Abstract
Purpose
Fast cine displacement encoding with stimulated echoes (DENSE) MR has higher spatial resolution and enables rapid post-processing. Thus we compared the accuracy of regional strains computation by DENSE with tagged MR in healthy and non-ischemic, non-valvular dilated cardiomyopathy (DCM) subjects.
Materials and Methods
Validation of 3D regional strains computed with DENSE was conducted in reference to standard tagged MRI (TMRI) in healthy subjects and patients with DCM. Additional repeatability studies in healthy subjects were conducted to increase confidence in DENSE. A meshfree multiquadrics radial point interpolation method (RPIM) was used for computing Lagrange strains in sixteen left ventricular segments. Bland-Altman analysis and Student's t-tests were conducted to observe similarities in regional strains between sequences and in DENSE repeatability studies.
Results
Regional circumferential strains ranged from -0.21 ± 0.07 (Lateral-Apex) to -0.11 ± 0.05 (Posterorseptal-Base) in healthy subjects and -0.15 ± 0.04 (Anterior-Apex) to -0.02 ± 0.08 (Posterorseptal-Base) in DCM patients. Computed mean differences in regional circumferential strain from the DENSE-TMRI comparison study was 0.01 ± 0.03 (95% limits of agreement) in normal subjects, -0.01 ± 0.06 in DCM patients and 0.0 ± 0.02 in repeatability studies, with similar agreements in longitudinal and radial strains.
Conclusion
We found agreement between DENSE and tagged MR in patients and volunteers in terms of evaluation of regional strains.
Keywords: displacement encoding with stimulated echoes (DENSE), tagged MRI, Lagrange strain, radial point interpolation method (RPIM), multiquadrics, Bland-Altman analysis
Introduction
MRI is considered the most viable noninvasive technology for visualizing regional myocardial function (1). MR is characterized by higher spatial and temporal resolution than echocardiography (2). The higher resolution facilitates detailed quantitative assessment of cardiac wall motion and computation of transmural strains (3). Developing imaging sequences and efficient post-processing techniques that accurately predict both homogeneous and heterogeneous functionality (and strain patterns) in the myocardium are highly important for the medical community (4,5,6,7). This study investigated the relatively new MRI technique of Displacement ENcoding with Stimulated Echoes (DENSE) that directly encodes tissue displacement in the phases of complex images and provides fast-track determination of regional myocardial function (6). Cine DENSE is characterized by high spatiotemporal resolution, compared to more conventional techniques such as tagged MRI (TMRI), rapidly providing a time series of myocardial displacements using 3D quality-guided spatiotemporal phase unwrapping (7-14).
DENSE is a relatively new imaging sequence for quantifying 3D cardiac contractile dysfunction (7). Hence, this study was conducted with the specific aims of validating 3D regional myocardial strains computed with DENSE in normal subjects and patients with DCM.
Materials and Methods
Human Subjects
Twelve normal subjects were imaged in a 1.5 T Avanto (Siemens, Erlangen, Germany) for DENSE-TMRI comparison and ten healthy subjects for DENSE repeatability studies. Six subjects were recruited for the DENSE-TMRI comparison studies in non-ischemic, non-valvular dilated cardiomyopathy (DCM). All subjects signed informed consents in accordance with the university's Institutional Review Board (IRB) guidelines. Average age of the twenty-two normal subjects was 45.9 ± 13.8 years and average mass was 73.4 ± 13.4 kg. Normal subjects were recruited as part of an ongoing effort to build a database of 3D DENSE strains in healthy subjects who have no prior history of cardiac dysfunction. All normal subjects were recruited without exclusions based on age, sex or ethnicity or racial background. Six female and six male subjects were recruited for the comparison study and six females and four males recruited for the repeatability study. Subjects for repeatability studies were removed from the scanner after the first scan and repositioned in approximately the same position during the second scan held on the same day. Average age of the six DCM patients was 60.3 ± 13.1 years and average mass was 93.4 ± 21.6 kg and the male to female recruitment ratio in DCM was 5:1. The clinical criteria for recruiting DCM patients were based on diagnosis of heart failure which included New York Heart Association (NYHA) heart failure class ranging II-III and LV ejection fraction (LVEF) less than 35%. Heart rates (HR) and blood pressures (BP) were recorded in both normal subjects, as detailed in a preceding 2D validation study (14), and in DCM patients.
Imaging Protocols
Cine DENSE data was acquired with displacement encoding applied in two orthogonal in-plane directions and one through plane direction and a four-channel body phased array radio-frequency (RF) coil was used for receiving signals. Typical imaging parameters included FOV of 359 × 269 mm, ETL of nine, TE of 8 ms, TR of 32.5 ms, matrix size of 112 × 84 pixels, 2.81 mm pixel size, 8 mm slice thickness and 12-21 cardiac phases using an echo planar imaging (EPI) cine DENSE sequence (8,9,14). TMRI images were acquired using established cine tagging protocols (4,5,15). Typical parameters included FOV of 359 × 359 mm, ETL of one, effective TE 1.5 ms, 32 ms TR, a matrix of 256 × 256 pixels, 1.37 mm squared pixel size, 8 mm slice thickness and 16-21 cardiac phases per subject, using a Spatial Modulation of Magnetization (SPAMM) TrueFISP sequence (15). More details of these protocols can be found in the preceding validation study (14). In addition to ECG-gated short-axis TMRI images acquired in multiple parallel planes between the mitral valve and the LV apex, four sets of radially oriented long-axis images at 45° of separation and intersecting at the LV centroid were also obtained from different breath-holds (4,5).
Computing Cardiac Mechanics from Cine DENSE and TMRI Images
Offline segmentation of the myocardium, phase unwrapping in DENSE, tracking tag deformations in TMRI and computation of specific mechanical parameters were accomplished with a custom C++ utility developed in this laboratory with more details outlined in a preceding publication (14). Dividing the geometry of the LV into sixteen American Heart Association (AHA) recommended segments was a part of the 3D reconstruction process and the segmental strains later used for comparison between the sequences and repeatability studies (4). Finding DENSE displacements by phase unwrapping consisted of computing and averaging phase angles of a series of Fourier transforms in an image that employed suppression of unwanted phase-wrap artifacts imposed on image resolution (8-17). Details of the quality guided spatiotemporal phase unwrapping algorithm can be found in the preceding study which was extended to 3D by phase unwrapping in the slice select (SS) or longitudinal direction (10-14). TMRI displacements were computed using a tag-finding algorithm based on pixel intensity that identified deforming dual sets of tag lines (oriented at 90° to each other) in short axis images and single set of tag lines in long axis (longitudinal) images (4,5). Figure 1a shows typical (uninterpolated) deformed tag lines in end-systole in a normal subject and Figure 1b shows the flood-filled quality image in the same subject that guide displacement finding in DENSE. Figures 1(c)-(e) show unwrapped smooth phases within the myocardium in vertical, horizontal and longitudinal orientations, respectively, in the same patient. Computation of myocardial strain was performed offline in MATLAB (Mathworks Inc., Natick, MA, United States) using a customized RPIM algorithm. The time taken for segmentation and combined displacement-strain analysis between reference and deformed timeframes was measured for both sequences. The Computation method for determining Signal to Noise Ratio (SNR) is detailed in a later section.
Figure 1.
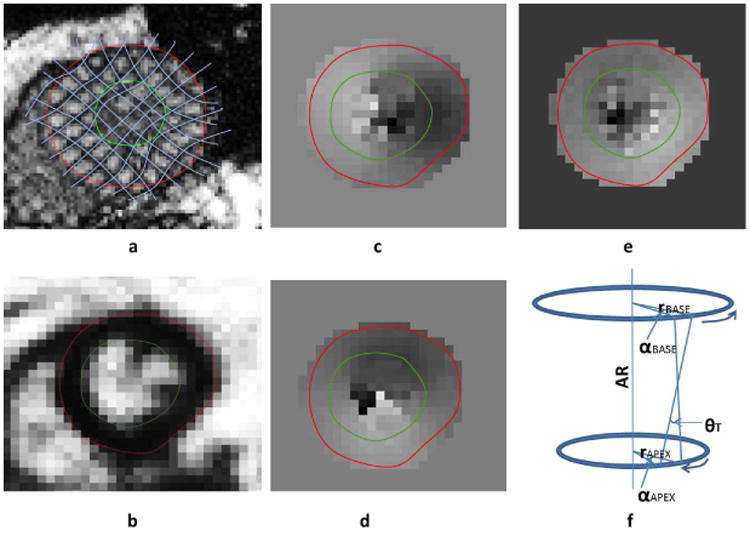
(a) Uninterpolated deformed tag lines in tagged MRI using a semiautomated tag-finding algorithm (b) Flood-filled quality image that guide displacement finding in DENSE. Unwrapped (smooth) phases within the myocardium in (c) vertical, (d) horizontal and (e) longitudinal orientations. (f) Directions of normal rotation (α) at the base and apex and torsion (θ). AR: axis of rotation, r: radius.
Meshfree 3D Lagrange Strain Analysis
A strain analysis strategy, different from conventional FEA, introduced earlier as RPIM (Galerkin weak form), was implemented for this study (18-22). RPIM is a meshfree numerical analysis method for multidimensional strain analysis within a continuum where a set of nodes neighboring a point of integration (quadrature point) are selected for shape function constructions (21). The RPIM technique was applied in this study, instead of the NNFEM technique used in the preceding 2D study as it became apparent that the radial basis functions in RPIM were a more stable approach for avoiding singularities (19-22). The RPIM method is briefly described in the following.
A continuous displacement field function, u(x), passing through a group of scattered nodes within a domain is considered, in a similar manner to the preceding study (14),
| [1] |
where P(x) is the matrix of monomial bases and b is vector of coefficients to which radial basis functions, B(x), with a as the coefficient vector, are added. The second component of Eqn. 1 passed through each scattered node point, xk, to generate a polynomial function. Mathematicians have proved the existence of B-1 for arbitrary scattered nodes and consider it a major advantage of radial basis over using only polynomial bases (21). There are a number of types of radial basis functions and the RPIM used for this study was of the multiquadrics (MQ) type (18,21), given by,
| [2] |
where ri is the distance between point xi and node x. R and q are parameter generally determined by numerical trials or values recommended in literature. With the introduction of radial basis functions additional polynomial terms can now be added without the risk of singularities (Eqn. 1). Additionally, a constraint of the form given below (21) must also be imposed,
| [3] |
where pk(x) are linear monomial terms with the limit m ≪ n. Eqn. 1 and Eqn. 3 can now be assembled into a point-wise global basis function G0,
| [4] |
with ue = [u1, u2,…,un]. The final interpolation displacement field function then becomes,
| [5] |
When complete k order polynomial terms are included in the basis, k order polynomial can be reproduced. Thus, including polynomial terms in the basis can be expected to obtain good approximation accuracy (21). The above function in Eqn. 5 can then be used to define the strain deformation tensor, F, as,
| [6] |
Lagrange strain can then be computed from F (14).RBFs have several advantages of which the first is approximation functions passing through each node point in the influence domain and secondly the ease of developing shape functions and derivatives with an arbitrary distribution of nodes (18,19,21). The continuously differentiable and integrable MQ RBFs used in this study provided true element free schemes of approximation (21) with individual patient-specific strain grid points undergoing strain computation from phase unwrapped displacements in DENSE and original and interpolated tagged data in TMRI. Furthermore RPIM also enabled computation of temporal strain trajectories for both sequences for all phases between and including end-diastole and end-systole.
The method of transformation of 2D endocardial and epicardial boundaries, displacements vectors and other landmarks into global 3D space was accomplished using a 4D Euclidean transformation given by (23),
| [7] |
where (x,y,z)3D and (x,y)2D are the spatial coordinates, nx,y,z is a normal vector to the imaging plane, cx,y,z and rx,y,z are unit column and row vectors to the imaging plane and tx,y,z is a translation vector corresponding to the origin of the imaging plane.
Computation of Twist and Torsion Angles
The twist (or rotation) angle was computed as the angle between the directions of end-diastolic radial and circumferential strains and in successive cardiac phases measured from the axis of rotation as shown in Figure 1f. A positive twist was defined as counterclockwise rotation when viewed from the LV apex. 3D torsion angles or global approximations of circumferential-longitudinal shear angles were computed from the differences in apical and basal rotation angles multiplied by the ratio of average LV radius and LV length (10). The axis of rotation for 3D torsion angles was computed from the 3D centers of each short-axis slice which approximated to a line between the most apical and basal slices. The equation for estimating torsion was given by,
| [8] |
where θA and θB are the apical and basal twist angles, RM is the average radius and L is the length of the LV.
Computation of Signal to Noise Ratio (SNR)
SNR was computed by first averaging the signal in a region of interest (ROI) in an image followed by sampling from an ROI of background air pixels (24). Care was taken to avoid placing the background region over artifacts. SNR (dimensionless) was given by,
| [9] |
where S is the mean signal in the image ROI and SDair is the standard deviation from the background air ROI. The correction factor of 0.655 is due to background noises in magnitude images that tend toward a Rayleigh distribution as SNR goes to zero (24). Three magnitude images (basal, mid-ventricular and apical sections) in three orthogonal directions (total of nine) from each subject's earliest and latest phases were used for SNR analysis.
Statistical Analysis
Bland-Altman statistical analyses were used for establishing agreement between the sequences and also to assess for repeatability (25). Bland-Altman evaluates the agreement between a new measurement technique and an existing ‘gold-standard’ one by estimating the bias which is their mean difference. The standard deviation (SD) of the difference is used to compute 95% limits of agreement (1.96SD) as an indicator of expected variations. The Bland-Altman equation for a given sample S(x,y) with two different measurements, S1 and S2, is given by,
| [10] |
where x and y are the abscissa and ordinate of the Cartesian coordinate system. Limits of agreement were found for differences between the sequences and repeatability studies. For regional Bland-Altman analysis, measurements were taken from six regions in each of the basal and mid-ventricular levels and four from the apex making it a total of sixteen regions per subject according to AHA recommended segmentation guidelines (14). Paired Student's t-tests were used to investigate the likelihood of similar mean strains between the sequences and in repeatability studies.
Results
Average HR monitored in normal subjects was 68.0 ± 8.1 bpm and average BP was 128.4 ± 18.7/79.7 ± 13.1 mmHg from comparison studies. Average HR monitored during repeatability studies was 68.7 ± 7.2 bpm and average BP was 124.3 ± 11.2/76.5 ± 7.3 mmHg. Average HR monitored in DCM patients was 75.9 ± 10.5 bpm and average BP was 117.7 ± 9.8/72.3 ± 8.8 mmHg. SNR computed from DENSE magnitude images were 35 ± 9 and 29 ± 4 at earliest and latest cardiac phases in healthy subjects and 26 ± 9 and 24 ± 5 in DCM patients. Time taken for 3D myocardial segmentation in both DENSE and TMRI was approximately 10-15 minutes per subject (or patient). Total time taken for phase unwrapping for displacements and strain analysis in DENSE was 290 ± 160 seconds per subject. In comparison, time taken for tissue tagging and strain analysis in TMRI averaged more than thirty minutes per subject. Figures 2 and 3 show Bland-Altman agreements between DENSE and TMRI studies and DENSE comparison studies, respectively, in normal subjects in regional circumferential, longitudinal and radial strains. Figure 4 shows Bland-Altman agreements between DENSE and TMRI studies in DCM patients for the above regional strains. Figure 5 shows 3D contour maps of the circumferential, longitudinal and radial strains computed with RPIM and DENSE in the myocardial walls and in individual slices between the LV apex and base in a normal subject. Figure 6 shows DENSE 3D strain contours in the myocardial walls in a DCM patient. Figure 7 shows temporal comparisons of strain between DENSE and TMRI in the sixteen AHA recommended regions in (i) normal subjects (ii) DCM patients spanning approximately two thirds of the cardiac cycle. The differences obtained between sequences in normal subjects in regional strain were 0.01 ± 0.03 in circumferential, 0.02 ± 0.07 in longitudinal and -0.01 ± 0.10 in radial strains. The differences in regional strains in the repeatability studies were 0.0 ± 0.02 in circumferential, -0.02 ± 0.04 in longitudinal and -0.02 ± 0.08 in radial strains. The differences (95% limits of agreement) in regional strains obtained between sequences in DCM patients were -0.01 ± 0.05 in circumferential, 0.02 ± 0.07 in longitudinal and -0.01 ± 0.04 in radial directions. Significant differences in means were found between DENSE and TMRI in all three circumferential longitudinal and radial strains (p < 0.01) in both normal subjects and DCM patients. DENSE repeatability studies showed significantly different means in longitudinal and radial strains (p < 0.05) but not in circumferential strain (p = 0.09). Table 1 show regional strain averages and twist and torsion angles computed with DENSE-RPIM in normal subjects and Table 2 shows those in DCM patients. Substantial reductions in all three strain components (circumferential, longitudinal and radial) can be seen in DCM patients (Table 1) in comparison to normal subjects (Table 1). Table 2 shows reduced apical and more clockwise basal direction of net rotations (twist) in DCM patients compared to normal subjects (Table 1). Similar differences in torsion can be seen between DCM patients and normal subjects.
Figure 2.
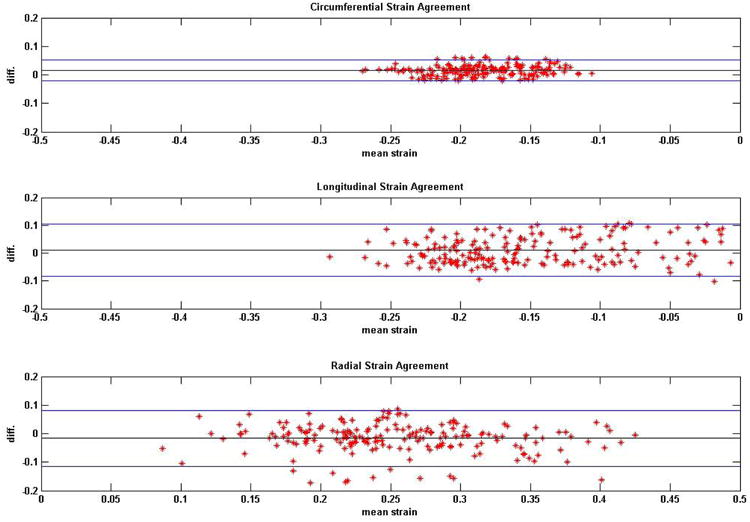
Bland-Altman plots on the agreements between DENSE and TMRI regional strains in normal (healthy) subjects. Analyses of agreements were conducted in circumferential, longitudinal and radial directions.
Figure 3.
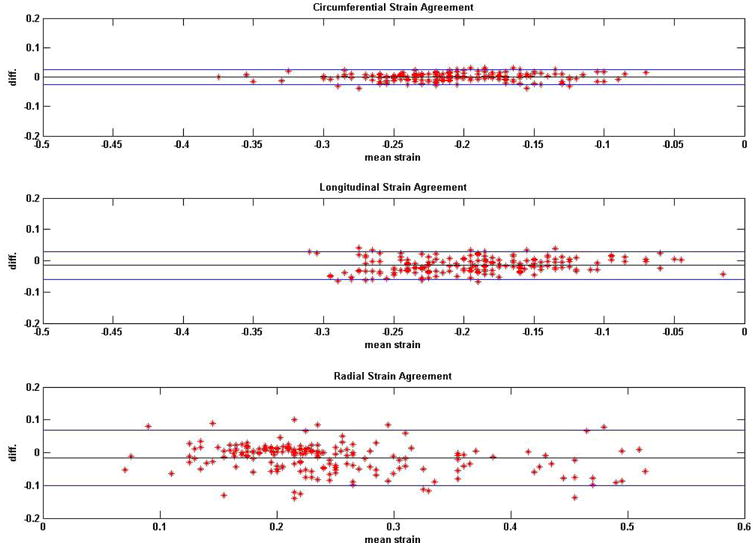
Bland-Altman plots on the agreements between DENSE repeatability studies in regional strains in normal (healthy) subjects. Analyses of agreements were conducted in circumferential, longitudinal and radial directions.
Figure 4.
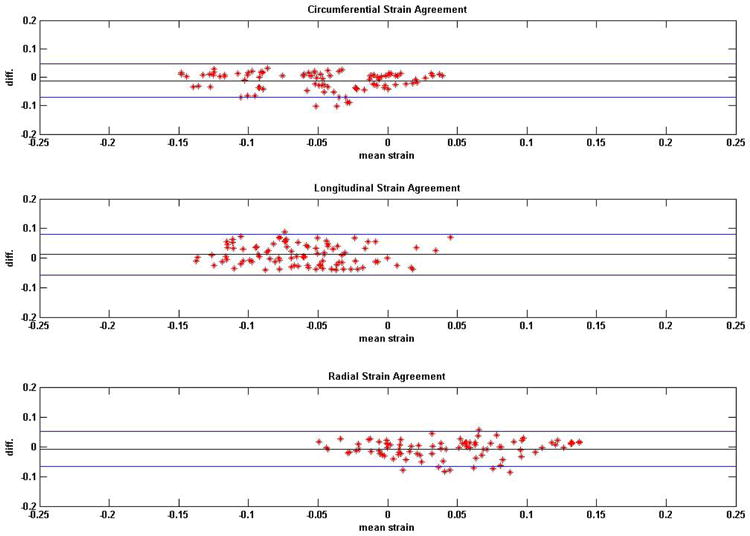
Bland-Altman plots on the agreements between DENSE and TMRI regional strains in non-ischemic, non-valvular dilated cardiomyopathy patients. Analyses of agreements were conducted in circumferential, longitudinal and radial directions.
Figure 5.
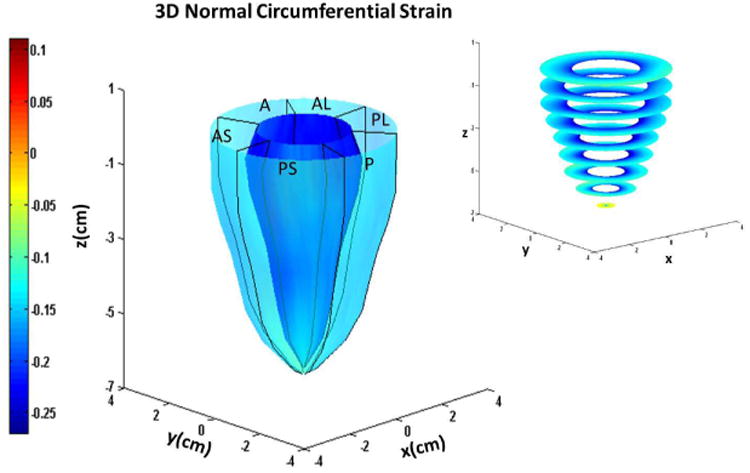
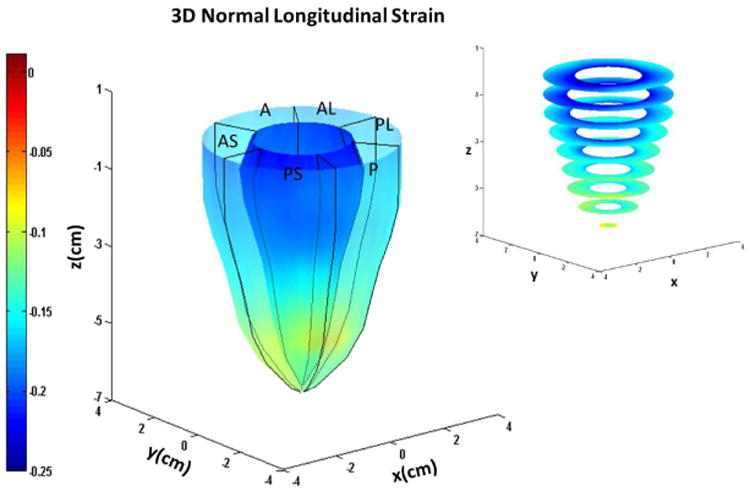
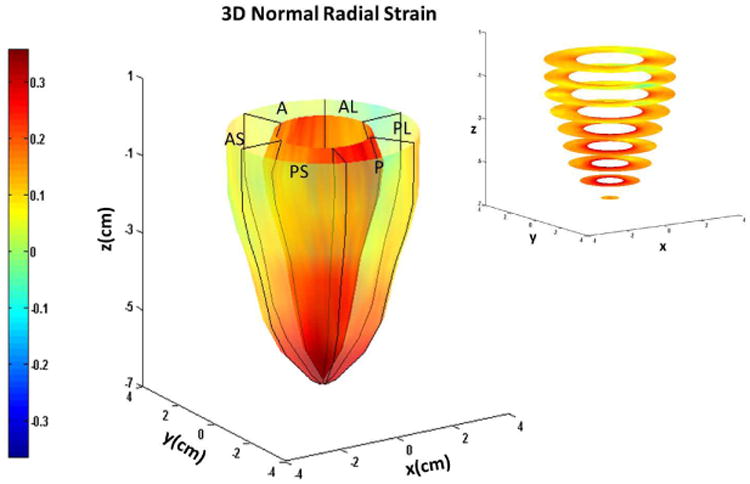
Systolic endocardial and epicardial (a) circumferential (b) longitudinal and (c) radial strains estimated with DENSE displacement fields and meshfree RPIM strain analysis methodology in normal subjects. Abbreviations: A: anterior, AL: anterolateral, AS: anteroseptal, P: posterior, PL: posterolateral, PS: posteroseptal.
Figure 6.
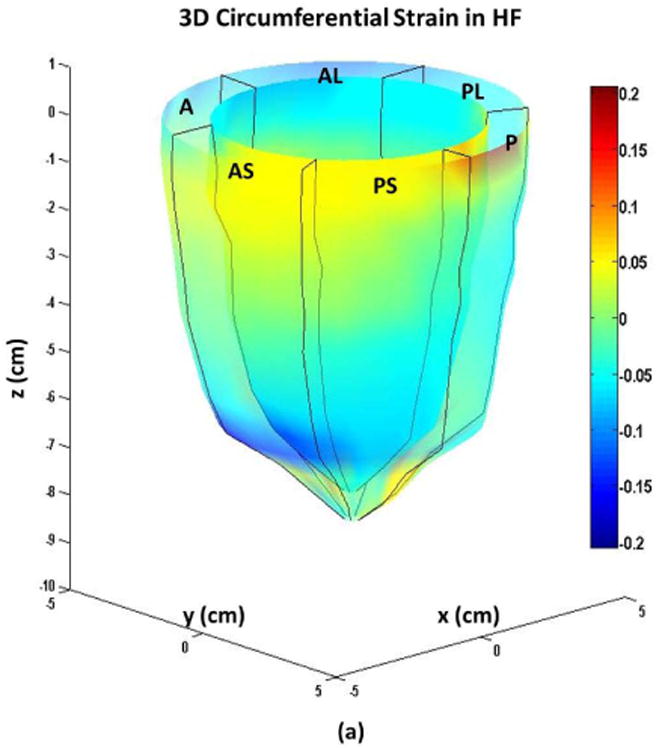
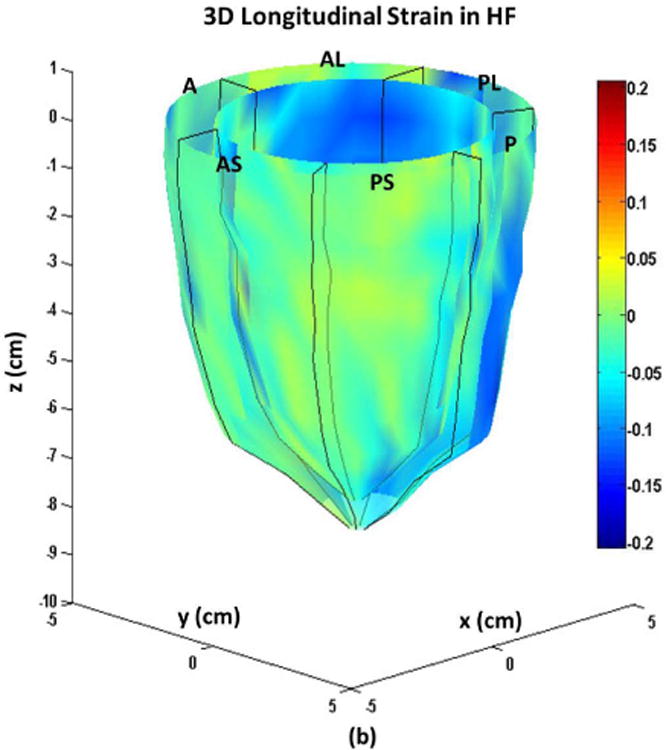
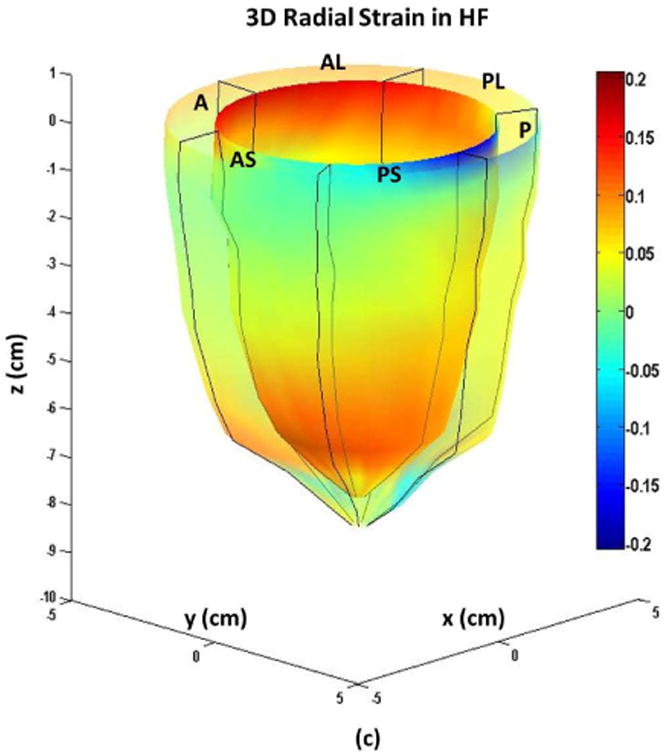
Systolic endocardial and epicardial (a) circumferential (b) longitudinal (c) and radial strains estimated from DENSE displacement fields and meshfree RPIM strain analysis methodology in non-ischemic, non-valvular dilated cardiomyopathy patients. Abbreviations: A: anterior, AL: anterolateral, AS: anteroseptal, P: posterior, PL: posterolateral, PS: posteroseptal.
Figure 7.
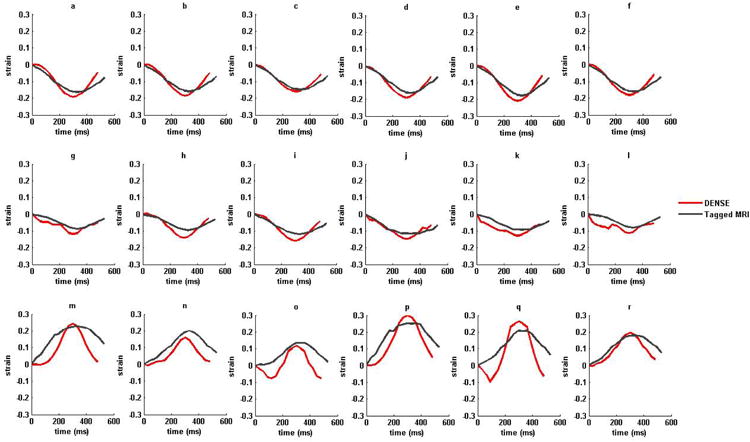
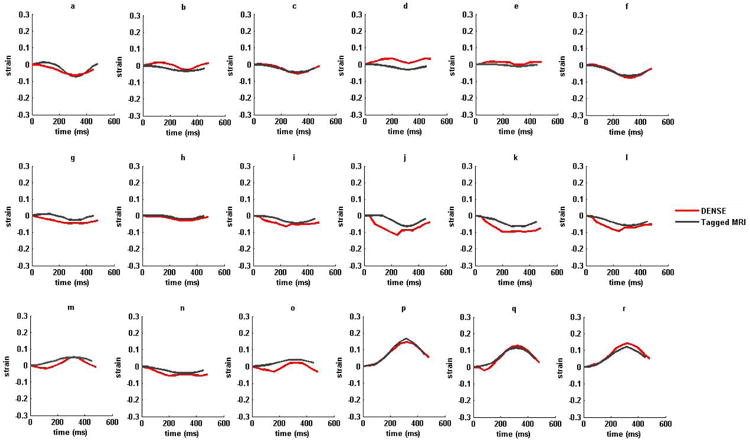
(i) Strain versus time plots for sixteen AHA recommended LV segments from a normal subject in (a)–(f) circumferential (g)-(l) longitudinal and (m)-(r) radial strains. (ii) Strain versus time plots for sixteen AHA recommended LV segments from a non-ischemic, non-valvular dilated cardiomyopathy (HF) patient in (a)–(f) circumferential (g)-(l) longitudinal and (m)-(r) radial strains. Strains estimated with DENSE are shown in red and tagged MRI in dark-gray. The six LV regions by column from left to right are anterior, anteroseptal, posteroseptal, posterior, posterolateral and anterolateral in both DENSE and TMRI.
Table 1.
Regional multi-parametric strains in the left ventricle in normal subjects, in sixteen AHA recommended segments, computed from DENSE displacements and meshfree RPIM numerical analysis technique.
| Region | Circ. | Long. | Rad. | Twist (°) | Tors. (°) |
|---|---|---|---|---|---|
| Anteroseptal-Basal | -0.14±0.05 | -0.13±0.06 | 0.19±0.10 | -13.1±8.3 | - |
| Anterior-Basal | -0.14±0.05 | -0.13±0.04 | 0.19±0.10 | -10.2±6.1 | - |
| Anterolateral-Basal | -0.15±0.03 | -0.15±0.04 | 0.19±0.13 | -10.2±7.3 | - |
| Posterolateral-Basal | -0.15±0.06 | -0.16±0.05 | 0.24±0.13 | -14.1±8.2 | - |
| Posterior-Basal | -0.12±0.05 | -0.16±0.05 | 0.23±0.12 | -11.5±7.0 | - |
| Posterorseptal-Basal | -0.11±0.05 | -0.14±0.05 | 0.18±0.12 | -12.3±10.2 | - |
| Anteroseptal-MidLV | -0.13±0.05 | -0.13±0.06 | 0.20±0.12 | 10.9±8.1 | 14.4±5.5 |
| Anterior-MidLV | -0.14±0.05 | -0.12±0.05 | 0.17±0.07 | 12.9±7.7 | 13.8±6.2 |
| Anterolateral-MidLV | -0.18±0.04 | -0.14±0.04 | 0.22±0.12 | 8.5±7.7 | 11.3±5.0 |
| Posterolateral-MidLV | -0.20±0.07 | -0.15±0.04 | 0.24±0.12 | 7.5±8.9 | 13.0±6.6 |
| Posterior-MidLV | -0.16±0.05 | -0.15±0.04 | 0.21±0.10 | 9.5±10.7 | 12.6±6.7 |
| Posterorseptal-MidLV | -0.14±0.06 | -0.13±0.05 | 0.20±0.12 | 16.5±9.7 | 15.2±7.6 |
| Septal-Apex | -0.15±0.07 | -0.12±0.06 | 0.17±0.09 | 13.7±6.1 | 7.9±3.8 |
| Anterior-Apex | -0.16±0.07 | -0.10±0.06 | 0.20±0.09 | 15.2±4.7 | 7.6±2.6 |
| Lateral-Apex | -0.21±0.07 | -0.12±0.04 | 0.22±0.12 | 8.1±7.2 | 6.1±3.6 |
| Posterior-Apex | -0.17±0.04 | -0.13±0.04 | 0.17±0.11 | 17.3±12.9 | 8.6±3.9 |
Table 2.
Regional multi-parametric strains in the left ventricle in dilated cardiomyopathic HF patients in sixteen AHA recommended segments, computed from DENSE displacements and meshfree RPIM numerical analysis technique.
| Region | Circ. | Long. | Rad. | Twist (°) | Tors. (°) |
|---|---|---|---|---|---|
| Anteroseptal-Basal | -0.04±0.06 | 0.03±0.04 | 0.01±0.05 | 0.4±1.7 | - |
| Anterior-Basal | -0.14±0.04 | -0.01±0.05 | 0.05±0.04 | 2.1±0.7 | - |
| Anterolateral-Basal | -0.10±0.03 | -0.07±0.03 | 0.10±0.05 | 3.7±1.1 | - |
| Posterolateral-Basal | -0.09±0.01 | -0.02±0.03 | 0.07±0.03 | 2.2±1.3 | - |
| Posterior-Basal | -0.02±0.05 | 0.03±0.04 | 0.05±0.04 | 0.4±0.9 | - |
| Posterorseptal-Basal | -0.02±0.08 | 0.03±0.06 | -0.01±0.03 | 1.1±1.7 | - |
| Anteroseptal-MidLV | -0.09±0.07 | -0.01±0.01 | 0.01±0.04 | -0.4±0.8 | -0.3±0.6 |
| Anterior-MidLV | -0.03±0.04 | -0.05±0.03 | 0.05±0.04 | 1.7±0.5 | -0.2±0.1 |
| Anterolateral-MidLV | -0.10±0.02 | -0.06±0.02 | 0.09±0.04 | 2.2±0.7 | -0.6±0.2 |
| Posterolateral-MidLV | -0.09±0.03 | -0.03±0.01 | 0.06±0.03 | 1.4±1.2 | -0.4±0.1 |
| Posterior-MidLV | -0.02±0.06 | 0.00±0.01 | 0.04±0.05 | 0.8±0.7 | 0.2±0.1 |
| Posterorseptal-MidLV | -0.02±0.08 | 0.01±0.03 | -0.02±0.03 | 0.3±0.8 | -0.3±0.6 |
| Septal-Apex | -0.09±0.08 | -0.02±0.02 | 0.01±0.03 | 0.1±0.4 | -0.2±0.6 |
| Anterior-Apex | -0.15±0.04 | -0.07±0.03 | 0.04±0.02 | 1.6±0.3 | -0.2±0.2 |
| Lateral-Apex | -0.10±0.03 | -0.07±0.02 | 0.06±0.03 | 0.1±0.9 | -0.9±0.1 |
| Posterior-Apex | -0.02±0.06 | -0.03±0.03 | 0.05±0.05 | 1.0±0.6 | 0.2±0.1 |
Discussion
Peak end-systolic strains were measured in sixteen regions within reconstructed 3D LV geometries in human subject experiments and Bland-Altman analysis conducted to show there is agreement between the sequences and in repeatability studies. It was shown that real time computation of 3D regional strain is possible using rapid unwrapping of phases and the RPIM framework. In comparison, this rapid post-processing technique is twice more fast than TMRI-HARP post-processing that use phase unwrapping like the Strain from Unwrapped Phases (SUP) technique (26). The regional circumferential, longitudinal and radial strains measured in normal subjects were found similar to previous literature in DENSE (10-12,14,27), TMRI (28-30) and HARP (31). Additionally, the regional distribution of circumferential strains computed with DENSE with highest values in the lateral region, followed by posterior, anterior, and septal regions is a distinguishable pattern shown in past studies with normal subjects (23,28-30). The plots of circumferential strain distribution in a healthy subject show prominent endocardial to epicardial strain gradients that were observed in previous studies (29). The mean difference in circumferential strain between DENSE and TMRI reciprocate a range found in similar 2D comparison studies (14,8,27). More precise limits of agreement were found for radial strain agreements than the preceding 2D study despite historically observed patterns of higher variations in radial strains (3,10,14,27,29). Radial strain variations between the sequences can be propagated by naturally varying transmural kinematics of the myocardium as explained in the preceding study (14) or technical differences like the real time demodulation of RF signals in DENSE versus TMRI (or HARP) post-processing techniques (17). However, variations in strain computations are also highly influenced by the technique used for numerical analysis. Better agreement between radial strains in this study is due to the RBFs added to the RPIM technique which introduced the major advantage of avoiding singularities associated with only polynomial bases (18,21).
This study also confirms the results of past studies regarding contractile function in patients with DCM that show abnormal values of both circumferential and longitudinal shortening in the septal walls when compared with relatively normal values in the lateral walls (5,32,33). It can also be seen that while symmetry was not compromised in remodeling the 3D LV geometries with DCM, regional and transmural myocardial contractile function occur heterogeneously in DCM patients. Similar strain-time plots of DENSE and TMRI clearly demonstrate that DENSE, which encodes displacement of discrete points as phase shifts, can be reliably used for tracking the progression of regional myocardial strains for two-thirds of the cardiac cycle in both healthy subjects and DCM patients. Reduced radial strains corresponding to reduced torsions in DCM were found in this study which was also coincidental with better agreement between DENSE and TMRI radial strains in DCM. Additionally, low magnitude (compared to normal) and almost identical radial strain trajectories with DENSE and TMRI was shown in a DCM patient. Given the above trends, it is postulated that impaired transmural mechanics (evidenced by reduced twist and torsion) in DCM reduced radial strain magnitudes and apparently contributed to better agreement in radial strains between sequences. Past studies have also investigated similar relationships between radial strains (displacements) and torsions (twists) with one study showing a linear correlation between torsion and radial displacement in both healthy subjects and elite athletes and another study in ionotropic stimulants showing increases in radial strains with augmented torsion (34,35).
Most studies define the peak end-systole direction of rotation (twist) as counterclockwise at the LV apex and clockwise at the base when viewed from the LV apex (36,37). This difference in base negative to apical positive peak rotation was observed in normal subjects during this study and our results are similar to regional averages from other studies in DENSE, HARP and TMRI (10,36,29,37). The reduced torsions in DCM patients is due to reductions in twists since computation of torsion is directly proportional to the difference between a twist angle in a particular segment and its corresponding one at the base. These reductions in twist and torsion angles in have also been shown in previous DCM studies (37-39).
There were several limitations to this study of which the first was not conducting rigorous interobserver variability tests on twist and torsion angles computed with DENSE. Computation of twist and torsion angles can be highly sensitive to factors such as the accurate location of the axis of rotation which depend on the delineation of myocardial walls by the observer (36). Hence, interobserver variability can contribute significantly toward establishing accurate ranges for twist and torsion angles. In future, more detailed investigations are required to establish the accuracy of predicting both normal and abnormal twists and torsions with DENSE. A second limitation pertained to not using a more state-of-the-art DENSE sequence such as a free-breathing navigator-gated method where matched respiratory phases in patients are used to synchronize image data acquisition and reduces respiration induced artifacts (40). A previous navigator-gated study combined with a spiral k-space trajectory to minimize echo time reported SNR values of ∼ 81 - 34 (earliest to latest cardiac phases) which was far superior to those achieved during this study (40). Nonetheless, a non-free-breathing DENSE sequence did not cause breath-hold difficulties or other respiratory issues in DCM patients. Some gating difficulties related to poor R-wave propagation were encountered with DCM patients but in all cases image acquisition was resumed after temporary cessation of breath-holds. Significant limitations in relation to the meshfree RPIM method should be mentioned of which the first is RPIM shape functions generally requiring more support nodes to achieve higher accuracy (18,21). This is not desirable, as it produces a non-sparse stiffness matrix which can directly affect computational efficiency. A second limitation of RPIM was using constant shape parameters (q and R) for strain computation where an iterative optimization scheme for estimating these parameters could have been used. The influential effect of shape parameters on radial basis functions have been minimally investigated and previous literature show that q < 1 and R ≈ 1.0 - 2.0 produce well-conditioned results (18,21). These shape parameters were duly followed during this study and no extra effort made to optimize them. The use of constant shape parameters remain a highly debated and under-investigated issue in computational science and its optimization for the purposes of computing 3D myocardial strains with RPIM should be addressed in future studies (18,19,21). A final limitation of this study was not depicting septal dominance in abnormal wall strains in DCM patients with a standardized (statistical) scoring system. A more elegant method of demonstrating dysfunction in DCM would be the use of composite multiparametric Z-score where point-by-point standard deviations from averages of expected norms can be used to color contour regions of dysfunction in a dense grid of patient specific LV geometry (4).
In conclusion this study shows that DENSE is a higher resolution technique that produces reliable estimates of regional strains (circumferential, longitudinal and radial) and twist and torsion angles that are important to the understanding of normal cardiac biomechanics and the etiology of cardiac disease. Cine DENSE is a promising non-invasive and high resolution diagnostic tool for quantifying regional myocardial biomechanics which can be a feasible tool for early detection and management of a wide range of myocardial diseases.
Acknowledgments
We thank Ms. Beckah Brady and Ms. Lina Reynolds for their role in recruiting healthy subjects and patients. This study was partly funded by the National Institute of Health (NIH) grant R01 HL112804. This study was also funded in part by the BJH Foundation at Washington University School of Medicine, St Louis, MO, USA.
References
- 1.Axel L, Dougherty L. MR imaging of motion with spatial modulation of magnetization. Radiology. 1989;171:841–845. doi: 10.1148/radiology.171.3.2717762. [DOI] [PubMed] [Google Scholar]
- 2.Sheheta ML, Cheng S, Osman NF, Bluemke DA, Lima JAC. Myocardial Tissue Tagging with Cardiovascular Magnetic Resonance. J Cardiovasc Magn R. 2009;11:55. doi: 10.1186/1532-429X-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal HK, Abd-Elmoniem KZ, Prince JL. Truharp: Single Breath-Hold Mri For High Resolution Cardiac Motion And Strain Quantification. Proceedings of IEEE Int Symp Biomed Imaging. 2009;2009:458–461. doi: 10.1109/ISBI.2009.5193083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cupps BP, Taggar AK, Reynolds LM, Lawton JS, Pasque MK. Regional Myocardial Contractile Function: Multiparametric Strain Mapping. Interact Cardiovasc Thorac Surg. 2010;10:953–957. doi: 10.1510/icvts.2009.220384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joseph S, Moazami N, Cupps BP, et al. Magnetic Resonance Imaging– based Multiparametric Systolic Strain Analysis and Regional Contractile Heterogeneity in Patients With Dilated Cardiomyopathy. The Journal of Heart and Lung Transplantation. 2009;28:388–394. doi: 10.1016/j.healun.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spottiswoode BS, Zhong X, Lorenz CH, Mayosi BM, Meintjes EM, Epstein FH. 3D Myocardial Tissue Tracking With Slice Followed Cine DENSE MRI. J Magn Reson Imag. 2008;27:1019–1027. doi: 10.1002/jmri.21317. [DOI] [PubMed] [Google Scholar]
- 7.Aletras AH, Ding S, Balaban RS, Wen H. DENSE: displacement encoding with stimulated echoes in cardiac functional MRI. J Magn Reson. 1999;137:247–252. doi: 10.1006/jmre.1998.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D, Gilson WD, Kramer CM, Epstein FH. Myocardial tissue tracking with 2D cine displacement-encoded MRI – development and initial evaluation. Radiology. 2004;230:862–871. doi: 10.1148/radiol.2303021213. [DOI] [PubMed] [Google Scholar]
- 9.Spottiswoode BS, Zhong X, Hess AT, et al. Tracking myocardial motion from cine DENSE images using spatiotemporal phase unwrapping and temporal fitting. IEEE Trans Med Imaging. 2007;26:15–30. doi: 10.1109/TMI.2006.884215. [DOI] [PubMed] [Google Scholar]
- 10.Zhong X, Spottiswoode BS, Meyer CH, Kramer MK, Epstein FH. Imaging three-dimensional myocardial mechanics using navigator-gated volumetric spiral cine DENSE MRI. Magn Reson Med. 2010;64:1089–1097. doi: 10.1002/mrm.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Wen H, Gorman RC, et al. Reconstruction of myocardial tissue motion and strain fields from displacement-encoded MR imaging. Am J Physiol Heart Circ Physiol. 2009;297:H1151–H1162. doi: 10.1152/ajpheart.00074.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong X, Gibberman LB, Spottiswoode BS, et al. Comprehensive Cardiovascular magnetic resonance of myocardial mechanics in mice using three-dimensional cine DENSE. J Cardiovasc Magn R. 2011;13:83–92. doi: 10.1186/1532-429X-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilliam AD, Zhong X, Epstein FH. Automated cardiac motion estimation from 3D Cine DENSE MRI. J Cardiovasc Magn R. 2012;14:W16. [Google Scholar]
- 14.Kar J, Knutsen AK, Cupps BP, Pasque MK. A Validation of Two-Dimensional In Vivo Regional Strain Computed from Displacement Encoding with Stimulated Echoes (DENSE), in Reference to Tagged Magnetic Resonance Imaging and Studies in Repeatability. doi: 10.1007/s10439-013-0931-2. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zwanenburg JJ, Kuijer JP, Marcus JT, Heethaar RM. Steady-state free precession with myocardial tagging: CSPAMM in a single breathhold. Magn Reson Med. 2003;49:722–30. doi: 10.1002/mrm.10422. [DOI] [PubMed] [Google Scholar]
- 16.Stoeck CT, Kozerke S, Maredia N, Crean A, Greenwood JP, Sven P. In vivo comparison of DENSE and CSPAMM for cardiac motion analysis. Proceedings of 12th Annual SCMR Scientific Sessions. 2009:142–143. [Google Scholar]
- 17.Kuijer JP, Hofman MBM, Zwanenburg JJ, Marcus JT, van Rossum AC, Heethaar RM. DENSE and HARP: Two Views on the Same Technique of Phase-Based Strain Imaging. J Magn Reson Imaging. 2006;24:1432–1438. doi: 10.1002/jmri.20749. [DOI] [PubMed] [Google Scholar]
- 18.Wang JG, Liu GR. A point interpolation meshless method based on radial basis functions. Int J Numer Meth Eng. 2002;54:1623–1648. [Google Scholar]
- 19.Wang JG, Liu GR. On the optimal shape parameters of radial basis functions used for 2-D meshlesss methods. Comput Methods Appl M. 2002;191:2611–2630. [Google Scholar]
- 20.Liu GR, Gu YT. A point interpolation method for two-dimensional solid. Int J Numer Meth Eng. 2001;50:937–951. [Google Scholar]
- 21.Liu GR. Meshfree Methods: Moving Beyond the Finite Element Method. 2nd. Boca Raton: CRC Press; 2009. p. 792. [Google Scholar]
- 22.Wendland H. Meshless Galerkin method using radial basis functions. Mathematics of Computation. 1999;68:1521–1531. [Google Scholar]
- 23.Moulton MJ, Creswell LL, Downing SW, et al. Spline surface interpolation for calculating 3-D ventricular strains from MRI tissue tagging. Am J Physiol. 1996;270:H281–97. doi: 10.1152/ajpheart.1996.270.1.H281. [DOI] [PubMed] [Google Scholar]
- 24.Firbank MJ, Coulthard A, Harrison RM, Williams ED. A comparison of two methods for measuring the signal to noise ratio on MR images. Phys Med Biol. 1999;44:N261–N264. doi: 10.1088/0031-9155/44/12/403. [DOI] [PubMed] [Google Scholar]
- 25.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;8476:307–310. [PubMed] [Google Scholar]
- 26.Venkateesh BA, Gupta H, Lloyd SG, Italia LD, Denny TS., Jr 3D Left Ventricular Strain from Unwrapped Harmonic Phase Measurements. J Magn Reson Imag. 2010;31:854–862. doi: 10.1002/jmri.22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young AA, Li B, Kirton RS, Cowan BR. Generalized Spatiotemporal Myocardial Strain Analysis for DENSE and SPAMM Imaging. Magn Reson Med. 2012;67:1590–1599. doi: 10.1002/mrm.23142. [DOI] [PubMed] [Google Scholar]
- 28.Hillenbrand HB, Lima JAC, Bluemke DA, Beache GM, McVeigh ER. Assessment of Myocardial Systolic Function by Tagged Magnetic Resonance Imaging. J Cardiovasc Magn R. 2000;2:57–66. doi: 10.3109/10976640009148674. [DOI] [PubMed] [Google Scholar]
- 29.Moore CC, Olivieri CH, McVeigh ER, Zerhouni EA. Three Dimensional Systolic Strain Patterns in the Normal Human Left Ventricle: Characterizing with Tagged MR Imaging. Radiology. 2000;214:453–466. doi: 10.1148/radiology.214.2.r00fe17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young AA, Kramer CM, Ferrari VA, Axel L, Reichek N. Three-dimensional left ventricular deformation in hypertrophic cardiomyopathy. Circulation. 1994;90:854–67. doi: 10.1161/01.cir.90.2.854. [DOI] [PubMed] [Google Scholar]
- 31.Osman NF, Kerwin WS, McVeigh ER, Prince JL. Cardiac motion tracking using CINE harmonic phase (HARP) magnetic resonance imaging. Magn Reson Med. 1999;42:1048–1060. doi: 10.1002/(sici)1522-2594(199912)42:6<1048::aid-mrm9>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potter DD, Araoz PA, Ng LL, et al. Cardiotropin-1 and myocardial strain change heterogeneously in cardiomyopathy. J Surg Res. 2007;141:277–83. doi: 10.1016/j.jss.2006.12.539. [DOI] [PubMed] [Google Scholar]
- 33.Young AA, Dokos S, Powell KA, et al. Regional heterogeneity of function in nonischemic dilated cardiomyopathy. Cardiovasc Res. 2001;49:308–18. doi: 10.1016/s0008-6363(00)00248-0. [DOI] [PubMed] [Google Scholar]
- 34.Nottin S, Doucende G, Schuster-Beck I, Dauzat M, Obert P. Alteration in left ventricular normal and shear strains evaluated by 2D-strain echocardiography in the athlete's heart. J Physiol. 2008;586:4721–4733. doi: 10.1113/jphysiol.2008.156323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akagawa E, Murata K, Nobuaki Tanaka N, et al. Augmentation of Left Ventricular Apical Endocardial Rotation With Inotropic Stimulation Contributes to Increased Left Ventricular Torsion and Radial Strain in Normal Subjects: Quantitative Assessment Utilizing a Novel Automated Tissue Tracking Technique. Circ J. 2007;71:661–668. doi: 10.1253/circj.71.661. [DOI] [PubMed] [Google Scholar]
- 36.Russel IK, Gotte MJW, Kuijer JP, Marcus JT. Regional assessment of left ventricular torsion by CMR tagging. J Cardiovasc Magn R. 2008;10:26–37. doi: 10.1186/1532-429X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russel IK, Gotte MJW, Bronwaer JG, et al. Left Ventricular Torsion: An Expanding Role in the Analysis of Myocardial Dysfunction. JACC: Cardiovascular Imaging. 2009;2:648–655. doi: 10.1016/j.jcmg.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 38.MacGowan GA, Shapiro EP, Azhari H, et al. Noninvasive measurement of shortening in the fiber and cross-fiber directions in the normal human left ventricle and in idiopathic dilated cardiomyopathy. Circulation. 1997;96:535–41. doi: 10.1161/01.cir.96.2.535. [DOI] [PubMed] [Google Scholar]
- 39.Sengupta PP, Tajik AJ, Chandrasekaran K, Khandheria BK. Twist Mechanics of the Left Ventricle Principles and Application. JACC: Cardiovascular Imaging. 2008;1:366–376. doi: 10.1016/j.jcmg.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Zhong X, Spottiswoode BS, Meyer CH, Kramer M, Epstein FH. Imaging Three-Dimensional Myocardial Mechanics Using Navigator-Gated Volumetric Spiral Cine DENSE MRI. Magn Res Med. 2010;64:1089–1097. doi: 10.1002/mrm.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]


