Abstract
Photoperiod, or the duration of light in a given day, is a critical cue that flowering plants utilize to effectively assess seasonal information and coordinate their reproductive development in synchrony with the external environment. The use of the model plant, Arabidopsis thaliana, has greatly improved our understanding of the molecular mechanisms that determine how plants process and utilize photoperiodic information to coordinate a flowering response. This mechanism is typified by the transcriptional activation of FLOWERING LOCUS T (FT) gene by the transcription factor CONSTANS (CO) under inductive long-day conditions in Arabidopsis. FT protein then moves from the leaves to the shoot apex, where floral meristem development can be initiated. As a point of integration from a variety of environmental factors in the context of a larger system of regulatory pathways that affect flowering, the importance of photoreceptors and the circadian clock in CO regulation throughout the day has been a key feature of the photoperiodic flowering pathway. In addition to these established mechanisms, the recent discovery of a photosynthate derivative trehalose-6-phosphate as an activator of FT in leaves has interesting implications for the involvement of photosynthesis in the photoperiodic flowering response that were suggested from previous physiological experiments in flowering induction.
Keywords: Photoperiodism, Flowering, Phenology, Circadian Clock, Florigen
I. INTRODUCTION
Seasonal variation in climate has selected for the ability of organisms to predict future environmental conditions and use this information to complete necessary adjustments to thrive. The tilt of the earth’s axis relative to the sun throughout the solar year can lead to radical changes in weather patterns and temperature, especially in non-equatorial regions (Thomas and Vince-Prue, 1996). Survival often depends on the development of strategies to cope with suboptimal conditions and to use optimal ones as fully as possible. Precise timing of key events in the span of a life cycle is a key trait for organisms faced with a seasonally shifting environment. The timing of the reproductive cycle is a good example of this phenomenon; in a substandard environment premature flowering can have severe implications for relative fitness. For plants dependent on pollinators for reproduction, flowering also must to be timed with the seasonal availability of other organisms (Hegland et al., 2009). As an irreversible process in most species, the timing of the reproductive transition in plants is especially critical (Kobayashi and Weigel, 2007).
The topic of how plants are able to recognize what constitutes optimal conditions for flowering has been an active area of research for almost a century. Wightman Garner and Henry Allard, two researchers at the USDA, were the first to empirically describe that the duration of light in a 24-hour period is a key cue for the induction of flowering in many plant species. Originally interested in explaining why soybeans planted sequentially over the summer decreased in days to flower as they were planted later in the season, they sought to find the casual variable behind the phenomenon. Over the course of two years from 1918 to 1920, they experimentally manipulated exposure of plants to light and dark cycles by moving plants from a common outdoor plot into darkened sheds. Through the careful control of light and dark duration to simulate different seasonal light conditions, they were able to determine critical durations of light or darkness that are required for induction of flowering in over 12 plant species and many different cultivars. This general principle of an exhibited response triggered by a change in day length, they coined “photoperiodism” (Garner and Allard, 1920). This revolutionary idea changed the thinking about seasonal responses by suggesting that the mechanism for sensing seasonal changes could be tied specifically to the sensing of duration of light in a given day. In addition, they found that plants could be grouped into three different groups groups by their flowering response. Some plants flower as day length increases in late spring (long-day plants), some flower as day length wanes as autumn begins (short-day plants), and some plants flower at certain times regardless of the photoperiods (day-neutral plants) (Garner and Allard, 1920).
The determination of day-length as a critical regulator of flowering time left several questions with regard to the physiology of the flowering response. Where was day-length sensed in the plant and how is the signal for floral induction carried throughout the organism? Elegant grafting experiments performed first by the Russian physiologist Mikhail Chailakhyan determined that a mobile signal from leaf scions exposed to inductive photoperiods could induce flowering in non-induced graft stock (Chailakhyan, 1937; Chailakhyan, 1968). Experimental evidence suggested that the transmissible signal could be universal or nearly universal among flowering plants, for instance grafts in which leaves from induced short-day Kalanchoë blossfeldiana (stock) and long-day Sedum spectabile plants (scion) were able were able to induce flowering when grafted to plants of the opposite response type (Wellensi, 1967; Zeevaart, 2006). Grafts between different species were also often found to lead to flowering induction (Zeevaart, 1976; 2006). These observations led Denis Carr and Lloyd Evans to propose a model for two-step floral induction (Carr, 1967; Evans, 1971). The first stimulus would be involved in the sensing of photoperiod and the incorporation of other endogenous and environmental factors, which would induce the secondary stimulus that was potentially universal and transmitted from the leaf.
The search for the chemical basis of florigen remained elusive and gradually fell out of favor until contributions from Arabidopsis that facilitated the discovery of FT protein as a key candidate. The discovery FT as a mobile signal in Arabidopsis along with recognition that its function is conserved in a range of distantly related plant species (Corbesier et al., 2007), has cemented the role of FT as a universal florigen (Abe et al., 2005; Kobayashi and Weigel, 2007; Kojima et al., 2002; Tamaki et al., 2007; Wigge et al., 2005). Increasingly, as our understanding of the photoperiodic sensing mechanism has expanded, we have found that similar regulatory networks govern flowering plant species other than Arabidopsis, and that the mechanism of photoperiodic flowering induction is highly conserved (Song et al., 2010).
In the following article, we will review developments in understanding the molecular mechanism of the photoperiodic flowering response through the model organism Arabidopsis thaliana, as well as recent discoveries that highlight the modulation of the photoperiodic sensing mechanism to accommodate both external environmental factors such as light quality through the action of photoreceptor proteins as well as internal physiological status through the sensing of internal photosynthetic accumulation.
II. PHOTOPERIODIC FLOWERING AND THE EXTERNAL COINCIDENCE MODEL
The key question that emerged with the discovery of photoperiodic flowering responses was the mechanism for how photoperiod was sensed. Since the early 18th century with the experiments of the astronomer De Mairan, plants have been known to have oscillatory leaf movements that occur in 24-hour cycles even in the absence of light, as if a light stimulus was present (De Mairan, 1729). These rhythms, which show a period of around 24 hours (hence circadian), show an inherited entrainment to the rotation of the earth that persists even after many generations of exposure to alternative day lengths in the laboratory (Bünning, 1960). This internal “clock” has extreme selective value in the regulation of daily output changes to the internal biochemical processes of the cell and the organism, which we can now appreciate given the advances in molecular biology in the last decades (Baudry and Kay, 2008). The connection between the internal clock and photoperiodic responses, however, was not immediately clear. First proposed by Erwin Bünning in 1936, and refined later by Colin Pittendrigh the “external coincidence” model, as it came to be known, proposed that photoperiodic phenomena could be explained by the interaction of light stimuli and the clock (Bünning, 1936; Pittendrigh and Minis, 1964). The clock would set the pace of the 24-hour rhythm, and define a period of photosensitivity to which exposure to light would be inductive for a photoperiodic response (Pittendrigh, 1972). In non-inductive photoperiods, the presence of darkness during the sensitive period of the response would result in no elicited reaction. In contrast, the encroachment of light into the photosensitive part of the circadian cycle, brought upon by longer inductive photoperiods, would cause a physiological response (see Figure 1).
Fig. 1.
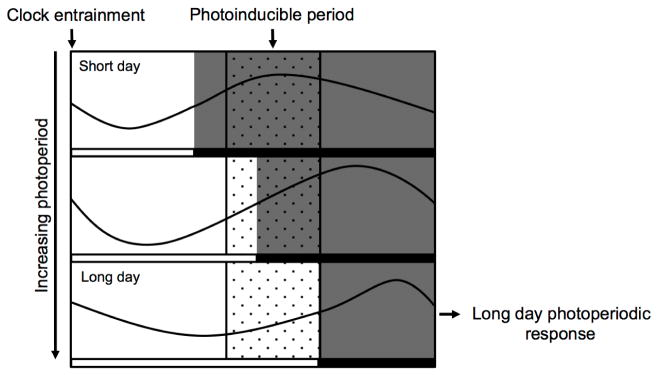
The external coincidence model for photoperiodic phenomena:
The following example represents a photoperiodic response that occurs in the afternoon of long days, as in photoperiodic flowering in Arabidopsis. The circadian clock generates a rhythm that determines a specific period of the day in which a light signal can induce the response. This period is similar regardless of day length. In short day conditions the photoinducible period does not coincide with a light signal, so no response occurs. As days lengthen with the coming of spring and summer, light begins to encroach on the photoinducible period, eliciting the photoperiodic response. Light serves a dual purpose; to reset the clock at dawn and dusk and to be present or absent during the photoinducible phase, to promote or halt the response.
For more than thirty years, it remained controversial that the endogenous circadian clock regulated the photoperiodic flowering response. Key experiments that unequivocally linked flowering to the clock were performed by Murray Coulter and Karl Hamner on the short-day plant Glycine max in 1964, by giving light pulses at different time points after transfer of plants into continuous darkness. One of the prevailing counter-hypotheses of the time posited that night duration was the primary cue for the photoperiodic response, and that this was mediated by the turnover kinetics of the photoreceptor phytochrome. According to this hypothesis, for short-day plants, in which photoperiods below a certain threshold are inductive, directing light pulses at different times of night should affect the photoperiodic flowering response equally as long as a certain night length was prevented. It was found, instead, that light pulses during the night (referred to as night breaks) affected the flowering response in a rhythmic fashion (Carpenter and Hamner, 1964; Coulter and Hamner, 1964). Additional experiments performed by Halaban in 1968 in the short day plant Coleus frederici showed that the phases in which flowering was inhibited by night break pulses of plants always correlated with leaf movement position rather than the duration of night (Halaban, 1968a; b). This was true for plants placed under several different photoperiods. These early findings helped to cement the clock as a crucial component in determining photoperiodic flowering responses.
A. GENETICS OF PHOTOPERIODIC FLOWERING IN ARABIDOPSIS
Most Arabidopsis accessions that were initially collected for use in the laboratory belong to the summer annual class of wild Arabidopsis, mainly due to the ease of flowering without vernalization treatment and compact stature. Interestingly, some of the earliest mutations described in Arabidopsis are part of the regulatory framework that determines the photoperiodic flowering response, as mutations in these genes often convert compact summer annual accessions into phenotypes with long vegetative phases of growth. Mutagenic screens performed by Gyorgy Redei in 1962 isolated gigantia (gi) and constans (co) (as supervital mutants), far earlier than the forward genetic screens that would later more clearly define the regulatory networks that govern the flowering response (Rédei, 1962).
The advent of molecular markers in Arabidopsis in the late 1980’s by Maarten Koorneef and colleagues enabled the systematic categorization of genes involved in the regulation of flowering time and mapping of their associated loci. Initial genetics of late flowering mutants of Arabidopsis found that CO, GI, and FT were likely components of the same regulatory pathway (Koornneef et al., 1991).
B. CO-FT MODULE IN ARABIDOPSIS
The co and gi mutant phenotype initially interested researchers studying the genetics basis of flowering time because these mutants exhibited a “day neutral” phenotype (Park et al., 1999; Putterill et al., 1995). Under inductive long day conditions, they flowered much later than wild type plants, but flowered about the same as wild type under non-inductive short day conditions. Additional phenotypic analysis led to the conclusion that CO is a limiting factor of flowering under short day conditions and that CO can promote flowering in a dose dependent manner under inductive photoperiods (Putterill et al., 1995). Transgenic analysis of plants expressing CO under a dexamethasone inducible construct found that plants could be induced to flower regardless of the external photoperiod when CO is highly expressed (Simon et al., 1996). Generation of mutants involved in the regulation of the circadian clock and light signaling also commonly affected the photoperiodic flowering response. Mutations in LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), EARLY FLOWERING 3 (ELF3), TIMING OF CAB EXPRESSION 1 (TOC1), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1), PSEUDO-RESPONSE REGULATOR 5 (PRR5), PRR7, PRR9, and CRYPTOCHROME 2 (CRY2) all displayed aberrant flowering phenotypes, which suggested that the clock on a molecular level was key to the proper induction of a photoperiodic response (El-Din El-Assal et al., 2001; Hicks et al., 1996; Ito et al., 2008; Nelson et al., 2000; Park et al., 1999; Sato et al., 2002; Schaffer et al., 1998; Somers et al., 2000). CO mRNA abundance was found to show a pronounced circadian oscillation in long day conditions, and was found to continue to occur after plants entrained to long day conditions were transferred to continuous light (Yanovsky and Kay, 2002). This suggested that the circadian clock regulated CO transcription. Additionally, the CO transcriptional pattern was significantly affected by mutations in clock components such as toc1-1, resulting in early flowering. toc1-1 mutants have a shortened circadian period to about 21 hours; when circadian periods were shortened to compensate for the short period defect in toc1-1, however, proper CO expression and function was restored. CO transcripts continue to oscillate in short day conditions, but CO protein was initially shown to be highly unstable and actively degraded in the dark (Valverde et al., 2004; Yanovsky and Kay, 2002). This discrepancy between transcript abundance and protein stability explains how the constriction of active CO protein to the afternoon of long days enables a photoperiodic response, and fits nicely with our understanding of the external coincidence model in reference to photoperiodic phenomena (Figure 1). Coupled with experimental evidence that CO was a transcriptional activator of FT (Kardailsky et al., 1999; Kobayashi et al., 1999; Onouchi et al., 2000; Samach et al., 2000) and that FT was directly involved in signaling the activation of floral meristem differentiation, a CO-FT module in which clock and light regulated CO would perceive photoperiodic information and signal for the induction of downstream flowering response through the activation of FT transcription began to take shape.
Thus in line with earlier experimental data from the 1960’s and 1970’s, molecular evidence suggested that the circadian clock could regulate the photoperiodic response, in this case through the transcriptional and post translational regulation of CO, and that this could lead to flowering under inductive conditions. Since this discovery, the regulation of photoperiodic flowering pathway has become increasing complex, and many factors have been shown to regulate CO and FT through a variety of mechanisms (Andres and Coupland, 2012).
III. CURRENT MOLECULAR MECHANISM OF PHOTOPERIODIC FLOWERING IN ARABIDOPSIS
In Arabidopsis, long days promote flowering through the function of FT protein (Andres and Coupland, 2012; Wigge, 2011). The protein, a mobile florigen, is synthesized in phloem companion cells of leaves and translocated to the shoot apical meristem where the floral primordia are formed (Corbesier et al., 2007). The timing of flowering is strongly correlated with the relative amount of FT; the high levels of FT transcript in longer photoperiods influence more rapid flowering compared with the low levels in shorter photoperiods (Kobayashi et al., 1999). The transcriptional activator CONSTANS (CO) protein directly induces the expression of FT gene in a day length-dependent manner (Samach et al., 2000). CO gene expression is controlled by the circadian clock (Suárez-López et al., 2001), and CO protein abundance is modulated by light signaling, which CO protein is stabilized in the afternoon of long days (Song et al., 2012b; Valverde et al., 2004). Together, these processes explain how day length is determined and how the floral transition is mediated under inductive photoperiod.
A. REGULATION OF CO TRANSCRIPTION
To accurately control the timing of seasonal flowering in Arabidopsis, the circadian clock-regulated CO expression is a crucial mechanism to precisely measure the difference in day length. CO transcription is controlled by many circadian clock proteins, such as CCA1, LHY, and PRRs. These clock proteins directly or indirectly regulate the gene expression of CYCLING DOF FACTORs (CDFs), transcriptional repressors of CO (Song et al., 2010). CDF1 directly binds to the CO promoter and represses its transcription in the morning redundantly with other CDF proteins, CDF2, CDF3, and CDF5 (Fornara et al., 2009; Imaizumi et al., 2005; Sawa et al., 2007). The expression level of CDF1 gene is positively regulated by CCA1 and LHY proteins (Nakamichi et al., 2007), which are most abundant at dawn (Schaffer et al., 1998; Wang and Tobin, 1998). Consequently, the expression level of CDF1 transcript remains high during the morning (Imaizumi et al., 2005). In the afternoon, the abundance of CDF transcripts is reduced through the function of four PRR family members, TOC1, PRR5, PRR7, and PRR9. These PRR proteins physically associate with the CCA1 and LHY loci and repress CCA1 and LHY gene expression (Huang et al., 2012; Nakamichi et al., 2010). TOC1, PRR5, PRR7, and PRR9 proteins also negatively regulate the expression of CDF1 gene (Ito et al., 2008; Nakamichi et al., 2007). In addition, PRR5 and PRR7 directly binds to the CDF2 and CDF5 loci to and repress their transcription (Liu et al., 2013b; Nakamichi et al., 2012) Coincident with their similar roles to CDF1 in CO regulation CDF2, CDF3, and CDF5 transcripts are also high in the morning (Fornara et al., 2009). Clock regulation of CDF expression, which keeps CO expression low in the morning, lays the groundwork for determining the photosensitive period later in the afternoon of long days, preventing early flowering in shorter photoperiods.
In long days, the repression of CO gene expression by CDF proteins is released through the function of FKF1-GI complex in the afternoon (Sawa et al., 2007). FKF1 protein is a blue light photoreceptor (Imaizumi et al., 2003; Sawa et al., 2007) and possesses an E3 ubiquitin ligase activity that mediates proteasome-dependent degradation of target proteins (Imaizumi et al., 2005). Once the expression patterns of FKF1 and GI proteins coincide with light in the afternoon, FKF1 absorbs blue light and is activated. Then, the blue light-activated FKF1 forms a protein complex with GI. The FKF1-GI complex recognizes CO repressors, the CDF proteins, and removes those repressors by ubiquitin-dependent degradation on the CO promoter (Sawa et al., 2007). FKF1 homologs, ZEITLUPE (ZTL) and LOV KELCH PROTEIN2 (LKP2) proteins, both of which interact with FKF1 and GI proteins, are also involved in the destabilization of CDF2 protein (Fornara et al., 2009). Removal of CDF proteins by the function of FKF1 protein constricts the action of CDF repressors to the morning of long days and facilitates the CO gene to be expressed during the late afternoon, while light still remains in the day (Figure 2). Maintaining this window of CO expression to the late afternoon allows for the subsequent peak of activation FT at dusk of long days, enabling the photoperiodic flowering response.
Fig. 2.
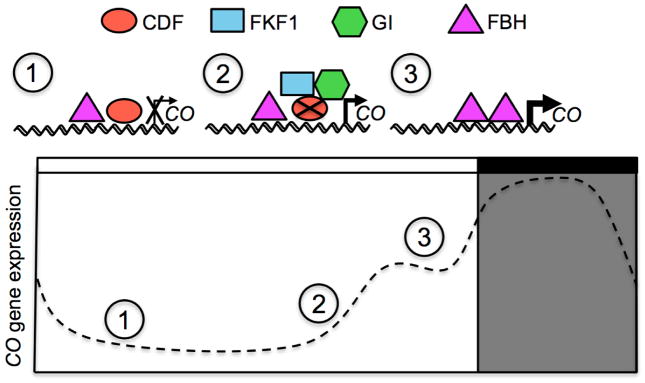
CONSTANS (CO) oscillatory transcription is dependent on multiple factors throughout the day: CO gene expression changes throughout the day. In inductive long-day conditions for flowering in Arabidopsis, the peak of CO expression is constrained to the afternoon before dusk. In the morning, CDF family transcription factors bind to the CO promoter to repress transcription. Beginning in the afternoon, FKF1 and GI form a protein complex that ubiquitinates CDFs through an F-box protein function on FKF1 and targets them for proteasomal degradation, freeing the CO promoter from repression. FBH transcriptional activators are then recruited to the CO genomic locus, resulting in increased transcription of CO before dusk. Constraining CO mRNA expression to the late afternoon, and stabilization of resultant CO protein results in FT expression at dusk and promotes flowering in long days.
In contrast to long days, the expression of FKF1 and GI proteins is out of phase in short day conditions. No functional complex between the proteins exits in the daytime under these conditions, which results in the accumulation of CO transcripts only during the dark period, which subsequently causes an extremely low level of FT expression throughout the day. Transcriptional regulation of CO gene expression thus is critical for sensing day length and differentiating between inductive and non-inductive photoperiods to coordinate the flowering response (Sawa et al., 2007).
Once CO repression by CDF proteins is relieved, four bHLH transcription factors, FLOWERING BHLH 1 (FBH1), FBH2, FBH3, and FBH4, activate CO expression (Ito et al., 2012). These FBH proteins directly bind to E-box elements in the CO promoter and redundantly induce CO expression in the late afternoon and the dark under both long and short day conditions (Figure 2). It is proposed that FBH mediated CO activation is conserved in other plant species because overexpression of FBH homologue genes in rice and poplar highly upregulates CO transcripts in Arabidopsis (Ito et al., 2012). To date, our knowledge of transcriptional repression of CO is much more developed than it’s activation (Song et al., 2013), and more work needs to be done to determine additional factors involved as well as time dependent impacts of CO activators on CO transcription.
B. POSTTRANSLATIONAL REGULATION OF CO PROTEIN
Along with the transcriptional regulation of CO gene, the posttranslational regulation of CO protein is crucial for the day length-dependent FT activation. In both long and short day conditions, the highest accumulation of CO mRNA occurs in the dark (Suárez-López et al., 2001). However, the expression of FT peaks at dusk in long days (Suárez-López et al., 2001). Various light signaling and proteasome-dependent protein degradation mechanisms have been shown to control CO protein stability and allow the protein to accumulate only in the late afternoon of long days, which accounts for day length-dependent FT expression (Jang et al., 2008; Lazaro et al., 2012; Liu et al., 2008b; Song et al., 2012b; Valverde et al., 2004). Red light delays flowering through the destabilization of CO protein, and far-red and blue light promote flowering through the stabilization of the protein (Valverde et al., 2004). PHYTOCHROME A (PHYA) and PHYB mediate far-red and red light responses, respectively, and CRY2 and FKF1 mediate blue light responses. Two RING finger E3 ubiquitin ligases, CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1), negatively regulate CO protein stability (Jang et al., 2008; Lazaro et al., 2012; Liu et al., 2008b).
CO protein is stable under far-red light and unstable under red light in wild-type Arabidopsis plants. In addition, the amount of the protein is reduced in a phyA mutant background throughout the daytime and, by contrast, increased in a phyB mutant background especially in the morning (Valverde et al., 2004). In natural conditions, the ratio of red to far-red light is high during the daytime and relatively low at dusk. Reflecting this, the levels of CO protein are reduced in the morning and high in the late afternoon; this seems to indicate that PHYA and PHYB antagonistically modulate the stability of the protein.
Recent evidence has suggested that the PHYB dependent regulation of CO protein stability is quite complex, and may contain several factors that both positively and negatively affect CO. Mutations in PHYTOCHROME-DEPENDENT LATE FLOWERING (PHL), cause a late flowering phenotype in long days, similar to other photoperiodic flowering pathway components (Endo et al., 2013). Double mutant combinations with phyB abolish the late flowering phenotype, suggesting that PHL affects the ability of PHYB to repress flowering. PHL does not appear to regulate CO transcription, but CO protein and PHL interact. PHL protein thus a likely factor involved in sheltering CO from PHYB dependent degradation (Endo et al., 2013). Similarly, it has been found that the VASCULAR PLANT ONE ZINC-FINGER1 (VOZ1) and VOZ2, two NAC domain transcription factors, interact with PHYB, and positively regulate flowering in long days. Like PHYB, VOZ1 and VOZ2 are expressed in the cytoplasm and are translocated into the nucleus (Yasui et al., 2012). Their expression is also vascular specific, together with other photoperiodic flowering components (Yasui et al., 2012). The discovery of these factors adds a new layer of complexity with regard to PHYB regulation of photoperiodic flowering, but exact mechanisms for how PHYB destabilizes CO protein remain to be determined. How these PHYB dependent positive regulators of flowering fit into the larger framework of antagonistic PHYB and PHYA signaling with regard to CO will have important implications for light quality dynamics and their impact on the photoperiodic response.
The HOS1 E3 ubiquitin ligase mediates degradation of CO protein in the morning by directly interacting with CO (Lazaro et al., 2012). Another E3 ubiquitin ligase COP1 forms a protein complex with SUPPRESSOR OF PHYA-105 1 (SPA1). The COP1-SPA1 complex binds to CO protein and degrades the protein in the night. Other SPA proteins, SPA2, SPA3, and SPA4, physically interact with CO protein and redundantly regulate the destabilization CO protein(Jang et al., 2008; Laubinger et al., 2006; Saijo et al., 2003). CRY2 is also involved in CO stabilization through protein complex formation with SPA1 (Zuo et al., 2011). The binding of photoactivated-CRY2 to SPA1 enhances the interaction between CRY2 and COP1 in response to blue light, resulting in the suppression of COP1-SPA1 activity and in turn the accumulation of CO in the daytime (Zuo et al., 2011). This function of CRY2 partially explains how blue light accelerates flowering through the stabilization of CO protein and induction of FT transcripts.
While we have a good idea of which factors contribute to CO protein stabilization and destabilization, the relationship between these factors throughout the day and how they compete or interact dynamically for CO protein needs to be further clarified. As has been discussed, three photoreceptors, PHYA, PHYB, and CRY2 and two E3 ubiquitin ligases, HOS1 and COP1, regulate CO protein stability. However, functions of the photoreceptors cannot fully account for the question about how CO protein is stabilized only at the end of day in long day conditions because those photoreceptors are constitutively expressed through the day (Mockler et al., 2003). The function of another blue light photoreceptor FKF1 provides a clue to answer the question. FKF1 protein physically interacts with CO protein in a blue light enhanced manner, and the FKF1-CO interaction increases CO stability at a specific time of day, in the afternoon, under long day conditions (Song et al., 2012b). Together with the similar expression profile of those proteins (Imaizumi et al., 2003; Valverde et al., 2004), the blue light enhanced FKF1-CO interaction supports the notion that FKF1 determines the timing of CO stabilization and that the expression of CO gene under light in long day condition is crucial for FT induction through CO protein accumulation, in which both gene expression and the protein accumulation are regulated by FKF1 function. As the core clock components CCA1 and LHY regulate the timing of FKF1 (Imaizumi et al., 2003), the circadian regulation of the FKF1 photoreceptor function is likely the molecular basis of the photosensitivity phase proposed in the external coincidence model in Arabidopsis.
C. TRANSCRIPTIONAL REGULATION OF THE FT GENE
The photoperiodic flowering pathway serves as a conduit for a large variety of environmental parameters that convert external information and integrate it into FT signal. These environmental signals merge to control the FT expression through numbers of transcription factors (cite Song TiPS 2013). Several classes of transcriptional repressors exist in the regulation of FT gene expression. SCHLAFMÜTZE (SMZ) gene encodes an APETALA2 (AP2)-related transcription factor that binds to the 3′-UTR of FT locus and represses FT transcription (Mathieu et al., 2009), and the expression of the gene is negatively regulated by GI function mediated through a microRNA pathway (Jung et al., 2007). GI protein positively regulates miRNA172 (miR172) accumulation in long days. The miR172 targets SMZ transcripts and decreases amount of the transcripts (Mathieu et al., 2009). TEMPRANILLO 1 (TEM1) protein directly associates with the 5′-UTR of FT gene and represses the gene expression throughout the day in long day conditions. TEM2 is involved in the regulation of FT expression redundantly with TEM1 (Castillejo and Pelaz, 2008). In addition, GI protein interacts with TEM1 and TEM2 in the nucleus in tobacco cells and probably changes activities of TEM proteins (Sawa and Kay, 2011).
Interestingly, the CO transcriptional regulator CDF1 also associates with the FT promoter near the transcriptional start site and represses FT transcription in the morning(Song et al., 2012b). Other CDF proteins (CDF2, CDF3, and CDF5) also likely regulate FT gene expression. The repression of FT transcription by CDF1 is released by the function of FKF1-GI complex on the FT promoter in the afternoon (Song et al., 2012b), concomitantly with the removal of CDF1 repression on the CO promoter. Together with CO protein stabilization, these observations suggest that FKF1 protein controls FT induction through a multiple-feed forward motif, which allow strong activation of flowering signals in long day conditions.
In the activation of FT transcription, two classes of transcription factors play major roles. A member of B-box transcription factor family, CO, acts as a strong activator of FT expression (Putterill et al., 1995; Robson et al., 2001; Tiwari et al., 2010). CO protein contains two functional motifs; two B-box domains at the N-terminus and the CCT (CONSTANS, CONSTANS-like, and TOC1) domain at the C-terminus (Robson et al., 2001). The protein associates with the FT promoter and activates FT gene expression through two modes of action (Song et al., 2012a; Song et al., 2012b; Tiwari et al., 2010; Wenkel et al., 2006); one is that CO directly binds to the CONSTANS (CO) responsive element (CORE) via the CCT motif (Tiwari et al., 2010), and the other is that CO is recruited by the CCAAT box-binding proteins including selected subunits of Nuclear Factor-Y (NF-Y) and ASYMMETRIC LEAVES 1 (AS1) that both physically interact with CO protein (Song et al., 2012a; Wenkel et al., 2006). FT induction is largely CO-dependent; the relative abundance of FT highly accumulates when CO expression is constitutive, regardless of day length (Valverde et al., 2004). Another transcription factor family that contains basic helix-loop-helix (bHLH) domain including CRYPTOCHROME-INTERACTING BASIC HELIX–LOOP–HELIX 1 (CIB1), CIB2, CIB4, and CIB5, are involved in FT induction (Liu et al., 2008a; Liu et al., 2013c). CIB1 protein forms a complex with CRY2 protein in a blue light-dependent manner and acts as a FT activator through directly binding to the FT promoter (Liu et al., 2008a). The blue light-dependent CIB1 accumulation is positively regulated by function of ZTL and LKP2, but not by FKF1 (Liu et al., 2013a). All other CIB proteins also interact with CRY2 in vitro but only CIB2 and CIB5 form complexes with CRY2 in vivo (Liu et al., 2013c). CIB proteins redundantly regulate FT transcription. CIB1 protein forms hetero-dimer complexes with other CIBs, and the hetero-dimerization increases the DNA-binding affinity of CIB1 protein to the specific cis-element in the FT promoter (Liu et al., 2013c). As described above, blue light signaling play a pivotal role in the regulation of FT induction through degradation of FT repressors and stabilization of FT activators in Arabidopsis.
D. MOVEMENT OF FT PROTEIN
Where a florigen is synthesized differs from where it functions; therefore, understanding how the florigen moves is also of great interest in the photoperiodic flowering pathway. FT protein, once synthesized in companion cells in the leaves, is loaded into the phloem and migrates towards its eventual destination at the shoot apex. Initial debate upon the discovery of FT as a primary component of the florigen occurred over whether the mobile signal was FT mRNA or FT protein (Corbesier et al., 2007; Huang et al., 2005; Jaeger and Wigge, 2007; Yoo et al., 2013b). Multiple studies have since confirmed that the movement of FT protein explains the florigenic signal. Grafting experiments in Cucurbita moschata in particular have proved a useful system for the study of FT movment. RT-PCR and mass spectrometry analysis on phloem sap detected no FT transcript but observed FT protein (Lin et al., 2007). Cross species grafting experiments using Cucurbita moschata and Cucurbita maxima, and subsequent analysis also showed that FT peptides belonging to the induced scion were detected in the phloem sap, but not FT mRNA (Yoo et al., 2013a). Additional work in this system has given a picture in which FT movement is regulated in different ways as it moves. Mutations in FT that prevent movement into the shoot apex have been shown to have the capacity to move through the companion cell to sieve tube element barrier. This is supported by evidence that protein size affects the ability of tagged FT to enter the phloem and that specific regions of FT protein are important for movement out of the phloem and into the shoot apex (Yoo et al., 2013a). This suggests a combination of FT movement by diffusion through the companion cell and into the phloem stream and a more active transport mechanism through the plasmodesmata of cells to move FT protein into the cells of the shoot apex (Yoo et al., 2013a). Several candidate proteins for interaction or facilitated movement of FT have been identified, but their roles need to be further clarified and a more nuanced model for FT movement at each step needs to be elucidated (Liu et al., 2012; Yoo et al., 2013a).
Once FT reaches the shoot apex, a complex cascade of interactions occurs that leads to the activation of downstream developmental patterning arrays that give rise to floral meristem initiation. FT protein interacts with the bZIP transcription factor FD and 14-3-3 to activate transcription of downstream floral targets such as APETALA1 (AP1) and LEAFY (LFY) (Abe et al., 2005; Kardailsky et al., 1999; Taoka et al., 2011; Wigge, 2011). Modeling of the interactors at the shoot apex has shown that maintenance of steady state levels of FT and other interactors at the shoot apex are necessary to maintain and push the reprogramming of the vegetative meristem forward into the inflorescence meristem (Jaeger et al., 2013). This mechanism is reminiscent of classical feed-forward genetic mechanisms found in Drosphila development (Thuringer and Bienz, 1993). This suggests that threshold levels of FT movement may be critical for the reproductive transition, and it will be interesting to see experimentally the quantitative effects of FT protein on the floral transition. Classical grafting experiments have shown that cross species grafts for floral induction can induce some partners but be insufficient for others, suggesting that threshold levels of FT may be different between species (Evans, 1971). Modeling at the shoot apex of these interactions in other species may have interesting implications for the dynamics of the reproductive transition across evolutionary lines.
IV. PHOTOSYNTHATES AS A COMPONENT OF THE PHOTOPERIODIC FLOWERING STIMULUS
Photosynthesis and photosynthetic assimilates have been also recognized in historical experiments to be involved in seasonal flowering, but determining the relationship between inductive photoperiods, the florigenic signal and the photosynthetic status of the plant could not easily be disentangled in the past and is far from concrete in the present (see (Evans, 1971; Zeevaart, 1976) for review of historical work). Recent molecular genetics evidence suggests that photosynthetic components can act in leaves in a photoperiodic manner to contribute in tandem to the known photoperiodic signaling pathway. This new information sheds light on older experimental data demonstrating that photosynthetic status may alter ability to respond to optimum photoperiod in long days in Arabidopsis.
A. EARLY EVIDENCE FOR THE INVOLVEMENT OF PHOTOSYNTHESIS IN THE PHOTOPERIODIC FLOWERING RESPONSE
Many experiments have been performed historically to determine the effect of changes in photosynthetic activity on the transition from vegetative to reproductive development. Although photoperiod remains a strict determinant of flowering in many species, the capacity of a plant to respond to an introduced inductive photoperiodic signal can depend on other factors. Experiments that use the application of DCMU, an inhibitor of photosynthesis, showed that flowering could be severely delayed in Lollium temulentum, a long-day grass (Evans, 1966). However, DCMU seemed to have no effect on many short-day species, but not universally (Evans, 1971). Prolonged growth in elevated CO2 coupled with inductive day lengths has been observed to accelerate flowering in several long-day species (Reekie et al., 1994). In contrast to these results, however, experiments utilizing albino Arabidopsis mutants grown on 1% glucose could still be induced to flower, suggesting that carbon availability rather that photosynthesis influences the flowering response (Brown and Klein, 1971). Recently, DCMU treatment and removal of CO2 have also been shown to influence the period of the circadian clock under free running conditions. Thus photosynthetic output could presumably effect downstream pathways such as photoperiodic components to change gene expression, in a manner similar to the photoperiodic flowering response in several circadian clock mutant backgrounds (Haydon et al., 2013).
Although inhibition and increase in photosynthetic activity seemed to be involved in flowering induction, it was not clear where in the plant photosynthates were acting, nor was it clear whether they were acting through the same mechanism or separately from the floral stimulus. During the 1980s and 1990s when the idea of a universal transmissible signal had fallen out of favor (Zeevaart, 2006), several studies demonstrated a marked increase in sucrose or glucose at the shoot apex of both long-day and short-day species around the time of the flowering induction (Lejeune, 1993; Milyaeva, 1996; Mirolo, 1985; Perilleux, 1997). Yet, in Sinapsis alba during a single displaced short day (8 hrs of light at the end of a subjective 16-hr day) and in Lolium tementulum in a single long day, an appreciable mobilization of carbohydrates to the shoot apex did not occur until after the floral stimulus left the leaf (Bodson et al., 1977; Perilleux, 1997). This lead Bodson and colleagues to speculate whether photosynthates could lead to floral induction in the leaves rather than at the shoot apex.
Arabidopsis, like Sinapsis alba, can be induced to flower in a single long day or in a displaced short day. Mutations in PHOSPHOGLYCERATE/BIS PHOSPHO-GLYCERATE MUTASE (PGM) result in the inability to accumulate starch. pgm mutants have low rates of floral induction and no increase in sucrose exudates from the leaves in a single displaced short day treatment compared to wild type plants or pgm exposed to one long day. This flowering repression of pgm in displaced short days, however, could be partially restored by application of sucrose at their apices (Corbesier et al., 1998). Laurent Corbesier and colleagues concluded that sufficient sucrose mobilization from the leaves was needed for flowering induction, and that both a florigenic signal as well as a photosynthetic component was required for the proper photoperiodic flowering response.
B. PHOTOSYNTHATES ACT IN THE LEAVES TO PROMOTE FLOWERING
Recent work regarding trehalose-6-phosphate has provided more detailed insight into the involvement of photosynthates in the leaf. Trehalose-6-phosphate (T6P) increases parallel to sucrose in the leaves and its levels correlate to increasing starch synthesis (Ponnu et al., 2011). It has been implicated as a signal for carbohydrate status in the plant; although new research has shown substantial starch accumulation cannot be induced by T6P alone (Martins et al., 2013). In Arabidopsis, T6P increases at dusk similar to the pattern FT displays in long days (Imaizumi et al., 2003; Wahl et al., 2013). Loss of TREHELOSE-6-PHOSPHATE SYNTHASE 1 (TPS1) markedly reduces the dusk peak of FT and delays flowering in long days. Together this evidence suggests a link between photosynthetic assimilation, long day induction of FT, and flowering. Expression of CO was only minimally altered in tps1 mutants, suggesting that increase in FT transcripts is CO independent (see Figure 4) (Wahl et al., 2013).
Fig. 4.
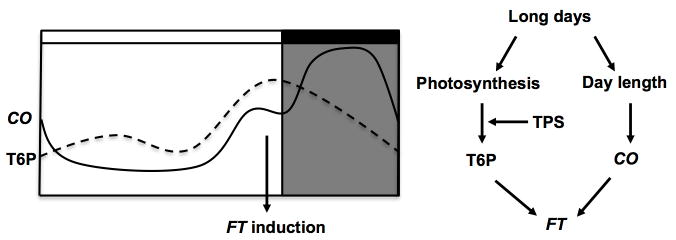
Trehalose-6-phospate regulates FT expression in long days: T6P levels peak in the afternoon of days in Arabidopsis, which coincides with the afternoon peak of FT transcription. Loss of function of tps1, the enzyme which produces T6P, results in a significant reduction in the dusk peak of FT expression. CO expression levels are unaltered by the tps1 mutation, suggesting that T6P regulation of FT in leaves occurs in a CO independent manner. Inductive long days may thus produce a photoperiodic flowering response through FT via multiple regulatory pathways.
Experimental evidence from Sinapsis alba, which is closely related to Arabidopsis, has also shown that photosynthate production during phases of the day can influence the flowering response. High light intensity provided by fluorescent lamps coupled with removal of CO2 from the air failed to promote flowering when the treatment occurred during the first eight hours of a long day cycle. However, flowering was strongly induced when the treatment occurred during the last eight hours of the daytime. To our knowledge parallel work has not been done in Arabidopsis, however, removal of CO2 throughout the entire day from Arabidopsis plants transferred from short days to long days resulted in a significant down regulation of FT transcription compared to normal CO2 controls (King et al., 2008). If T6P indeed interacts with the photoperiodic pathway to induce flowering in leaves, a time dependent sensitivity to photosynthate accumulation could explain how T6P promotes FT transcription only at dusk. It will be interesting to see if T6P levels are strongly reduced by removal of CO2 early in the day; however, this remains to be tested.
King and colleagues demonstrated that high-intensity (270 μmol m−2 s−1) fluorescent light, presumably increasing photosynthetic intake, led to relatively early flowering in short days compared to normal (100 μmol m−2 s−1) light intensity in ft mutants. This indicates that photosynthesis possibly can override the lack of FT signal under short day conditions (King et al., 2008). Clarifying a possible mechanism, Wahl and colleagues found that unlike the ft mutant, a TPS1 deficiency delayed flowering in Arabidopsis in short days as well as long days, indicating that T6P could interact with floral signals besides FT (Wahl et al., 2013). Further, loss of TPS1 resulted in reduced expression of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) SPL3, SPL4 and SPL5 at the shoot apex. The SPL protein family is the known component of the age-dependent flowering pathway in Arabidopsis. Reduced SPL expression appeared to be accomplished partially through and partially independently of miR156, which delays the vegetative-reproductive phase transition. Mature miR156 was initially higher in tps1 mutants compared to wild type, and although it declined to wild type levels over time, SPL3, 4, and 5 accumulated more slowly in tps1 mutants (Wahl et al., 2013). Finally, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and FRUITFUL (FUL) were not altered in the tps1 mutants, although they have been implicated as inducing FT downstream of the SPL proteins in the leaves (Wahl et al., 2013). It seems that T6P acts to regulate FT in the leaves mainly independently of the age-dependent pathways, while it acts to induce flowering directly at the shoot apex in response to plant age (Samach et al., 2000; Teper-Bamnolker and Samach, 2005). Because of this, T6P probably occupies a role as a stimulus of flowering in both a photoperiodic and non-photoperiodic context based on tissue specificity.
The FT protein is still the primary component of the transmissible signal in the lengthening days of spring and summer in Arabidopsis. Now, it is becoming clear that photosynthetic bi-products can lead to induction of FT at the leaf level, additively with the established photoperiod sensing mechanism through CO. This synergy appears to be long-day specific. Under short day conditions, it is common to increase the light intensity in photoperiodic studies to normalize the radiative energy received by plants when comparing plants grown in long days and short days to around 120 μmol m−2 s−1. In these conditions, FT induction does not occur. Therefore, greater accumulation of photosynthates from high-intensity light in short days seems not to over-ride the requirement of late-afternoon light for FT induction. Further, it appears that photosynthesis enhances the photoperiodic response, but cannot completely abrogate it, as FT did not decline to short-day levels when CO2 was removed from the air (King et al., 2008). By what mechanism do photosynthesis and photosynthates interact with the photoperiodic pathway to induce FT and flowering? Because FT induction through T6P is likely CO independent, a heretofore-unknown factor or pathway must be involved in FT transcriptional regulation in response to photosynthetic accumulation. Clearly, more work to determine the effects of T6P on photoperiodic pathway components is needed.
Earlier studies into the mechanisms of photoperiodic flowering and photosynthetic involvement in the flowering response highlight interactions between age and photoperiod that we do not fully understand. Many early experiments that were able to induce flowering by a single inductive long day did so by first growing their plants in short days for several weeks (Corbesier et al., 1998; Evans and Wardlaw, 1966; King et al., 2008). It appears, therefore, that age or carbohydrate status increases the amount or reduces the threshold requirement of the floral stimulus, or both. The activity of T6P at the shoot apex, proposed as a fail-safe to ensure flowering will occur even in the absence of inductive conditions (Wahl et al., 2013), suggests one mechanism. The decline of miR156 in the leaves over time resulting in up regulation of FT suggests another (Srikanth and Schmid, 2011). Although T6P seems to act independently of FT at the shoot apex, it is possible these two pathways act in parallel to modify plant response. A better understanding of how age and the carbohydrate statuses of the plants interact with photoperiodic induction either at the leaves or the shoot apex is critical to determine the threshold FT necessary to promote flowering under different timescales and spatial contexts.
V. CONCLUSIONS
While our current understanding of the underlying mechanisms that confer a photoperiodic flowering response in Arabidopsis are now better understood, the number of factors that are involved in the process makes it a very complex system of interactors. Circadian clock control of a variety of CO and FT regulators, light perception through photoreceptors, as well as photosynthetic status through T6P can affect the photoperiodic response synergistically to promote flowering in Arabidopsis as days get longer in the springtime. Brought into a larger context of all of flowering time regulation, there are a maze of players whose roles and domains may not be easily defined and whose outputs affect feedback within the system. One of the greater challenges in the future will be understanding how the plant is able to assimilate information regarding day length, light quality, temperature, precipitation, photosynthetic status, developmental age and other external and physiological characteristics and to incorporate this information in a way that is meaningful towards timing the floral transition. To this end, systems level approaches will be necessary to untangle the influence of so many factors on one output, and this will be critical if we are to understand how flowering time functions under natural conditions. At a surface level, we assume the large amount of redundancy, overlap, and crosstalk within and among flowering pathway regulators must be necessary and of selective value in coordinating the flowering response; but is this the case? As our mechanistic knowledge of flowering improves, we should continue to look out among natural populations in Arabidopsis and other species to see whether what we presume is indeed the case. Can we see that these factors affect fitness as plants expand and contract across geographic ranges, climates, and latitudes?
As detailed, work in Arabidopsis has established that the CO-FT module is critical for day length sensing, and recent developments have confirmed the highly conserved nature of this mechanism for flowering and it’s co-option for other photoperiodic outputs across the angiosperm lineage (Böhlenius et al., 2006; Kloosterman et al., 2013; Song et al., 2010). While the limited information we have on other species points to this similarity, much work is needed to better characterize mechanisms of photoperiodic sensing in other plants. With the improved genomic and functional systems at our disposal, hopefully these will shed light on the commonalities and divergence of seasonal adaptation and how plants utilize that information to survive, and hopefully how we can use that knowledge to better adapt the plants which we depend on to flourish in changing habitats.
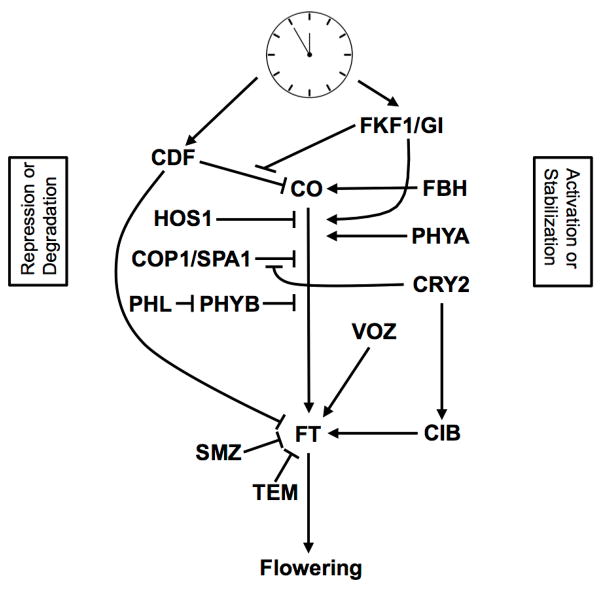
Fig. 3.
Regulators of the photoperiodic flowering pathway: Flowering under inductive long days requires a peak of FT expression in the late afternoon. CO transcription, CO protein stability, and FT transcription are critical to the photoperiodic flowering response. Blue light promotes flowering through FKF1 dependent degradation of CDFs and stabilization of CO protein, direct activation of FT through CIB transcription factors, and stabilization of the COP1/SPA1 complex by CRY2, which normally destabilizes CO protein in the dark. Red light inhibits flowering through destabilization of CO protein by PHYB. Far-red light promotes flowering through increased stability of CO protein by PHYA. Low temperature destabilizes CO protein through HOS1. The promotion or inhibition of each respective component can affect the flowering output, and thus serves to integrate multiple environmental signals such as day length, light quality, and temperature.
Acknowledgments
This work was supported by a Pre-Doctoral Developmental Biology Training Grant (5T32HD007183) from the National Institutes of Health to G.S.G., the National Science Foundation Graduate Research Fellowship Program to H.K.S., funding from the Next-Generation BioGreen 21 Program (SSAC, PJ009495) to Y.H.S., and the National Institutes of Health (GM079712) to T.I.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–6. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Baudry A, Kay S. Clock control over plant gene expression. Advances in Botanical Research. 2008;48:69–105. [Google Scholar]
- Bodson M, King RW, Evans LT, Bernier G. The role of photosynthesis in flowering of the long-day plant Sinapis alba. Funct Plant Biol. 1977;4:467–478. [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–3. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Brown JA, Klein WH. Photomorphogenesis in Arabidopsis thaliana (L.): threshold intensities of and blue-far-red synergism in floral induction. Plant Physiol. 1971;47:393–399. doi: 10.1104/pp.47.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünning E. Die endogene tagesrhythmik als grundlage der photoperiodischen reaktion. Berichte der Deutschen Botanischen Gesellschaft. 1936;54:590–607. [Google Scholar]
- Bünning E. Opening address - biological clocks. Cold Spring Harb Sym Quant Biol. 1960;25:1–9. [Google Scholar]
- Carpenter BH, Hamner KC. The effect of dual perturbations on the rhythmic flowering response of Biloxi soybean. Plant Physiol. 1964;39:884–9. doi: 10.1104/pp.39.6.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr DJ. Relationship between florigen and flower hormones. Annals of the New York Academy of Sciences. 1967;144:305. [Google Scholar]
- Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–43. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Chailakhyan MK. Concerning the hormonal nature of plant development process. CR (Dokl) Acad Sci URSS. 1937;16:227. [Google Scholar]
- Chailakhyan MK. Internal factors of plant flowering. Annu Rev Plant Physiol. 1968;19:1–36. [Google Scholar]
- Corbesier L, Lejeune P, Bernier G. The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta. 1998;206:131–137. doi: 10.1007/s004250050383. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–3. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Coulter MW, Hamner KC. Photoperiodic flowering response of Biloxi soybean in 72-hour cycles. Plant Physiol. 1964;39:848–56. doi: 10.1104/pp.39.5.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mairan JJ. Histoire de l’ Academie Royale des Sciences. 1729. Observation botanique. [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29:435–40. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- Endo M, Tanigawa Y, Murakami T, Araki T, Nagatani A. PHYTOCHROME-DEPENDENT LATE-FLOWERING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc Natl Acad Sci U S A. 2013;110:18017–22. doi: 10.1073/pnas.1310631110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LT. Abscisin II: Inhibitory Effect on Flower Induction in a Long-Day Plant. Science. 1966;151:107–8. doi: 10.1126/science.151.3706.107. [DOI] [PubMed] [Google Scholar]
- Evans LT. Flower induction and florigen concept. Ann Rev Plant Physiol. 1971;22:365. [Google Scholar]
- Evans LT, Wardlaw IF. Independent translocation of C14-labelled assimilates and of the floral stimulus in Lolium Temulentum. Planta. 1966;68:310–326. doi: 10.1007/BF00386331. [DOI] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. Journal of agricultural research. 1920;18:553–606. [Google Scholar]
- Halaban R. Circadian rhythm of leaf movement of Coleus blumei x C frederici a short day plant: effects of light and temperature signals. Plant Physiol. 1968a;43:1887. doi: 10.1104/pp.43.12.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R. Flowering response of Coleus in relation to photoperiod and circadian rhythm of leaf movement. Plant Physiol. 1968b;43:1894. doi: 10.1104/pp.43.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AA. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502:689–92. doi: 10.1038/nature12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegland SJ, Nielsen A, Lázaro A, Bjerknes AL, Totland Ø. How does climate warming affect plant-pollinator interactions? Ecol Lett. 2009;12:184–195. doi: 10.1111/j.1461-0248.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–2. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- Huang T, Böhlenius H, Eriksson S, Parcy F, Nilsson O. The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science. 2005;309:1694–6. doi: 10.1126/science.1117768. [DOI] [PubMed] [Google Scholar]
- Huang W, Perez-Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–9. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–7. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–6. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- Ito S, Niwa Y, Nakamichi N, Kawamura H, Yamashino T, Mizuno T. Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:201–13. doi: 10.1093/pcp/pcm178. [DOI] [PubMed] [Google Scholar]
- Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:3582–7. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 2013;25:820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. FT Protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007 doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. Embo J. 2008;27:1277–88. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19:2736–48. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–5. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- King RW, Hisamatsu T, Goldschmidt EE, Blundell C. The nature of floral signals in Arabidopsis. I. Photosynthesis and a far-red photoresponse independently regulate flowering by increasing expression of FLOWERING LOCUS T (FT) J Exp Bot. 2008;59:3811–3820. doi: 10.1093/jxb/ern231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman B, Abelenda JA, Gomez Mdel M, Oortwijn M, de Boer JM, Kowitwanich K, Horvath BM, van Eck HJ, Smaczniak C, Prat S, Visser RG, Bachem CW. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature. 2013;495:246–50. doi: 10.1038/nature11912. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–2. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D. Move on up, it’s time for change--mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–84. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–22. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Pineiro M, Jarillo JA. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell. 2012;24:982–99. doi: 10.1105/tpc.110.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune PB, Requier G, Kinet MJ. Sucrose increase during floral induction in the phloem sap collected at the apical part of the shoot of the long day plant Sinapis alba L. Planta. 1993;90:71–74. [Google Scholar]
- Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka K, Miura E, Xoconostle-Cazares B, Gendler K, Jorgensen RA, Phinney B, Lough TJ, Lucas WJ. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the Cucurbits. Plant Cell. 2007;19:1488–506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Q, Liu Y, Zhao X, Imaizumi T, Somers DE, Tobin EM, Lin C. Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc Natl Acad Sci U S A. 2013a;110:17582–7. doi: 10.1073/pnas.1308987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008a;322:1535–9. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H. FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 2012;10:e1001313. doi: 10.1371/journal.pbio.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008b;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. The Plant J. 2013b;76:101–114. doi: 10.1111/tpj.12276. [DOI] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013c;9:e1003861. doi: 10.1371/journal.pgen.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins MC, Hejazi M, Fettke J, Steup M, Feil R, Krause U, Arrivault S, Vosloh D, Figueroa CM, Ivakov A, Yadav UP, Piques M, Metzner D, Stitt M, Lunn JE. Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant Physiol. 2013;163:1142–63. doi: 10.1104/pp.113.226787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009:7. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milyaeva ELK, EN Changes in the sugar content in stem apices of the short-day plant Perilla nankinensis at floral transition. Russ Journal of Plant Physl. 1996;43:149–154. [Google Scholar]
- Mirolo CB, Bernier MG. Effects of flower induction on the import of C-14 assimilates into the apical bud of Xanthium. Archives internationales de physiologie et de biochemie. 1985;93:13–13. [Google Scholar]
- Mockler T, Yang H, Yu X, Parikh D, Cheng YC, Dolan S, Lin C. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci U S A. 2003;100:2140–5. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010 doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci U S A. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niimura K, Ito S, Yamashino T, Mizoguchi T, Mizuno T. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–32. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101:331–40. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell. 2000;12:885–900. doi: 10.1105/tpc.12.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–82. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- Perilleux CBG. Leaf carbohydrate status in Lolium temulentum during the induction of flowering. New Phytol. 1997;135:59–66. doi: 10.1046/j.1469-8137.1997.00629.x. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS. Circadian surfaces and the diversity of possible roles of circadian organization in photoperiodic induction. Proc Natl Acad Sci U S A. 1972:2734–2737. doi: 10.1073/pnas.69.9.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Minis DH. The entrainment of circadian oscillations by light and their role as photoperiodic clocks. American Naturalist. 1964:261–294. [Google Scholar]
- Ponnu J, Wahl V, Schmid M. Trehalose-6-Phosphate: connecting plant metabolism and development. Front Plant Sci. 2011:2. doi: 10.3389/fpls.2011.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–57. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Rédei GP. Supervital mutants of Arabidopsis. Genetics. 1962;47:443–60. doi: 10.1093/genetics/47.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reekie JYC, Hicklenton PR, Reekie EG. Effects of elevated CO2 on time of flowering in four short-day and four long-day species. Can J Bot. 1994;72:533–538. [Google Scholar]
- Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28:619–31. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–7. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–6. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Sato E, Nakamichi N, Yamashino T, Mizuno T. Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 2002;43:1374–85. doi: 10.1093/pcp/pcf166. [DOI] [PubMed] [Google Scholar]
- Sawa M, Kay SA. GIGANTEA directly activates FLOWERING LOCUS T in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2011;108:11698–703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–5. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–29. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Simon R, Igeno MI, Coupland G. Activation of floral meristem identity genes in Arabidopsis. Nature. 1996;384:59–62. doi: 10.1038/384059a0. [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–29. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T. Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol. 2010;13:594–603. doi: 10.1016/j.pbi.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18:575–83. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Lee I, Lee SY, Imaizumi T, Hong JC. CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J. 2012a;69:332–42. doi: 10.1111/j.1365-313X.2011.04793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012b;336:1045–9. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci. 2011;68:2013–37. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–6. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14–3–3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–5. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell. 2005;17:2661–75. doi: 10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in plants. Academic Press; 1996. [Google Scholar]
- Thuringer F, Bienz M. Indirect autoregulation of a homeotic Drosophila gene mediated by extracellular signaling. Proc Natl Acad Sci U S A. 1993;90:3899–903. doi: 10.1073/pnas.90.9.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, Belachew A, Repetti PP, Reuber TL, Ratcliffe OJ. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–6. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339:704–7. doi: 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–17. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Wellensi S. Relations between flower inducing factors in Silene Armeria L. Zeitschrift Fur Pflanzenphysiologie. 1967;56:33. [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–84. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge Philip A. FT, a mobile developmental signal in plants. Curr Biol. 2011;21:R374–R378. doi: 10.1016/j.cub.2011.03.038. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–9. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–12. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- Yasui Y, Mukougawa K, Uemoto M, Yokofuji A, Suzuri R, Nishitani A, Kohchi T. The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell. 2012;24:3248–63. doi: 10.1105/tpc.112.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SC, Chen C, Rojas M, Daimon Y, Ham BK, Araki T, Lucas WJ. Phloem long-distance delivery of FLOWERING LOCUS T (FT) to the apex. Plant J. 2013a;75:456–68. doi: 10.1111/tpj.12213. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Hong SM, Jung HS, Ahn JH. The cotyledons produce sufficient FT protein to induce flowering: evidence from cotyledon micrografting in Arabidopsis. Plant Cell Physiol. 2013b;54:119–28. doi: 10.1093/pcp/pcs158. [DOI] [PubMed] [Google Scholar]
- Zeevaart JA. Physiology of flower formation. Annu Rev Plant Physiol. 1976;27:321–48. [Google Scholar]
- Zeevaart JA. Florigen coming of age after 70 years. Plant Cell. 2006;18:1783–9. doi: 10.1105/tpc.106.043513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 2011;21:841–7. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]