Abstract
The geographic origins of populations can be identified by their maternally inherited mitochondrial DNA (mtDNA) haplogroups. This study compared human cybrids (cytoplasmic hybrids), which are cell lines with identical nuclei but mitochondria from different individuals with mtDNA from either the H haplogroup or L haplogroup backgrounds. The most common European haplogroup is H while individuals of maternal African origin are of the L haplogroup. Despite lower mtDNA copy numbers, L cybrids had higher expression levels for nine mtDNA-encoded respiratory complex genes, decreased ATP turnover rates and lower levels of ROS production, parameters which are consistent with more efficient oxidative phosphorylation. Surprisingly, GeneChip arrays showed that the L and H cybrids had major differences in expression of genes of the canonical complement system (5 genes), dermatan/chondroitin sulfate biosynthesis (5 genes) and CCR3 signaling (9 genes). Quantitative nuclear gene expression studies confirmed that L cybrids had (a) lower expression levels of complement pathway and innate immunity genes and (b) increased levels of inflammation-related signaling genes, which are critical in human diseases. Our data support the hypothesis that mtDNA haplogroups representing populations from different geographic origins may play a role in differential susceptibilities to diseases.
Keywords: Mitochondrial haplogroups, transmitochondrial cybrids, inflammation, complement, mitochondria, complement activation, innate immunity, haplogroups, cybrids, retina
1. Introduction
Mitochondria (mt) have their own unique, circular DNA that can be categorized into haplogroups defined by single nucleotide polymorphism (SNP) variants, and these haplogroups represent populations of different ancestral origin. Recent studies have shown that mtDNA haplogroups can be associated with human aging and diseases [1]. The coding region of mtDNA encodes for 37 genes including 13 protein subunits that are essential for oxidative phosphorylation (OXPHOS), 2 ribosomal RNAs and 22 transfer RNAs [2–4]. The non-coding mtDNA D-loop (also called the Control Region) contains 1121 nucleotides and is important for replication and transcription. It is known that mtDNA is critical for OXPHOS but recent studies have provided evidence that mtDNA haplogroups can also influence expression of genes related to oxidative stress [5, 6] and the clinical severity of diseases [7, 8]. Most importantly mtDNA haplogroups have been associated with various diseases, including Alzheimer’s disease (AD) [9], Parkinson’s disease [10–12], osteoarthritis [13], type 2 diabetes (T2D) [14] and various cancers [15].
In medicine, it has long been recognized that certain diseases are more prevalent in specific racial/ethnic populations [16–21]. For example, there are differences in the prevalence of Alzheimer’s disease (AD) depending upon the ethnic/racial groups. Proportionately to the size of their population, older African-Americans are ~2-times more likely and Hispanics are approximately 1.5-times more likely to have AD or dementia compared to older non-Hispanic whites [22–24]. Another example of racial/ethnic differences can be found in the incidence of diabetes. Compared to non-Hispanic white adults, Asian-Americans have an 18% rate, Hispanic/Latinos have 66%, and non-Hispanic blacks a 77% higher risk of diabetes (http://www.diabetes.niddk.nih.gov/dm/pubs/statistics). With systemic lupus erythematosus (SLE), there is higher incidence within the African-American community compared to white subjects [20, 21]. While some have suggested that epigenetic changes may contribute to these ethnic/racial differences [25], we suspect that non-synonymous mtDNA variants (causing amino acid changes)associated with the different haplogroups may contribute to altered functions and disease susceptibilities. In addition, the SNP variants within the mt-D-loop region can cause changes in replication and transcription rates, leading to lower levels of mtDNA and mtRNA [26].
In the past it has been difficult to evaluate the contribution that mtDNA variants might have to molecular processes or cellular behavior. However, with the development and use of the novel cybrid (cytoplasmic hybrid) model, many questions related to the functional importance of the mtDNA haplogroup variants and mitochondrial-nuclear interactions can be addressed. These cybrid cell lines are created by fusing mitochondrial-free (Rho0) cells with mitochondria-rich platelets from different individuals so the resultant cells have identical nuclei but vary in their mtDNA haplogroups.
Our initial interests have been related to age-related macular degeneration and other retinal diseases, so we elected to use a well characterized retinal pigment epithelial cell line, ARPE-19, for our cybrids. Our findings with this novel cybrid model show that surprisingly the L haplogroup mtDNA variants (African origins) can differentially mediate the expression of nuclear genes involved in the complement, inflammatory and innate immunity pathways, which are critical in human diseases. These data also support the hypothesis that the differential susceptibilities to diseases found in ethnic/racial populations may be related in part to their mtDNA haplogroup backgrounds, which can influence nuclear gene expression, cellular functions and induce variable phenotypic severity of diseases.
2. Material and methods
2.1. Transmitochondrial cybrids and culture conditions
Institutional review board approval was obtained from the University of California, Irvine (#2003–3131). For DNA analyses, 10mls of peripheral blood were collected via venipuncture in tubes containing 10mM EDTA from normal volunteers (H haplogroup, n=3, average age 35.3±7.3 years; L haplogroup, n=3, average age 44.6±4.8 years, p=0.5). DNA was isolated with a DNA extraction kit (PUREGENE, Qiagen, Valencia, CA). Platelets were isolated by a series of centrifugation steps and final pellets were suspended in Tris-buffered saline. The ARPE-19 cells deficient in mtDNA (Rho0) were created by serial passage in low dose ethidium bromide [27]. Cybrids were produced by polyethylene glycol fusion of platelets with Rho0 ARPE-19 cells according to modified procedures of Chomyn [28]. Verification of transfer of the mitochondria into the Rho0 ARPE-19 cells was accomplished by polymerase chain reaction (PCR), restriction enzyme digestion, and sequencing of the mtDNA to identify the mitochondrial haplogroup of each cybrid [29].
2.2. Identification of cybrid haplogroups
Cybrid DNA was extracted from cell pellets using a spin column kit (DNeasy Blood and Tissue Kit, Qiagen) and quantified using the Nanodrop 1000 (Thermo Scientific, Wilmington, DE). PCR and restriction enzyme digests [29] allelic discrimination and sequencing of the MT-Dloop were performed to determine mitochondrial haplogroups. The H defining SNPs were T7028C, G73A, G2706A, A11719G and T14766C. The samples were further sequenced and identified to be H, H and H5a for the three cybrids (Figure 1a). The L cybrids were further sequenced using primers to L9611-H12111 and to the MT-Dloop. The L samples were identified to be L0a1′4, L1b, and L2bas defined by the SNP variants (Figure 1b).
Figure 1.
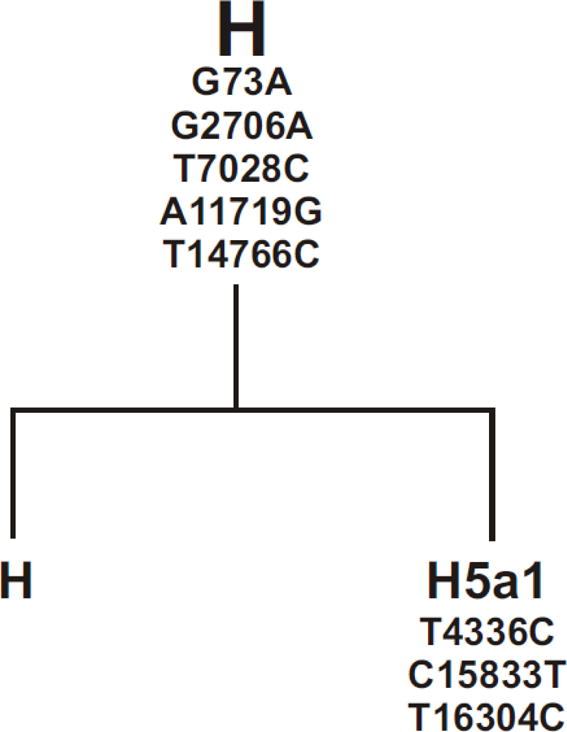
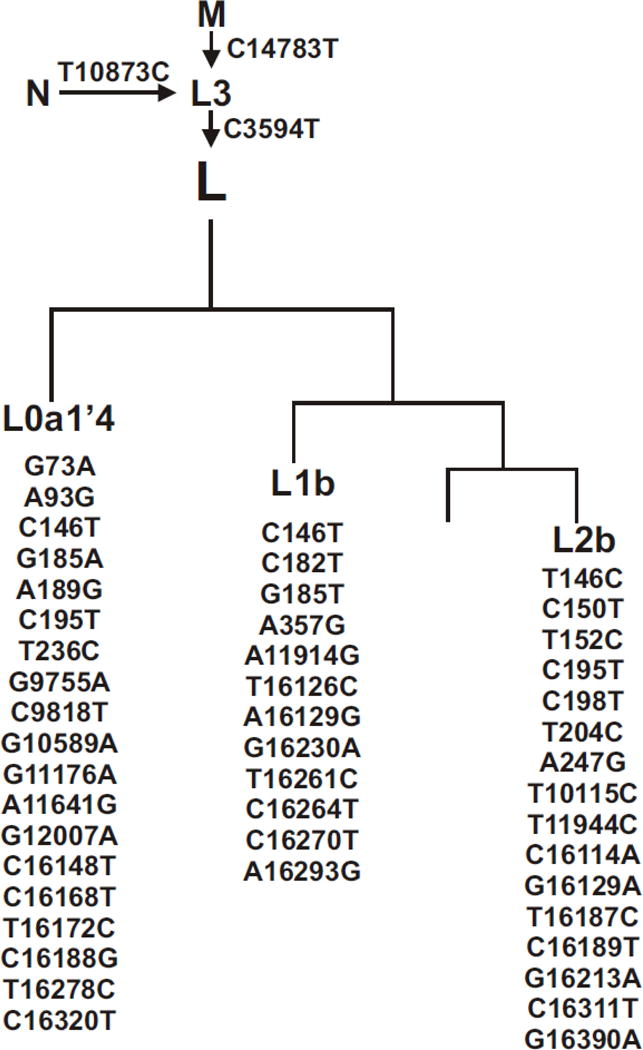
a and b. Diagrams of haplogroups trees showing the subsets of L cybrids and H cybrids used in this study. The mtDNA from the individual L cybrids were analyzed and the SNPs which define the Loa1′4, L1b, and L2b haplogroup subsets are listed. The SNPs listed were all verified by sequencing and comparison to The H haplogroup represents the Cambridge reference standard used to define the mtDNA sequences.
Allelic discrimination was also performed to confirm the haplogroups. The primers for allelic discrimination were synthesized by ABI Assay-by-Design. The samples were run at GenoSeq, the UCLA Genotyping and Sequencing Core, on an ABI 7900HT. Data were analyzed with Sequence Detection Systems software from ABI. All experiments used passage 5 cybrid cells for the assays described below.
2.3. ROS production
Cybrids from different individuals with either H (n=3) or L (n=3) haplogroups were incubated in 24 well plates (10×105 cells/well). After 24 hours, the cells were exposed to fluorescent dye, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Invitrogen-Molecular Probes) and ROS production was measured by the FMBio III instrument at 490 nM. The values represent results combined from different cybrids with H (n=3) or L (n=3) haplogroups, each experiment was repeated two times, and the assays run in quadruplicate. For comparison, cybrids containing J haplogroups (n=3) were also analyzed for ROS production levels.
2.4. ATP production assay
Intracellular ATP levels were measured for H and L cybrids using the luminescence ATP detection assay (ATPlite Perkin Elmer Inc., Waltham, MA USA) as per the supplier’s instructions. Cybrid lines (H cybrids n=3, L cybrids n=3) were cultured 24 hours on a 96 well plate at 2 different concentrations, 100K and 50K cells per well with a final volume of 100μl/well. Luminescence was measured using a Synergy HT Multi-Mode microplate reader and Gen5 Data Analysis software (BioTek instruments, Winooski, VT USA). All experiments were repeated twice and assayed in quadruplicate.
2.5. Lactate assay
Lactate concentrations in the H (n=3) and L (n=3) cybrid samples were measured by the Lactate Assay Kit (Eton Bioscience Inc., San Diego, CA). Cells were plated at 100K and 50K in 96-well plates and incubated overnight. Lactate levels were measured according to the manufacturer’s protocol. Standards and samples were set up as duplicates and quadruplicates and experiments were repeated twice.
2.6. Growth curve assay
The growth curves of three different H cybrids were compared to three different L cybrids over six days, under similar environmental conditions. The different H cybrids and L cybrids were grown to passage 5 using methods described above. 300,000 cells per well were plated onto six-well plates, incubated in standard conditions and culture medium was changed every other day. The cell numbers were measured using a Cell Viability Analyzer (ViCell, Beckman Coulter, Miami, FL). The numbers of cells plated at time-point 0 were designated as 100% and the percentage increase in growth for each cybrid at Days 2, 4, and 6 were calculated. A mean percentage increase value of all three H cybrids and three L cybrids were compared by nonlinear regression analysis (Prism, version 5.0; GraphPad Software Inc., San Diego, CA). Within each experiment the assays were run in duplicate and the experiments repeated twice.
2.7. Extracellular flux analysis
The oxygen consumption rate (OCR) and bioenergetic profiles for the cybrids were measured at 37°C using a Seahorse XF Extracellular Flux Analyzer (Seahorse Bioscience, Billerica, MA). Cybrids with haplogroups H (n=3) and L (n=3) were plated at 30,000 cells/well and cultured overnight at 37°C under 5% CO2. Samples were run in triplicate and experiments repeated three times. Plates were then washed and placed 1 hour in a 37°C incubator under air in 500 μl of unbuffered DMEM (Dulbecco’s modified Eagle’s medium, pH 7.4), supplemented with 17.5mM Glucose (Sigma, St Louis, MO), 200 mM L-glutamine (Invitrogen-Molecular Probes, Carlsbad, CA) and10 mM sodium pyruvate (Invitrogen-Molecular Probes). There was sequential injection into the wells of Oligomycin (1 μM final concentration, which blocks ATP synthase to assess respiration required for ATP turnover), FCCP (1 μM final concentration, a proton ionophore which induces chemical uncoupling and maximal respiration), and Rotenone plus Antimycin A (1 μM final concentration of each, completely inhibits electron transport to measure non-mitochondrial respiration). Data from each well were normalized by measuring total protein. Total protein was isolated with Ripa lysis buffer (Millipore, Billerica, MA) containing protease inhibitor (Sigma, St. Louis, MO) and phosphatase arrest (Gbiosciences, St. Louis, MO). Isolated protein was mixed with Qubit buffer and measured with Qubit 2.0 fluorometer (Invitrogen, Grand Island, NY).
All data from XF24 assays were collected using the XF Reader software from Seahorse Bioscience. The OCR is determined by measuring the drop in O2 partial pressure over time followed by linear regression to find the slope. The ECAR is determined by measuring the change in pH levels over time followed by linear regression to find the slope of the line which represents ECAR. The percentage ATP Turnover Rate is calculated by the following formula: 100 − (ATP coupler response/basal respiration ×100). The percentage Spare Respiratory Capacity represents a bioenergetic value for cells needing high amounts of ATP in response to demands placed upon them. This is calculated by the formula: electron transport chain (ETC) accelerator response/basal respiration × 100. The percentage Proton Leak equals the ATP coupler response - non-mitochondrial respiration. Data from these experiments were exported to GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA) where they were analyzed, normalized and graphed. Statistical significance was determined by performing two-tailed Student t tests and p≤0.05 was considered significant in all experiments.
2.8. Isolation of RNA and Amplification of cDNA
Cells from cybrid cultures (H cybrids, n=3 and L cybrids, n=3) were pelleted, and RNA isolated using the RNeasy Mini-Extraction kit (Qiagen, Inc.) following the manufacturer’s protocol. The RNA was quantified using a NanoDrop1000 (ThermoScientific). For Q-PCR analyses, 100 ng of individual RNA samples were reverse transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen).
2.8. Gene expression arrays and statistical analyses
For the GeneChip array analyses, equal amounts of RNAs (250 ng/μl per sample) from H cybrids (n=3) were combined into a single sample. This was compared to the sample of combined RNAs from L cybrids (250 ng/μl per sample, n=3). The RNA samples were sent to the UCLA Clinical MicroArray Core Lab for analyses with the Affymetrix Human U133 Plus 2.0 Array. Gene expression results were analyzed with IPA summary pathway analysis software (INGENUITY Systems, Redwood City, CA). The array analyses showed that genes related to inflammation, complement activation and cell signaling pathways were differentially expressed in the H versus the L cybrids. The raw and processed data files can be accessed through NCBI Gene Expression Omnibus (GEO) Series.
2.10. Quantitative PCR (Q-PCR) analyses
Gene expression changes identified by the GeneCHip array were verified by Q-PCR using 19 different primers (QuantiTect Primer Assay, Qiagen) for genes associated with complement pathway (CFH, CFHR4, CFP, CFP-var1, CD55/DAF, C3, CD59, C1QC, C1S, C1R, C4B, C4BPB) and inflammatory genes (TGFA, TGFB2, IL-33, IL-6, NFκB2, MAPK8, MAPK10). In addition, twelve mtDNA-encoded genes were also analyzed by Q-PCR using primers obtained from Drs. Marquis Vawter and Nitin Udar (Integrated DNA Technologies, Inc., Coralville, IA). The Q-PCR analyses were performed on individual H or L cybrids and not on combined samples. Total RNA was isolated from individual pellets of cultured cells of haplogroup H cybrids (n=3) and haplogroup L cybrids (n=3) as described above. These cDNA samples were not pooled but rather analyzed individually. Q-PCR was performed on individual samples using a QuantiFast SYBR Green PCR Kit (Qiagen) on a Bio-Rad iCycleriQ 500 detection system and expression levels were standardized for all primers using TATA box binding protein (TBP) as the reference gene. The C1QC, C1S, C4BPB primers were run with the housekeeping gene HMBS. The C4B primers were run with the housekeeping gene HPRT1. All others were run with the housekeeping gene TBP. The analyses were performed in triplicate. Statistical analyses of gene expression levels were performed to measure difference between haplogroups using Prism, version 5.0; (GraphPad Software Inc.).
2.11. Statistical analyses
Data were subjected to statistical analysis by ANOVA and GraphPad Prism (version 5.0). Newman-Keuls multiple-comparison test was done to compare the data within each experiment. P<0.05 (two-sided) was considered statistically significant. Error bars in the graphs represent SEM (standard error mean).
3. Results
For the following series of experiments there were three H cybrids from different individuals (subsets H, H and H5a) and three L cybrids from different individuals (subsets L0a1′4, L1b and L2b). In the various assays, all samples were analyzed independently except for the Affymetrix GeneChip experiment, which had equal amounts of RNA from the three H cybrids or the three L cybrids combined for the assay. However, based upon the GeneChip results, we ran Q-PCR for specific genes with individual RNA samples from each of the different cybrids.
3.1. ROS, ATP and Lactate production levels and growth curve analysis
The ATP and lactate levels were similar in the H (n=3) and L (n=3) cybrid cultures (Figures 2A and 2B). We compared the H and L cybrids plated at concentrations of 100K cells or 50K cells per well for the relative ATP and lactate levels. The ATP production levels were similar in the H cybrids compared to the L cybrids in the cultures with 100K cells (100% versus 98.86 ± 0.75, p=0.27) and 50K cells (94.13 ± 10.72 versus 77.24 ± 5.10, p=0.4). Lactate levels were similar in the H cybrids and L cybrids in the 100K/well cultures (99.95 ± 0.05 versus 95.6 ± 1.9, p=0.22) and 50K/well cultures, (73.3 ± 12.0 versus 71.2 ± 8.8, p=0.89).
Figure 2.
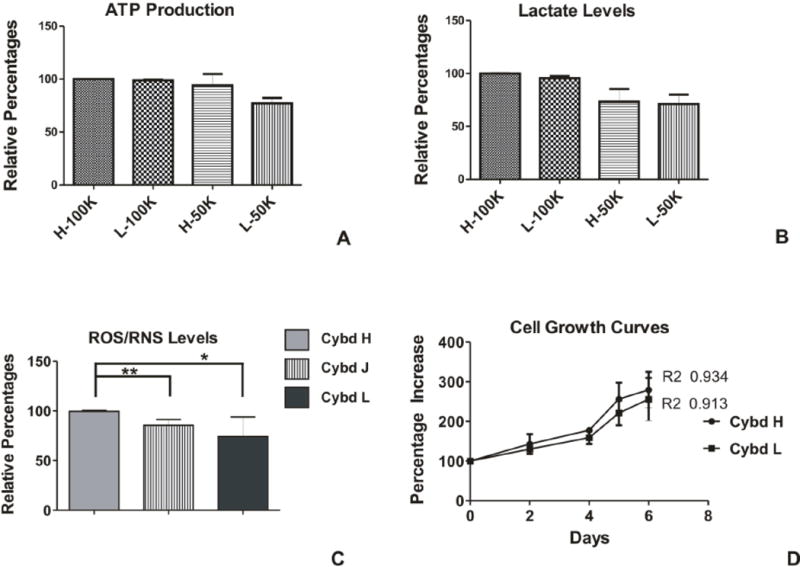
A) H and L cybrids that were plated at 100K cells or 50K cells per well showed similar levels of ATP production after 24 hours incubation. All experiments were repeated twice and assayed in quadruplicate. B) The lactate production levels were similar in the H and L cybrids when plated at concentrations of 100K cells and 50K cells per well and cultured 24 hours. All experiments were repeated twice and assayed in quadruplicate. C) After being cultured 24 hours, the H cybrids showed increased production levels of ROS compared to the J cybrids (**p<0.01) and the L cybrids (*p<0.05). D) At Day 0, 30k cells per well were plated and the cell viabilities measured at Days 2, 4, and 6. The graph shows that the L cybrids grew at a similar growth rate pattern to the H cybrids. ROS/RNS reactive oxygen/nitrogen species; Cybd, cybrid.
The ROS production for the L cybrids was significantly lower than the H cybrids (Figure 2C). As oxygen consumption occurs, electron leakage from the electron transport chain (ETC) can cause increased production of endogenous ROS. The L cybrids (74.21% ± 6.21%, p=0.007) had lower ROS production levels compared to the H cybrids (99.5% ± 0.29) which was unexpected because of the production levels of ATP were similar in the H and L cybrid cultures. In order to verify the ROS assay, we also measured the ROS production of J cybrids, which are known to have lower levels than H cybrids [30]. We found that J-cybrid ROS levels were significantly lower than the H cybrids (83.46% ± 1.84%, p=0.006) but were not significantly different from the L cybrids (p=0.13).
Since the J cybrids have been shown to have a higher growth rate than H cybrids[30] we wanted to compared the growth rates of the L cybrids and H cybrids over a six day incubation period (Figure 2D). The L and H cybrids had similar slopes to their growth rate curves at all time periods examined (Day 0, 100% versus 100%; Day 2, 130% versus 143%; Day 4, 159% versus 178%, Day 6, 256% versus 279%).
3.2. Measurements of the bioenergetic profiles for cybrids
The bioenergetic profiles in the H (n=3) and L (n=3) cybrids were measured after sequential treatments with Oligomycin, FCCP and Rotenone/Antimycin A using values generated by the Seahorse XF24 flux analyzer. Figure 3A shows the profile representing basal respiration, ATP turnover, maximal respiration, spare respiratory capacity, proton leak and non-mitochondrial respiration. The percentage ATP turnover rate was significantly higher for the H cybrids compared to the L cybrids (49.42 ± 1.3 versus 41.3 ± 3.1, p=0.024, Figure 3B). The percentage spare respiratory capacity of the H cybrids (166.5 ± 4.07) was significantly higher than the L cybrids (143.4 ± 4.7, p=0.02, Figure 3C). The percentage proton leak values for the H cybrids (12.43 ± 1.27) were similar to the L cybrids (15.13 ± 0.8, p=0.09).
Figure 3.
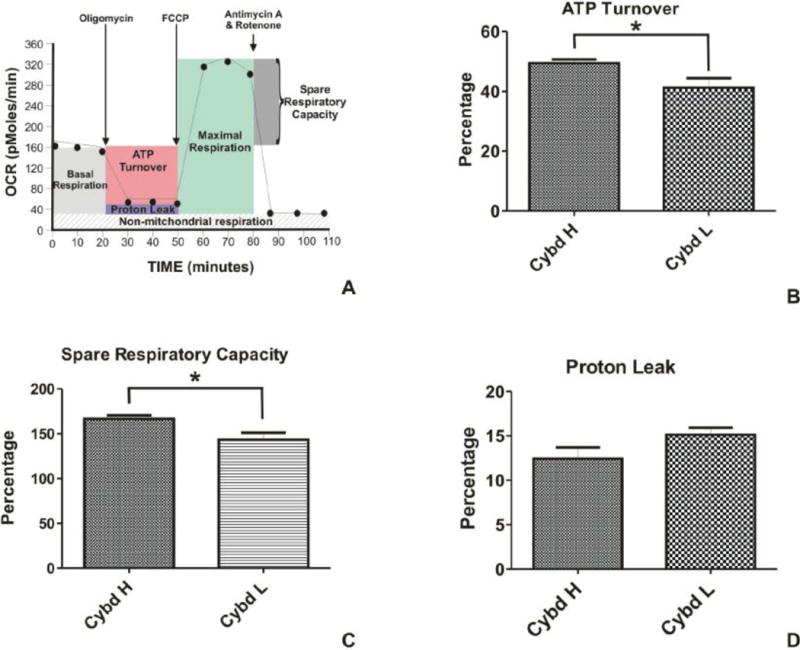
A) Schematic representation showing regions that define the basal aerobic respiration, ATP turnover, maximal respiration, spare respiratory capacity, proton lead and non-mitochondrial respiration as measured by Seahorse XF24 flux analyzer. B) In response to Oligomycin treatment, the L cybrids showed significantly lower ATP turnover compared to the H cybrids (*p=0.05). C) After treatment with Antimycin A plus Rotenone, the L cybrids showed significantly lower spare respiratory capacity compared to the H cybrids (*p=0.05). D) The percentage proton leak levels were similar in the H and L cybrids (p=0.09). Cybd, cybrid.
The Seahorse XF24 flux analyzer allows for real-time measurement in the individual wells of the oxygen consumption rates (OCR) representing the basal aerobic respiration of the cells and extracellular acidification rates (ECAR) representing glycolysis. The OCR to ECAR ratios for the H and L cybrids were not significantly different from each other (23.85 ± 2.50 versus 17.17 ± 2.44, p=0.1).
3.3. Levels of expression for mtDNA encoded genes
The H and L haplogroups are defined by specific patterns of SNPs, some of which are non-synonymous SNPs that lead to amino acid changes within the OXPHOS respiratory complexes. Using the cybrid model (H cybrids, n=3 and L cybrids, n=3), we measured the expression levels of the mtDNA encoded genes in respiratory complexes I, III, IV, and V of the OXPHOS pathway and normalized the H cybrids values to 1 (Table 1). The L cybrids showed significantly higher gene expression levels of nine of the genes compared to H cybrids: Complex I, mt-ND1 (p=0.047), mt-ND4/ND4L (p=0.005), mt-ND5 (p=0.007); mt-ND6 (p=0.019); Complex III, mt-CYB (p=0.006); Complex IV, (mt-CO1 (p=0.004), mt-CO2 (p=0.033), mt-CO3 (p=0.025); Complex V, mt-ATP6 (p=0.015); and mt-ATP8 (p=0.013). Only the mt-ND4/ND4L expression levels in the L cybrids were lower than the H cybrids (p=0.005).
Table 1.
Q-PCR Expression Levels for mtDNA Encoded Genes from Complexes I, III, IV, and V Found in the L versus H Cybrids
| Symbol | Gene Name | L vs. H p-value | L vs. H ΔΔCT/Fold |
|---|---|---|---|
| MT-ND1 | NADH Dehydrogenase subunit 1 | 0.047 | 0.65±0.30/1.57 |
| MT-ND2 | NADH Dehydrogenase subunit 2 | 0.338 | 0.37±0.37/1.29 |
| MT-ND3 | NADH Dehydrogenase subunit 3 | 0.337 | 0.40±0.41/1.32 |
| MT-ND4/ND4L | NADH Dehydrogenase subunit 4/4L | 0.005 | −2.64±0.81/0.16 |
| MT-ND5 | NADH Dehydrogenase subunit 5 | 0.007 | 1.07±0.34/2.10 |
| MT-ND6 | NADH Dehydrogenase subunit 6 | 0.019 | 0.90±0.35/1.87 |
| MT-CYB | Cytochrome b | 0.006 | 1.08±0.34/2.11 |
| MT-CO1 | Cytochrome c oxidase subunit I | 0.004 | 0.95±0.29/1.93 |
| MT-CO2 | Cytochrome c oxidase subunit II | 0.033 | 0.83±0.36/1.78 |
| MT-CO3 | Cytochrome c oxidase subunit III | 0.025 | 0.79±0.32/1.73 |
| MT-ATP6 | ATP synthase F0 subunit 6 | 0.015 | 0.85±0.31/1.80 |
| MT-ATP8 | ATP synthase F0 subunit 8 | 0.013 | 0.86±0.31/1.82 |
N=3 with six values for each sample.
Fold values greater than 1 indicate up regulation of the gene compared to H cybrids. H cybrids are assigned a value of 1.
Fold values less than 1 indicate down regulation of the gene compared to H cybrids. H cybrids are assigned a value of 1.
Fold = 2−ΔΔCT
3.4. Levels of expression of nuclear genes related to complement activation, inflammation and apoptosis
Based upon differences in the expression of mtDNA-encoded OXPHOS complexes found in H and L cybrids, we hypothesized that the cybrids might also have different gene expression patterns for other pathways. Therefore, we isolated the RNA of three H cybrids and three L cybrids, pooled equal quantities from each sample and then analyzed them using the GeneChip array that characterizes gene expression for over 40K genes. These data were then analyzed with the Ingenuity software program which showed that the pooled RNA from the L cybrids had statistically significant differences in the Complement System, CCR3 Signaling in Eosinophils, Dermatan Sulfate Biosynthesis (late stages), and Chondroitin Sulfate Biosynthesis (late stages) compared to the pooled RNA from the H cybrids (Table 2). We were surprised that differences in major pathways of nuclear genes unrelated to energy production were found in these cybrids with the identical nuclei, cytoplasm and culture conditions but varying only in the haplotype of their mitochondria.
Table 2.
| a. Comparison of Gene Expression Differences in Canonical Pathways for L Cybrids versus H Cybrids as Determined by IPA Summary Program Analyses of Affymetrix Human U133 Plus 2.0 Array | |||
|---|---|---|---|
| Ingenuity Canonical Pathways | P-value | Ratio | Molecules |
| Complement System | 0.00298 | 5/35 (0.143) | C1R, CFD, C3, C1S, CFH |
| CCR3 Signaling in Eosinophils | 0.00688 | 9/126 (0.071) | GNAI3, PLA2G4A, GNB4, MPRIP, PIK3C2A, PLA2G4C, GNB1L, PRKD3, LIMK1 |
| Dermatan Sulfate Biosynthesis (Late Stages) | 0.00794 | 5/47 (0.106) | CHST2, CHST5, SULT1C2, HS3ST1, CHST15 |
| Chondroitin Sulfate Biosynthesis (Late Stages) | 0.00975 | 5/54 (0.093) | CHST2, CHST5, SULT1C2, HS3ST1, CHST15 |
| b. Details of the Genes Most Differentially Expressed in the Canonical Pathways as Identified by the Affymetrix Human U133 Plus 2.0 Array (see Table 2a) | ||||||
|---|---|---|---|---|---|---|
| Probe Set ID | Represent ative Public ID | Gene Title | Gene Symbol | CYBD -L Signal | CYBD -H Signal | L vs H (Fold) |
| Complement Pathway | ||||||
| 205382_s_at | NM_00192 8 | complement factor D (adipsin) | CFD | 110.22 28 | 221.97 49 | −2.014 |
| 217767_at | NM_00006 4 | complement component 3 | C3 | 47.807 12 | 266.43 35 | −5.573 |
| 208747_s_at | M18767 | complement component 1, s subcomponent | C1S | 373.55 14 | 1077.6 74 | −2.885 |
| 213800_at | X04697 | complement factor H | CFH | 374.22 68 | 731.56 13 | −1.955 |
| 233645_s_at | AK024084 | complement component 1, r subcomponent-like | C1RL | 34.922 33 | 80.276 31 | −2.299 |
| 212067_s_at | AL573058 | complement component 1, r subcomponent | C1R | 462.33 73 | 921.95 | −1.994 |
| CCR3 Signaling in Eosinophils | ||||||
| 201179_s_at | J03005 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | GNAI 3 | 1984.7 61 | 964.71 7 | 2.057 |
| 210145_at | M68874 | phospholipase A2, group IVA (cytosolic, calcium-dependent) | PLA2 G4A | 508.06 08 | 239.03 16 | 2.125 |
| 225710_at | H99792 | guanine nucleotide binding protein (G protein), beta polypeptide 4 | GNB 4 | 2083.3 21 | 1604.4 51 | 1.298 |
| 238330_s_at | BE545235 | Myosin phosphatase Rho interacting protein | MPRI P | 45.435 6 | 38.304 87 | 1.186 |
| 213070_at | AV682436 | phosphoinositide-3-kinase, class 2, alpha polypeptide | PIK3 C2A | 3398.1 76 | 3108.4 79 | 1.093 |
| 1554810_at | BC017956 | phospholipase A2, group IVC (cytosolic, calcium-independent) | PLA2 G4C | 2.8145 04 | 3.7450 29 | −1.331 |
| 204357_s_at | NM_00231 4 | LIM domain kinase 1 | LIMK 1 | 364.53 68 | 172.30 25 | 2.116 |
| Dermatan Sulfate Biosynthesis (late stages) | ||||||
| 203921_at | NM_00426 7 | carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 | CHST 2 | 172.48 77 | 574.86 35 | −3.333 |
| 221164_x_at | NM_01212 6 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 | CHST 5 | 166.99 94 | 66.774 11 | 2.501 |
| 205342_s_at | AF026303 | sulfotransferase family, cytosolic, 1C, member 2 | SULT 1C2 | 1550.9 49 | 727.04 83 | 2.133 |
| 213991_s_at | BF940710 | heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | HS3S T1 | 8.1710 75 | 11.568 58 | −1.416 |
| 203066_at | NM_01486 3 | carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 | CHST 15 | 123.38 05 | 259.39 76 | −2.102 |
| Chondroitin Sulfate Biosynthesis (late stages) | ||||||
| 203921_at | NM_00426 7 | carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 | CHST 2 | 172.48 77 | 574.86 35 | −3.333 |
| 221164_x_at | NM_01212 6 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 | CHST 5 | 166.99 94 | 66.774 11 | 2.501 |
| 205342_s_at | AF026303 | sulfotransferase family, cytosolic, 1C, member 2 | SULT 1C2 | 1550.9 49 | 727.04 83 | 2.133 |
| 213991_s_at | BF940710 | heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | HS3S T1 | 8.1710 75 | 11.568 58 | −1.416 |
| 203066_at | NM_01486 3 | carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 | CHST 15 | 123.38 05 | 259.39 76 | −2.102 |
The significance of our Affymetrix gene array studies is that by IPA analyses, the four pathways with the greatest changes are related to complement activation, inflammation and autoimmunity. This strongly suggests that mtDNA can greatly influence the expression of nuclear gene pathways associated with diseases which are more prevalent in African populations (e.g., SLE, diabetes, asthma). The next series of experiments were to verify the genes of the complement pathway but in the future we will need to perform Q-PCR to measure expression levels for the CCR3 and sulfotransferase pathways.
We used Q-PCR to analyze twelve nuclear genes associated with the complement pathway and seven nuclear genes related to cell signaling and inflammation (Table 3). In the Q-PCR studies, we analyzed the RNA from individual cybrids (H haplogroups, n=3 and L haplogroups, n=3). We demonstrated that the L cybrids showed decreased levels for C1QC (0.46 fold, p= 0.0002), C1R (0.16 fold, p=0.025), C3 (0.17 fold, p=0.0004), and CFH (0.58 fold, p=0.009) compared to H cybrids. There was a trend for decreased values for the C1S gene but it did not quite reach significance (0.21 fold, p=0.063). The inflammation-related (type 2 immunity) genes, TGFA and IL-33, were expressed at lower levels in the L cybrids (0.4 fold, p=0.02 and 0.3 fold, p=0.002) compared to the H cybrids. The L cybrids had higher expression levels for NFκB2 (1.36 fold, p=0.0013) and MAPK10 (1.62 fold, p=0.004). The H and L cybrids had similar expression levels of the IL-6, TGFB2, and MAPK8 genes.
Table 3.
Q-PCR Analyses for Differential Gene Expressions Found in the L versus H Cybrids
| Symbol | Gene Name | GenBank Accession No. | L vs. H p-value | L vs. H ΔΔCT/Fold | Pathways/Functions |
|---|---|---|---|---|---|
| C1QC* | Complement component 1, q subcomponent, C chain |
NM_001114101 NM_172369 |
0.0002 | −1.13±0.23/0.46 | Classical Complement |
| C1S* | Complement component 1, s subcomponent |
NM_001734 NM_201442 |
0.063 | −2.28±1.14/0.21 | ” |
| C1R* | Complement Factor H | NM_000186 | 0.025 | −2.65±1.07/0.16 | ” |
| C4B** | Complement component 4B (Chido blood group) |
NM_001002029 NM_000592 |
0.35 | −0.89 ± 0.39/0.54 | ” |
| C4BPB* | Complement component 4 binding protein, beta |
NM_000716 NM_001017364 NM_001017365 NM_001017366 NM_001017367 |
0.06 | −1.19±0.57/0.44 | ” |
| CFHR4 | Complement factor H-related 4 |
NM_006684 NM_001201550 NM_001201551 |
0.09 | −0.84 ± 0.46/0.56 | ” |
| C3 | Complement Component 3 | NM_000064 | 0.0004 | −2.6±0.6/0.17 | Classical and Alternative |
| CFH | Complement Factor H | NM_000186 | 0.009 | −0.8±0.3/0.58 | Alternative Complement |
| CFP* | Complement Factor Properdin |
NM_001145252 NM_002621 |
0.1 | −0.38±0.22/0.77 | ” |
| CFP-var 1* | Complement Factor Properdin | NM_002621 | 0.52 | −0.51 ± 0.77 | ” |
| CD59 | CD59 molecule, Complement regulatory protein |
NM_000611 NM_203329 NM_203331 NM_001127223 NM_001127225 NM_001127226 NM_001127227 |
0.17 | 0.19±0.13/1.14 | |
| CD55/DAF | Decay accelerating factor for complement |
NM_000574 NM_001114543 NM_001114544 NM_001114752 |
0.14 | −0.37±0.24/0.77 | ” |
| TGFA | Transforming growth factor, alpha |
NM_003236 NM_001099691 |
0.02 | −1.31±0.53/0.40 | Activates signaling pathway for cell proliferation, differentiation |
| TGFB2 | Transforming growth factor, beta 2 |
NM_003238 NM_001135599 |
0.27 | −0.28±0.25/0.82 | Regulates proliferation, differentiation, adhesion, migration |
| IL-33 | Interleukin 33 |
NM_033439 NM_001199640 NM_001127180 |
0.002 | −1.74±0.46/0.30 | Proinflammatory, Involved in production of T helper cytokines |
| IL-6 | Interleukin 6 | NM_000600 | 0.11 | −1.37±0.81/0.39 | Involved in inflammation and maturation of B cells. |
| NFKB2 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 |
NM_001077494 NM_001077493 NM_002502 |
0.0013 | 0.44±0.13/1.36 | Central activator of inflammation and immune function genes |
| MAPK8 | Mitogen-activated protein kinase 8 |
NM_139046 NM_002750 NM_139047 NM_139049 |
0.7 | −0.08±0.21/0.95 | Targets specific transcription factors involved with apoptosis |
| MAPK10 | Mitogen-activated protein kinase 10/JNK3 |
NM_002753 NM_138980 NM_138982 |
0.004 | 0.7±0.21/1.62 | Regulatory role in signaling pathways during apoptosis |
N=3 with six values for each sample.
Fold values greater than 1 indicate up regulation of the gene compared to H cybrids. H cybrids are assigned a value of 1.
Fold values less than 1 indicate down regulation of the gene compared to H cybrids. H cybrids are assigned a value of 1.
Fold = 2−ΔΔCT
These primers were run with the housekeeping gene HMBS.
These primers were run with the housekeeping gene HPRT1. All others were run with the housekeeping gene TBP.
3.5. mtDNA copy numbers
The mtDNA copy numbers in the H and L cybrid cultures were measure by Q-PCR using 18S to represent nDNA and mt-ND2 to represent mtDNA. An average of 3 independent representatives was used for each haplogroup and run in triplicate. At Day 1 the L cybrids had nDNA:mtDNA ratios of 1.26±0.007 (p<0.0001) and at Day 7 the ratio was 1.18±0.06 (p<0.036), which were significantly lower than the H haplogroup cybrids (H cybrids at Day 1 were 1.38±0.016 and day 7 were 1.31±0.022). Therefore, our results showed that the L haplogroup cybrids had decreased mtDNA copy numbers compared to the H cybrids.
4. Discussion
Over the past few decades there have been tremendous advances in the identification of nuclear genes that are associated with human diseases but there are still many questions with respect to population susceptibilities to diseases. We hypothesize that mtDNA variants contribute to this susceptibility for the following reasons: 1) Single mtDNA mutations can have profound effects on tissues and lead to debilitating and lethal diseases [1]. Within the mtDNA haplogroups, there are large accumulations of SNPs that have occurred over thousands of years, some of which cause amino acid and functional changes, while others cause changes in the rates of replication and transcription of the mtDNA. These SNP variations can influence levels of ATP and ROS, which are signaling elements for pathways that can affect cellular behavior. 2) Our laboratory, along with others, have used the cybrid model to provide evidence that mtDNA haplogroups can mediate expression of nuclear genes, rates of cell growth and cell behavior [5, 6, 30]. In addition, clinical studies have shown that mtDNA haplogroups can influence the severity and penetrance of diseases [7, 8]. It is reasonable to speculate that if an ethnic/racial population has a mtDNA haplogroup profile which causes major alterations in gene expression within a disease-related pathway (i.e., complement or innate immunity), that this group may respond uniquely to nuclear genetic patterns or environmental patterns compared to a population with a totally different mtDNA haplogroup SNP profile (Figure 4). In other words, we propose that when we look at diseases, we not only examine the nuclear genetics and environmental factors, but also include the mtDNA haplogroup backgrounds because the mtDNA profiles seem to greatly influence the nuclear genes expressed for major pathways related to diseases.
Figure 4.
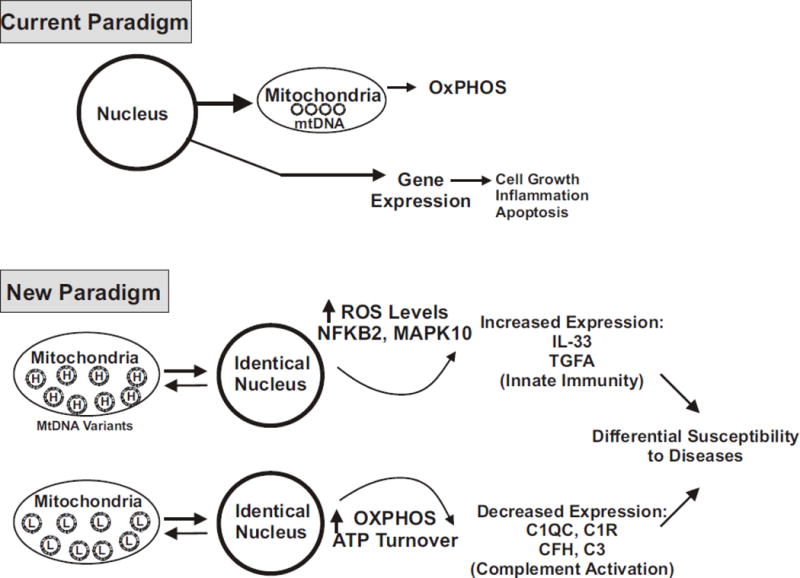
The upper panel is a schematic of the current paradigm in which nuclear genes dominating cellular functions. The lower panel is a schematic which describes a new paradigm, based upon our results with the model of cybrids, which have identical nuclei but differ in the mtDNA haplogroups (H versus L). We propose that mtDNA, which mediates energy production pathways and ROS formation, can also greatly influence the expression of nuclear genes related to complement activation, inflammation and signaling pathways. As a result, the cells with different mtDNA haplogroups (e.g., European versus African) will have different baseline expressions of major functional pathways. Therefore, since the cells will have unique complement/inflammation backgrounds, then responses of these cells to identical nuclear genes or environmental factors may then contribute to differential susceptibilities to diseases for the different ethnic/racial haplogroups.
Attempts were made to correlate the results from the ROS, ATP, Lactate and growth curve assays for each L-subtype and H-subtype. We found that within the L-subtypes, the values trended together while the H values were similar to each other. In other words, there was not a particular subtype, either H or L, that stood out from the others and this is reflected in that the error bars were relatively low and that there were no differences in the ATP, Lactate and growth curve assays. In addition, based upon the information presently at hand, it was not possible to assign the differences found in the gene expression levels to individual SNPs. However, when additional cybrids are generated, we should be able to analyze the data with programs to help us understand the potential consequences of the particular mitochondrial variants.
4.1. Respiration rates and mtDNA encoded genes for complexes I, III, IV, and V
The XF24 Flux analyzer allows for simultaneous, real-time measurements of bioenergetic profiles for multiple cybrid samples. In our study, measurements were taken after different compounds were added sequentially to metabolically alter the bioenergetic profile of the cybrids. Oligomycin acts as an ATP coupler which inhibits ATP synthase (Complex V). FCCP, an ETC accelerator, acts as an uncoupling agent as it transports hydrogen ions across the mitochondrial membrane and disrupts ATP synthesis associated with Complex V activity. The third compound is a combination of Rotenone, a Complex I inhibitor, plus Antimycin A, a Complex III inhibitor, which together shut down mitochondrial respiration.
Most studies have focused on European mtDNA haplogroups. To date there are very few cybrid model studies using the L haplogroups, which are individuals with African maternal origins. In our study, the L cybrids have similar levels of lactate and ATP production, suggesting similar glycolysis and OXPHOS levels as compared to the European H cybrids. We were surprised to find that ROS levels in the L cybrids were significantly less than the H cybrids, because usually the ATP production and ROS levels parallel each other, as was seen when H cybrids were compared to J cybrids [30]. Our findings suggest that in the L haplogroup variants, there may not be a direct correlation between ROS production and mtDNA gene expression within the respiratory Complexes I and III. One possible explanation for the dissimilarity between the ATP and ROS levels in L cybrids maybe that the L mtDNA haplogroups are more efficient in OXPHOS so electrons leakage is lower, resulting in diminished ROS formation.
Our Q-PCR studies demonstrate that although the L cybrids have lower mtDNA copy numbers, they show higher expression levels for nine of the mtDNA-encoded respiratory complex genes, which were associated with Complexes I, III, IV and V, which suggests that these mitochondria may be more efficient in their aerobic respiration. In contrast, while mtRNA expression for L cybrids were 1.57-fold to 2.11-fold higher than H cybrids, the cybrids that contained J haplogroup mtDNA showed high levels of glycolysis and also significantly lower expression for the mtDNA-encoded OXPHOS genes (data not shown). Our data demonstrate that the mtDNA haplogroup variants can greatly influence the efficiency of respiration, irrespective the nuclei, since in our cybrid system all nuclei are identical.
The Seahorse Flux bioenergetic analyses showed that compared to the European H cybrids, the L cybrids have lower ATP turnover and spare respiratory capacity, which suggests that they may not be able to respond to stress as readily as the H cybrids. The relationships of elevated mtRNA expression levels but lower ATP turnover rates and ROS production may be supportive of the theory that African mtDNA variants are associated with endurance performance in elite athletes. Scott and coworkers reported L0 haplogroups were found in higher numbers of Kenyan athletes while L3 haplogroups were significantly reduced [31]. However, no correlation between haplogroups and athletic performance was found in Ethiopian athletes [32]. Further studies will be required to clarify in L cybrids the relationship between ATP production, mtRNA expression levels and ROS production.
The H and L cybrids were similar to each other in their growth rates patterns and lactate levels, but both cybrids differ considerably from the European J cybrids, which showed preference toward use of glycolysis and very rapid growth rates [30]. In summary, although H and L cybrids had identical nuclei, the bioenergetic parameters and ROS levels were different, reflecting the influence of the mtDNA on the cybrid “nuclear machinery”. The mechanisms of nuclear-mitochondrial interactions in mammalian cells are not understood but deserve much closer scrutiny.
4.2. Differences in Gene Expression Patterns Identified by Affymetrix GeneChip Array
Individuals with African background are at a higher risk for developing asthma and other inflammation-related diseases. The IPA summary of the GeneChip array data (Table 3) showed that the H and L cybrids possess differences in the pathways related to Complement System and the CCR3 Signaling in Eosinophils. The CCR3 pathway is important in neovascularization and inflammation, with the CCR3 receptor (also known as eotaxin receptor) being expressed on eosinophils, airway epithelial cells, basophils, mast cells and T2 helper cells [71]. The CCR3 receptors can be found in the human choroidal neovascular membrane and various cancers [72–74]. In a mouse model, when CCR3 is blocked then choroidal neovascular is decreased [75]. Investigators suggest that CCR3 plays a significant role in the development of neovascularization by promoting endothelial cell migration and activation of VEGFR2 [76] and may be a target for therapeutic intervention [77]. In either case, the finding that expression of genes in the CCR3 pathway are significantly different between the H and L cybrids is notable because the cybrids had identical nuclei but differed only in the mtDNA haplogroups, suggesting that the mtDNA can mediate this major inflammatory pathway.
The IPA program also identified five genes (CHST2, CHST5, SULT1C2, HS3ST1 AND CHST15) associated with the late stages of the Dermatan sulfate (DS) and Chondroitin sulfate (CS) pathways as being significantly different between the H cybrids and L cybrids. All five enzymes are sulfotransferases, which catalyze the transfer of sulfate onto DS, CS or keratan sulfate (KS). This may be significant for cellular behavior because the degree of sulfation of GAGs can mediate biological functions [33], which include cellular proliferation, migration, differentiation, angiogenesis and activation of cytokine/growth factors.
Studies show that DS and CS play a role in autoimmune diseases as they can be targets for auto-antigen formation and responses of specific auto-reactive B-1a cells [34]. For example, in SLE patients, CS can cross-react with anti-DNA antibodies and in autoimmune thyroid disease and diabetes, the urinary DS and CS levels correlate with disease severity [35]. In patients with SLE and kidney disease, the ratio of chondroitin sulfate to heparan sulfate can be used to identify the degree of disease activity [36]. Campo and coworkers showed that chondroitin 4-sulfate (C4S) have antioxidant properties by inhibiting lipid peroxidation and lowering ROS damage, and suggested that GAGs can act as immunomodulators [37]. As part of the extracellular matrix (ECM) network, the CS proteoglycans can influence the functions of immune cells during pathologic central nervous system diseases [38]. At a molecular level, CS may have antioxidant and anti-inflammatory properties because it can decreased activation of NFκB, which in turn leads to lower expression of proinflammatory cytokines [39–41].
A recent study showed that mutations in genes for biosynthetic enzymes that sulfate CS and DS are associated with various connective tissue diseases [42], which indicates that the sulfation patterns of GAGs are very important for cellular behavior. Our findings demonstrate that the expression of five sulfotransferase enzymes in the cybrids are different based solely upon the mtDNA within the cell. Weiser and coworkers has shown that auto-antibodies in SLE patients have selective binding to cell surface GAGs [43]. One can speculate that if there are different levels of the negatively charged sulfates on DS and CS, such as might be seen with the different expression levels of sulfotransferase genes, this might influence the autoantibody-binding capacity to the cells.
Finally, C1q inhibitor (C1q INH) is a chondroitin 4-sulfate proteoglycan, which inhibits C1q activation by binding and precipitating the molecule. C1q is critical for the first steps of activation of the complement pathway, another pathway different in H cybrids compared to L cybrids. This demonstrates a possible connection between the DS/CS pathway and complement pathway, all of which are altered in the L cybrids.
In summary, our GeneChip array data shows that the mtDNA can influence the expression levels for key sulfotransferase enzymes of DS and CS, molecules which are involved with autoimmune diseases. One can speculate that if the DS and CS have different sulfation patterns, then cells may have different signaling and inflammation pathways activated. This could lead to different susceptibilities of European H haplogroup and African L haplogroup individuals to high risk nuclear genes and/or environmental oxidative stressors. Therefore, when examining the pathogenesis of diseases, we believe it is important to evaluate the mtDNA background because some mtDNA patterns may make a person more likely to get a disease than other haplogroup patterns.
The mechanisms by which mtDNA can mediate the sulfotransferase enzymes is unknown but deserves further studies. The mtDNA encodes only 37 genes, no transcription factors and does not encode for any of the genes that are altered in the Affymetrix Human U133 Plus 2.0 array. Perhaps the ROS levels may influence the genes because the H cybrids have higher ROS production than the L cybrids. It is less likely that the ATP levels are involved since they have similar levels in the H and L cybrids. Using the osteosacroma cybrid model, D’Aquila et al reported that the SIRT3 gene contributes to the mitochondria-nuclear cross talk after oxidative stress [78]. Furthermore, experiments with Drosophila simulans which measured the catalytic capacity of the electron transport chain also showed the functional differences of mitochondrial metabolism were related to mtDNA haplogroups, and were not mediated via mitochondrial-nuclear interactions or nuclear DNA properties [79]. In any case, there is enough evidence accumulating which supports a paradigm shift in thinking about mitochondrial-nuclear interactions [80].
4.3. Gene expression for complement pathway genes
The C1QC and C1R gene expressions were significantly lower in the L cybrids compared to H cybrids. C1q binds with C1r and C1s to become C1 complex of the classical pathway. As immunoglobulins bind to the C1 complex then the proteases C1r and C1s are activated, which leads to further activation of the classical pathway. Complete genetic deficiency of C1q is highly predictive of systemic lupus erythematosus (SLE) disease and low C1q serum levels have been associated with SLE flare-ups [44]. Studies show that C1q has a regulatory role on interferon-alpha, interleukin (IL)-6, IL-8 and tumor necrosis factor (TNF)-alpha [45]. Deficiency of C1S is also associated with SLE-like syndromes and glomerulonephritis [46]. These data indicate that mtDNA variants can mediate gene expression for two components of the classical pathway, which are important in inflammatory diseases.
Abnormalities of the alternative complement pathway are strongly associated with various diseases and our cybrids model shows significant effects of the haplogroups on that activity. The L cybrids have 0.6-fold lower levels of the CFH inhibitor of complement compared to H cybrids (p=0.009). In a previous study, we reported that the J cybrids had significantly lower expression levels of two alternative pathway inhibitors, CFH (0.5-fold lower, p=0.0001) and CD55/DAF (0.6-fold lower, p=0.03) compared to the H cybrids [30]. Activation of the alternative complement pathway and altered levels of CFH are linked with AD, obesity and T2D [47–49]. If we extrapolate our cybrid findings to individuals, then the H haplogroup subjects would have two inhibitors of the alternative pathways (CFH and CD55), the L haplogroup subjects would have decreased CFH inhibitor and J haplogroups would be deficient in two inhibitors. Then theoretically, the H individual would be the most protected from complement activation, the L would be intermediate and the J individuals would be highest risk. This scenario may be a mechanism by which ethnic/racial origins could influence development of diseases which had major inflammation components.
Our findings that mtDNA haplogroups mediate gene expression have been supported by other using the osteosarcoma cybrid model, which reported the influence of mtDNA variants upon stress response genes including heat shock protein (HSP)60, HSP75, IL-6, IL-1 and TNF-receptor2 [5, 6]. In addition, cybrids from patients with Huntington’s disease and Leber hereditary optic neuropathy (LHON) have shown that mtDNA mutations can affect various cellular functions [50–53]. Therefore, our findings, along with cybrid studies using different background Rho0 cells and different mtDNA, demonstrate that mtDNA variants can affect gene expression of major molecular pathways and cell behavior.
4.4. Inflammation Pathway Genes
IL-33 was identified by both GeneChip array and Q-PCR to be differentially expressed in the L cybrids compared to H cybrids. This recently discovered cytokine has generated tremendous interest because it plays a key role in immune-inflammatory diseases and angiogenesis [54–56]. IL-33 seems to act as a “switch” cytokine which can alter expression of IL-4, IL-5, IL-13, IgA, IgE and IgG levels [57–62] and also plays a role in the induction of naïve T cells to become cytokine-producing Th2 cells [63–65]. In mice, IL-33 increases IgE serum levels and triggers degranulation of mast cells [65].
IL-33 has a role in asthma, arthritis, cardiovascular disease, clearance of parasites, central nervous system disease and various types of infections [66]. Milovanovic reports that the IL-33/ST2 complex plays a role in animal models of diabetes, autoimmune diseases and cancer, and that IL-33 can be anti-inflammatory for T cell responses [67]. Ali and coworkers have shown that the full-length IL-33 interacts with NF-kB by reducing its DNA binding capacity, which they suggest lowers gene transcription and pro-inflammatory signaling [68]. If IL-33 acts as a potent activator of the immune system, then differing levels could greatly influence allergic and inflammatory responses. One can speculate that this might play a role in the increased prevalence of disease such as asthma and SLE in populations with maternal African origins. This is important because if cybrids with different mtDNA haplogroups show disparity in the IL-33 expression levels, it may be a mechanism by which innate immunity is involved in population susceptibilities of various diseases. In the future, Il-33 should be investigated as a potential trigger and therapeutic target site to diminish the inflammatory component of human diseases.
The L cybrids have lower expression for TGF-alpha, a growth factor with 40% homology to epidermal growth factor (EGF) that binds to the EGF receptor and induces c-fos production and functions in cell proliferation and differentiation [69, 70]. TGF-alpha has been found in retina, neural tissues and tumors [71–73]. Abnormal interactions between TGF-alpha and the transcription factor IRF6 (interferon regulatory factor-6) are associated with tooth agenesis and cleft lip-palate development [74]. Vieira has suggested that TGF-alpha is a genetic modifier associated with developmental diseases [75]. It has been suggested that serum concentrations of TGF-alpha can be predictive for the response to EGFR-tyrosine kinase inhibitors for the treatment of non-small cell lung cancer [76]. TGF-alpha plays a role in the circadian sleep-wake cycles and neuronal-retinal cell behavior through the retinohypothalamic tract [77]. TGF-alpha is likely an important growth factor that has a functional impact on diseases, and it is significant that expression levels vary based upon the mtDNA content of cells.
Using both the GeneChip expression array and Q-PCR methods, we found that the NFκB2 and MAPK10 gene expression levels for L cybrids were higher than in H cybrids. Activation of NFκB2 has been associated with inflammation and its inhibition has been linked to delayed growth and apoptosis. MAPK10 (also known as JNK3) is responsible for activating the Jun transcription factor and usually is distributed in neurons, heart, testis and pancreatic islet cells [78, 79]. MAPK10/JNK3 activation is elevated in human Alzheimer’s disease and an AD mouse model and JNK3 deletion improves cognition in familial AD mice [80]. In a retinal ganglion cell mouse model, JNK3 is involved with cell death, which is a key feature of glaucoma [81]. Our findings are significant because we show that in cells with identical nuclei, changing the mtDNA variants can lead to differential expression levels of NFκB2 and MAPK10/JNK3, important signaling molecules for many cellular functions.
5. Conclusions
This study uses a cybrid model to study mitochondrial-nuclear interactions and shows that although H and L cybrids had identical nuclei, the bioenergetic parameters and ROS levels were distinctly different from each other. In addition, Q-PCR analyses show differential expression of genes associated with the cell signaling, complement activation, inflammation and innate immunity in the L cybrids compared to H cybrids. Our findings demonstrate that mtDNA variants can mediate nuclear gene expression and alter major functional pathways, which is extremely important as different ethnic/racial populations have different susceptibilities to diseases associated with complement activation and inflammation. The cybrid model may offer insights into mechanisms by which these disease susceptibilities may occur and help us to identify novel targets for treatments.
Highlights.
H (European) and L (African) cybrids have identical nuclei but different mtDNA
L cybrids have lower mtDNA copy numbers, ROS production and ATP turnover rates
L cybrids have higher expression levels of mtDNA-encoded respiratory complex genes
H and L cybrids differentially express genes from major inflammatory pathways
mtDNA haplogroups may play a role in differential susceptibilities to diseases
Acknowledgments
The authors wish to thank the participants in the study. This work was supported by Discovery Eye Foundation, Guenther Foundation, Beckman Macular Research Initiative, Polly and Michael Smith Foundation, Max Factor Family Foundation, Skirball Foundation, Lincy Foundation, Iris and B. Gerald Cantor Foundation, and unrestricted grant from Research to Prevent Blindness. National Institute on Aging grant (AG006168) to SMJ. National Institutes of Mental Health grant (R01-MH-085801) awarded to MPV.
Abbreviations
- ABI
Applied Biosystems
- ARPE-19
Retinal pigmented epithelium cell line
- ATP
Adenosine triphosphate
- CFH
Complement factor H
- C1s
Complement component 1, s subcomponent
- C3
Complement component 3
- C4B
Complement component 4B
- DMEM
Dulbecco’s modified Eagle’s medium
- DNA
Deoxyribonucleic acid
- ECAR
Extracellular acidification rate
- EDTA
Ethylenediaminetetracetic acid
- ETC
Electron transport chain
- FCCP
Carbonyl Cyanide 4-trifluoromethoxy-phenylhydrazone
- μM
MicroMolar
- MT-CYB
Mitochondria encoded cytochrome B
- MT-ND1
Mitochondria encoded NADH dehydrogenase 1
- MT-ND3
Mitochondria encoded NADH dehydrogenase 3
- MT-ND5
Mitochondria encoded NADH dehydrogenase 5
- MT-CO1
Mitochondria encoded cytochrome oxidase 1
- MT-CO2
Mitochondria encoded cytochrome oxidase 2
- MT-CO3
Mitochondria encoded cytochrome oxidase 3
- OCR
Oxygen consumption rate
- OXPHOS
Oxidative phosphorylation
- Q-PCR
Quantitative polymerase chain reaction
- SEM
Standard error mean
- SNPs
Single nucleotide polymorphisms
- UCLA
University of California, Los Angeles
- VO2max
Maximal oxygen uptake
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallace DC, Lott MT, Procaccio V. Mitochondrial genes in degenerative diseases, cancer and aging. 5. Churchill Livingstonr Elsevier; Philadelphis, PA: 2007. [Google Scholar]
- 2.Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. Mitochondrial DNA mutations in diseases of energy metabolism. J Bioenerg Biomembr. 1994;26:241–250. doi: 10.1007/BF00763096. [DOI] [PubMed] [Google Scholar]
- 4.McFarland R, Turnbull DM. Batteries not included: diagnosis and management of mitochondrial disease. J Intern Med. 2009;265:210–228. doi: 10.1111/j.1365-2796.2008.02066.x. [DOI] [PubMed] [Google Scholar]
- 5.Bellizzi D, Taverna D, D’Aquila P, De Blasi S, De Benedictis G. Mitochondrial DNA variability modulates mRNA and intra-mitochondrial protein levels of HSP60 and HSP75: experimental evidence from cybrid lines. Cell Stress Chaperones. 2009;14:265–271. doi: 10.1007/s12192-008-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellizzi D, Cavalcante P, Taverna D, Rose G, Passarino G, Salvioli S, Franceschi C, De Benedictis G. Gene expression of cytokines and cytokine receptors is modulated by the common variability of the mitochondrial DNA in cybrid cell lines. Genes Cells. 2006;11:883–891. doi: 10.1111/j.1365-2443.2006.00986.x. [DOI] [PubMed] [Google Scholar]
- 7.Hudson G, Carelli V, Spruijt L, Gerards M, Mowbray C, Achilli A, Pyle A, Elson J, Howell N, La Morgia C, Valentino ML, Huoponen K, Savontaus ML, Nikoskelainen E, Sadun AA, Salomao SR, Belfort R, Jr, Griffiths P, Man PY, de Coo RF, Horvath R, Zeviani M, Smeets HJ, Torroni A, Chinnery PF. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet. 2007;81:228–233. doi: 10.1086/519394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strauss KA, DuBiner L, Simon M, Zaragoza M, Sengupta PP, Li P, Narula N, Dreike S, Platt J, Procaccio V, Ortiz-Gonzalez XR, Puffenberger EG, Kelley RI, Morton DH, Narula J, Wallace DC. Severity of cardiomyopathy associated with adenine nucleotide translocator-1 deficiency correlates with mtDNA haplogroup. Proc Natl Acad Sci U S A. 2013;110:3453–3458. doi: 10.1073/pnas.1300690110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, Schmechel DE, Murali Doraiswamy P, Gilbert JR, Haines JL, Vance JM, Pericak-Vance MA. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 10.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Mastaglia F, Stajich JM, McLaurin AC, Middleton LT, Scott BL, Schmechel DE, Pericak-Vance MA, Vance JM. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huerta C, Castro MG, Coto E, Blazquez M, Ribacoba R, Guisasola LM, Salvador C, Martinez C, Lahoz CH, Alvarez V. Mitochondrial DNA polymorphisms and risk of Parkinson’s disease in Spanish population. J Neurol Sci. 2005;236:49–54. doi: 10.1016/j.jns.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Coskun P, Wyrembak J, Schriner S, Chen HW, Marciniack C, Laferla F, Wallace DC. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Moreno M, Soto-Hermida A, Oreiro N, Pertega S, Fenandez-Lopez C, Rego-Perez I, Blanco FJ. Mitochondrial haplogroups define two phenotypes of osteoarthritis. Frontiers Physiol. 2012;3:129. doi: 10.3389/fphys.2012.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Achilli A, Olivieri A, Pala M, Hooshiar Kashani B, Carossa V, Perego UA, Gandini F, Santoro A, Battaglia V, Grugni V, Lancioni H, Sirolla C, Bonfigli AR, Cormio A, Boemi M, Testa I, Semino O, Ceriello A, Spazzafumo L, Gadaleta MN, Marra M, Testa R, Franceschi C, Torroni A. Mitochondrial DNA backgrounds might modulate diabetes complications rather than T2DM as a whole. PLoS ONE. 2011;6:e21029. doi: 10.1371/journal.pone.0021029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czarnecka AM, Bartnik E. The role of the mitochondrial genome in ageing and carcinogenesis. J Aging Res. 2011;2011:136435. doi: 10.4061/2011/136435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzfeld JJ, LaVeist TA, Gaston-Johansson FG. Racial/ethnic disparities in the prevalence of selected chronic diseases among US Air Force members, 2008. Preventing chronic disease. 2012;9:E112. doi: 10.5888/pcd9.110136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mensah GA, Mokdad AH, Ford ES, Greenlund KJ, Croft JB. State of disparities in cardiovascular health in the United States. Circulation. 2005;111:1233–1241. doi: 10.1161/01.CIR.0000158136.76824.04. [DOI] [PubMed] [Google Scholar]
- 18.Kurian AK, Cardarelli KM. Racial and ethnic differences in cardiovascular disease risk factors: a systematic review. Ethnicity & Disease. 2007;17:143–152. [PubMed] [Google Scholar]
- 19.Peek ME, Cargill A, Huang ES. Diabetes health disparities: a systematic review of health care interventions. Medical care research and review: MCRR. 2007;64:101S–156S. doi: 10.1177/1077558707305409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldman CH, Hiraki LT, Liu J, Fischer MA, Solomon DH, Alarcon GS, Winkelmayer WC, Costenbader KH. Epidemiology and sociodemographics of systemic lupus erythematosus and lupus nephritis among US adults with Medicaid coverage, 2000–2004. Arthritis Rheum. 2013;65:753–763. doi: 10.1002/art.37795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiraki LT, Feldman CH, Liu J, Alarcon GS, Fischer MA, Winkelmayer WC, Costenbader KH. Prevalence, incidence, and demographics of systemic lupus erythematosus and lupus nephritis from 2000 to 2004 among children in the US Medicaid beneficiary population. Arthritis Rheum. 2012;64:2669–2676. doi: 10.1002/art.34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MR Manly JJ. Ethnic differences in dementia and Alzheimer’s disease. Natl Acad Press; Washington, DC: 2004. [Google Scholar]
- 23.Dilworth-Anderson P, Hendrie HC, Manly JJ, Khachaturian AS, Fazio S. Diagnosis and assessment of Alzheimer’s disease in diverse populations. Alzheimer’s & dementia: J Alzheimer’s Assoc. 2008;4:305–309. doi: 10.1016/j.jalz.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Potter GG, Plassman BL, Burke JR, Kabeto MU, Langa KM, Llewellyn DJ, Rogers MA, Steffens DC. Cognitive performance and informant reports in the diagnosis of cognitive impairment and dementia in African Americans and whites. Alzheimer’s & dementia: J Alzheimer’s Assoc. 2009;5:445–453. doi: 10.1016/j.jalz.2009.04.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiley KL, Treadwell E, Manigaba K, Word B, Lyn-Cook BD. Ethnic differences in DNA methyltransferases expression in patients with systemic lupus erythematosus. J Clinical Immunol. 2013;33:342–348. doi: 10.1007/s10875-012-9803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Duran A, Pacheu-Grau D, Martinez-Romero I, Lopez-Gallardo E, Lopez-Perez MJ, Montoya J, Ruiz-Pesini E. Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber’s hereditary optic neuropathy. Biochim Biophys Acta. 2012;1822:1216–1222. doi: 10.1016/j.bbadis.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Miceli MV, Jazwinski SM. Nuclear gene expression changes due to mitochondrial dysfunction in ARPE-19 cells: implications for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2005;46:1765–1773. doi: 10.1167/iovs.04-1327. [DOI] [PubMed] [Google Scholar]
- 28.Chomyn A. Platelet-mediated transformation of human mitochondrial DNA-less cells. Academic Press, Inc.; Salt Lake City, UT: 1996. [DOI] [PubMed] [Google Scholar]
- 29.Udar N, Atilano SR, Memarzadeh M, Boyer D, Chwa M, Lu S, Maguen B, Langberg J, Coskun P, Wallace DC, Nesburn AB, Khatibi N, Hertzog D, Le K, Hwang DW, Kenney MC. Mitochondrial DNA haplogroups associated with Age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:2966–2974. doi: 10.1167/iovs.08-2646. [DOI] [PubMed] [Google Scholar]
- 30.Kenney MC, Chwa M, Atilano SR, Pavlis JM, Falatoonzadeh P, Ramirez C, Malik D, Hsu T, Woo G, Soe K, Nesburn AB, Boyer DS, Kuppermann BD, Jazwinski SM, Miceli MV, Wallace DC, Udar N. Mitochondrial DNA variants mediate energy production and expression levels for CFH, C3, and EFEMP1 genes: Implications for age-related macular degeneration. PLoS ONE. 2013;8:e54339–e54348. doi: 10.1371/journal.pone.0054339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott RA, Fuku N, Onywera VO, Boit M, Wilson RH, Tanaka M, W HG, Pitsiladis YP. Mitochondrial haplogroups associated with elite Kenyan athlete status. Med Sci Sports Exercise. 2009;41:123–128. doi: 10.1249/MSS.0b013e31818313a2. [DOI] [PubMed] [Google Scholar]
- 32.Scott RA, Wilson RH, Goodwin WH, Moran CN, Georgiades E, Wolde B, Pitsiladis YP. Mitochondrial DNA lineages of elite Ethiopian athletes, Comparative biochemistry and physiology. Part B, Biochemistry & Molecular Biology. 2005;140:497–503. doi: 10.1016/j.cbpc.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Schnabelrauch M, Scharnweber D, Schiller J. Sulfated glycosaminoglycans as promising artificial extracellular matrix components to improve the regeneration of tissues. Curr Med Chem. 2013;20:2501–2523. doi: 10.2174/0929867311320200001. [DOI] [PubMed] [Google Scholar]
- 34.Rho JH, Zhang W, Murali M, Roehrl MH, Wang JY. Human proteins with affinity for dermatan sulfate have the propensity to become autoantigens. Am J Pathol. 2011;178:2177–2190. doi: 10.1016/j.ajpath.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen C, Otto E, Kuhlemann K, Forster G, Kahaly GJ. Glycosaminoglycans in autoimmunity. Clinical and Exp Rheum. 1996;14(Suppl 15):S59–67. [PubMed] [Google Scholar]
- 36.De Muro P, Faedda R, Formato M, Re F, Satta A, Cherchi GM, Carcassi A. Urinary glycosaminoglycans in patients with systemic lupus erythematosus. Clinical and Exp Rheum. 2001;19:125–130. [PubMed] [Google Scholar]
- 37.Campo GM, Avenoso A, Campo S, D’Ascola A, Traina P, Sama D, Calatroni A. Purified human plasma glycosaminoglycans reduced NF-kappaB activation, pro-inflammatory cytokine production and apoptosis in LPS-treated chondrocytes. Innate Immunity. 2008;14:233–246. doi: 10.1177/1753425908094725. [DOI] [PubMed] [Google Scholar]
- 38.Haylock-Jacobs S, Keough MB, Lau L, Yong VW. Chondroitin sulphate proteoglycans: extracellular matrix proteins that regulate immunity of the central nervous system. Autoimmun Rev. 2011;10:766–772. doi: 10.1016/j.autrev.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 39.Vallieres M, du Souich P. Modulation of inflammation by chondroitin sulfate. Osteoarthritis Cartilage. 2010;18(Suppl 1):S1–6. doi: 10.1016/j.joca.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 40.du Souich P, Garcia AG, Verges J, Montell E. Immunomodulatory and anti-inflammatory effects of chondroitin sulphate. Journal of Cellular and Molecular Medicine. 2009;13:1451–1463. doi: 10.1111/j.1582-4934.2009.00826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egea J, Garcia AG, Verges J, Montell E, Lopez MG. Antioxidant, antiinflammatory and neuroprotective actions of chondroitin sulfate and proteoglycans. Osteoarthritis Cartilage. 2010;18(Suppl 1):S24–27. doi: 10.1016/j.joca.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Mizumoto S, Ikegawa S, Sugahara K. Human genetic disorders caused by mutations in genes encoding biosynthetic enzymes for sulfated glycosaminoglycans. J Biol Chem. 2013;288:10953–10961. doi: 10.1074/jbc.R112.437038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiser P, Qian Y, Pan J, Zhou X, Lu H, Studelska DR, Shih FF, Zhang L. Activated contact system and abnormal glycosaminoglycans in lupus and other auto- and non-autoimmune diseases. Progress in Mole Biol and Translational Sci. 2010;93:443–472. doi: 10.1016/S1877-1173(10)93019-6. [DOI] [PubMed] [Google Scholar]
- 44.Robson MG, Walport MJ. Pathogenesis of systemic lupus erythematosus (SLE) Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2001;31:678–685. doi: 10.1046/j.1365-2222.2001.01147.x. [DOI] [PubMed] [Google Scholar]
- 45.Lood C, Gullstrand B, Truedsson L, Olin AI, Alm GV, Ronnblom L, Sturfelt G, Eloranta ML, Bengtsson AA. C1q inhibits immune complex-induced interferon-alpha production in plasmacytoid dendritic cells: a novel link between C1q deficiency and systemic lupus erythematosus pathogenesis. Arthritis Rheum. 2009;60:3081–3090. doi: 10.1002/art.24852. [DOI] [PubMed] [Google Scholar]
- 46.Inoue N, Saito T, Masuda R, Suzuki Y, Ohtomi M, Sakiyama H. Selective complement C1s deficiency caused by homozygous four-base deletion in the C1s gene. Hum Genet. 1998;103:415–418. doi: 10.1007/s004390050843. [DOI] [PubMed] [Google Scholar]
- 47.Ohno-Matsui K. Parallel findings in age-related macular degeneration and Alzheimer’s disease. Prog Retin Eye Res. 2011;30:217–238. doi: 10.1016/j.preteyeres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Moreno-Navarrete JM, Martinez-Barricarte R, Catalan V, Sabater M, Gomez-Ambrosi J, Ortega FJ, Ricart W, Bluher M, Fruhbeck G, Rodriguez de Cordoba S, Fernandez-Real JM. Complement factor H is expressed in adipose tissue in association with insulin resistance. Diabetes. 2010;59:200–209. doi: 10.2337/db09-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujita T, Hemmi S, Kajiwara M, Yabuki M, Fuke Y, Satomura A, Soma M. Complement-mediated chronic inflammation is associated with diabetic microvascular complication. Diabetes/metabolism Research and Reviews. 2012 doi: 10.1002/dmrr.2380. [DOI] [PubMed] [Google Scholar]
- 50.Ferreira IL, Nascimento MV, Ribeiro M, Almeida S, Cardoso SM, Grazina M, Pratas J, Santos MJ, Januario C, Oliveira CR, Rego AC. Mitochondrial-dependent apoptosis in Huntington’s disease human cybrids. Exp Neurol. 2010;222:243–255. doi: 10.1016/j.expneurol.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Jun AS, Trounce IA, Brown MD, Shoffner JM, Wallace DC. Use of transmitochondrial cybrids to assign a complex I defect to the mitochondrial DNA-encoded NADH dehydrogenase subunit 6 gene mutation at nucleotide pair 14459 that causes Leber hereditary optic neuropathy and dystonia. Mol Cell Biol. 1996;16:771–777. doi: 10.1128/mcb.16.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown MD, Trounce IA, Jun AS, Allen JC, Wallace DC. Functional analysis of lymphoblast and cybrid mitochondria containing the 3460, 11778, or 14484 Leber’s hereditary optic neuropathy mitochondrial DNA mutation. J Biol Chem. 2000;275:39831–39836. doi: 10.1074/jbc.M006476200. [DOI] [PubMed] [Google Scholar]
- 53.Arning L, Haghikia A, Taherzadeh-Fard E, Saft C, Andrich J, Pula B, Hoxtermann S, Wieczorek S, Akkad DA, Perrech M, Gold R, Epplen JT, Chan A. Mitochondrial haplogroup H correlates with ATP levels and age at onset in Huntington disease. J Mol Med (Berl) 2010;88:431–436. doi: 10.1007/s00109-010-0589-2. [DOI] [PubMed] [Google Scholar]
- 54.Murphy GE, Xu D, Liew FY, McInnes IB. Role of interleukin 33 in human immunopathology. Ann Rheum Dis. 2010;69(Suppl 1):i43–47. doi: 10.1136/ard.2009.120113. [DOI] [PubMed] [Google Scholar]
- 55.Liew FY. IL-33: a Janus cytokine. Ann Rheum Dis. 2012;71(Suppl 2):i101–104. doi: 10.1136/annrheumdis-2011-200589. [DOI] [PubMed] [Google Scholar]
- 56.Kuchler AM, Pollheimer J, Balogh J, Sponheim J, Manley L, Sorensen DR, De Angelis PM, Scott H, Haraldsen G. Nuclear interleukin-33 is generally expressed in resting endothelium but rapidly lost upon angiogenic or proinflammatory activation. Am J Pathol. 2008;173:1229–1242. doi: 10.2353/ajpath.2008.080014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller AM, Xu D, Asquith DL, Denby L, Li Y, Sattar N, Baker AH, McInnes IB, Liew FY. IL-33 reduces the development of atherosclerosis. J Exp Med. 2008;205:339–346. doi: 10.1084/jem.20071868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rank MA, Kobayashi T, Kozaki H, Bartemes KR, Squillace DL, Kita H. IL-33-activated dendritic cells induce an atypical TH2-type response. J Allergy Clin Immunol. 2009;123:1047–1054. doi: 10.1016/j.jaci.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, Bouche G, Girard JP. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci U S A. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leung BP, Xu D, Culshaw S, McInnes IB, Liew FY. A novel therapy of murine collagen-induced arthritis with soluble T1/ST2. J Immunol. 2004;173:145–150. doi: 10.4049/jimmunol.173.1.145. [DOI] [PubMed] [Google Scholar]
- 61.Seidelin JB, Bjerrum JT, Coskun M, Widjaya B, Vainer B, Nielsen OH. IL-33 is upregulated in colonocytes of ulcerative colitis. Immunol Letters. 2010;128:80–85. doi: 10.1016/j.imlet.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Kurowska-Stolarska M, Stolarski B, Kewin P, Murphy G, Corrigan CJ, Ying S, Pitman N, Mirchandani A, Rana B, van Rooijen N, Shepherd M, McSharry C, McInnes IB, Xu D, Liew FY. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183:6469–6477. doi: 10.4049/jimmunol.0901575. [DOI] [PubMed] [Google Scholar]
- 63.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 64.Smithgall MD, Comeau MR, Yoon BR, Kaufman D, Armitage R, Smith DE. IL-33 amplifies both Th1-and Th2-type responses through its activity on human basophils, allergen-reactive Th2 cells, iNKT and NK cells. International Immunology. 2008;20:1019–1030. doi: 10.1093/intimm/dxn060. [DOI] [PubMed] [Google Scholar]
- 65.Komai-Koma M, Xu D, Li Y, McKenzie AN, McInnes IB, Liew FY. IL-33 is a chemoattractant for human Th2 cells. Eur J of Immunol. 2007;37:2779–2786. doi: 10.1002/eji.200737547. [DOI] [PubMed] [Google Scholar]
- 66.Mirchandani AS, Salmond RJ, Liew FY. Interleukin-33 and the function of innate lymphoid cells. Trends in Immunol. 2012;33:389–396. doi: 10.1016/j.it.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 67.Milovanovic M, Volarevic V, Radosavljevic G, Jovanovic I, Pejnovic N, Arsenijevic N, Lukic ML. IL-33/ST2 axis in inflammation and immunopathology. Immunologic Res. 2012;52:89–99. doi: 10.1007/s12026-012-8283-9. [DOI] [PubMed] [Google Scholar]
- 68.Ali S, Mohs A, Thomas M, Klare J, Ross R, Schmitz ML, Martin MU. The dual function cytokine IL-33 interacts with the transcription factor NF-kappaB to dampen NF-kappaB-stimulated gene transcription. J Immunol. 2011;187:1609–1616. doi: 10.4049/jimmunol.1003080. [DOI] [PubMed] [Google Scholar]
- 69.Lillien L, Cepko C. Control of proliferation in the retina: temporal changes in responsiveness to FGF and TGF alpha. Development. 1992;115:253–266. doi: 10.1242/dev.115.1.253. [DOI] [PubMed] [Google Scholar]
- 70.Sagar SM, Edwards RH, Sharp FR. Epidermal growth factor and transforming growth factor alpha induce c-fos gene expression in retinal Muller cells in vivo. J Neurosci Res. 1991;29:549–559. doi: 10.1002/jnr.490290416. [DOI] [PubMed] [Google Scholar]
- 71.Fassio JB, Brockman EB, Jumblatt M, Greaton C, Henry JL, Geoghegan TE, Barr C, Schultz GS. Transforming growth factor alpha and its receptor in neural retina. Invest Ophthalmol Vis Sci. 1989;30:1916–1922. [PubMed] [Google Scholar]
- 72.Brissenden JE, Derynck R, Francke U. Mapping of transforming growth factor alpha gene on human chromosome 2 close to the breakpoint of the Burkitt’s lymphoma t(2;8) variant translocation. Cancer Res. 1985;45:5593–5597. [PubMed] [Google Scholar]
- 73.Ellis DL, Kafka SP, Chow JC, Nanney LB, Inman WH, McCadden ME, King LE., Jr Melanoma, growth factors, acanthosis nigricans, the sign of Leser-Trelat, and multiple acrochordons. A possible role for alpha-transforming growth factor in cutaneous paraneoplastic syndromes. N Engl J Med. 1987;317:1582–1587. doi: 10.1056/NEJM198712173172506. [DOI] [PubMed] [Google Scholar]
- 74.Letra A, Fakhouri W, Fonseca RF, Menezes R, Kempa I, Prasad JL, McHenry TG, Lidral AC, Moreno L, Murray JC, Daack-Hirsch S, Marazita ML, Castilla EE, Lace B, Orioli IM, Granjeiro JM, Schutte BC, Vieira AR. Interaction between IRF6 and TGFA genes contribute to the risk of nonsyndromic cleft lip/palate. PLoS ONE. 2012;7:e45441. doi: 10.1371/journal.pone.0045441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vieira AR. Association between the transforming growth factor alpha gene and nonsyndromic oral clefts: a HuGE review. Am J Epidemiol. 2006;163:790–810. doi: 10.1093/aje/kwj103. [DOI] [PubMed] [Google Scholar]
- 76.Vollebergh MA, Kappers I, Klomp HM, Buning-Kager JC, Korse CM, Hauptmann M, de Visser KE, van den Heuvel MM, Linn SC. Ligands of epidermal growth factor receptor and the insulin-like growth factor family as serum biomarkers for response to epidermal growth factor receptor inhibitors in patients with advanced non-small cell lung cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2010;5:1939–1948. doi: 10.1097/JTO.0b013e3181f77a39. [DOI] [PubMed] [Google Scholar]
- 77.Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–2515. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 78.Resnick L, Fennell M. Targeting JNK3 for the treatment of neurodegenerative disorders. Drug Discovery Today. 2004;9:932–939. doi: 10.1016/S1359-6446(04)03251-9. [DOI] [PubMed] [Google Scholar]
- 79.Abdelli S, Puyal J, Bielmann C, Buchillier V, Abderrahmani A, Clarke PG, Beckmann JS, Bonny C. JNK3 is abundant in insulin-secreting cells and protects against cytokine-induced apoptosis. Diabetologia. 2009;52:1871–1880. doi: 10.1007/s00125-009-1431-7. [DOI] [PubMed] [Google Scholar]
- 80.Yoon SO, Park DJ, Ryu JC, Ozer HG, Tep C, Shin YJ, Lim TH, Pastorino L, Kunwar AJ, Walton JC, Nagahara AH, Lu KP, Nelson RJ, Tuszynski MH, Huang K. JNK3 perpetuates metabolic stress induced by Abeta peptides. Neuron. 2012;75:824–837. doi: 10.1016/j.neuron.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fernandes KA, Harder JM, Fornarola LB, Freeman RS, Clark AF, Pang IH, John SW, Libby RT. JNK2 and JNK3 are major regulators of axonal injury-induced retinal ganglion cell death. Neurobiol Dis. 2012;46:393–401. doi: 10.1016/j.nbd.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


