrows show the donor-specific PCR primers used for PCR analysis of CRISPR-treated, selected, and expanded clones. (C) Representative gel image of PCR screening approach for the left (“Lt”) and right (“Rt”) HDR using the donor-specific and locus-specific primers from (A) and (B). (D) Number of gene-corrected clones obtained. Numbers indicate the number of clonally expanded cells that showed a positive HDR PCR product. Two independent experiments were performed, and the data pooled together to obtain the total number of clones positive for HDR. (E) HDR-mediated c.456+4A>T mutation correction. Representative Sanger sequence data of the c.456+4A>T locus in untreated cells (top) and gene-corrected clones (bottom). Blue shading indicates the mutant thymine or corrected adenine base. Arrows on bottom sequence file show the donor-derived silent mutations present in the corrected clones.
Introduction of the gene-editing reagent with the donor followed by puromycin selection allowed us to screen the treatment pools in bulk with an HDR-specific PCR strategy that uses a donor-specific primer and locus-specific primer located outside the donor arms (Fig. 6A and B). Once a bulk population showed evidence of HDR by PCR (Fig. 6C), it was plated at low density in order to isolate and expand single-cell-derived clones. The expansion of individual clones allowed us to then quantitate the rate of HDR in a pool using the HDR PCR strategy. Nearly 20 individual clones for each of the CRISPR/Cas9 nuclease and nickase treatment groups were able to be expanded and assessed for HDR by PCR. Each resulted in at least one clone that showed HDR, with the nickase treatment resulting in the derivation of the most clones exhibiting donor-derived repair (Fig. 6D). Sanger sequencing of these HDR PCR products showed the presence of donor-derived polymorphisms, as well as correction of the c.456+4A>T mutation (Fig. 6E and Supplementary Fig. S4). These data document the ability of CRISPR/Cas9 reagents to mediate precise correction of FANCC c.456+4A>T mutation by HDR.
Restoration of FANCC gene expression and assessment in hematopoietic stem cells
The result of the c.456+4A>T mutation is the skipping of exon 4 (Fig. 7A and B and Supplementary Fig. S5). To determine whether genome editing by CRISPR/Cas9 resulted in restoration of exon 4 expression, we performed a corrected transcript/allele-specific RT-PCR using a forward primer that recognizes unique donor-derived bases and a reverse primer in exon 8 that is, importantly, several kilobases downstream of the terminus of the donor arm (Fig. 7B and C). CRISPR/Cas9 nuclease and nickase clones each showed the presence of the modified transcript, while untreated FA-C and wild-type cells did not show a product, thus confirming the specificity of the assay (Fig. 7C). To conclusively demonstrate seamless continuity of exon 4 with downstream exons, we sequenced the amplicons and showed the presence of polymorphisms present from HDR and intact exon–exon junctions (Fig. 7D and Supplementary Fig. S6).
FIG. 7.
CRISPR-mediated restoration of FANCC. (A) The FANCC locus with mutation is indicated with a red asterisk. The mutation results in aberrant splicing (red lines) that cause exon 4 (green box) skipping. Normal splicing is indicated by the green dashed lines. Tan box represents exon 3, green box is exon 4, purple box is exon 5, and orange box is exon 8. (B) FANCC transcripts. The c.456+4A>T mutation-induced exon skipping results in deletion of exon 4. Gene correction results in restoration of exon 4 in the transcript. The green arrow indicates an allele-specific primer for the silent base changes that were introduced by donor-derived HDR. The blue primer is an exon 8-specific primer. (C) Allele-specific PCR of nickase- and nuclease-corrected cell clones. A representative gel of an allele-specific PCR showing normalized transcripts in the nuclease and nickase clones. The specificity of the primer set is evident because of absence of amplification in FA-C (FC) or wild-type (WT) cells. To ensure that cDNA was amplification grade, samples were subjected to PCR with GAPDH primers (bottom). Mw, molecular weight standards. (D) Sanger sequencing of gene-modified allele. At left is the start of exon 4, with blue arrows indicating the silent polymorphisms that were incorporated into the genome-targeting donor. At right is the junction (shaded in blue) of the restored exon 4 contiguous with exon 5. (E) FANCC activity. Graph is a representation of four experiments of the nuclease and two nickase clones utilizing flow cytometric analysis of phosphorylated γ-H2AX in FA cells that are untreated or treated with 2 mM hydroxyurea. Nuclease or nickase clones were assessed simultaneously and data are presented as the mean fluorescence intensity (MFI) of the phospho-γ-H2AX antibody signal. FC cells (red bar) were stained with an isotype control. Mean±SD are graphed and * is gene-corrected clones vs. controls with p<0.05.
These data confirm the ability of CRISPR/Cas9-mediated HDR of the c.456+4A>T mutation to restore proper expression of FANCC exon 4 in cells from an individual with FA-C. The positioning of our exogenous sequences (i.e., puromycin and FANCC cDNA) within the donor construct resulted in their insertion by HDR into intronic sequence >400 bp away from an exon and thus did not result in perturbation of splicing. However, to assess whether the gene correction we observed at the DNA and mRNA levels extended to functional rescue, we utilized cre-recombinase to remove the floxed sequences (Supplementary Fig. S7). In agreement with a previous report that documented an inability of FA-C fibroblasts to phosphorylate γ-H2AX in response to DNA-damaging agents in vitro, we show that untreated FA-C cells do not phosphorylate γ-H2AX (Fig. 7E).42 The clones that were corrected by the nickase or the nuclease showed restored ability to phosphorylate γ-H2AX (Fig. 7E). In totality, at the DNA, mRNA, and protein levels, we show correction of the c.456+4A>T mutation in fibroblasts.
Discussion
We generated a CRISPR/Cas9 reagent for the FANCC locus and showed this system's usefulness in repairing the FANCC c.456+4A>T mutation in patient-derived, transformed fibroblasts utilizing a donor that contained a floxed puromycin and FANCC cDNA flanked by arms of homology to the FANCC locus. We found that our format of donor drastically impacted the outcome of gene repair and our ability to derive corrected clones. The ideal strategy, and our initial work, focused on delivering either a single-stranded or double-stranded DNA donor to FA cells that would correct the mutation and allow for preferential expansion of the cells because of the restored DNA repair ability and resistance to agents such as mitomycin C (MMC). However, untreated or CRISPR-treated normal and FA fibroblasts were prone to senesce/arrest in response to MMC rather than a high degree of cell death, preventing a selective outgrowth of gene-corrected cells (unpublished observations). Moreover, our results using the TLR assay showed maximal correction rates of ∼2% (Fig. 2), which, while comparatively high to canonical gene targeting without genome-editing nucleases, are still sufficiently low and necessitated the use of a drug selection-based strategy.
We utilized a floxed puromycin-based selection strategy that has proven reliable in previous gene correction studies.31,43 Our initial attempts focused on a donor similar to that depicted in Fig. 6B except lacking the FANCC cDNA component. Using this first-generation donor, we were unable to expand clones to meaningful numbers. Therefore, we reformatted the donor to allow for transient FANCC correction by virtue of the FANCC cDNA expression cassette. Using this donor we were able to achieve gene correction with the highest frequency observed when utilizing the D10A nickase version of Cas9 (Fig. 6D). This resulted in restoration of proper splicing and functional rescue of the FA phenotype (Figs. 6 and 7).
To assess the preferred pathway of DNA repair for the CRISPR/Cas9 system, we employed the TLR system. Directly comparing the two versions of Cas9 showed that the HDR rates for the nuclease were higher than the nickase (Fig. 2B–H). However, this was offset by a correspondingly high rate of nuclease-induced NHEJ that was essentially absent from nickase-treated cells (Fig. 2). As such, expressing the outcome of DNA cleavage as a ratio of HDR versus NHEJ showed that the nickase possesses a strong bias toward faithful gene repair by HDR (Fig. 2I). These findings are consistent with other studies in the field showing that DNA nicking is associated with preferential repair by HDR.35,37,44–47 We postulate that the phenotype of FA may make nickases especially valuable based on a recent study by Davis and Maizels that shows that DNA nicks can be resolved by an alternative HDR (altHDR) pathway that proceeds when BRCA2 or RAD51 are downregulated.44 Given the intimate connection of FANCC and other FA proteins with BRCA2 and RAD51 for mediating HDR following a DSB,48 FA cells may preferentially employ altHDR. A second key finding from the Davis study showed that targeting the nontemplate strand as we have done promotes higher levels of HDR.44 Our results support these findings and show that Cas9 nickase promotes HDR and minimizes NHEJ (Figs. 2, 6, and 7).38,44 This resulted in correction at the genomic locus, restoration of proper mRNA splicing, and phenotypic rescue in patient-derived fibroblasts (Figs. 6 and 7).
The primary utility of the fibroblasts in our study was to establish proof of principle for gene editing in the FA class of disorders. The ideal cell type from a clinical perspective is the correction of HSCs. An elegant new study reports the use of ZFN for the first ever correction of patient-derived HSCs.49 A key finding from the Genovese report is the importance of optimizing the cellular culture conditions and donor and nuclease delivery platforms. They delivered their donor as an IDLV followed by ZFN mRNA 1 day later to cells cultured in the aryl hydrocarbon antagonist StemRegenin 1 (SR1) plus/minus 16,16-dimethyl-prostaglandin E2 (dmPGE2).49 SR1 promotes self-renewal and expansion of CD34+progenitors and dmPGE2 enhances engraftment rates.34,50 Importantly, the foundation for these studies was based on multiple previous reports describing the optimization and activity of these ZFNs as part of an additive approach to ultimate HSC application.28,30,51–54 We have initiated a similar stepwise and progressive experimental approach and report here proof of concept for the ability of CRISPR/Cas9 to mediate a gene correction event in FA fibroblasts. These data will synergize with optimized HSC culture, expansion, and gene transfer protocols as part of the formation of next-generation therapies.
A second crucial determinant for clinical use of gene-editing reagents is a detailed analysis of OT effects. Our in silico analysis identified five OT sites within coding regions that shared significant sequence homology to the FANCC target site (Fig. 3A): HERC2 encodes a large protein believed to function as a ubiquitin ligase,55 RLF and HNF4G are predicted to be transcriptional regulators,56,57 ERC2 is involved in neurotransmitter release,58 and LOC399715 is an uncharacterized RNA gene. We assessed these sites for evidence of CRISPR OT activity by the Surveyor method and observed no demonstrable OT activity at the predicted OT sites (Fig. 3). To identify OT sites that may exist within the genome but that are not predicted by in silico methods, we utilized the ultrasensitive, unbiased, genome-wide LAM PCR methodology.30,32,33,40 Greater than 100,000 sequence reads were evaluable for each of the reagents and control samples (Fig. 5B). The numerous IS observed in control (i.e., IDLV alone)-treated cells were consistent with our previous studies showing IDLV capture at genomic fragile spots that occur endogenously and independent of nuclease activity.30,31 Importantly, only cells treated with Cas9 contained IDLV CLIS indicative of site-specific nuclease activity. The CLIS were solely localized to the FANCC locus, showing that the reagents employed in our study are exquisitely specific. Concerns for poor CRISPR/Cas9 specificity have been detailed in two prior studies.39,59
One of these studies showed that CCR5 CRISPR/Cas9, similar to CCR5 ZFNs and TALENs, also exhibits OT activity at CCR2.59 These studies and ours highlight the importance of rigorously designing CRISPR/Cas9 candidates for loci of sufficient sequence complexity in order to minimize OT effects. Doing so, as we have shown here and is in agreement with others, can result in a highly specific gene-editing reagent.41
The cell type utilized for the OT effects was carefully considered, and our intention was to perform the OT screens in the 293T cell line to identify candidates that could then be validated in patient-derived cells. The rationale for this is that the rapidly proliferating nature of 293Ts would facilitate dilution of episomal IDLV, thus decreasing background and minimizing the number of ectopic IDLV integration events at genomic fragile sites. The 293s rapidly diluted the unintegrated IDLV (Fig. 4B) and, because of their open chromatin profile, we predicted that OT events would manifest to the highest possible degree representing the most thorough and stringent screening procedure. Moreover, previous studies have shown that laboratory cell lines employed for IDLV gene mapping prove a useful predictor for gene-editing OT site analysis in primary cells.30 As such, the lack of OT sites in 293Ts suggests a highly specific reagent. What role the phenotype of FA plays in the targeting profile of gene-editing proteins is under investigation because of the possibility of a higher degree of DNA breaks in FA cells that may lead to a higher prevalence of OT effects. These data will be essential for clinical application and are being assessed on the genome level.
In summary, we show that while both the CRISPR/Cas9 nuclease and nickase mediated direct c.456+4A>T mutation repair that resulted in normalization of the FANCC gene, the nickase was more efficient. Further, we provide support for a favorable safety profile using these synthetic molecules for correction of genetic disease in human cells. The observation that CRISPR/Cas9 mediates HDR in FA establishes further proof of principle for the application of genome editing for human genetic disorders, including those with defects in the DNA repair pathway.
Supplementary Material
Acknowledgments
We are grateful to Nancy Griggs Morgan for expert assistance in article preparation and editing. We are also thankful for the generosity of the Kidz 1st Fund, the Fanconi Anemia Research Fund, the Children's Cancer Research Fund, the Lindahl Family & the Corrigan Family. MJO is supported by 8UL1TR000114-02. JT is supported in part by R01 AR063070 and P01 CA065493. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR000114 (MJO). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Disclosure Statement
J.K.J. has a financial interest in Transposagen Biopharmaceuticals. J.K.J.'s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. No competing financial interests exist for the remaining authors.
References
- 1.Huard CC, Tremblay CS, Helsper K, et al. . Fanconi anemia proteins interact with CtBP1 and modulate the expression of the Wnt antagonist Dickkopf-1. Blood 2013;121:1729–1739 [DOI] [PubMed] [Google Scholar]
- 2.Whitney MA, Jakobs P, Kaback M, et al. . The Ashkenazi Jewish Fanconi anemia mutation: incidence among patients and carrier frequency in the at-risk population. Hum Mutat 1994;3:339–341 [DOI] [PubMed] [Google Scholar]
- 3.Whitney MA, Saito H, Jakobs PM, et al. . A common mutation in the FACC gene causes Fanconi anaemia in Ashkenazi Jews. Nat Genet 1993;4:202–205 [DOI] [PubMed] [Google Scholar]
- 4.Grompe M, D'Andrea A. Fanconi anemia and DNA repair. Hum Mol Genet 2001;10:2253–2259 [DOI] [PubMed] [Google Scholar]
- 5.Pickering A, Zhang J, Panneerselvam J, Fei P. Advances in the understanding of Fanconi anemia tumor suppressor pathway. Cancer Biol Ther 2013;14:1089–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacMillan ML, Wagner JE. Haematopoeitic cell transplantation for Fanconi anaemia—when and how? Br J Haematol 2010;149:14–21 [DOI] [PubMed] [Google Scholar]
- 7.Barker JN, Davies SM, Defor T, et al. . Survival after transplantation of unrelated donor umbilical cord blood is comparable to that of human leukocyte antigen-matched unrelated donor bone marrow: results of a matched-pair analysis. Blood 2001;97:2957–2961 [DOI] [PubMed] [Google Scholar]
- 8.Davies SM, Khan S, Wagner JE, et al. . Unrelated donor bone marrow transplantation for Fanconi anemia. Bone Marrow Transplant 1996;17:43–47 [PubMed] [Google Scholar]
- 9.Fagerlie SR, Diaz J, Christianson TA, et al. . Functional correction of FA-C cells with FANCC suppresses the expression of interferon gamma-inducible genes. Blood 2001;97:3017–3024 [DOI] [PubMed] [Google Scholar]
- 10.Hyland KA, Olson ER, Clark KJ, et al. . Sleeping Beauty-mediated correction of Fanconi anemia type C. J Gene Med 2011;13:462–469 [DOI] [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. . Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 2008;118:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. . LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003;302:415–419 [DOI] [PubMed] [Google Scholar]
- 13.Maeder ML, Thibodeau-Beganny S, Osiak A, et al. . Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 2008;31:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cermak T, Doyle EL, Christian M, et al. . Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 2011;39:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christian M, Cermak T, Doyle EL, et al. . Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010;186:757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, Ran FA, Cox D, et al. . Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P, Yang L, Esvelt KM, et al. . RNA-guided human genome engineering via Cas9. Science 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, East A, Cheng A, et al. . RNA-programmed genome editing in human cells. Elife 2013;2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotin RM, Linden RM, Berns KI. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J 1992;11:5071–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer 2011;12:51–58 [DOI] [PubMed] [Google Scholar]
- 21.Tan I, Ng CH, Lim L, Leung T. Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J Biol Chem 2001;276:21209–21216 [DOI] [PubMed] [Google Scholar]
- 22.DeKelver RC, Choi VM, Moehle EA, et al. . Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res 2010;20:1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hockemeyer D, Soldner F, Beard C, et al. . Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 2009;27:851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cumming RC, Liu JM, Youssoufian H, Buchwald M. Suppression of apoptosis in hematopoietic factor-dependent progenitor cell lines by expression of the FAC gene. Blood 1996;88:4558–4567 [PubMed] [Google Scholar]
- 25.Pang Q, Christianson TA, Keeble W, et al. . The Fanconi anemia complementation group C gene product: structural evidence of multifunctionality. Blood 2001;98:1392–1401 [DOI] [PubMed] [Google Scholar]
- 26.Doyon Y, Choi VM, Xia DF, et al. . Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods 2010;7:459–460 [DOI] [PubMed] [Google Scholar]
- 27.Guschin DY, Waite AJ, Katibah GE, et al. . A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 2010;649:247–256 [DOI] [PubMed] [Google Scholar]
- 28.Lombardo A, Genovese P, Beausejour CM, et al. . Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol 2007;25:1298–1306 [DOI] [PubMed] [Google Scholar]
- 29.Matrai J, Cantore A, Bartholomae CC, et al. . Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology 2011;53:1696–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabriel R, Lombardo A, Arens A, et al. . An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 2011;29:816–823 [DOI] [PubMed] [Google Scholar]
- 31.Osborn MJ, Starker CG, Mcelroy AN, et al. . TALEN-based gene correction for epidermolysis bullosa. Mol Ther 2013;21:1151–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paruzynski A, Arens A, Gabriel R, et al. . Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc 2010;5:1379–1395 [DOI] [PubMed] [Google Scholar]
- 33.Arens A, Appelt JU, Bartholomae CC, et al. . Bioinformatic clonality analysis of next-generation sequencing-derived viral vector integration sites. Hum Gene Ther Methods 2012;23:111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boitano AE, Wang J, Romeo R, et al. . Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 2010;329:1345–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu PD, Scott DA, Weinstein JA, et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 2012;109:E2579–E2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Certo MT, Ryu BY, Annis JE, et al. . Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods 2011;8:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho SW, Kim S, Kim Y, et al. . Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014;24:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, Foden JA, Khayter C, et al. . High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013;31:822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabriel R, Eckenberg R, Paruzynski A, et al. . Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med 2009;15:1431–1436 [DOI] [PubMed] [Google Scholar]
- 41.Smith C, Gore A, Yan W, et al. . Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 2014;15:12–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brozovic A, Damrot J, Tsaryk R, et al. . Cisplatin sensitivity is related to late DNA damage processing and checkpoint control rather than to the early DNA damage response. Mutat Res 2009;670:32–41 [DOI] [PubMed] [Google Scholar]
- 43.Zou J, Maeder ML, Mali P, et al. . Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell 2009;5:97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis L, Maizels N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc Natl Acad Sci USA 2014;111:E924–E932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metzger MJ, Mcconnell-Smith A, Stoddard BL, Miller AD. Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Res 2011;39:926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez CL, Certo MT, Mussolino C, et al. . Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res 2012;40:5560–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilber A, Frandsen JL, Wangensteen KJ, et al. . Dynamic gene expression after systemic delivery of plasmid DNA as determined by in vivo bioluminescence imaging. Hum Gene Ther 2005;16:1325–1332 [DOI] [PubMed] [Google Scholar]
- 48.Kitao H, Yamamoto K, Matsushita N, et al. . Functional interplay between BRCA2/FancD1 and FancC in DNA repair. J Biol Chem 2006;281:21312–21320 [DOI] [PubMed] [Google Scholar]
- 49.Genovese P, Schiroli G, Escobar G, et al. . Targeted genome editing in human repopulating haematopoietic stem cells. Nature 2014;510:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goessling W, Allen RS, Guan X, et al. . Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell 2011;8:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doyon Y, Vo TD, Mendel MC, et al. . Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 2011;8:74–79 [DOI] [PubMed] [Google Scholar]
- 52.Holt N, Wang J, Kim K, et al. . Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 2010;28:839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lombardo A, Cesana D, Genovese P, et al. . Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods 2011;8:861–869 [DOI] [PubMed] [Google Scholar]
- 54.Orlando SJ, Santiago Y, Dekelver RC, et al. . Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res 2010;38:e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harlalka GV, Baple EL, Cross H, et al. . Mutation of HERC2 causes developmental delay with Angelman-like features. J Med Genet 2013;50:65–73 [DOI] [PubMed] [Google Scholar]
- 56.Gerdin AK, Surve VV, Jonsson M, et al. . Phenotypic screening of hepatocyte nuclear factor (HNF) 4-gamma receptor knockout mice. Biochem Biophys Res Commun 2006;349:825–832 [DOI] [PubMed] [Google Scholar]
- 57.Makela TP, Hellsten E, Vesa J, et al. . The rearranged L-myc fusion gene (RLF) encodes a Zn-15 related zinc finger protein. Oncogene 1995;11:2699–2704 [PubMed] [Google Scholar]
- 58.Kiyonaka S, Nakajima H, Takada Y, et al. . Physical and functional interaction of the active zone protein CAST/ERC2 and the beta-subunit of the voltage-dependent Ca(2+) channel. J Biochem 2012;152:149–159 [DOI] [PubMed] [Google Scholar]
- 59.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 2013;41:9584–9592 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.