Abstract
This article considers the problem whereby, given two metabolic networks N1 and N2, a set of source compounds, and a set of target compounds, we must find the minimum set of reactions whose removal (knockout) ensures that the target compounds are not producible in N1 but are producible in N2. Similar studies exist for the problem of finding the minimum knockout with the smallest side effect for a single network. However, if technologies of external perturbations are advanced in the near future, it may be important to develop methods of computing the minimum knockout for multiple networks (MKMN). Flux balance analysis (FBA) is efficient if a well-polished model is available. However, that is not always the case. Therefore, in this article, we study MKMN in Boolean models and an elementary mode (EM)-based model. Integer linear programming (ILP)-based methods are developed for these models, since MKMN is NP-complete for both the Boolean model and the EM-based model. Computer experiments are conducted with metabolic networks of clostridium perfringens SM101 and bifidobacterium longum DJO10A, respectively known as bad bacteria and good bacteria for the human intestine. The results show that larger networks are more likely to have MKMN solutions. However, solving for these larger networks takes a very long time, and often the computation cannot be completed. This is reasonable, because small networks do not have many alternative pathways, making it difficult to satisfy the MKMN condition, whereas in large networks the number of candidate solutions explodes. Our developed software minFvskO is available online.
Key words: : algorithm, Boolean model, elementary mode, integer linear programming, metabolic network, NP-complete
1. Introduction
Metabolic networks represent relations between biochemical reactions and metabolites in living cells. The removal (or knockout) of metabolism-related genes is often simulated in metabolic networks, as the perturbation of genes frequently corresponds to the inhibition of certain reactions in metabolic networks.
Many types of mathematical models have been developed for this purpose. For small size networks, models using the ordinary differential equations (ODEs) are often used. Although ODEs have a detailed explanatory power, their applicability is limited by the difficulty of obtaining the necessary kinetic parameters, and their limited scalability. On the other hand, less detailed approaches like Boolean models and constraint-based models have been used in larger networks (Gonçalves et al., 2013).
Flux balance analysis (FBA) (Raman and Chandra, 2009; Varma and Palsson, 1994) is a constraint-based mathematical model of metabolic networks in which the stoichiometry and the biomass objective functions are used to predict cell growth rates (Kauffman et al., 2003). Although the standard FBA simply maximizes the biomass objective function, the minimization of metabolic adjustment method (MOMA) (Segre et al., 2002) seeks to minimize the difference between the wild and the knocked-out flows. Flux variability analysis (FVA) assesses the range of this difference (Shlomi et al., 2009), and the optimal knockout strategies have been developed based on such flow models. Optknock (Burgard et al., 2003) determines which reactions should be knocked-out to maximize the biomass objective function. Using bilevel programming, the biomass objective function is first maximized for each knockout(s), and then a reaction set is chosen from the resulting candidates. In contrast, although RobustKnock (Tepper and Shlomi, 2010) is also based on bilevel programming, it maximizes the minimized biomass objective functions by selecting different reactions. Optorf (Kim and Reed, 2010) integrates transcriptional regulatory networks and metabolic networks. The above-mentioned flow models need to define the biomass objective function, which often involves a linear combination of more than 100 metabolites (Raman and Chandra, 2009). Furthermore, it is difficult to define the biomass objective function for higher organisms such as humans, and the essential assumption concerning growth-optimal behavior is not always fulfilled (Schuster et al., 2008).
Another flow model approach for metabolic networks is based on the elementary mode (EM) concept, which does not require a biomass objective function. EM is a minimal set of reactions in a steady state, in which all irreversible reactions are used in the appropriate direction (Schuster and Hilgetag, 1994; Schuster et al., 2000). Stelling et al. (2002) estimated the effect of knockouts by the number of elementary modes (EMs) that include the knocked-out reaction. Based on the idea of topological flux balance (TFB) (Smart et al., 2008), the topological impact degree (TID) calculates the number of reactions that have an EM in common with the knocked-out reactions (Jiang et al., 2009; Tamura et al., 2011). The flux balance impact degree (FBID) is the number of reactions that are not included in any EMs that do not include the knocked-out reactions (Zhao et al., 2013). A minimal cut set (MCS) is the minimal set of reactions whose inactivation leads to a failure of the specified reactions (Klamt and Gilles, 2004; Klamt, 2006). Acuña et al. (2009) proved that computing the MCS is NP-hard. MCSs in a metabolic network are EMs in a dual network (Ballerstein et al., 2012). EMs and MCSs can be calculated by FluxAnalyzer (Klamt and Gilles, 2004) and CellNetAnalyzer (CNA) (Klamt et al., 2007), MATLAB-based tools that can be applied to many network-based problems. Because knockouts may induce side effects that disables the desired functionality, the constrained MCS allows additional constraints that preserve a set of desired modes (Hädicke and Klamt, 2011). Based on an EM-based analysis and the idea of minimal metabolic functionality, optimal knockout strategies were analyzed to maximize industrial production, and the effect was confirmed by biological experiments (Trinh et al., 2006; Unrean et al., 2010).
Another mathematical model of metabolic networks is the Boolean model. Although not suitable for analyzing the mass flow of metabolic networks, the logical analysis of this model is relatively solid as it needs less information than the flow-based models. The synthetic accessibility is defined as the number of reactions required to transform a set of source metabolites into a set of target metabolites (Wunderlich and Mirny, 2006). For a given set of source nodes (called seeds), the scope is defined as the set of producible compounds (Handorf et al., 2005). The damage is the number of reactions that are affected by the knocked-out reactions (Lemke et al., 2004). To take side effects into account, Sridhar et al. (2008) developed a branch-and-bound-based algorithm, OPMET, which minimizes damage to nontarget nodes. Tamura et al. (2010) developed an integer linear programming (ILP)-based method for the Boolean reaction cut (BRC) problem and analyzed the computational complexity of BRC (Tamura and Akutsu, 2010), in which the number of inhibited reactions is minimized to make target compounds nonproducible (Tamura et al., 2010). In the Boolean model of metabolic networks, reactions and compounds can be represented by “AND” and “OR” nodes, respectively, and the network can then be considered as a bipartite graph. Lu et al. (2014) considered the problem of adding a minimum reaction set so that the target compound becomes producible in a Boolean metabolic network. Although the above research focuses on metabolic networks, the basic framework of the Boolean model can be extended to other types of biological networks. For example, Flöttmann et al. formalized signal transduction networks using reaction contingency-based bipartite Boolean modeling (Flöttmann et al., 2013), and Samaga et al. (2010) studied minimal intervention sets (MISs) for Boolean signaling networks.
Thus, in many cases, the effects of reaction inhibition are first estimated in each metabolic network model, and then a certain optimization problem is defined for each estimation model. Furthermore, in addition to maximizing the desired functions, some problems minimize undesirable functions as side effects in the same network.
As a reasonable extension of the above research, this article considers the situation in which different types of cells exist in the same place. For example, many types of bacteria exist in our intestine. Some of them are good bacteria, others are bad bacteria, and they have different metabolic networks. As another example, normal cells and cancer cells may exist in the same organ. In such cases, it may be useful to compute the optimal knockout strategies that would cause the bad cells to lose some essential functionality but allow the good cells to survive. Therefore, we consider the following minimal knockout for multiple networks (MKMN) problem: given two different metabolic networks with source and target compounds, obtain the minimum number of reactions whose inhibition induces the target compounds to become nonproducible in bad cells but producible in good cells.
Although we focus in this article on the case in which two networks are given, it is straightforward to extend the proposed methods to the case in which more than two networks are given.
We analyze this problem using the Boolean model (MKMN-B) and the EM-based model (MKMN-EM). As both MKMN-B and MKMN-EM are NP-complete (as shown in the “Theoretical results” section), we develop methods based on ILP (Schrijver, 1998; Li et al., 2007) for MKMN-B and MKMN-EM. ILP is often used to formalize NP-complete problems, and there is an efficient free ILP solver called CPLEX (IBM, 2010). In MKMN-B, to properly account for the effect of cycles, we utilize the notion of the maximal valid assignment (MaxVA) (Tamura et al., 2010) and the minimal valid assignment (MinVA) (Lu et al., 2014). To obtain faster ILP-based methods by reducing the number of variables, we develop IP-FVS1 and IP-FVS2, which utilize the idea based on feedback vertex sets (FVS) (Tamura et al., 2010), MaxVA is strictly applied in IP-FVS1 but is not applied to the nodes detected by FVS in IP-FVS2. We also develop faster algorithms IP-FVS1-approx and IP-FVS2-approx by limiting the number of time steps, although the optimality of the resulting solutions is not ensured. For MKMN-EM, we develop IP-EM, in which CNA is used to obtain the EMs, and then formalize MKMN-EM using ILP, since MKMN-EM is NP-complete, even when EMs are given.
We apply our developed methods to the metabolic networks data of clostridium perfringens SM101 (CPR) and bifidobacterium longum DJO10A (BLJ), as downloaded from the KEGG database (Kanehisa and Goto, 2000). The CPR network is denoted by N1, and the BLJ network is denoted by N2. Dataset 1 consists of only the central metabolism, and N1 and N2 consist of 73 and 82 nodes, respectively. Dataset 2 consists of the carbon metabolism, fatty acid metabolism, and biosynthesis of amino acids, and N1 and N2 consist of 251 and 328 nodes, respectively. Dataset 3 consists of whole metabolic networks, and N1 and N2 consist of 1231 and 1881 nodes, respectively. We apply IP-FVS1, IP-FVS2, and IP-EM to datasets 1, 2, and 3, where the target compounds are pyruvate, acetyl-CoA, acetate, oxaloacetate, and phosphoenolpyruvate. For most cases in dataset 3, IP-FVS1, IP-FVS2, and IP-EM could not complete the computation in a provisional time limit of 2 hr. Hence, we applied IP-FVS1-approx and IP-FVS2-approx to this dataset and limited the number of time steps to 10. For dataset 1, IP-FVS1, IP-FVS2, and IP-EM took at most 3 sec for every target compound, but in most cases there are no solutions. For dataset 2, IP-FVS1, IP-FVS2, and IP-EM took at most 20 sec for every target compound, and solutions were obtained in about half of the cases. Finally, for dataset 3, IP-FVS1-approx(10) and IP-FVS2-approx(10) finished their calculations within 15 sec for every target compound. We examine the relations between the obtained solutions and predecessors of the target compound in N1 and N2.
2. Materials and Methods
2.1. Main problems
In this subsection, the main problem minimal knockout for multiple networks, (MKMN) is explained using examples. Mathematical definitions are given in the next subsection. The MKMN for the Boolean model and the EM-based model are called MKMN-B and MKMN-EM respectively.
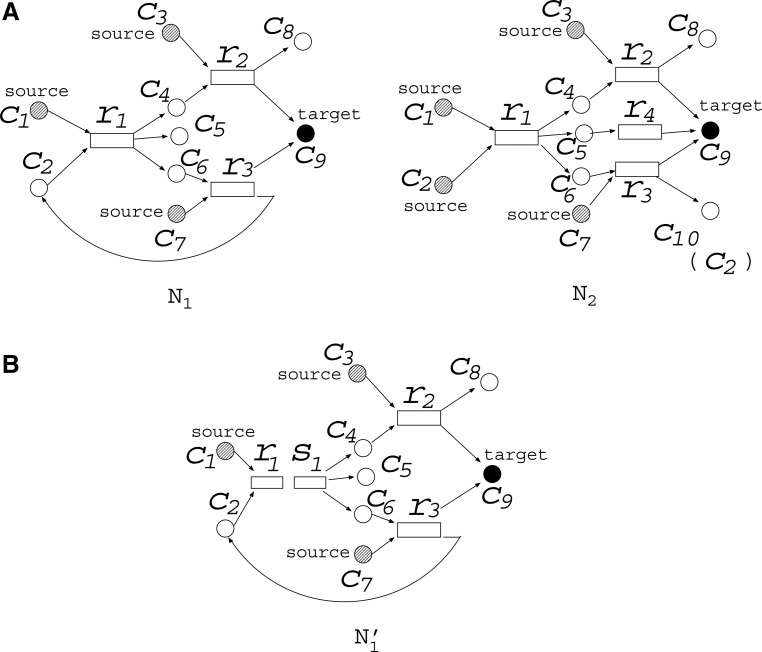
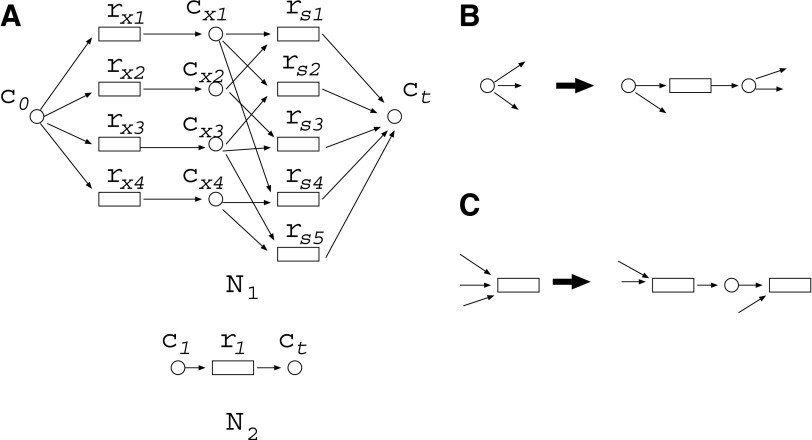
Suppose we have two metabolic networks N1 and N2, as shown in Figure 1A. Rectangles and circles represent reactions and compounds, respectively. For example, reaction r1 has the substrates (reactants) {c1, c2}, and products {c4, c5, c6} for both N1 and N2. However, since the topologies of N1 and N2 are slightly different, N1 does not include reaction r4. For N1, {c1, c3, c7} are called source nodes and are assumed to be supplied by the external environment, whereas {c1, c2, c3, c7} are source nodes for N2. The purpose of MKMN is to find the minimum number of reactions whose inhibition (deletion) induces the target compound to be nonproducible in N1 but producible in N2. In Figure 1A, c9 is the target compound. This should be nonproducible in N1 but producible in N2 after the minimum number of reaction deletions.
FIG. 1.
An example of minimum knockout for multiple networks (MKMN) problem. MKMN is a problem to find the minimum number of reactions whose inhibition makes the target compound nonproducible in N1 but producible in N2. (A) The MKMN solution for this example is {r3}, whose inhibition prevents production of vc9 in N1 but not in N2; (B) r1 of N1 in Figure 1A is decomposed into r1 and s1.
MKMN-B is defined as a problem to solve MKMN in a Boolean model. In MKMN-B, if all substrates exist, the reaction occurs. For example, in Figure 1A, r1 occurs if both c1 and c2 exist. Note that the existence of c2 is not always the same in N1 and N2. In this example, the solution of MKMN-B is to inhibit {r3}, as c2 becomes nonproducible, r1 cannot take place, and as c4 becomes nonproducible, r2 cannot take place, and then c9 becomes non producible in N1 but producible in N2 via r2 or r4. Note that directed cycles often play important roles. On the other hand, the inhibition of {r1} is not a solution as this makes c9 non producible in both N1 and N2. Similarly, inhibiting {r2} is not a solution either, since c9 would remain producible in both N1 and N2.
The problem setting of MKMN-B for N1 is the same as that for the Boolean reaction cut in Tamura et al. (2010). To properly account for the effect of cycles, Tamura et al. (2010) defined the maximal valid assignment (MaxVA). For example, although the optimal solution of MKMN-B for N1 and N2 as in Figure 1A is {r2}, if neither c2 nor c6 initially exists in N1, then neither r1 nor r3 can occur, and c9 becomes nonproducible. Therefore, “inhibiting no reactions” looks like the optimal solution of MKMN-B if c2 is not supplied to N1 from the external environment. To avoid such ambiguity, MaxVA is defined as the 0-1 assignment that satisfies the Boolean constraint for each node, and the number of 1's is maximal among such 0-1 assignments. In Tamura et al. (2010), it was proved that MaxVA can be calculated by initially assigning a value of 1 to all nodes; the effect of reaction inhibition gradually reaches other nodes, and finally converges to some 0-1 assignment, which corresponds to the MaxVA. In MKMN-B, the assumption of MaxVA is applied to N1.
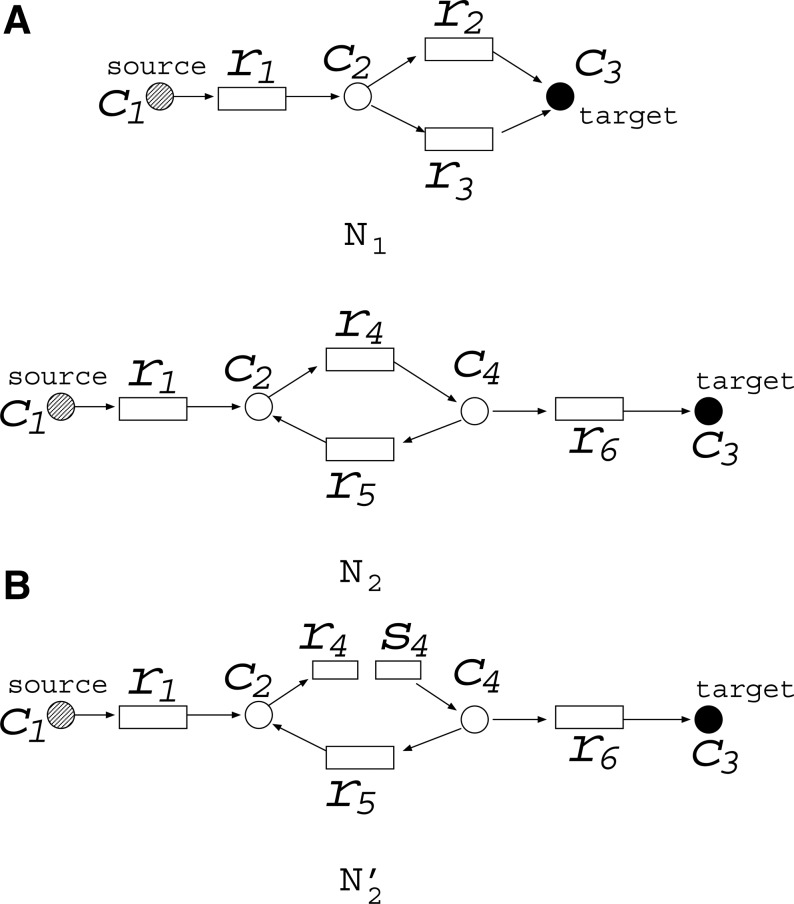
However, the assumption of MaxVA is not appropriate for N2 in MKMN-B. For example, suppose that N1 and N2 in Figure 2A are given. If the assumption of MaxVA is applied to both N1 and N2, {r1} is obtained as the optimal solution of MKMN-B, since (c1, c2, c3, c4, r1, r4, r5, r6) = (1, 1, 1, 1, 0, 1, 1, 1) is the MaxVA of N2 if {r1} is inhibited. However, {r2, r3} seems to be a more appropriate solution of MKMN-B, as the inhibition of {r1} disconnects the source node c1 and the target compound c3 in N2. To account for the effect of cycles, and make the target compound producible, the MinVA notion has been shown to be efficient (Lu et al., 2014). Similar to MaxVA, MinVA is a valid assignment, in which the number of 1's is minimal. In MKMN-B, MaxVA and MinVA are applied to N1 and N2, respectively.
FIG. 2.
The minimal valid assignment (MinVA) is applied to N2. The FVS-based operation for N2 is different from that of N1; (A) {r2, r3} is the optimal solution of MKMN-B for N1 and N2; (B) However, if s4 is initially assigned a value of 1 in 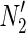 , {r1} is obtained as the solution.
, {r1} is obtained as the solution.
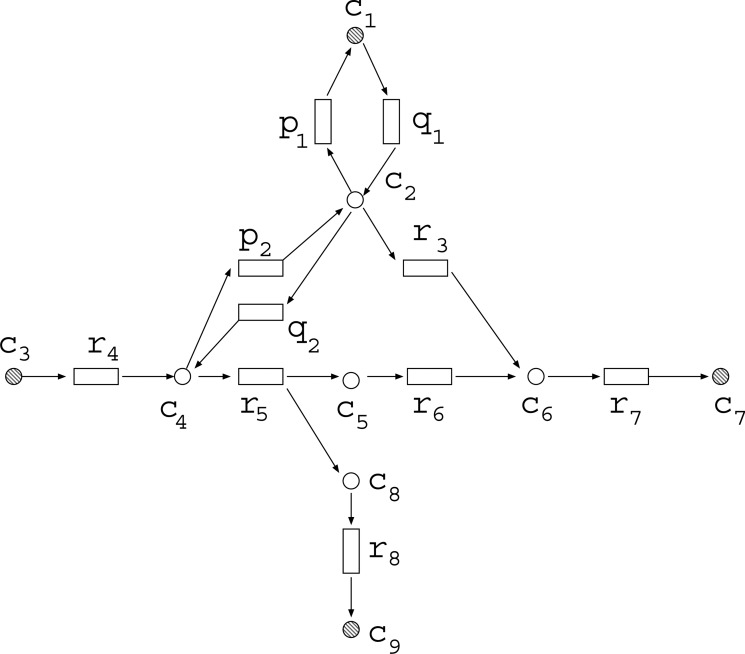
MKMN-EM is defined as a problem to solve MKMN in the elementary mode (EM)-based model. An EM describes a feasible and balanced (steady-state) flux distribution through the network, which is minimal with respect to the utilized reactions (enzymes) (Klamt and Gilles, 2004). Note that {r3} is not the solution of MKMN-EM in Figure 1A, since the target compound c9 becomes nonproducible in both N1 and N2. In MKMN-EM, we consider only the topology of each EM, and do not consider the magnitude of each flow. Therefore, an EM can be represented by a 0-1 assignment of reaction and compound nodes. For example, Table 1 shows the 0-1 assignments of the EMs for the example network shown in Figure 3, where {c1, c3, c7, c9} is a set of external compounds. Suppose that the target compound c7 should become nonproducible, where {c1, c3, c9} is a set of source nodes. As EM1, EM2, EM3, and EM5 can produce c7, the set of reactions to be inhibited must include at least one from each of these four EMs. For example, inhibiting {r3,r5} makes c7 nonproducible since r3 is included in EM2 and EM5, and r5 is included in EM1 and EM3.
Table 1.
The 0-1 Assignments Corresponding to the Elementary Modes of the Example Network of Figure 3, Where {c1, c3, c7, c9} is a Set of External Compounds
| p1 | q1 | p2 | q2 | r3 | r4 | r5 | r6 | r7 | r8 | c1 | c2 | c3 | c4 | c5 | c6 | c7 | c8 | c9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| EM2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| EM3 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| EM4 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| EM5 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
FIG. 3.
The 0-1 assignments of elementary modes (EMs) of this network are listed in Table 1.
2.2. Mathematical definitions of main problems
In this subsection, the main problems of this article are mathematically defined. A metabolic network is defined as a directed bipartite network N = (Vc, Vr, E), where Vc is a set of compound nodes, and Vr is a set of reaction nodes. E is a set of edges connecting a compound node and a reaction node. Therefore, neighbors of compound nodes are always reaction nodes, and neighbors of reaction nodes are always compound nodes. In MKMN, two metabolic networks N1 = (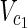 ,
, 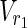 , E1) and N2 = (
, E1) and N2 = (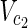 ,
, 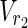 , E2) are given, where |
, E2) are given, where |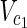 | = m1, |
| = m1, |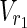 | = n1, |
| = n1, |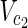 | = m2, |
| = m2, |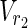 | = n2, m = m1 + m2, and n = n1 + n2 hold. For example, in Figure 1A,
| = n2, m = m1 + m2, and n = n1 + n2 hold. For example, in Figure 1A, 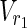 = {r1, r2, r3},
= {r1, r2, r3}, 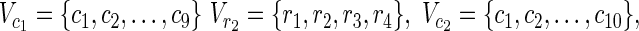 m1 = 9, n1 = 3, m2 = 10, and n2 = 4 hold.
m1 = 9, n1 = 3, m2 = 10, and n2 = 4 hold.
Vex is a set of external nodes, that are affected by the external environment not described in N. 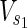 ⊂
⊂ 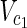 and
and 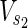 ⊂
⊂ 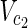 are sets of source nodes in N1 and N2, respectively. Similarly,
are sets of source nodes in N1 and N2, respectively. Similarly, 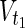 ⊂
⊂ 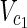 and
and 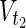 ⊂
⊂ 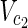 are sets of target nodes in N1 and N2, respectively. It holds that
are sets of target nodes in N1 and N2, respectively. It holds that 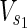 ,
, 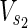 ,
, 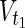 ,
,
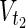 ⊂ Vex. In Figure 1A,
⊂ Vex. In Figure 1A, 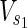 = {c1, c3, c7},
= {c1, c3, c7}, 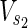 = {c1, c2, c3
c7}, and
= {c1, c2, c3
c7}, and 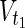 =
= 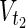 = {c9} hold. Note that some
= {c9} hold. Note that some 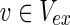 may not be included by either Vs or Vt in the EM-based analysis. In this article, we assume that source nodes do not have incoming edges but that target nodes are allowed to have outgoing edges to take the multiple target compounds into account.
may not be included by either Vs or Vt in the EM-based analysis. In this article, we assume that source nodes do not have incoming edges but that target nodes are allowed to have outgoing edges to take the multiple target compounds into account.
In both the Boolean model and the EM-based model, values of either “0” or “1” are assigned to each node as the magnitude of each flow is not taken into account in MKMN-EM. Suppose that “1” is assigned to a node. In the Boolean model, this means that either the reaction occurs or the compound exists. In the EM-based model, it means that the corresponding node is included in some EM. If “0” is assigned to a node, then either the reaction does not occur, or the compound does not exist in the Boolean model. In the EM-based model, it means that the corresponding node is not included in the EM. Let Va ⊆ 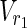 ∪
∪ 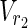 be perturbed reactions. If
be perturbed reactions. If 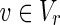 is included in Va, v = 0 always holds whatever values are assigned to other nodes.
is included in Va, v = 0 always holds whatever values are assigned to other nodes.
Let AB be such an assignment. In the Boolean model, AB is called a valid assignment (VA) if it satisfies each of the following:
(i) For each
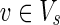 , v = 1 holds;
, v = 1 holds;(ii) for each
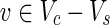 , v = 1 if and only if there is some
, v = 1 if and only if there is some 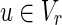 such that
such that 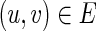 and u = 1 hold;
and u = 1 hold;(iii) for each
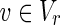 , v = 1 holds if and only if
, v = 1 holds if and only if 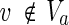 holds and u = 1 holds for all
holds and u = 1 holds for all 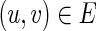 .
.
Therefore, in the Boolean model, each reaction node corresponds to an “AND” node, and each compound node corresponds to an “OR” node. A is the MaxVA for Va, if A is a valid assignment for the given Va, and the number of 1's is the maximal. Similarly, A is the MinVA for Va, if it is a valid assignment for the given Va and the number of 1's is minimal.
MKMN-B is mathematically defined as follows:
Problem: MKMN-B (minimum knockout for multiple networks in the Boolean model)
• Input: Metabolic networks N1 = (
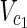 ,
, 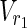 , E1), N2 = (
, E1), N2 = (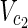 ,
, 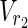 , E2), source nodes
, E2), source nodes 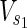 ⊂
⊂ 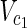
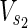 ⊂
⊂ 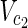 , and target nodes
, and target nodes 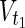 ⊂
⊂ 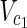 ,
, 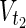 ⊂
⊂ 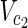 .
.• Output: The minimum cardinality set Va ⊆
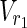 ∪
∪ 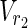 such that MaxVA for N1 ensures every
such that MaxVA for N1 ensures every 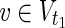 is 0, and MinVA for N2 ensures every
is 0, and MinVA for N2 ensures every 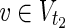 is 1.
is 1.
Note again that the MaxVA and the MinVA are applied to N1 and N2, respectively. (See also the examples of Figs. 1 and 2.)
On the other hand, an EM describes a feasible and balanced (steady-state) flux distribution through the network, which is minimal with respect to utilized reactions (enzymes) (Klamt and Gilles, 2004). An EM is said to be a relevant EM if v1 = v2 = 1 holds for some 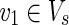 and
and 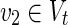 in its 0-1 assignment. Then, MKMN-EM is mathematically defined as follows:
in its 0-1 assignment. Then, MKMN-EM is mathematically defined as follows:
Problem: MKMN-EM (minimum knockout for multiple networks in the elementary mode model)
• Input: Metabolic networks N1 = (
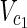 ,
, 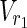 , E1), N2 = (
, E1), N2 = (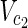 ,
, 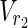 , E2), external nodes Vex1 ⊂
, E2), external nodes Vex1 ⊂ 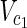 ∪
∪ 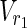 , Vex2 ⊂
, Vex2 ⊂ 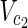 ∪
∪ 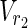 , target nodes
, target nodes 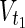 ⊂
⊂ 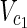 ,
, 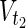 ⊂
⊂ 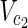 , and a stoichiometry matrix S.
, and a stoichiometry matrix S.-
• Output: The minimum cardinality set Va ⊆
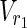 ∪
∪ 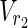 , which satisfies each of the following:
, which satisfies each of the following:–For all
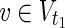 , v = 0 holds for any relevant EM on N1 = (
, v = 0 holds for any relevant EM on N1 = (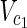 ,
, 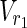 \Va, E1).
\Va, E1).–For all
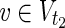 , v = 1 holds for some relevant EM on N2 = (
, v = 1 holds for some relevant EM on N2 = (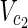 ,
, 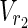 \Va, E2).
\Va, E2).
2.3. Integer linear programming–based method for MKMN-B
Since MKMN-B is NP-hard as discussed in “Theoretical results,” we develop an integer linear programming (ILP)-based method for its solution. In ILP, every Boolean constraint must be represented by linear equations or inequalities. In this article, we use two linear representations of Boolean constraints:
LP1 (Tamura et al., 2010): Since the Boolean “AND” relation 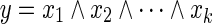 can be converted into
can be converted into 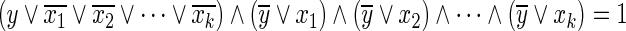 , it can be represented by the following linear inequalities:
, it can be represented by the following linear inequalities:
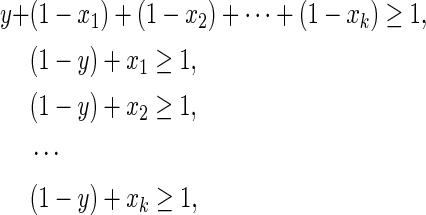 |
where all variables are binary.
Similarly, as the Boolean “OR” relation 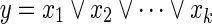 can be converted into
can be converted into 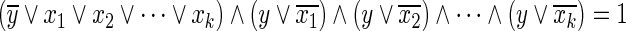 , it can be represented by the following linear inequalities:
, it can be represented by the following linear inequalities:
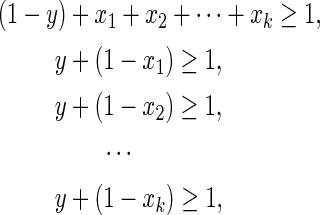 |
where all variables are binary.
LP2 (Akutsu et al., 2012): Another type of linear function representation of Boolean functions is as follows: The Boolean “AND” can be represented by the following linear inequalities:
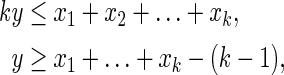 |
where all variables are binary.
Similarly, the Boolean “OR” can be represented by the following linear inequalities:
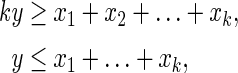 |
where all variables are binary.
As described in Tamura et al. (2010), we should introduce notion of time for calculating the MaxVA. A naive method for calculating the MaxVA is as follows. Each node is initially assigned 1, specified reactions are inhibited, effects of the inhibition affect neighbor nodes in the next time step, and the finally converged 0-1 assignment is the MaxVA. Similarly, as described in Lu et al. (2014), the MinVA can be calculated by initially assigning 0 to each node other than the source nodes. However, in this naive method, the number of time steps should be the same as the total number of nodes to ensure that the 0-1 assignment converges. Thus, the number of variables needed for the ILP formalization is O((m + n)2).
By utilizing an FVS, the number of time steps is reduced to f, and then the number of variables in the ILP formalization becomes O(f (m + n)). [The FVS is a set of nodes whose removal makes the graph acyclic, and f is the size of the FVS (Tamura et al., 2010).] For example, r1 of N1 in Figure 1A can be decomposed into r1 and s1, as shown in  of Figure 1B. Different from the naive method, the time step advances by one only when the value of the “r” node is copied to the “s” node. Suppose that r3 is inhibited in Figure 1B. Because we consider the MaxVA for N1 (and
of Figure 1B. Different from the naive method, the time step advances by one only when the value of the “r” node is copied to the “s” node. Suppose that r3 is inhibited in Figure 1B. Because we consider the MaxVA for N1 (and  ), all source nodes and “s” nodes are assigned 1 at t = 0. If r3 is not inhibited, all nodes become 1 at t = 0 since the time step advances only when the value of r1 is copied to s1. However, since r3 is inhibited, (r3, c2, r1) = (0, 0, 0) holds at t = 0, whereas (s1, c6) = (1, 1) holds at t = 0. Then, r1 = 0 at t = 0 is copied to s1 at t = 1. Thus, all nodes other than c1, c3, and c7 become 0 when r3 is inhibited. Note that the necessary time steps for calculating this is 2 (t = 0 and t = 1). This is the size of FVS + 1. In the naive method, the time step advances whenever 0 affects its adjacent nodes. Therefore, the number of necessary time steps is as large as the number of nodes to ensure that the 0-1 assignment converges.
), all source nodes and “s” nodes are assigned 1 at t = 0. If r3 is not inhibited, all nodes become 1 at t = 0 since the time step advances only when the value of r1 is copied to s1. However, since r3 is inhibited, (r3, c2, r1) = (0, 0, 0) holds at t = 0, whereas (s1, c6) = (1, 1) holds at t = 0. Then, r1 = 0 at t = 0 is copied to s1 at t = 1. Thus, all nodes other than c1, c3, and c7 become 0 when r3 is inhibited. Note that the necessary time steps for calculating this is 2 (t = 0 and t = 1). This is the size of FVS + 1. In the naive method, the time step advances whenever 0 affects its adjacent nodes. Therefore, the number of necessary time steps is as large as the number of nodes to ensure that the 0-1 assignment converges.
Based on this idea, we formalize an ILP-based method to solve MKMK-B. In the following, we explain our proposed methods using examples. It is straightforward to extend the examples to a general case.
When N1 and N2 in Figure 1A are given, IP-FVS1-LP1 is formalized as follows.
Maximize
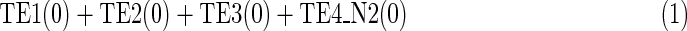 |
Subject to
 |
 |
/* Definitions of N1
for all
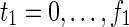
/* Reactions
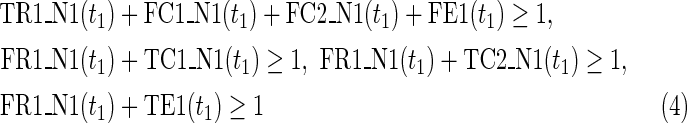 |
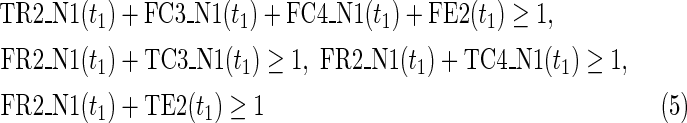 |
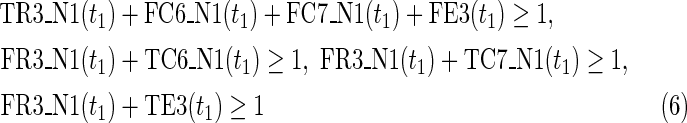 |
/* Compounds
 |
 |
 |
 |
 |
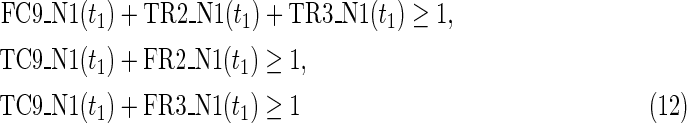 |
/* SR1
 |
/* Es
 |
 |
 |
/* Source compounds
 |
 |
 |
/* Definitions of N2
for all
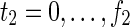
/* Reactions
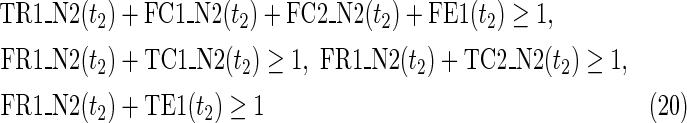 |
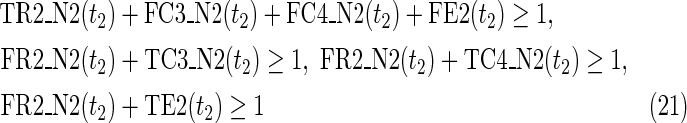 |
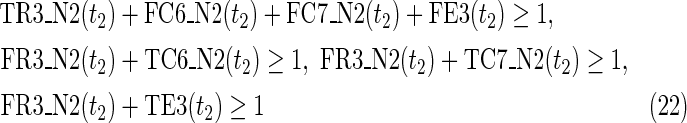 |
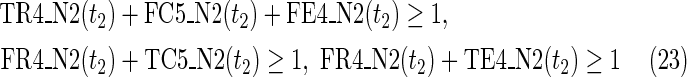 |
/* Compounds
 |
 |
 |
 |
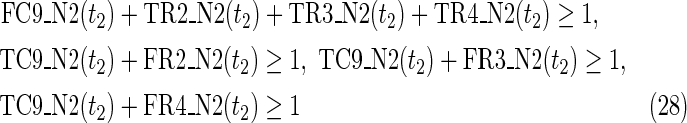 |
 |
/* Es
 |
 |
 |
 |
/* Source compounds
 |
 |
 |
 |
/* SR1
 |
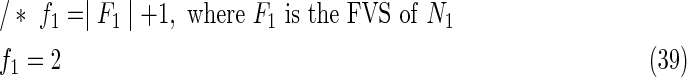 |
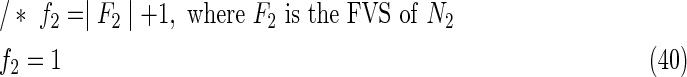 |
 |
where every variable takes a value of either 0 or 1. “T” and “F” stand for true (1) and false (0), respectively. “R”, “C,” and “E” stand for reaction, compound, and enzyme, respectively. For each reaction node, TRi(t) = 1 (resp., FRi(t) = 1) represents ri = 1 (resp., ri = 0). Therefore, TRi(t) + FRi(t) = 1 holds for any reaction node ri at time step t; “t = 0” means the initial time step. For example, if TR2(1) = 0, which means r2 = 0 (the reaction node r2 does not occur) at t = 1, FR2(1) = 1 automatically holds at the same time. The use of FRi(t) simplifies this illustration. In the implementation, FRi(t) is replaced by 1-TRi(t) to reduce the number of variables. Similarly, each compound ci is represented by TCi(t) and FCi(t), where TCi(t) + FCi(t) = 1. For example, TC3(1) = 1 means that c3 = 1 (the compound c3 exists) at t = 1, and FC3(1) = 1 means that c3 = 0 (the compound c3 does not exist) at t = 1.
The maximal number of time steps f1 and f2 can be calculated by f1 = |F1| +1 and f2 = |F2| +1, where |F1| and |F2| are the sizes of the FVS for N1 and N2, respectively. Note that “ +1” is necessary because at least 1 time step is necessary even if there is no FVS. Furthermore, since the number of time steps of N1 and N2 may be different, the ILP formalization is defined for each network. In the above example for IP-FVS1-LP1, (2), (4)–(19), (38), and (39) give the ILP formalization of N1, whereas (3), (20)–(37), and (40) give the ILP formalization of N2. To distinguish each node, “_N1” (resp., “_N2”) is appended to each variable name to denote which network the node belongs to. For example, TR1_N1(1) = 1 means that r1 = 1 (the reaction node r1 occurs) in N1 at t = 1, and FR1_N2(3) = 1 means that r1 = 0 (the reaction r1 does not occur) in N2 at t = 3. “_N1” and “_N2” are not appended to the variables of nodes that are common to both N1 and N2.
In IP-FVS1-LP1, the Boolean “AND” relation of a reaction node is converted into LP1 type linear inequalities as shown in (5). The relation in (5) represents the constraints between the reaction node r2 and each incoming compound node (c3 and c4) in N1. Two additional variables TE2(t1) and FE2(t1) are also included in (5), since we add a virtual predecessor node ei to each reaction node ri in both networks. TEi(t) and FEi(t) denote whether ri is inhibited. Since ri is represented by an “AND” node, ei = 0 can ensure that ri remains inactive, even if all other predecessors of ri are 1. Furthermore, the same TEi(t) and FEi(t) are used for each common reaction node ri between N1 and N2. In the above example for IP-FVS1-LP1, the common reaction nodes r2 of the two networks are connected by e2 [TE2 and FE2 in (5) and (21)].
The Boolean “OR” relation of a compound node is converted into LP1-type linear inequalities, as shown in (12), when the indegree of the compound node is more than 1. If the indegree of the compound is 1, we need only copy the value from the variable of the predecessor reaction node as shown in (7).
For N1, the values of each source node and each node newly created by si at t = 0 are set to 1 to realize the MaxVA as shown in (17)–(19) and (38). The value of the target node is set to 0 at t = f1 to ensure that the target node eventually cannot be produced in N1. For N2, only the source nodes and target node values at t = f1 are set to 1, as shown in (3) and (34)–(37), to realize MinVA.
The MKMN result can be calculated by maximizing TEi(t), as shown in (1), since ei represents whether the reaction node is inhibited. As mentioned above, FEi(t) is replaced by 1-TEi(t). Relation (1) implies that the minimal number of reaction knockouts is calculated by maximizing the number of reaction nodes that are not knocked out. In addition, (2) indicates that the target compound c9 should eventually not be produced in N1, whereas (3) suggests that the target compound c9 should eventually be produced in N2.
Relations (4)–(19) are the node constraints for N1; {r1} is chosen as the FVS F1. The maximal time step f1 of N1 is 2, where f1 = |F1| +1 = 1 + 1 = 2. Relations (4)–(6) represent the constraints on reactions r1–r3, respectively, and (7)–(12) represent the constraints on compounds c2, c4, c5, c6, c8, and c9, respectively. In (8)–(10), the predecessor reaction node r1 of compounds c4–c6 is replaced by s1, where TSR1_N1(t1) = 1 represents s1 = 1 (s1 is active) at t1 in N1, and TSR1_N1(t1) = 0 represents s1 = 0 (s1 is inactive) at t1 in N1. Relation (13) increases the time step by 1 when the value of r1 is copied to s1. Relations (14)–(16) ensure that the state of each reaction (inhibited or uninhibited) does not change during the time transition, and (17)–(19) define c1, c3, and c7 as the source nodes of N1.
Relations (20)–(37) are the node constraints for N2. Since the FVS F2 = {}, the maximal time step f2 of N2 is 1, where f2 = |F2| +1 = 0 + 1 = 1. Relations (20)–(23) represent the constraints on reactions r1–r4, respectively, and (24)–(29) give the constraints on compounds c4, c5, c6, c8, c9, and c10, respectively. Relations (30)–(33) ensure that the state of each reaction (inhibited or uninhibited) does not change during the time transition. Reaction nodes r1, r2, and r3 are common to both N1 and N2, whereas r4 exists only in N2. Relations (34)–(37) define c1, c2, c3, and c7 as the source nodes of N2.
Relation (38) states that the initial value of s1 is set to 1 (s1 = 1 at t1 = 0) for the MaxVA in N1, and (39)–(40) give the maximal time steps of N1 and N2, respectively. Relation (41) denotes that “T … ” represents “true (1)” and “F … ” represents “false (0).”
Since MinVA is calculated for N2, IP-FVS1-LP2 for MKMN-B shown in Figure 2 is as follows:
Maximize
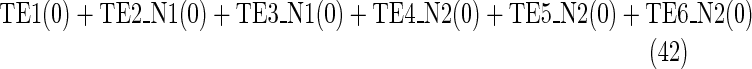 |
Subject to
 |
 |
/* Definitions of N1
for all
/* Reactions
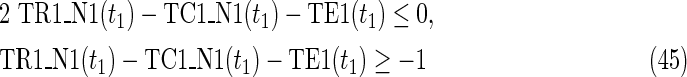 |
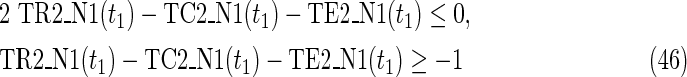 |
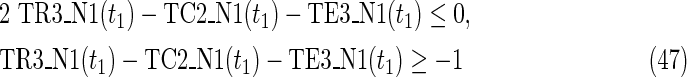 |
/* Compounds
 |
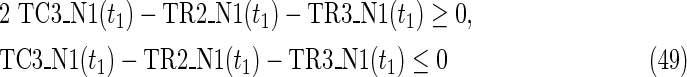 |
/* Es
 |
 |
 |
/* Source compounds
 |
/* Definitions of N2
for all
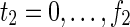
/* Reactions
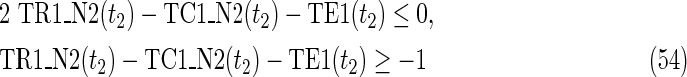 |
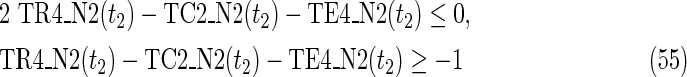 |
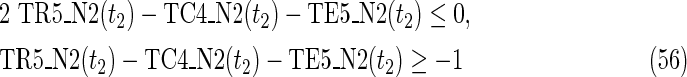 |
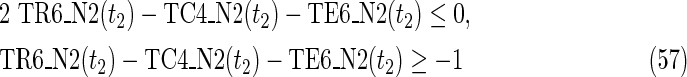 |
/* Compounds
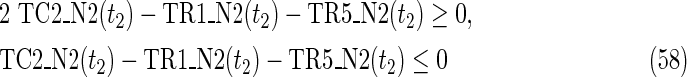 |
 |
 |
/* SR4
 |
/* Es
 |
 |
 |
 |
/* Source compounds
 |
/* SR4
 |
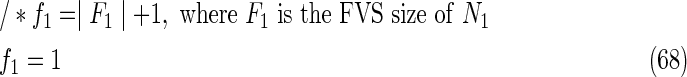 |
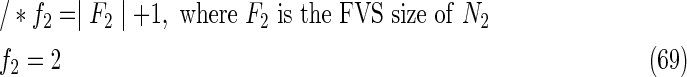 |
where each variable takes a value of either 0 or 1. The Boolean relations of the reaction nodes and compound nodes are converted into LP2 type linear inequalities here. As (42) maximizes the number of reactions that are not inhibited, it corresponds to minimizing the number of inhibited reactions. Relation (43) ensures that the target compound c3 finally becomes nonproducible in N1, whereas (44) guarantees that c3 remains producible in N2. Relations (45)–(47) and (54)–(57) represent the Boolean relations of reactions in N1 and N2, respectively. Similarly, (48)–(49) and (58)–(60) represent the Boolean relations of compounds in N1 and N2, respectively. Relations (50)–(52) and (62)–(65) ensure that the state of each reaction (inhibited or uninhibited) does not change in that time transition. Relations (53) and (66) mean that c1 is the source compound in both N1 and N2, whereas (68)–(69) give the numbers of time steps for N1 and N2, which are calculated from the sizes of the FVS. Here, {r4} is chosen as the FVS. In (60), the predecessor reaction node r4 of the compound c4 is replaced by s4. Relation (67) states that the initial value of s4 is set to 0 (s4 = 0 at t = 0) as MinVA is applied to N2. All variables represented by “F … ” are replaced with “T … ” variables in the implementation.
Although the above ILP-formalizations do not include cycles in both N1 and N2, in the general case, MaxVA and MinVA are applied to N1 and N2, respectively.
2.4. Exception of the maximal valid assignment
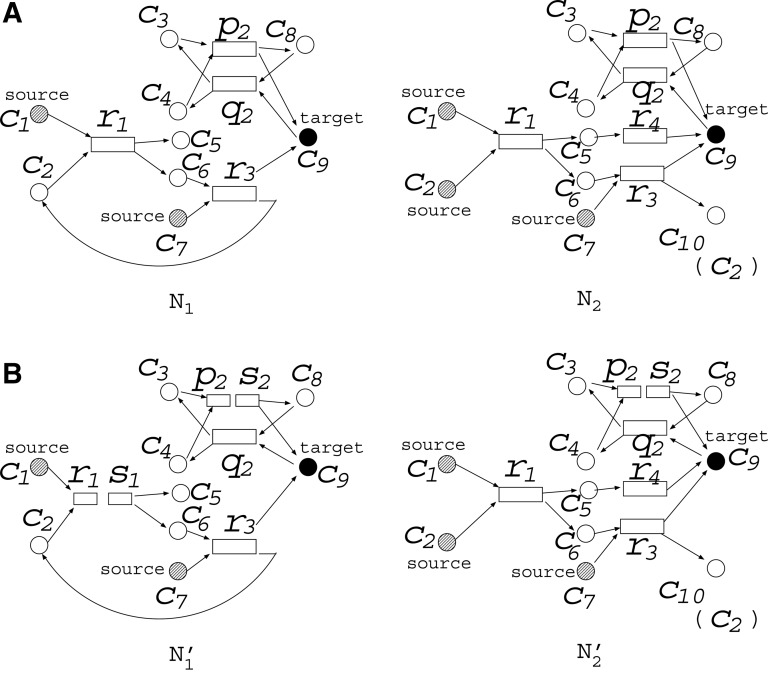
MKMN-B can be solved by IP-FVS1. However, the problem setting of MKMN-B, in which MaxVA and MinVA are applied to N1 and N2, respectively, may be inappropriate for some cases. For example, suppose that N1 and N2 are as in Figure 4A. Originally, p2 and q2 are from one reversible reaction (r2), which is decomposed into two irreversible reactions. Both p2 and q2 can be inhibited by the inhibition for r2. If MaxVA is assumed in N1, neither p2 nor q2 becomes 0 unless r2 is inhibited. Thus, there is a case where an original reversible reaction works as if it were a source node in N1.
FIG. 4.
A reversible reaction r2 decomposed into p2 and q2. The r1 is decomposed in N1, and p2 is decomposed in N1 and N2 so that there is no directed cycle in the resulting networks. Additionally, s1(0) = 1 since the detected cycle in (A) includes an irreversible reaction. However, s2(0) = 0 since the detected cycle in (A) does not include an irreversible reaction.
To handle such reversible reactions more appropriately, we define a variant of MaxVA by modifying the FVS-based method as follows: if the cycle detected by the FVS-based method does not include an original irreversible reaction, 0 is assigned to the newly created node at t = 0.
For example, suppose that N1 and N2 are as in Figure 4A. Since the cycle consisting of {r1, c6, r3, c2} includes an original irreversible reaction, after decomposing r1 of N1 into r1 and s1 (see Fig. 4B), s1(0) is assigned a value of 1. Furthermore, cycles consisting of {c3, p2, c8, q2}, {c3, p2, c9, q2}, {c4, p2, c8, q2}, and {c4, p2, c9, q2} are also decomposed. Suppose that p2 is decomposed as shown in N1 and N2 in Figure 4B. In this case, as the cycle does not include an original irreversible reaction, s2(0) is assigned a value of 0 for both N1 and N2 (see also Table 2). Note that if s2(0) = 1, the values of c9 in N1 are fixed to 1, and r2 must be inhibited to make c9 = 0, as it operates as if a reversible reaction was a kind of source node.
Table 2.
Initial Values of Nodes Included by the FVS
| IP-FVS1 | IP-FVS2 | |
|---|---|---|
| s2(0) of N1 | 1 | 0 |
| s2(0) of N2 | 0 | 0 |
In IP-FVS1, N1 and N2 are based on MaxVA and MinVA, respectively. However, in IP-FVS2, in addition to the above assumption, nodes included by the FVS consisting of only reversible reactions are assigned an initial value of 0. IP-FVS1 and IP-FVS2, integer linear programming-based methods; MaxVA, maximal valid assignment; MinVA, minimal valid assignment.
2.5. Fast approximation algorithm for MKMN-B with large networks
Although IP-FVS1 and IP-FVS2 successfully reduce the number of variables in the ILP formalization from O((m + n)2) to O(f (m + n)), O(f (m + n)) may be still too large if the network is very big. One reasonable strategy for handling this problem is to limit the number of time steps to some small constant. IP-FVS1 and IP-FVS2 need f time steps to ensure that the MaxVA and MinVA are always calculated, and hence the optimal solution of MKMN-B is always obtained. However, as the number of time steps necessary to obtain the optimal solution of MKMN-B depends on the topology of the network obtained after removing the FVS, we generally require fewer than f time steps. Although the proposed method does not ensure that the solution is optimal (as f time steps are necessary for the worst case), it is possible to confirm that MaxVA and MinVA in the obtained solution satisfy the condition of MKMN-B; that is, the target compound is not producible in N1 but is producible in N2. Note that this validation process can be conducted in polynomial time, since MKMN-B is NP-complete (see “Theoretical results” section). Let IP-FVS1-approx(t) and IP-FVS2-approx(t) be the IP-FVS1 and IP-FVS2 algorithms in which the number of time steps is limited to t. If t is not large, the numbers of variables used in IP-FVS1-approx(t) and IP-FVS2-approx(t) are O(m + n).
2.6. Elementary mode–based method for MKMN-EM
To solve MKMN-EM, we utilize CNA to calculate the EMs. Since MKMN-EM is NP-complete even if the EMs are given (see “Theoretical results” section), we develop an ILP-based method, IP-EM, to solve MKMN-EM for given EMs. For example, suppose that N1 and N2 have four and three relevant EMs, respectively, as shown in Table 3.
Table 3.
Example for MKMN-EM
| N1 | N2 | ||
|---|---|---|---|
| EM1 | r1, r2, r4 | EM5 | r1, r2, r7 |
| EM2 | r1, r3, r6 | EM6 | r2, r3, r4 |
| EM3 | r2, r4, r5 | EM7 | r4, r5 |
| EM4 | r5, r7 | ||
After obtaining the EMs that produce the target compound using CellNetAnalyzer, MKMN-EM can be formalized by ILP. MKMN-EM, minimum knockout for multiple reactions in the elementary mode model; EM, elementary mode; ILP, integer linear programming.
Then IP-EM can be formulated as follows:
maximize r1 + r2 + r3 + r4 + r5 + r6 + r7
such that
 |
 |
 |
 |
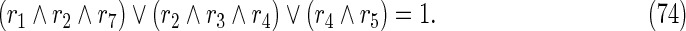 |
where all variables are binary, and Boolean relations should be converted into linear inequalities. Constraints (70)–(73) are converted into
 |
 |
 |
 |
For example, since (70) requires that at least one of {r1, r2, r4} must be 0, it can be represented by (75). Furthermore, the constraint (74) is converted into
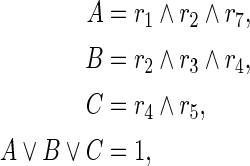 |
and then converted into
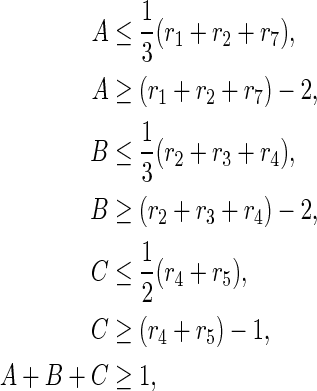 |
using LP2, where all variables are binary. In this example, inhibiting {r1, r5} is an optimal solution, since either r1 or r5 is included in each of EM1, EM2, EM3, and EM4, and neither r1 nor r5 is included in EM6.
3. Computational Experiments
We conducted computational experiments on an Intel(R) Xeon(R) CPU E5-2690 0, 2.90GHz with cache size 20.48 MB, operating system on SUSE Linux Enterprise Server 11 SP3 (x86 64). CPLEX version 12.4.0.0 was used as the ILP solver to obtain the global optimality (IBM, 2010).
Metabolic networks of clostridium perfringens SM101 (CPR) and bifidobacterium longum DJO10A (BLJ) were represented by N1 and N2, respectively, in the MKMN. KGML KEGG Markup Language (KGML) of BLJ and CPR were downloaded from the KEGG PATHWAY database (Kanehisa and Goto, 2000). Datasets 1, 2, and 3 were extracted and used as small, medium, and large networks, respectively. Details of datasets 1, 2, and 3 are given in Tables 4, 5, and 6, respectively. We considered the target compounds to be C00022 (pyruvate), C00024 (acetyl-CoA), C00033 (acetate), C00036 (oxaloacetate), and C00074 (phosphoenolpyruvate). In addition, “5 compounds” indicates the problem in which none of the above five compounds are producible in N1 but all are producible in N2.
Table 4.
Details of the Networks of Dataset 1
| N1 | N2 | Total (N1 + N2) | |
|---|---|---|---|
| #compound | 36 | 43 | 79 |
| #irreversible reaction | 11 | 8 | 19 |
| #reversible reaction | 26 | 31 | 57 |
| #node | 73 | 82 | 155 |
N1 and N2 are the central metabolism of bifidobacterium longum DJO10A (BLJ) and clostridium perfringens SM101 (CPR), respectively. N1 consists of {cpr00010.xml cpr00030.xml}, and N2 consists of {blj00010.xml,blj00020.xml,blj00030.xml} in the KEGG database.
Table 5.
Details of the Networks of dataset 2
| N1 | N2 | Total (N1 + N2) | |
|---|---|---|---|
| #compound | 132 | 157 | 289 |
| #irreversible reaction | 86 | 133 | 219 |
| #reversible reaction | 33 | 38 | 71 |
| #node | 251 | 328 | 579 |
N1 and N2 consist of carbon metabolism, fatty acid metabolism, and biosynthesis of amino acids of BLJ and CPR, respectively. N1 is {cpr01200.xml, cpr01212.xml cpr01230.xml}, and N2 is {blj01200.xml,blj01212.xml,blj01230.xml} in the KEGG database.
Table 6.
Details of the Networks of Dataset 3
| N1 | N2 | Total (N1 + N2) | |
|---|---|---|---|
| #compound | 622 | 567 | 1189 |
| #irreversible reaction | 400 | 337 | 737 |
| #reversible reaction | 209 | 205 | 414 |
| #node | 1231 | 1881 | 2340 |
N1 and N2 consist of whole metabolic networks of BLJ and CPR, respectively, downloaded from KEGG markup language (KGML) in the KEGG database.
3.1. Datasets
In dataset 1, N1 and N2 correspond to the central metabolism of BLJ and CPR, respectively. N1 includes 36 compounds and 37 reactions (26 reversible reactions) and consists of glycolysis, gluconeogenesis, and pentose phosphate pathway. N2 includes 43 compounds and 39 reactions (31 reversible) and consists of glycolysis, gluconeogenesis, citrate (TCA) cycle, and pentose phosphate pathway. Thus, there are a total of 155 nodes for dataset 1; increasing to 212 nodes after decomposing each reversible reaction into two irreversible reactions.
In dataset 2, N1 and N2 correspond to the carbon metabolism, fatty acid metabolism, and biosynthesis of amino acids of BLJ and CPR, respectively. N1 includes 132 compounds and 119 reactions (33 reversible), whereas N2 includes 157 compounds and 171 reactions (38 reversible). Thus, there are a total of 579 nodes for dataset 2, rising to 650 nodes after decomposing each reversible reaction into two irreversible reactions.
In dataset 3, N1 and N2 correspond to the whole pathways of BLJ and CPR, respectively. N1 includes 622 compounds and 609 reactions (209 reversible), whereas N2 includes 1189 compounds and 1151 reactions (414 reversible). Thus, there are a total of 2340 nodes for dataset 3, increasing to 2714 nodes after decomposing each reversible reaction into two irreversible reactions.
3.2. results for dataset 1
Tables 7 and 8 show the computation time and size of the solutions obtained for each target compound by each MKMN method for dataset 1. As dataset 1 has small networks, the computation time is less than 2 sec for every case, and there is generally no MKMN solution.
Table 7.
The Computation Time of IP-FVS1, IP-FVS2, and IP-EM for Each Target Compound with Dataset 1
| IP-FVS1 | IP-FVS1 | IP-FVS2 | ||
|---|---|---|---|---|
| target compound\type | (with LP1) | (with LP2) | (with LP2) | IP-EM |
| C00022 (Pyruvate) | 2.07 sec | 1.45 sec | 0.65 sec | 0.47 sec |
| C00024 (Acetyl-CoA) | <0.01 sec | <0.01 sec | <0.01 sec | <0.01 sec |
| C00033 (Acetate) | 0.21 sec | 0.18 sec | 0.19 sec | 0.27 sec |
| C00036 (Oxaloacetate) | <0.01 sec | <0.01 sec | <0.01 sec | <0.01 sec |
| C00074 (Phosphoenolpyruvate) | 0.18 sec | 0.18 sec | 0.57 sec | 0.49 sec |
| All the above 5 compounds | 0.18 sec | 0.18 sec | 0.18 sec | 0.49 sec |
Table 8.
Size of the Solutions Given by IP-FVS1, IP-FVS2, and IP-EM for Each Target Compound with Dataset 1
| Target compound\type | IP-FVS1 | IP-FVS2 | IP-EM |
|---|---|---|---|
| C00022 (Pyruvate) | no solution | 2 | 2 |
| C00024 (Acetyl-CoA) | no solution | no solution | no solution |
| C00033 (Acetate) | no solution | no solution | no solution |
| C00036 (Oxaloacetate) | no solution | no solution | no solution |
| C00074 (Phosphoenolpyruvate) | no solution | 2 | 1 |
| All the above 5 compounds | no solution | no solution | 2 |
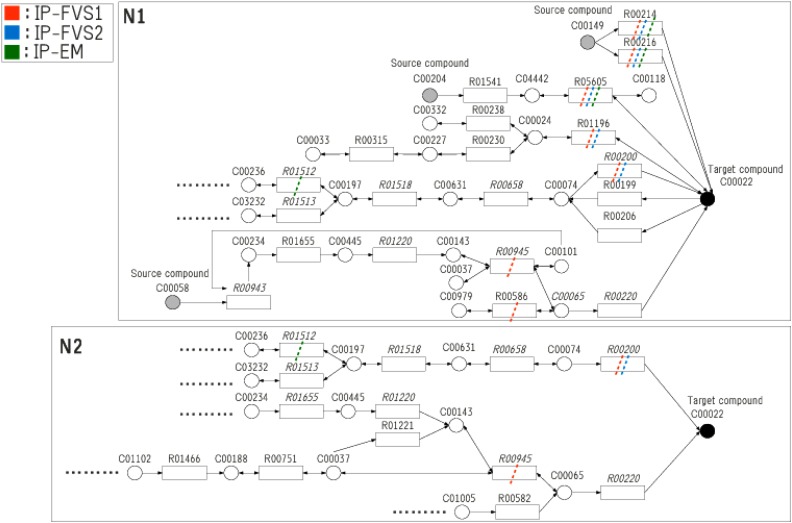
However, when the target compound is C00022 (pyruvate), {R01541, R04779} and {R04779, R05605} are solutions for IP-FVS2 and IP-EM, respectively, as shown in Table 9, where RXXXXX ( ) are the IDs in KEGG. R04779 is a reaction from beta-D-Fructose 6-phosphate to beta-D-Fructose 1,6-bisphosphate located seven steps before pyruvate in the pathway of glycolysis. R01541 is a reaction from 2-Dehydro-3-deoxy-D-gluconate to 2-Dehydro-3-deoxy-6-phospho-D-gluconate, locating two steps before pyruvate in the pentose phosphate pathway, and R05605 is a reaction between 2-Dehydro-3-deoxy-6-phospho-D-gluconate and pyruvate in the pentose phosphate pathway.
) are the IDs in KEGG. R04779 is a reaction from beta-D-Fructose 6-phosphate to beta-D-Fructose 1,6-bisphosphate located seven steps before pyruvate in the pathway of glycolysis. R01541 is a reaction from 2-Dehydro-3-deoxy-D-gluconate to 2-Dehydro-3-deoxy-6-phospho-D-gluconate, locating two steps before pyruvate in the pentose phosphate pathway, and R05605 is a reaction between 2-Dehydro-3-deoxy-6-phospho-D-gluconate and pyruvate in the pentose phosphate pathway.
Table 9.
Solutions Given by IP-FVS1, IP-FVS2, and IP-EM for Each Target Compound with Dataset 1
| Target compound | Type | The obtained solution |
|---|---|---|
| C00022 | IP-FVS1 | no solution |
| IP-FVS2 | R01541(N1), R04779(N1) | |
| IP-EM | R04779(N1), R05605(N1) | |
| C00074 | IP-FVS1 | no solution |
| IP-FVS2 | R04779(N1), R01541(N1) | |
| IP-EM | R04779(N1) | |
| 5 compounds | IP-FVS1 | no solution |
| IP-FVS2 | no solution | |
| IP-EM | R04779(N1), R05605(N1) |
“(N1)” indicates that the reaction appears in N1 but not in N2.
When the target compound is C00074 (phosphoenolpyruvate), {R04779, R01541} and {R04779} are solutions for IP-FVS2 and IP-EM, respectively. When all five compounds are targeted, only MKMN-EM has a solution {R04779, R05605}.
Supplementary Tables S1–S3 the Supplementary Material (available online at www.liebertpub. Com/cmb) show the relation among the target compound, predecessors of the target compound in N1 and N2, and solutions given by each MKMN method for each target compound. For example, Supplementary Table S1 shows that R00200 and R00703 are predecessors of the target compound C00022 for both N1 and N2. R05605 is a predecessor of C00022 in N1 but not in N2. R04779 (or R01541) is not a predecessor of C00022 in either N1 or N2. In the tables of supporting information, “ko” indicates reactions included in the solution by each MKMN method. We can see that solutions for MKMN are not trivial, as they do not consist of only predecessors of the target compound.
3.3. Results for dataset 2
Tables 10 and 11 show the computation time and size of the solutions obtained for each target compound by each MKMN method for dataset 2. As the networks are not very large, the computation time is generally less than 20 sec. MKMN-B-FVS1 only has solutions when the target compound is C00022 or C00024, and MKMN-B-FVS2 only has solutions for C00022, C00024, or C00033; in contrast, MKMN-EM has solutions for all cases.
Table 10.
Computation Time of IP-FVS1, IP-FVS2 and IP-EM for Each Target Compound with Dataset 2
| IP-FVS1 | IP-FVS1 | IP-FVS2 | ||
|---|---|---|---|---|
| Target compound\type | (with LP1) | (with LP2) | (with LP2) | IP-EM |
| C00022 (Pyruvate) | 8.95 sec | 11.19 sec | 10.5 sec | 14.59 sec |
| C00024 (Acetyl-CoA) | 8.57 sec | 11.31 sec | 11.91 sec | 4.62 sec |
| C00033 (Acetate) | 0.74 sec | 0.74 sec | 9.82 sec | 17.48 sec |
| C00036 (Oxaloacetate) | 8.17 sec | 7.05 sec | 5.32 sec | 10.92 sec |
| C00074 (Phosphoenolpyruvate) | 0.72 sec | 0.74 sec | 0.72 sec | 13.36 sec |
| All the above 5 compounds | 0.74 sec | 0.76 sec | 0.74 sec | 4.64 sec |
Table 11.
Size of the Solutions Given by IP-FVS1, IP-FVS2, and IP-EM for Each Target Compound with Dataset 2
When the target compound is C00022 (pyruvate), the solutions obtained for IP-FVS1, IP-FVS2, and IP-EM are {R00200, R00214, R00216, R00586, R00945, R01196, R05605}, {R00200, R00214, R00216, R01196, R05605}, and {R00214, R00216, R01512, R05605}, respectively listed in Table 12. As shown in Figure 5, among the above reactions, {R00200, R00220} are predecessors of C00022 in both N1 and N2, whereas {R00214, R00216, R001196, R05605} are predecessors of C00022 in N1 but not in N2 (Supplementary Table S4). However, {R00586, R00945, R01512} are not predecessors of C00022 in either N1 or N2. R00586 is a reaction between L-Serine and O-Acetyl-L-serine in the pathway of cysteine and methionine metabolism. R00945 is a reaction between L-Serine and glycine in the pathway of glycine, serine, and threonine metabolism. R01512 is a reaction between 3-Phospho-D-glyceroyl phosphate and 3-Phospho-D-glycerate in the pathway of glycolysis. The relations among the reactions included in the obtained solutions and predecessors of N1 and N2 are described in Supplementary Table S4.
Table 12.
Solutions Given by IP-FVS1, IP-FVS2, and IP-EM for Each Target Compound with Dataset 2
| Target compound | Type | The obtained solution |
|---|---|---|
| C00022 | IP-FVS1 | R00200, R00214(N1), R00216(N1), R00586(N1) R00945, R01196(N1), R05605(N1) |
| IP-FVS2 | R00200, R00214(N1), R00216(N1), R01196(N1), R05605(N1) | |
| IP-EM | R00214(N1), R00216(N1), R01512, R05605(N1) | |
| C00024 | IP-FVS1 | R00230, R00238(N1), R01196(N1) |
| IP-FVS2 | R01196(N1) | |
| IP-EM | R01196(N1) | |
| C00033 | IP-FVS1 | no solution |
| IP-FVS2 | R01196(N1) | |
| IP-EM | R01196(N1) | |
| C00036 | IP-FVS1 | no solution |
| IP-FVS2 | no solution | |
| IP-EM | R00345 | |
| C00074 | IP-FVS1 | no solution |
| IP-FVS2 | no solution | |
| IP-EM | R00199(N1), R00206(N1), R01015(N1), R01829(N1), R04533(N1) | |
| 5 compounds | IP-FVS1 | no solution |
| IP-FVS2 | no solution | |
| IP-EM | R00214(N1), R00216(N1), R01512, R05605(N1) |
“(N1)” indicates that the reaction appears in N1 but not in N2.
FIG. 5.
Solutions for dataset 2 with the target compound C00022 (Pyruvate). When the target compound is C00022 (Pyruvate), solutions of IP-FVS1, IP-FVS2, and IP-EM for dataset 2 are represented by red, blue, and green lines, respectively.
When the target compound is C00024 (Acetyl-CoA), the solutions for IP-FVS1, IP-FVS2, and IP-EM are {R00230, R00238, R01196}, {R01196}, and {R01196}, respectively (Table 12). R00230 is a predecessor of C00024 in both N1 and N2, whereas R00238 and R01196 are predecessor of C00024 in N1 but not in N2 (Supplementary Table S5).
When the target compound is C00033 (acetate), IP-FVS1 does not obtain a solution, but IP-FVS2 and IP-EM both give the solution {R00196}. As described in Supplementary Table S6, R00196 is not the predecessor of either N1 or N2. Next, when the target compound is C00036 (oxaloacetate), only MKMN-EM has a solution, {R00345}. R00345 is a predecessor of C00036 in both N1 and N2 as shown in Table S7 of supplemental data When the target compound is C00074(phosphoenolpyruvate), only MKMN-EM has a solution {R00199, R00206, R01015, R01829, R04533}. As described in Supplementary Table S8, R00199 and R00206 are predecessors of C00074 in N1 but not in N2. However, none of R01015, R01829, or R04533 are predecessors of C00074 in either N1 or N2.
Finally, when all five compounds are targeted, MKMN-B does not have the solution, but MKMN-EM has the solution {R00214, R00216, R01512, R05605}. As described in Supplementary Table S9, R00214, R00216 and R05605 are predecessors of at least one of the five target compounds in N1 but not in N2. R01512 is not the predecessor of any of the five target compounds in either N1 or N2, and is a reaction between 3-Phospho-D-glyceroyl phosphate and 3-Phospho-D-glycerate locating two steps before C00074 (phosphoenolpyruvate) in the glycolysis pathway.
3.4. Results for dataset 3
Tables 13 and 14 show the computation time and size of the solutions obtained for each target compound by each MKMN method for dataset 3. For IP-FVS1, although the calculation was completed for every target compound, no solutions were found when the target compound was either C00033 (acetate), C00074 (phosphoenolpyruvate), or “5 compounds.” For IP-FVS2, the calculation only completed when the target compound was C00074 (phosphoenolpyruvate) or “5 compounds.” CNA could not calculate the EMs for the EM-based method, as the networks are too large.
Table 13.
The Computation Time by IP-FVS1, IP-FVS2, and IP-EM for Each Target Compound with Dataset 3
| IP-FVS1 | IP-FVS1 | LP-type2 | IP-EM | |
|---|---|---|---|---|
| Target compound\type | (with LP1) | (with LP2) | IP-FVS2 | |
| C00022 (Pyruvate) | 26 min 37 sec | 54 min 56 sec | >2 hours | NA |
| C00024 (Acetyl-CoA) | 32 min 6 sec | 56 min 19 sec | >2 hours | NA |
| C00033 (Acetate) | 16 min 17 sec | 12 min 14 sec | >2 hours | NA |
| C00036 (Oxaloacetate) | 32 min 38 sec | 1 hr 8 min 35 sec | >2 hours | NA |
| C00074 (Phosphoenolpyruvate) | 21 min 1 sec | 12 min 34 sec | 5 min 29 sec | NA |
| All the above 5 compounds | 21 min 44 sec | 12 min 15 sec | 7 min 2 sec | NA |
NA: Not available.
Table 14.
Size of the Solutions Given by IP-FVS1, IP-FVS2, and IP-EM for Each Target Compound with Dataset 3
As listed in Table 13, IP-FVS1 can compute solutions with either LP1 or LP2 within 70 min for each target compound. However, IP-FVS2 requires a computation time of over 2 hr, and frequently cannot obtain a solution.
As the computation time is large and solutions are not always obtained, we applied IP-FVS1-approx(10) and IP-FVS2-approx(10) to dataset 3. Tables 15 and 16 show the computation time and size of the solutions given by IP-FVS1-approx(10) and IP-FVS2-approx(10) for each target compound for MKMN-B for dataset 3. As can be seen in Table 15, the computation time of IP-FVS1-approx(10) and IP-FVS2-approx(10) is at most 15sec, and computation is always completed although there is no solution for some cases. The relations among solutions of IP-FVS1, IP-FVS1-approx(10), IP-FVS2, and IP-FVS2-approx(10) are summarized in Table 17.
Table 15.
Computation Time of IP-FVS1-approx(10) and IP-FVS2-approx(10) for Each Target Compound with Dataset 3
| IP-FVS1-approx(10) | IP-FVS1-approx(10) | IP-FVS2-approx(10) | |
|---|---|---|---|
| Target compound \ type | (with LP1) | (with LP2) | (with LP2) |
| C00022 (Pyruvate) | 9.43 sec | 6.92 sec | 12.05 sec |
| C00024 (Acetyl-CoA) | 9.18 sec | 6.71 sec | 14.95 sec |
| C00033 (Acetate) | 4.46 sec | 1.22 sec | 14.01 sec |
| C00036 (Oxaloacetate) | 8.78 sec | 7.81 sec | 10.66 sec |
| C00074 (Phosphoenolpyruvate) | 2.81 sec | 0.97 sec | 1.32 sec |
| All the above 5 compounds | 2.72 sec | 0.96 sec | 1.37 sec |
Table 16.
The Size of the Obtained Solutions by IP-FVS1-approx(10) and IP-FVS2-approx(10) for Each Target Compound with Dataset 3
Table 17.
Solutions Given by IP-FVS1, IP-FVS1-approx(10), IP-FVS2, IP-FVS2-approx(10), and IP-EM for Each Target Compound with Dataset 3
| Target compound | Type | The obtained solution |
|---|---|---|
| C00022 | IP-FVS1 | R00200, R00212, R00214(N1), R00220, R00470(N1), R00703, R00704(N1), R00782, R00896, R02320, R05605(N1), R05636 |
| IP-FVS1-approx(10) | R03105 is chosen instead of R00896. | |
| IP-FVS2 | NA | |
| IP-FVS2-approx(10) | R00200, R00470(N1), R01220 | |
| IP-EM | NA | |
| C00024 | IP-FVS1 | R00212, R00228, R00230, R00238(N1), R01177(N1), R04386(N1), R05351 |
| IP-FVS1-approx(10) | the same as the above | |
| IP-FVS2 | NA | |
| IP-FVS2-approx(10) | R00212, R03026(N1), R05351 | |
| IP-EM | NA | |
| C00033 | IP-FVS1 | no solution |
| IP-FVS2 | NA | |
| IP-FVS2-approx(10) | R00212, R00238(N1), R05351 | |
| IP-EM | NA | |
| C00036 | IP-FVS1 | R00345, R00355, R00483(N1) |
| IP-FVS1-approx(10) | R00345, R00355, R00357 | |
| IP-FVS2 | NA | |
| IP-FVS2-approx(10) | R00345, R00485(N1) | |
| IP-EM | NA |
“(N1)” indicates that the reaction appears in N1 but not in N2.
When the target compound is C00022 (pyruvate), the solution obtained by IP-FVS1 is {R00200, R00212, R00214, R00220, R00470, R00703, R00704, R00782, R00896, R02320, R05605, R05636}; the solution given by IP-FVS1-approx(10) is almost the same, but R00896 is replaced by R03105. Although IP-FVS2 cannot obtain a solution, IP-FVS2-approx(10) obtains the solution {R00200, R00470, R01220}. The relations among solution obtained by each method and the predecessors of the target compound in N1 and N2 are described in Supplementary Table S10.
When the target compound is C00024 (acetyl-CoA), IP-FVS1 and IP-FVS1-approx(10) obtain the same solution {R00212, R00228, R00230, R00238, R01177, R04386, R05351}. Although IP-FVS2 cannot obtain a solution, IP-FVS2-approx(10) gives {R00212, R03026, R05351}. The relations among the selected reactions and predecessors of the target compound are described in Supplementary Table S11.
Next, when the target compound is C00033 (acetate), FVS-type1 has no solution and IP-FVS2 cannot complete the calculation within 2 hr, whereas IP-FVS2-approx(10) obtains the solution {R00212, R00238, R05351} (see also Supplementary Table S12). When the target compound is C00036 (oxaloacetate), the solutions given by IP-FVS1 and IP-FVS1-approx(10) are {R00345, R00355, R00483} and {R00345, R00355, R00357}, respectively. Although IP-FVS2 cannot complete the calculation, IP-FVS2-approx(10) obtains the solution {R00345, R00485}. The relation is summarized in Supplementary Table S13.
Finally, when the target compound is C00074 (phosphoenolpyruvate) or“5 compounds,” neither FVS-type1 nor FVS-type2 has solutions.
4. Theoretical Results
Solving ILP is NP-complete. However, it does not mean that a problem that can be formalized as ILP is NP-complete. Therefore, in this section, we prove that MKMN-B and MKMN-EM are NP-complete and show the appropriateness of formalizing MKMN-B and MKMN-EM as ILP.
Theorem 1
MKMN-B is NP-complete even if the maximum degree is limited to 2, and the substrates and products of a reaction in N1 are the same as those in N2.
Proof: The proof of the NP-hardness is similar to that of Theorem 1 in Tamura and Akutsu (2010). The NP-completeness of the decision version of MKMN-B is shown by the polynomial time reduction from the decision version of the hitting set problem (HSP). The former aims to determine whether there exists |Va| ≤ z, whose MaxVA satisfies that v1 = 0 for all 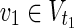 and v2 = 1 for all
and v2 = 1 for all 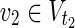 hold. Given a set of elements
hold. Given a set of elements 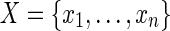 , a set of subsets
, a set of subsets 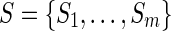 , and an integer z, the HSP determines whether Y (Y ⊂ X) exists such that Si ∩ Y ≠ φ for any
, and an integer z, the HSP determines whether Y (Y ⊂ X) exists such that Si ∩ Y ≠ φ for any 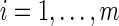 and |Y| ≤ z (Ausiello, 1999).
and |Y| ≤ z (Ausiello, 1999).
When an instance of HSP with X, S, and z is given, N1 and N2 of MKMN-B are constructed as follows.
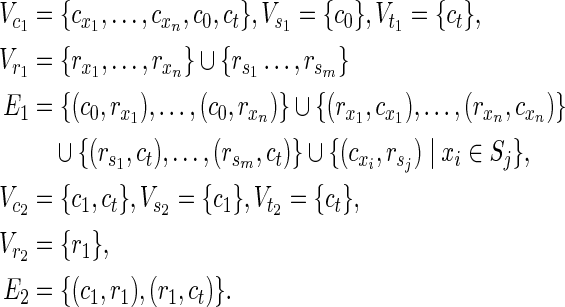 |
For example, if an instance of the HSP is given as X = {1, 2, 3, 4} and S = {{1, 2}, {1, 3}, {2, 3}, {1, 4}, {3, 4}}, then N1 and N2 of MKMN-B are constructed as shown in Figure 6A. This conversion can be conducted in polynomial time.
FIG. 6.
Example of the polynomial time reduction from the HSP. (A) N1 and N2 of MKMN-B are constructed from an instance of the HSP with X = {1, 2, 3, 4} and S = {{1, 2}, {1, 3}, {2, 3}, {1, 4}, {3, 4}}. (B) Compound nodes with outdegree greater than 2 can be converted to nodes with outdegree at most 2. (C) Reaction nodes with indegree greater than 2 can be converted to nodes with indegree at most 2.
In the following, we show that HSP has a solution with |Y| = z if and only if MKMN-B has a solution with |Va| = z. Suppose that HSP has a solution 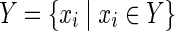 . Then,
. Then, 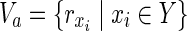 is a solution of MKMN-B since |Y| = |Va| holds. Next, suppose that MKMN-B has a solution Va. If
is a solution of MKMN-B since |Y| = |Va| holds. Next, suppose that MKMN-B has a solution Va. If 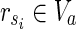 holds, then
holds, then 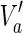 , where
, where 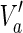 is obtained by replacing
is obtained by replacing 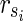 with
with 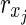 satisfying
satisfying 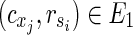 , is also a solution. Therefore, we can assume, without generality that Va does not include
, is also a solution. Therefore, we can assume, without generality that Va does not include 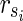 . Then,
. Then, 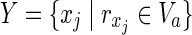 is a solution of the HSP. Since the decision version of MKMN-B is clearly in NP, it belongs to NP-complete.
is a solution of the HSP. Since the decision version of MKMN-B is clearly in NP, it belongs to NP-complete.
Each node with degree greater than 2 can be converted into nodes with degree at most 2 by the methods shown in Figure 6B and C. Since reactions in N1 and N2 created by this reduction do not intersect, MKMN-B is NP-hard, even when reactions common to both N1 and N2 have the same substrates and the same products. ■
Theorem 2
MKMN-EM is NP-hard even if the maximum degree is limited to 2, and the substrates and products of a reaction in N1 are the same as those in N2.
Proof: In N1 of the proof of Theorem 1 with 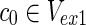 , there are m EMs
, there are m EMs 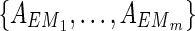 with ct = 1 where
with ct = 1 where 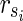 = 1 holds only for
= 1 holds only for 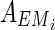 . If the HSP has a solution
. If the HSP has a solution 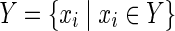 , then
, then 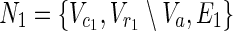 for
for 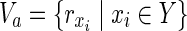 does not have any EM with ct = 1. On the other hand, suppose that
does not have any EM with ct = 1. On the other hand, suppose that 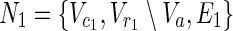 does not have any EM with ct = 1. If Va includes some
does not have any EM with ct = 1. If Va includes some 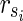 , then
, then 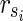 can be replaced with some
can be replaced with some 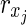 satisfying
satisfying 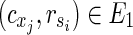 since N1 = {
since N1 = {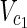 ,
, 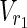 \{V
a\{
\{V
a\{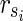 }}\ {
}}\ {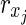 }, E1} does not have any EM with ct = 1 either. Therefore we can assume without generality that Va does not include any
}, E1} does not have any EM with ct = 1 either. Therefore we can assume without generality that Va does not include any 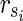 . Then,
. Then, 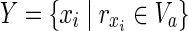 is the solution of HSP. The other parts of the proof are similar to Theorem 1 other than
is the solution of HSP. The other parts of the proof are similar to Theorem 1 other than 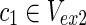 is assumed. ■
is assumed. ■
Theorem 3
MKMN-EM is NP-complete even if EMs of N1 and N2 are given.
Proof: If the sets of reactions of N1 and N2 have no intersections, then the problem can clearly be reduced to the HSP for EMs of N1. Note that the completeness holds for the number of EMs but not for the number of nodes since the number of EMs may be exponential to the number of nodes. ■
5. Discussion
In this article, we have studied the MKMN problem to determine the minimum set of reactions whose inhibition induces that the target compounds are not producible in N1 but are producible in N2. MKMN-B and MKMN-EM are the Boolean version and elementary mode (EM)-based version of MKMN, respectively.
For MKMN-B, we developed integer linear programming (ILP)-based methods, IP-FVS1 and IP-FVS2, utilizing the idea of feedback vertex sets (FVS) to reduce the number of variables present in the ILP formalizations. In IP-FVS1, MaxVA and MinVA are strictly applied to N1 and N2, respectively, to appropriately take the effect of cycles into account. However, since each node in FVS works as if it were a source node in the MaxVA, the solution obtained by IP-FVS1 is not always realistic. To avoid this problem, IP-FVS2 does not apply MaxVA to cycles consisting only of original reversible reactions in N1. To solve MKMN-B for large networks, we developed IP-FVS1-approx and IP-FVS2-approx. These algorithms are fast since the number of time steps is limited to a small constant, but the optimality of their solutions is not ensured. We also developed IP-EM, an ILP-based method for MKMN-EM in which every EM including a source node and a target compound is inhibited in N1, but at least one such EM remains in N2.
We implemented IP-FVS1, IP-FVS2, IP-FVS1-approx, IP-FVS2-approx, and IP-EM in computational experiments, using metabolic networks of bifidobacterium longum DJO10A (BLJ) and clostridium perfringens SM101 (CPR) for N1 and N2, respectively. The datasets 1, 2, and 3 include 155, 650, and 2340 nodes in total, respectively. From Tables 8, 11, 14, and 16, it is seen that MKMN tends to have no solution for smaller networks but has solutions for larger networks. However, Tables 13 and 14 show that solving MKMN for large networks is computationally very expensive. In particular, IP-FVS2 and the IP-EM could not finish the computation within 2 hr for most cases. Tables 15 and 16 show that IP-FVS1-approx and IP-FVS2-approx can solve MKMN-B very efficiently even for large networks. Although optimality is not ensured by IP-FVS1-approx and IP-FVS2-approx, the solutions they produced are optimal for most cases in the dataset 3 experiment as shown in Table 17. Furthermore, since MKMN-B belongs to NP, it is not difficult to confirm that the obtained solutions by IP-FVS1-approx and IP-FVS2-approx satisfy that the target compound is not producible in N1 but producible in N2 even for nonrestricted time steps. It remains as a future work to develop a method of finding the smallest number of the time steps to ensure the optimality of IP-FVS1-approx and IP-FVS2-approx. It is to be noted that some bilevel programming-based method seems to be necessary to define MKMN in an FBA model since FBA needs another objective function in addition to minimizing the number of reactions. Applying Petri-net-based methods (Jin et al., 2011) is also interesting since they may extract the good points of both Boolean-based methods and FBA-based methods.
We also analyzed the obtained solutions by checking the relation among the predecessors of N1 and N2, and the selected reactions. From Supplementary Tables S5, S10, and S11, it may be thought that the solutions given by IP-FVS1 and IP-FVS1-approx are trivial when the predecessors of N2 are not a subset of those of N1, since choosing all predecessors of N1 clearly satisfies the condition of MKMN-B. However, Supplementary Tables S3 and S7 show that this method cannot always obtain the correct solution. On the other hand, when the predecessors of N2 are a subset of those of N1, the obtained solution is complex as shown in Supplementary Table S4. It seems that the solutions given by IP-FVS2, IP-FVS2-approx, and IP-EM are somewhat involved, and it is difficult to infer solutions from these tables. Since a smaller set of inhibited reactions is preferable, solutions of IP-FVS2 and IP-EM are considered to be better than those from IP-FVS1. Moreover, for large networks, since IP-FVS2 and IP-EM are generally unable to complete the calculation, IP-FVS2-approx is considered to be the best method, although the optimality of its solution is not ensured in the worst-case scenario.
As theoretical results, we proved that MKMN-B is NP-complete for the number of nodes, MKMN-EM is NP-hard for the number of nodes, and MKMN-EM is NP-complete for the number of EMs. The software developed in this study and reported here is available online (minFvskO, 2015).
Supplementary Material
Acknowledgments
T.T. was partially supported by JSPS, Japan [Grant-in-Aid for Young Scientists (B) 25730005 and Grant-in-Aid for Scientific Research (A) 25250028]. J.S. is an Australian National Health and Medical Research Council (NHMRC) Peter Doherty fellow and a recipient of the Hundred Talents Program of the Chinese Academy of Sciences (CAS). T.A. was partly supported by the Chinese Academy of Sciences Visiting Professorship for Senior International Scientists, China, and Grant-in-Aid #22240009 from JSPS, Japan.
Disclosure Statement
No competing financial interests exist.
References
- Acuña V., Chierichetti F., Lacroix V., et al. 2009. Modes and cuts in metabolic networks: Complexity and algorithms. Biosystems 95, 51–60 [DOI] [PubMed] [Google Scholar]
- Akutsu T., Zhao Y., Hayashida M., et al. 2012. Integer programming-based approach to attractor detection and control of boolean networks. IEICE Trans. Inf. Syst. 95, 2960–2970 [Google Scholar]
- Ausiello G.1999. Complexity and approximation: Combinatorial optimization problems and their approximability properties. Springer, New York [Google Scholar]
- Ballerstein K., von Kamp A., Klamt S., et al. 2012. Minimal cut sets in a metabolic network are elementary modes in a dual network. Bioinformatics 28, 381–387 [DOI] [PubMed] [Google Scholar]
- Burgard A.P., Pharkya P., and Maranas C.D.2003. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84, 647–657 [DOI] [PubMed] [Google Scholar]
- Flöttmann M., Krause F., Klipp E., et al. 2013. Reaction-contingency based bipartite boolean modelling. BMC Syst. Biol. 7, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves E., Bucher J., Ryll A., et al. 2013. Bridging the layers: towards integration of signal transduction, regulation and metabolism into mathematical models. Mol. BioSyst. 9, 1576–1583 [DOI] [PubMed] [Google Scholar]
- Hädicke O., and Klamt S.2011. Computing complex metabolic intervention strategies using constrained minimal cut sets. Metab. Eng. 13, 204–213 [DOI] [PubMed] [Google Scholar]
- Handorf T., Ebenhöh O., and Heinrich R.2005. Expanding metabolic networks: scopes of compounds, robustness, and evolution. J. Mol. Evol. 61, 498–512 [DOI] [PubMed] [Google Scholar]
- IBM ILOG CPLEX Optimizer. 2010. www-01.ibm.com/software/integration/optimization/cplex-optimizer/
- Jiang D., Zhou S., and Chen Y.P.2009. Compensatory ability to null mutation in metabolic networks. Biotechnol. Bioeng. 103, 361–369 [DOI] [PubMed] [Google Scholar]
- Jin G., Zhao H., Zhou X., et al. 2011. An enhanced petri-net model to predict synergistic effects of pairwise drug combinations from gene microarray data. Bioinformatics 27, i310–i316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., and Goto S.2000. Kegg: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman K.J., Prakash P., and Edwards J.S., 2003. Advances in flux balance analysis. Curr. Opin. Biotechnol. 14, 491–496 [DOI] [PubMed] [Google Scholar]
- Kim J., and Reed J.L.2010. Optorf: Optimal metabolic and regulatory perturbations for metabolic engineering of microbial strains. BMC Syst. Biol. 4, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamt S.2006. Generalized concept of minimal cut sets in biochemical networks. Biosystems 83, 233–247 [DOI] [PubMed] [Google Scholar]
- Klamt S., and Gilles E.D.2004. Minimal cut sets in biochemical reaction networks. Bioinformatics 20, 226–234 [DOI] [PubMed] [Google Scholar]
- Klamt S., Saez-Rodriguez J., and Gilles E.D.2007. Structural and functional analysis of cellular networks with cellnetanalyzer. BMC Syst. Biol. 1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke N., Herédia F., Barcellos C.K., et al. 2004. Essentiality and damage in metabolic networks. Bioinformatics 20, 115–119 [DOI] [PubMed] [Google Scholar]
- Li Z., Zhang S., Wang Y., et al. 2007. Alignment of molecular networks by integer quadratic programming. Bioinformatics 23, 1631–1639 [DOI] [PubMed] [Google Scholar]
- Lu W., Tamura T., Song J., et al. 2014. Integer programming-based method for designing synthetic metabolic networks by minimum reaction insertion in a boolean model. PLOS ONE 9, e92637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- minFvskO. 2015. http://sunflower.kuicr.kyoto-u.ac.jp/∼rogi/minFvskO/minFvskO.html
- Raman K., and Chandra N., 2009. Flux balance analysis of biological systems: applications and challenges. Brief. Bioinform. 10, 435–449 [DOI] [PubMed] [Google Scholar]
- Samaga R., Von Kamp A., and Klamt S.2010. Computing combinatorial intervention strategies and failure modes in signaling networks. J. Comp. Biol. 17, 39–53 [DOI] [PubMed] [Google Scholar]
- Schrijver A.1998. Theory of linear and integer programming. John Wiley & Sons, Hoboker, NJ [Google Scholar]
- Schuster S., Fell D.A., and Dandekar T.2000. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 18, 326–332 [DOI] [PubMed] [Google Scholar]
- Schuster S., and Hilgetag C.1994. On elementary flux modes in biochemical reaction systems at steady state. Journal of Biological Systems 2, 165–182 [Google Scholar]
- Schuster S., Pfeiffer T., and Fell D.A.2008. Is maximization of molar yield in metabolic networks favoured by evolution? J. Theor. Biol. 252, 497–504 [DOI] [PubMed] [Google Scholar]
- Segre D., Vitkup D., and Church G.M.2002. Analysis of optimality in natural and perturbed metabolic networks. Proceedings of the National Academy of Sciences 99, 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T., Cabili M.N., and Ruppin E.2009. Predicting metabolic biomarkers of human inborn errors of metabolism. Mol. Syst. Biol. 5, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart A.G., Amaral L.A.N., and Ottino J.M.2008. Cascading failure and robustness in metabolic networks. Proceedings of the National Academy of Sciences 105, 13223–13228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar P., Song B., Kahveci T., et al. 2008. Mining metabolic networks for optimal drug targets. Pac. Symp. Biocomput. 13, 291–302 [PubMed] [Google Scholar]
- Stelling J., Klamt S., Bettenbrock K., et al. 2002. Metabolic network structure determines key aspects of functionality and regulation. Nature 420, 190–193 [DOI] [PubMed] [Google Scholar]
- Tamura T., and Akutsu T.2010. Exact algorithms for finding a minimum reaction cut under a boolean model of metabolic networks. IEICE Transactions on Fundamentals of Electronics, Communications and Computer Sciences 93, 1497–1507 [Google Scholar]
- Tamura T., Cong Y., Akutsu T., et al. 2011. An efficient method of computing impact degrees for multiple reactions in metabolic networks with cycles. IEICE Trans. Inf. Syst. 94, 2393–2399 [Google Scholar]
- Tamura T., Takemoto K., and Akutsu T.2010. Finding minimum reaction cuts of metabolic networks under a boolean model using integer programming and feedback vertex sets. International Journal of Knowledge Discovery in Bioinformatics (IJKDB) 1, 14–31 [Google Scholar]
- Tepper N., and Shlomi T.2010. Predicting metabolic engineering knockout strategies for chemical production: accounting for competing pathways. Bioinformatics 26, 536–543 [DOI] [PubMed] [Google Scholar]
- Trinh C.T., Carlson R., Wlaschin A., et al. 2006. Design, construction and performance of the most efficient biomass producing e. coli bacterium. Metab. Eng. 8, 628–638 [DOI] [PubMed] [Google Scholar]
- Unrean P., Trinh C.T., and Srienc F.2010. Rational design and construction of an efficient e.coli for production of diapolycopendioic acid. Metab. Eng. 12, 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A., and Palsson B.O.1994. Metabolic flux balancing: Basic concepts, scientific and practical use. Nat. Biotechnol. 12, 994–998 [Google Scholar]
- Wunderlich Z., and Mirny L.A.2006. Using the topology of metabolic networks to predict viability of mutant strains. Biophys. J. 91, 2304–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Tamura T., Akutsu T., et al. 2013. Flux balance impact degree: a new definition of impact degree to properly treat reversible reactions in metabolic networks. Bioinformatics 29, 2178–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.