Abstract
Lung cancer is the most common human malignancy and leads to about one-third of all cancer-related deaths. Lung adenocarcinomas harboring KRAS mutations, in contrast to those with EGFR and EML4-ALK mutations, have not yet been successfully targeted. Here, we describe a combination therapy for treating these malignancies using two agents: a lipophilic bisphosphonate and rapamycin. This drug combination is much more effective than either agent acting alone in the KRAS G12D induced mouse lung model. Lipophilic bisphosphonates inhibit both farnesyl and geranylgeranyldiphosphate synthases, effectively blocking prenylation of the KRAS and other small G-proteins critical for tumor growth and cell survival. Bisphosphonate treatment of cells initiated autophagy but was ultimately unsuccessful and led to p62 accumulation and concomitant NF-κB activation, resulting in dampened efficacy in vivo. However, we found that rapamycin, in addition to inhibiting the mTOR pathway, facilitated autophagy and prevented p62 accumulation-induced NF-κB activation and tumor cell proliferation. Overall, these results suggest that using lipophilic bisphosphonates in combination with rapamycin may provide an effective strategy for targeting lung adenocarcinomas harboring KRAS mutations.
Introduction
Lung adenocarcinomas account for about 50% of all non-small cell lung cancers (NSCLC), the most common type of human malignancy and a leading cause of cancer-related mortality worldwide. There has been rapid progress in developing targeted therapies for lung adenocarcinomas over the last decade, including gefitinib and erlotinib, which target EGF receptor mutations (1, 2) and crizotinib, which targets the transforming EML4-ALK fusion gene (3). However, KRAS mutations, which are commonly found in smokers and Caucasian patients, are not effectively targeted by currently available therapeutics and have low survival rates, as well as frequent drug resistance (4). KRAS mutations at amino acid positions 12, 13 or 61 are widely found in human pancreatic, thyroid, lung and colorectal cancers (5). They typically impair GTPase activity and lead to constitutive activation of downstream signaling pathways. It is therefore difficult to develop potent KRAS mutant-specific inhibitors that can directly restore intrinsic GTPase activity, although specific inhibitors of KRAS G12C have recently been reported (6), as have attempts to interfere with mutated KRAS function by altering its membrane localization; inhibiting its downstream effectors, as well as searching for synthetic lethality (7, 8).
Farnesylation and correct membrane localization are essential for the in vivo biological activity of RAS proteins (9, 10). CAAX peptido-mimetics; farnesyltransferase and geranylgeranyltransferase inhibitors (FTI/GGTIs); farnesylthiosalicylic acid (Salirasib) –which mimics farnesylcysteine, as well as small molecule inhibitors of KRAS-PDEδ interactions have all been developed to circumvent KRAS post-translational modification and membrane anchoring (7, 11–13). Knockout mouse models support the notion that disruption of protein prenylation severely impairs lung cancer development induced by KRAS mutations (14, 15). However, there has been little success in clinical trials with these small molecule inhibitors, probably due to the existence of “cross-prenylation”(16), in which a FTI can fail due to alternative KRAS geranylgeranylation, suggesting the need for combination therapies. In addition to the protein-prenyltransferase inhibitors, there is interest in the development of compounds, such as bisphosphonates (see fig. S1) that directly inhibit the biosynthesis of the two prenyldiphosphate substrates: farnesyldiphosphate (FPP) and geranylgeranyldiphosphate (GGPP), catalyzed by the respective synthases, FPP synthase (FPPS) and GGPP synthase (GGPPS). Bisphosphonates are used to treat a variety of bone resorption diseases and function by blocking FPPS activity in osteoclasts. In previous work, we developed “lipophilic” bisphosphonates in which hydrophobic side-chains were added to a pyridinium bisphosphonate. These compounds do not bind to bone mineral, but maintain inhibitory activity against both FPPS as well as GGPPS(17), both of which can provide membrane anchoring 15 and 20-carbon isoprenoid chains for Kras post-translational modification.
Impaired protein processing, folding and trafficking usually induce ER stress and autophagy if the protective unfolded protein response (UPR) is not sufficient to clear the incorrectly processed proteins (18) and indeed, inhibitors of FPP and GGPP biosynthesis such as bisphosphonates and statins have been reported to initiate autophagy in cells (19, 20). The role of autophagy in tumorigenesis has been considered as a “double-edged sword”, since it can either inhibit tumor initiation at an early stage or get adopted by tumor cells as survival mechanisms at an advanced stage (21). In this work, we sought a combination therapy that would ultimately stop KRAS prenylation and temporally modulate autophagy as an effective two-pronged approach against lung adenocarcinomas.
Results
Bisphosphonates inhibit FPPS and GGPPS activity
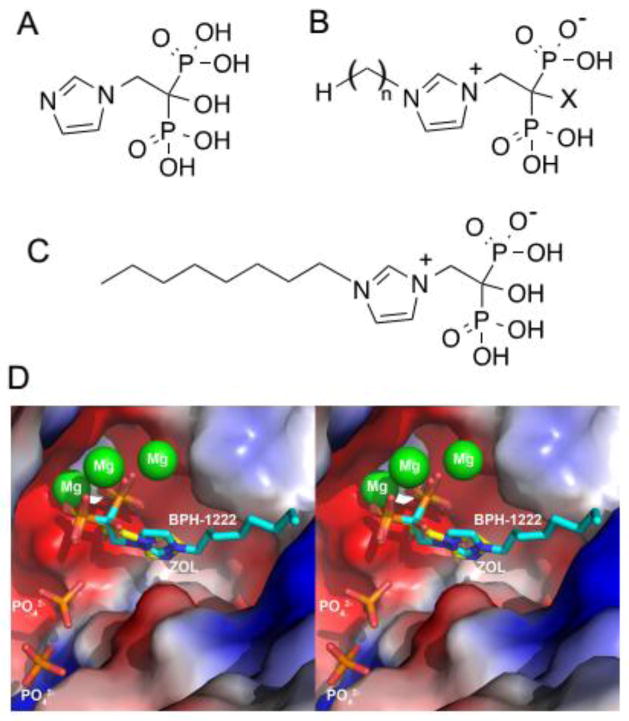
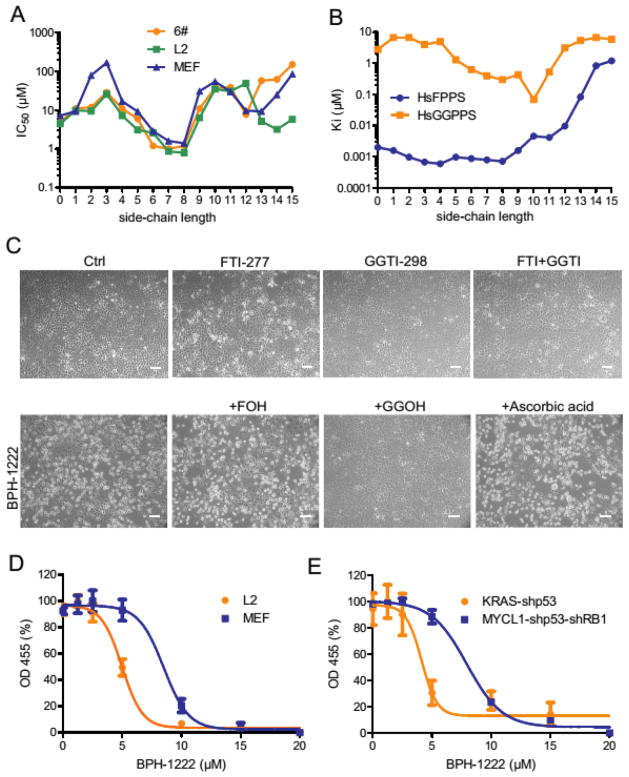
We tested a library of 30 synthetic analogs of zoledronate (Fig. 1) for growth inhibition of two KRAS mutant cell lines (6#, L2) and of control mouse embryonic fibroblasts (MEF). We found most anti-growth activity with BPH-1222, a zoledronate analog having a C8 side-chain and a 1-OH group (~1 μM IC50, Fig. 2A and fig. S2A, B). Compounds with very short or very long chains inhibited growth the least while BPH-1222 and other intermediate chain length compounds had the most activity. In vitro inhibitory activities against human FPPS (Ki as low as ~1 nM) correlated well with activities in inhibiting cell growth, suggesting FPPS as one possible in vivo target (Fig. 2B and table S1 and fig. S2C). To uncover how BPH-1222 binds and inhibits FPPS, we determined the structure of the BPH-1222-FPPS complex using single crystal X-ray crystallography. As shown in Fig. 1D, the bisphosphonate, head-group and imidazolium ring bind to human FPPS in essentially the same manner as does zoledronate (shown superimposed) with a 0.45 Å rmsd (root mean square deviation) for the common atoms (see table S2 for a summary of structural and refinement statistics).
Fig. 1. BPH-1222 and zoledronate bind to the same site of FPPS.
(A) Zoledronate. (B) A library of zoledronate analogs, side-chain length n= 0–15, X= H or OH. (C) BPH-1222. (D) Structure of BPH-1222 binding to FPPS was determined by single crystal X-ray crystallography. Zoledronate binding is shown superimposed with BPH-1222 (zoledronate in yellow and BPH-1222 in cyan).
Fig. 2. Bisphosphonates suppress cell growth by inhibiting FPPS and GGPPS.
(A) IC50 of zoledronate and its hydroxy analogs (see fig. S2 for desoxy analogs) was determined in cell lines derived from KRAS-shp53 mouse lung cancer model (6# and L2) and control mouse embryonic fibroblasts (MEF) using MTT assay. (B) Ki of compounds in (A) was measured in vitro against human FPPS or GGPPS. (C) Mouse lung cancer cells (6#) were treated with single drug (FTI-277, 15 μM; GGTI-298, 15 μM; BPH-1222, 10 μM; FOH, 10 μM; GGOH, 10 μM; ascorbic acid, 50 μM) or drug combinations as indicated for 48 h, and images were taken under phase-contrast microscope. Scale bars, 100 μm. (D) Mouse lung cancer cells (L2) and embryonic fibroblasts (MEF, matched genetic background), or (E) mouse embryonic fibroblasts transformed by KRAS-shp53 and MYCL1-shp53-shRB1 were treated with different concentrations of BPH-1222 for 3 days, and cell survival was measured using Cell Proliferation Reagent WST-1 from Roche. All data were collected from two independent experiments (each has triplicate wells) and raw ODs were normalized to a common scale with GraphPad Prism. Data are presented as means ± SE, n = 6.
Moreover, the lipophilic bisphosphonates also inhibit human GGPPS (Fig. 2B and fig. S2C and table S1). Zoledronate, as well as other bisphosphonates such as risedronate and alendronate, all inhibit FPPS, but do not inhibit GGPPS, since the latter lacks the third Asp required for [Mg2+] coordination to the bisphosphonate (22). With the lipophilic bisphosphonates binding to GGPPS, the loss of the additional binding interaction due to the absence of the third Asp residue is likely made up for, by increased van der Waals interactions in the hydrophobic tunnel that normally houses the allylic side-chains of the growing isoprenoid diphosphate products. The intermediate chain length species possess the best inhibitory activity for GGPPS, a Ki of ~300 nM for BPH-1222. There is little difference in GGPPS enzyme inhibition activity between the 1-hydroxy and 1-desoxy species (table S1), so in this work we focused on BPH-1222, the N-octyl analog of zoledronate, over its desoxy analog in order to eliminate the possibility of a retro-Michael reaction (in which the hydrophobic side-chain would be lost) in vivo.
As can be seen in table S1, the Kis for FPPS inhibition by zoledronate (ID 1) and BPH-1222 (ID 9) are quite similar (1–2 nM), but the IC50 values for cell growth inhibition are ~10–20x lower with BPH-1222. This likely correlates with the lower GGPPS Ki values (~3 μM for zoledronate; 300 nM for BPH-1222) and with the higher clogP (the computed logarithm of the oil/water partition coefficient) value for BPH-1222 (0.25) versus zoledronate (−3.9).
Inhibition of protein farnesylation (with FTI-277), geranylgeranylation (with GGTI-298) or both had little effect on cell proliferation (Fig. 2C, and fig S2D, E). However, there was massive cell death within 3 days with BPH-1222. Supplementation with geranylgeraniol (GGOH), but not farnesol (FOH) or anti-oxidants, substantially rescued cells from bisphosphonate-induced cell death (Fig. 2C). Similar rescue effects are seen with zoledronate (23) as well as the lipophilic pyridinium bisphosphonate BPH-714 ((17)) and with simvastatin, a potent HMG-CoA reductase inhibitor that shuts down all isoprenoid biosynthesis (fig. S2F). As noted by Goffinet et al.(23), the chemical target for zoledronate is FPPS (not GGPPS), but the main “biological effect” of zoledronate involves protein geranylgeranylation. So, dual FPPS/GGPPS-targeting bisphosphonates (such as BPH-1222) are expected to be particularly potent since formation of the FPP substrate for GGPP biosynthesis is blocked by FPPS inhibition, with GGPP production being particularly important for cell survival. While inhibition of FPPS is expected to have effects on diverse metabolic pathways (such as sterol and steroid biosynthesis), these effects cannot be major ones responsible for cell death since the BPH-1222 effects are reversible upon GGOH addition.
We found that BPH-1222 was more toxic to cells harboring KRAS mutations, such as cell lines derived from a mouse model of KRAS-induced lung adenocarcinoma, as well as in mouse embryonic fibroblasts transformed by KRAS in vitro (Fig. 2D, E). Taken together, these results suggest that blocking protein prenylation by lipophilic bisphosphonates can be used as a targeted therapy for cancer cells that carry KRAS mutations, since potentially both FPPS as well as GGPPS can be targeted; the lipophilic bisphosphonates have much better clogP values than do more conventional bisphosphonates; and furthermore, they do not bind to bone mineral, which rapidly removes them from the circulatory system (24).
Bisphosphonates block KRAS prenylation and induces its degradation
“KRAS addiction” has been shown in a mouse lung cancer model using inducible KRAS G12D (25), so given the observation that cells bearing KRAS mutations were more sensitive to bisphosphonate treatment, we were interested in determining if the lipophilic bisphosphonates blocked KRAS prenylation. We used a cell fractionation assay to check KRAS protein prenylation status, since unprenylated KRAS proteins lose their ability to avidly associate with cell membranes and, consequently, appear in the cytosolic fraction (26). BPH-1222 treatment robustly inhibited protein farnesylation as well as protein geranylgeranylation, as indicated by HRAS degradation and RAP1A dislodgement from the membrane fraction respectively (27). Other bisphosphonates, including BPH-714 and zoledronate, showed similar effects, but at higher concentrations (Fig. 3A). As expected, a substantial amount of KRAS was unprenylated and was also partially degraded. There was also a reduction of downstream AKT activity, induced by oncogenic KRAS expression (Fig. 3A). This result was further confirmed with HDJ2, RAP1A and KRAS electrophoretic mobility shift assay (Fig. 3B and table S3) (28). To further verify whether cytosolic KRAS represents the unprenylated form of the protein, we determined the molecular weight of KRAS expressed in U2OS cells after BPH-1222 treatment. As expected, the molecular weights of KRAS proteins from cytosolic fraction (Supernatant) and membrane-bound fraction (Pellet) fit well with unprenylated form and farnesylated form, respectively (see fig. S3A and table S4 for a summary). The effects of a lipophilic bisphosphonate on KRAS function was further tested in a mouse lung cancer (M3L2) and two human pancreatic cancer (Panc-1 and MiaPaCa2) cell lines carrying endogenous KRAS G12D or G12C mutations (Fig. 3C, D). BPH-1222 treatment greatly reduced the amount of GTP-bound KRAS (representing the active form of the KRAS protein), and led to down-regulation of the AKT pathway as well as the activation of apoptosis as shown by increase in Caspase-3 cleavage (Fig. 3C).
Fig. 3. Bisphosphonates inhibit KRAS prenylation and induce ER stress and autophagy.
(A) U2OS cells expressing Flag-KRAS G12D were treated with bisphosphonates (BPH-1222, 10 μM; BPH-714, 10 μM; zoledronate, 20 μM) or chloroquine (30 μM) for 48 h. Cellular distribution of proteins (HRAS, KRAS, and RAP1A) that require prenylation was examined by immunoblotting. p, pellet contains correctly prenylated proteins which bind avidly to membrane; s, supernatant contains unmodified proteins in the cytoplasm. *, HRAS signal left on the membrane. **, autophagy activation determined by the presence of LC3-II (bottom band, PE-conjugated form). (B) Same cells were treated with FTI-277 (15 μM), GGTI-298 (15 μM) or BPH-1222 (5, 10, 15 μM) for 48 h. Cell lysates were separated with 15-cm SDS-PAGE and blotted with indicated antibodies. *, KRAS mobility shift was observed. (C) Mouse lung cancer cells (M3L2) were treated with BPH-1222 for 48 h and analyzed for KRAS, AKT, caspase-3 activation, and LC3 conversion. KRAS-GTP was pulled down from whole cell lysate with RAF-1 RBD beads and immunoblotted with total KRAS antibody. (D) Human pancreatic cancer cells harboring KRAS mutations (Panc-1 and MiaPaCa2) were treated with BPH-1222 (10 μM) for 48 h and analyzed the same way as in (C). (E) BPH-1222 treatment (10 μM) for 1, 2, or 3 days induced ER stress (CHOP, BiP), autophagy (PE-conjugated LC3II) and apoptosis (caspase-3) in mouse lung cancer cells (6#) in a time-dependent manner.
Bisphosphonates enhance ER stress and initiate autophagy
Cancer cells usually exhibit a “stress phenotype” that consists of replicative stress, mitotic stress, metabolic stress, oxidative stress and proteotoxic or ER (endoplasmic reticulum) stress. They are, therefore, vulnerable to further enhancement of these stresses by chemotherapy (29). Bisphosphonates, as demonstrated above, potently block protein prenylation by eliminating the source of isoprenoid chains and lead to the accumulation of incorrectly folded proteins, inducing the so-called Unfolded Protein Response (UPR), or ER stress, as is also observed when cells are treated with HMG-CoA reductase inhibitors (30, 31). As shown in Fig. 3E, treatment of cells with the lipophilic bisphosphonate BPH-1222 potently induced C/EBP homologous protein (CHOP) and Binding immunoglobulin protein (BiP) protein levels, the two main markers for the ER stress response (18). Notably, the ER stress response induced by this bisphosphonate is specifically due to blockade of protein prenylation, since supplementation with GGOH, but not antioxidants, completely abolished the upregulation of CHOP, BiP as well as phospho-PERK induced by the bisphosphonate (fig. S3B). When cells are unable to handle excessive ER stress, they activate autophagy to eliminate incorrectly processed proteins as a defense mechanism for cell survival. In tumor cells treated with BPH-1222, autophagy was induced within 2–3 days, as indicated by the accumulation of the phosphatidylethanolamine (PE)-conjugated form of LC3 (Fig. 3A, E). Similar results have been observed in cells treated with other bisphosphonates and it has been proposed that the combination of inhibitors of autophagy with GGPPS inhibitors might be a therapeutic strategy, since as noted above, autophagy is a defense mechanism (19).
Rapamycin, but not chloroquine, sensitizes tumor cells to bisphosphonates in vivo
Recent reports have indicated that KRAS-driven tumor cells depend on autophagy to help reduce reactive oxygen species (ROS), as well as provide substrates to fuel cell metabolism (32, 33). We therefore tested the hypothesis that blocking autophagy might sensitize tumor cells to bisphosphonate treatment. We first determined the effects on cell survival of treatment with bisphosphonate and an autophagy inhibitor—chloroquine (34). As expected, essentially additive effects of the bisphosphonate and chloroquine were observed in all the lung cancer cell lines derived from the mouse KRAS lung cancer model (with KRAS G12D and p53 knockdown), as well as in human NSCLC cell lines with KRAS mutations (A549 and A427) (Fig. 4A and fig. S4A).
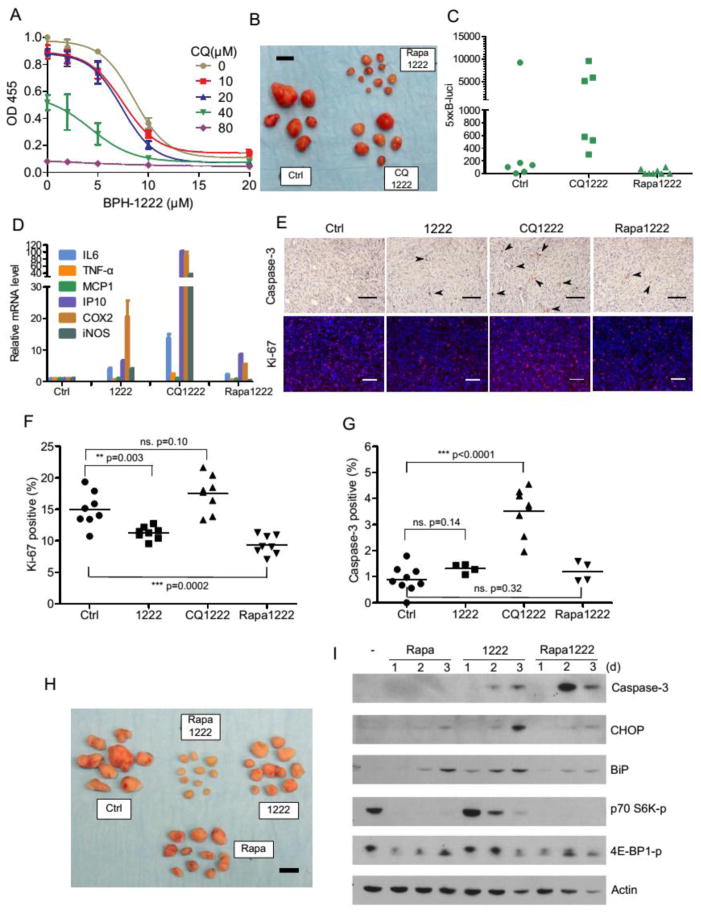
Fig. 4. Rapamycin but not chloroquine sensitizes tumor cells to BPH-1222 treatment in vivo.
(A) Mouse lung cancer cells (6#) were treated with BPH-1222 in the presence of different concentrations of chloroquine (CQ) for 3 days, and cell survival was examined with Cell Proliferation Reagent WST-1. All data were collected from triplicate wells and are presented as means ± SD, n = 3. (B) Mouse lung cancer (L2, infected with 5xκB-luci reporter) syngeneic grafts were treated with BPH-1222 (2 mg/kg) plus either chloroquine (60 mg/kg) or rapamycin (2.5 mg/kg) on alternating days for 3 weeks. Scale bar, 10 mm. NF-κB activity in tumor grafts was examined by in vivo luciferase imaging system (IVIS) immediately after the 3-week treatment (C). (D) Mouse lung cancer cells (6#) were treated with BPH-1222 (10 μM) or combinations with CQ (30 μM) or Rapa (0.1 μM) for 48 h, and NF-κB target genes were examined by quantitative RT-PCR. All data were collected from triplicate wells and are presented as means ± SD, n = 3. (E, F, G) L2 tumor samples treated with different drug combinations were sectioned and immuno-stained with Ki-67 and cleaved caspase-3 antibodies. Caspase-3 positive cells were marked with arrowheads. Scale bars, 100 μm. Percentages of Ki-67 and caspase-3 positive cells were quantified and plotted in (F, G). (H) Mouse lung cancer (L2) syngeneic grafts were treated with BPH-1222 (2 mg/kg), rapamycin (2.5 mg/kg), or the combination for 3 weeks. Scale bar, 10 mm. (I) Mouse lung cancer cells (6#) were treated with BPH-1222 (10 μM), rapamycin (0.1 μM), or the combination for 48 h and examined by immunoblotting.
We next tested this combination in a syngeneic graft model using L2 cells derived from a mouse lung adenocarcinoma (with KRAS G12D and p53 knockdown) developed in C57B mice. Treatment was started 3 weeks after subcutaneous transplantation when the tumor mass was palpable. Surprisingly, 24 days of BPH-1222 plus chloroquine therapy only minimally suppressed tumor growth in animals. In contrast, BPH-1222 plus rapamycin (a potent autophagy inducer) showed substantially better efficacy (Fig. 4B). Our results are more in line with those recently reported in pancreatic cancer (KRAS G12D and p53−/−) development where it was shown that another autophagy inhibitor, hydroxychloroquine, tended to accelerate tumor formation (35). We noticed that tumors treated with BPH-1222 plus chloroquine had elevated NF-κB activity as shown by in vivo imaging (Fig. 4C). This result was further confirmed in cultured cells treated with combination of drugs by quantitative RT-PCR of NF-κB target genes (Fig. 4D). The NF-κB activity was correlated with p62 protein accumulation in the cells, probably due to the blockage of autophagy flux by chloroquine (fig. S4B). Indeed, high p62 protein level in cells is known to activate NF-κB through TRAF6-TBK1 pathway (36). We have reported earlier that high NF-κB activity in tumors may stimulate cell proliferation (37), and this appeared to be the case in tumor samples treated with BPH-1222 plus chloroquine (Fig. 4E, F), although these tumors also showed increased number of apoptotic cells (Fig. 4E, G). We suggest that the elevated NF-κB activity and cell proliferation impaired the efficacy of bisphosphonate to reduce the size of tumors (Fig. 4B).
The results obtained in Fig. 4B and E suggested that a combination therapy using a lipophilic bisphosphonate together with the autophagy inducer rapamycin might be more effective in treating KRAS induced lung adenocarcinomas. We found that in the L2 cell syngeneic graft model, the combination of BPH-1222 with rapamycin was indeed much more effective than either single agent acting alone (Fig. 4H). Additionally, rapamycin also potently blocked phosphorylation of the two mTOR substrates (p70 S6 kinase and 4E-BP1), both of which are important for boosting metabolism in cancer cells (Fig. 4I).
Bisphosphonate and rapamycin combination therapy potently suppresses tumor growth in lung cancer models
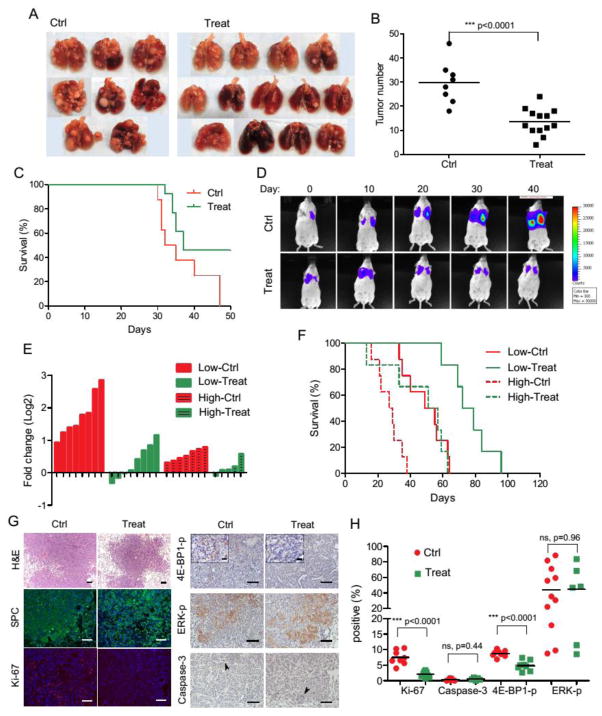
We next tested the BPH-1222 plus rapamycin combination in treating lung adenocarcinoma in both the syngeneic orthotopic graft model and the lentiviral vector-mediated model, which well represent the lung microenvironment. In the orthotopic model, tail veil injection of 2×105 M3L2 cells reproducibly generated massive lung tumors in FVB mice, resulting in a median survival of 33 days. For the treatment group, all mice (n=13) were given BPH-1222 and rapamycin alternately 16 days after the transplantation, for a total of 9 doses of each compound. BPH-1222 has good pharmacokinetic properties (fig. S4C) and this treatment potently suppressed tumor growth, though mice survival was not significantly prolonged (37 d, n = 13, compared to 33.5 d, n= 8 in the control group, p= 0.06) due to internal bleeding in the thoracic cavity in some of the mice under treatment, even those with small tumor burdens (Fig. 5A–C).
Fig. 5. Combination therapy inhibits tumor growth in orthotopic graft model and KRAS-shp53 lentiviral model.
(A–C) Mouse orthotopic grafts were induced by tail vein injection of M3L2 cells. Mice were left untreated or given treatment (BPH-1222 plus rapamycin) 16 days after the inoculation. Lungs were collected when mice reached the IACUC end-point (having severe breathing difficulty) (A). Tumor lesions larger than 1mm in diameter were counted. Ctrl, 29.8 average lesions per mouse, n=8; Treat, 13.5 average lesions per mouse, n=13 (B). Kaplan-Meier curves of mice from control group and treatment group. All surviving mice were collected on day 50. Median survival time: Ctrl, 33.5 d, n=8; Treat, 37 d, n=13 (C). (D–H) Mouse lung adenocarcinomas were induced by intra-tracheal infection with KRAS-shp53 lentiviral vectors. Mice were left untreated or given treatment when luciferase signals from tumors were detectable (Low-Ctrl and Low-Treat groups, luciferase signal: 103–105; High-Ctrl and High-Treat groups, luciferase signal: > 105). (D) Luciferase imaging results of one mouse from Low-Ctrl group and one mouse from Low-Treat group, to show the shrinkage of tumor after the combination therapy. See fig. S5 for results from all mice. (E) Fold changes of luciferase signal after 2-weeks of treatment. Negative value means shrinkage in tumor size. (F) Kaplan-Meier curves of mice from all groups. Median survival time: Low-Ctrl, 55 d, n=8; Low-Treat, 75.5 d, n=6; High-Ctrl, 28 d, n=8; High-Treat, 54 d, n=6. (G) Tumors from Low-Ctrl and Low-Treat groups were sectioned and immuno-stained with different antibodies and quantified in (H). Caspase-3 positive cells were marked with arrowheads. Scale bars, 100 μm (insets in 4E-BP1-p staining: scale bars, 20 μm).
In the lentiviral vector-mediated model, as we reported previously, lung tumors appeared “visible” in the luciferase imaging system at around two months after lentiviral vector infection. In the absence of any treatment, the tumors grew rapidly and all mice (n=8) died from full-blown lung cancer with a median survival of 55 days from when the tumors were first detected by imaging. A combination therapy of BPH-1222 and rapamycin was given to 12 mice around 3 months after lentiviral infection (counted as Day 1 in Fig. 5D and F), for a total of 16 doses of each compound. We monitored the tumor load of each mouse every 9–10 days throughout the whole treatment. The 12 mice were divided into two groups according to their tumor burden at the beginning of treatment—a high burden group (n=6, initial luciferase signal >105) and a low burden group (n=6, initial luciferase signal 103–105). The combination treatment substantially delayed tumor development and prolonged mice survival in both the low burden group (median survival of 75.5 days, compared to 55 days without treatment, p=0.0009), as well as the high burden group (median survival of 54 days, compared to 28 days without treatment, p=0.02, Fig. 5F). Tumor regression was observed in most of the mice during treatment, although tumor size did increase after treatment ceased (Fig. 5D, E and fig. S5). Tumors from treated mice showed a large reduction in cell proliferation as indicated by Ki-67 staining, although apoptotic cells were rarely found (Fig. 5G, H). Unlike the results we reported previously with an IKK2 inhibitor that suppressed tumor progression by reducing ERK signaling (37), there was no significant change of ERK phosphorylation after the bisphosphonate plus rapamycin treatment, in vivo (Fig. 5G, H). Notably, BPH-1222 (either alone or combined with other agents) slightly increased ERK phosphorylation in cell culture conditions (fig. S3C), although its mechanism of action here is unknown. Nevertheless, we tested if a combination with an ERK inhibitor might further improve treatment efficacy. However, ERK inhibitor U0126 increased KRAS protein, c-RAF, MEK and AKTphosphorylation levels under all conditions tested (fig. S3D), perhaps due to the interruption of the ERK negative feedback loop (38).
Interestingly, zoledronate itself has been shown to potentiate the killing of osteosarcoma cells by RAD001 (a rapamycin analogue) (39) and, although less potent than BPH-1222, zoledronate still synergizes with rapamycin in vivo for killing KRAS tumors (fig. S4D, E). This suggests the possibility of a more immediate translation into clinical trials, of zoledronate plus rapamycin (or an analog) for lung adenocarcinoma, given that both drugs have been prescribed for many years with good safety records.
Discussion
RAS mutations (including HRAS, KRAS and NRAS) are commonly found in a variety of human cancers including lung, colon and pancreatic cancers (5), and in this study, we investigated the efficacy of treating lung adenocarcinomas carrying a KRAS mutation with a combination of a lipophilic bisphosphonate, an analog of zoledronate, with rapamycin. Bisphosphonates are a class of drugs widely used for treating osteoporosis and for preventing bone metastasis of certain cancers (40). Mechanistically, they tightly bind to bone mineral and inhibit FPPS in osteoclasts. This results in impaired protein prenylation and function, inducing cell death of osteoclasts. However, this strong binding to bone mineral (24) – a desirable feature for a drug to treat bone resorption diseases – makes them less suited to treat solid tumors. Lipophilic bisphosphonates, on the other hand, do not bind to bone mineral and, in addition to inhibiting FPPS, they also target GGPPS. Compared with the most potent commercial bisphosphonate drug, zoledronate, the lipophilic bisphosphonate BPH-1222 is more efficient in killing tumor cells with KRAS mutations both in vitro and in vivo. This enhanced efficacy is due to a combination of factors: good FPPS and GGPPS inhibition, as well as greatly enhanced lipophilicity (clogP of 0.25 for BPH-1222 versus −3.9 for zoledronate), and BPH-1222 has good pharmacokinetic properties (41, 42).
Targeting protein prenylation (including farnesylation and geranylgeranylation) has been pursued for more than 20 years, ever since researchers first found that RAS requires post-translational prenylation for its malignancy-transforming activity. FTIs and GGTIs were developed early on to kill tumor cells in vitro, however, little success has been achieved using these compounds in animals. More interestingly, responses to these inhibitors do not always correspond to RAS mutation status. This observation strongly suggests the existence of other targets (27). Here, we show that KRAS prenylation and activity is largely inhibited by lipophilic bisphosphonate treatment and suggest that this is one of the major mechanisms of action of this class of compounds. However, other small G-proteins, such as RAL, RHO, RAC and CDC42, require exclusively geranylgeranylation, and all of these proteins have been shown to be involved in RAS induced transformation in a context-dependent manner (15, 43–45). Indeed, results from this as well as other studies all support the idea that suppression of protein geranylgeranylation is critical for bisphosphonate mediated cytotoxicity. Based on this observation, dual-target (FPPS+GGPPS) inhibitors should be particularly potent in inhibiting tumor cell growth. They might also be particularly toxic in vivo, so their use in combination therapies might be of more interest. For examples, it has been reported that high dose treatment with dual prenyltransferase inhibitor (DPI, 1000–2000 mg/kg) led to substantial lethality (11). However, in our experiments we haven’t observed any severe toxicity in animals even after treatments up to 6 weeks.
KRAS tumors cells have been shown to rely on autophagy for providing metabolic intermediates and clearing excess reactive oxygen species (ROS)(32, 33). In vitro, the additive effects of blocking protein prenylation and autophagy with chloroquine resulted in very potent tumor cell killing. However, in animals, the pro-proliferative effect induced by p62 accumulation and NF-κB activation became dominant and the combination was ineffective. In sharp contrast, the combination of a lipophilic bisphosphonate with rapamycin was far more effective because rapamycin not only facilitated autophagy, but also inhibited the mTOR pathway that is critical for the tumor cell survival. Thus, the combination of a lipophilic bisphosphonate plus rapamycin offers a promising therapeutic lead for treating KRAS-related lung cancers.
Materials and Methods
Study design
The objective of this study was to develop combination therapies of lipophilic bisphosphonates, analogs of zoledronate, in mouse lung cancer models. 30 bisphosphonates were screened for activity against their putative targets, farnesyl diphosphate synthase (FPPS) and geranylgeranyl diphosphate synthase (GGPPS), as well as in vitro against a panel of tumor cell lines. The most effective compounds were then tested in vivo in mouse lung cancer models. In monotherapy the bisphosphonates had only moderate activity. We then sought to determine if efficacy could be improved by using combination therapies with chloroquine or rapamycin in vivo, in both syngeneic transplantation (subcutaneous and orthotopic) and lentiviral models. Tumor size was monitored 1–2 times every week during the treatment, by either palpation or in vivo luciferase imaging. At least 5 mice were used for each group (either control or treatment). All the treatment experiments have been repeated at least twice independently. Mice used for control and treatment were randomly grouped from a pool of model mice.
Chemical reagents
Lipophilic bisphosphonates were synthesized as described before (17, 46). Protein farnesyltransferase and geranylgeranyltransferase inhibitors (FTI-277 and GGTI-298) were purchased from Calbiochem. MEK inhibitor U0126 was purchased from Cell Signaling. Farnesol (FOH) and geranylgeraniol (GGOH), ascorbic acid, Trolox, Lipoic acid, Morin hydrate and chloroquine diphosphate salt were purchased from Sigma. Simvastatin was purchased from Tokyo Chemical Industry Co. Rapamycin was purchased from Alfa Aesar.
Stable cell lines and cell survival assay
Mouse lung cancer cell lines (6#, L2 and M3L2) were derived from primary tumors of LSL-KrasG12D/+ mice infected with CA2Cre-shp53 lentiviral vector (37). L2 and M3L2 cells were derived from mouse tumors with pure C57B and pure FVB background respectively, so that they form syngeneic grafts in matched recipient mice. To monitor NF-κB activation in tumor grafts, L2 cells were stably infected with 5xκB-luci lentiviral vector. Human cancer cell lines (A549, A427, Panc-1 and MiaPaCa2) were purchased from ATCC. MEFs were prepared from mouse embryo with matched genetic background and immortalized by shRNAs against p53 and Rb1. Cell survival after drug treatment was measured using Cell Proliferation Reagent (Wst-1) from Roche or MTT Cell Proliferation Assay Kit (30-1010K) from ATCC. Dose-response curves and corresponding IC50s were fitted using GraphPad Prism.
Lentiviral vector mediated mouse lung cancer model and syngeneic graft model
LSL-KRASG12D/+ Rosa26luc/luc mice were used for the lentiviral vector (CA2Cre-shp53) mediated lung cancer model as described before (37). Tumor size was monitored by in vivo luciferase imaging system (IVIS 100) from Caliper Lifesciences. Syngeneic graft experiments were done by either subcutaneously transplanting 106 L2 tumor cells in the flank region of C57B mice, or tail vein injection of 2×105 M3L2 cells into FVB mice. Subcutaneous tumor size was measured every 7 days after the transplantation. For single drug treatment, animals were given the drug every day. For combination therapies, animals were given BPH-1222 on days 1, 3, 5 and the other drug on days 2, 4, 6, alternately. BPH-1222 (2 mg/kg) and chloroquine (60 mg/kg) were diluted in PBS while rapamycin (2.5 mg/kg) in 75% DMSO and 25% PBS. All drugs were given i.p. in a volume of 100 μl. All mice studies were carried out according to the protocols that were approved by the Institutional Animal Care and Use Committee of Salk Institute.
Cellular fractionation
106 Cells treated with drugs for 48 h were washed with PBS and resuspended in 0.5 ml 0.1M TrisHCl (pH 7.5) with protease inhibitors for 15 min. Supernatant from 10,000 g 30 min centrifugation was collected as cytosolic fraction. The pellet was resuspended in 0.5 ml lysis buffer containing 1% Triton X-100 for 20 min and centrifuged. The supernatant was collected as membrane-bound fraction.
Histology, immunofluorescence staining, and immunoblotting analysis
Mouse lung tumor and syngeneic graft samples were fixed with 10% formalin, paraffin-embedded and sectioned for haematoxylin and eosin staining (H&E) and immunofluorescence staining. Elite ABC system (Vector labs) was applied where staining signal was weak. Immunoblotting analysis was performed according to standard protocols. For detecting Hdj2, Rap1A and Kras prenylation with mobility shift assay, proteins were separated with 15-cm 8% and 13% SDS-PAGE, respectively. Antibodies were purchased from Millipore (SPC, 1:2000), Cell Signaling (phos-ERK, total ERK, phos-AKT, total AKT, phos-MEK, phos-c-RAF, Cleaved Caspase-3, CHOP, BiP, phos-PERK, LC3I/II, phos-p70 S6K, phos-4E-BP1, all 1:1000), Abgent (p62, clone 2C11, 1:2000), Vector Labs (Ki-67, 1:500), and Santa Cruz Biotechnology (KRAS, HRAS, RAP1A, HDJ2, ACTIN, all 1:1000).
Quantitative RT-PCR
Total RNA isolated from the treated cells was reverse transcribed using Superscript III system (Invitrogen) with random primers. Quantitative PCR was performed in triplicate using 7900HT Fast Real-Time PCR system with SYBR green method (Applied Biosystems). Results were analyzed for the relative expression of mRNAs normalized against GAPDH and cyclophilin. A list of primers used for PCR is in table S3.
KRAS MALDI-mass spectrum analysis
Purified protein samples were spotted onto a MALDI target using sinapinic acid as the matrix. Spectra (averages of 200 laser shots) were obtained on an Applied Biosystems Voyager DE-STR instrument in linear mode with delayed extraction. The accelerating voltage was set to 25,000V and the laser repletion rate was 20 Hz. Masses were corrected using human ACTIVIN-A (MH+ave = 25934.8) as an external standard.
FPPS and GGPPS inhibition assay and crystallization of FPPS with BPH-1222
Human FPPS and GGPPS proteins were prepared and used for enzymatic inhibition assay as described before (17). Crystallization of FPPS with BPH-1222 was carried out as reported (46). Diffraction data was collected at the Life Sciences Collaborative Access Team (LS-CAT) beamline 21ID-Gat Argonne National Laboratory. Data was collected at 100K with wavelength of 0.97857Å. Data was processed using HKL3000 and refined by using CCP4 and Coot (47–49). Crystallographic figures were drawn by using PyMOL (http://www.pymol.org). Data collection and refinement statistics are shown in table S2. After refinement, the Ramachandran statistics showed that the percentages of the most favorable and additional allowed regions are 97.6% and 2.4%, respectively.
Pharmacokinetics test
Pharmacokinetic studies were performed using 3 female SD rats (230–240 g body weight). Plasma concentrations were measured at 1min, 5 min, 15min, 30min, 1h, 2h, 4h, 8h, 24h, and 48h following a single i.v. injection of BPH-1222 at 5 mg/kg. 0.3 ml blood was taken each time. Data were analyzed using DAS2.0 software.
Statistical analysis
Statistical analyses were performed with GraphPad Prism Software. Statistical significance of the differences of tumor number and Ki-67, Cleaved Caspase-3 positive staining was evaluated using Student’s unpaired two-tailed t-test. The Kaplan-Meier curves were analyzed by Log-rank test. P values less than 0.05 were considered statistically significant.
Supplementary Material
Fig. S1. Structural diagram of bisphosphonates.
Fig. S2. FPPS/GGPPS activity and cell growth inhibited by bisphosphonates.
Fig. S3. ER stress induced by blocking protein prenylation.
Fig. S4. Comparison of drug combinations.
Fig. S5. Combination therapy in KRAS-shp53 lentiviral model.
Fig. S6. Working model of combination therapy.
Table S1. FPPS/GGPPS activity and cell growth inhibited by bisphosphonates.
Table S2. Data collection and refinement statistics.
Table S3. Summary of protein prenylation inhibited by various compounds.
Table S4. Summary of KRAS molecular weight (M.W.) measured by Mass spectrum.
Table S5. PCR primer sets used in the study.
Acknowledgments
We thank Drs. R. Alvarez Rodriguez, J. Moscat, M.T. Diaz-Meco and J.P. Noel for helpful discussions, and Liping Li for technical assistance.
Funding: I.M.V is an American Cancer Society Professor of Molecular Biology, and holds the Irwin and Joan Jacobs Chair in Exemplary Life Science. This work was supported in part by grants from the NIH (R01-AI048034 from the National Institute of Allergy and Infectious Diseases to I.M.V. and R01CA158191 from the National Cancer Institute to E.O.), Ipsen Biomeasure, the H.N. and Frances C. Berger Foundation, and the Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002 (I.V.M.), a Harriet A. Harlin Professorship (E.O.), and the 1000 Young Talents Program and start-up funds from Tsinghua University (Y.Z.). Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Y. Xie was supported by the post-doctoral fellowship from Tsinghua-Peking Center for Life Sciences.
Footnotes
Competing interests: The authors declare that they have no competing financial interests.
Data and materials availability: Coordinates and structure factors have been deposited in the PDB with access codes 4N1Z for human FPPS in complex with BPH-1222.
Author Contributions: Y.X. and Y.Z. designed, and Y.X. performed the experiments with the help of Y.Z., Y.L., Y. Xie, W.Z., F.G., X.Z. (compound synthesis, enzymatic assay, crystallization analysis, pharmacokinetics test and cell survival assay), S.S. (animal works), N.Y. (quantitative RT-PCR), and W.F., W.L. (Mass spec). I.M.V., E.O. and Y.Z. provided funding and supervised the project. Y.X., E.O. and I.M.V. wrote the paper.
References
- 1.Ansari J, Palmer DH, Rea DW, Hussain SA. Role of tyrosine kinase inhibitors in lung cancer. Anticancer Agents Med Chem. 2009;9:569–575. doi: 10.2174/187152009788451879. [DOI] [PubMed] [Google Scholar]
- 2.West H, Lilenbaum R, Harpole D, Wozniak A, Sequist L. Molecular analysis-based treatment strategies for the management of non-small cell lung cancer. J Thorac Oncol. 2009;4:S1029–1039. doi: 10.1097/JTO.0b013e3181b27170. quiz S1041–1022. [DOI] [PubMed] [Google Scholar]
- 3.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, Bekele BN, Herbst RS, Wistuba KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 5.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 6.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 8.Chan DA, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat Rev Drug Discov. 2011;10:351–364. doi: 10.1038/nrd3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casey PJ, Solski PA, Der CJ, Buss JE. p21ras is modified by a farnesyl isoprenoid. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:8323–8327. doi: 10.1073/pnas.86.21.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson JH, Cochrane CG, Bourne JR, Solski PA, Buss JE, Der CJ. Farnesol modification of Kirsten-ras exon 4B protein is essential for transformation. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:3042–3046. doi: 10.1073/pnas.87.8.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lobell RB, Omer CA, Abrams MT, Bhimnathwala HG, Brucker MJ, Buser CA, Davide JP, deSolms SJ, Dinsmore CJ, Ellis-Hutchings MS, Kral AM, Liu D, Lumma WC, Machotka SV, Rands E, Williams TM, Graham SL, Hartman GD, Oliff AI, Heimbrook DC, Kohl NE. Evaluation of farnesyl:protein transferase and geranylgeranyl:protein transferase inhibitor combinations in preclinical models. Cancer Res. 2001;61:8758–8768. [PubMed] [Google Scholar]
- 12.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, Waldmann H. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Sjogren AK, Karlsson C, Ibrahim MX, Andersson KM, Olofsson FJ, Wahlstrom AM, Dalin M, Yu H, Chen Z, Yang SH, Young SG, Bergo MO. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6471–6476. doi: 10.1073/pnas.0908396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sjogren AK, Andersson KM, Liu M, Cutts BA, Karlsson C, Wahlstrom AM, Dalin M, Weinbaum C, Casey PJ, Tarkowski A, Swolin B, Young SG, Bergo MO. GGTase-I deficiency reduces tumor formation and improves survival in mice with K-RAS-induced lung cancer. J Clin Invest. 2007;117:1294–1304. doi: 10.1172/JCI30868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sousa SF, Fernandes PA, Ramos MJ. Farnesyltransferase inhibitors: a detailed chemical view on an elusive biological problem. Curr Med Chem. 2008;15:1478–1492. doi: 10.2174/092986708784638825. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Cao R, Yin F, Hudock MP, Guo RT, Krysiak K, Mukherjee S, Gao YG, Robinson H, Song Y, No JH, Bergan K, Leon A, Cass L, Goddard A, Chang TK, Lin FY, Van Beek E, Papapoulos S, Wang AH, Kubo T, Ochi M, Mukkamala D, Oldfield E. Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation. J Am Chem Soc. 2009;131:5153–5162. doi: 10.1021/ja808285e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verfaillie T, Salazar M, Velasco G, Agostinis P. Linking ER Stress to Autophagy: Potential Implications for Cancer Therapy. Int J Cell Biol. 2010;2010:930509. doi: 10.1155/2010/930509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wasko BM, Dudakovic A, Hohl RJ. Bisphosphonates induce autophagy by depleting geranylgeranyl diphosphate. J Pharmacol Exp Ther. 2011;337:540–546. doi: 10.1124/jpet.110.175521. [DOI] [PubMed] [Google Scholar]
- 20.Wojtkowiak JW, Sane KM, Kleinman M, Sloane BF, Reiners JJ, Jr, Mattingly RR. Aborted autophagy and nonapoptotic death induced by farnesyl transferase inhibitor and lovastatin. J Pharmacol Exp Ther. 2011;337:65–74. doi: 10.1124/jpet.110.174573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Artz JD, Wernimont AK, Dunford JE, Schapira M, Dong A, Zhao Y, Lew J, Russell RG, Ebetino FH, Oppermann U, Hui R. Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites. The Journal of biological chemistry. 2011;286:3315–3322. doi: 10.1074/jbc.M109.027235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goffinet M, Thoulouzan M, Pradines A, Lajoie-Mazenc I, Weinbaum C, Faye JC, Seronie-Vivien S. Zoledronic acid treatment impairs protein geranyl-geranylation for biological effects in prostatic cells. BMC Cancer. 2006;6:60. doi: 10.1186/1471-2407-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee S, Huang C, Guerra F, Wang K, Oldfield E. Thermodynamics of bisphosphonates binding to human bone: a two-site model. J Am Chem Soc. 2009;131:8374–8375. doi: 10.1021/ja902895p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haklai R, Weisz MG, Elad G, Paz A, Marciano D, Egozi Y, Ben-Baruch G, Kloog Y. Dislodgment and accelerated degradation of Ras. Biochemistry. 1998;37:1306–1314. doi: 10.1021/bi972032d. [DOI] [PubMed] [Google Scholar]
- 27.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berndt N, Sebti SM. Measurement of protein farnesylation and geranylgeranylation in vitro, in cultured cells and in biopsies, and the effects of prenyl transferase inhibitors. Nat Protoc. 2011;6:1775–1791. doi: 10.1038/nprot.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morck C, Olsen L, Kurth C, Persson A, Storm NJ, Svensson E, Jansson JO, Hellqvist M, Enejder A, Faergeman NJ, Pilon M. Statins inhibit protein lipidation and induce the unfolded protein response in the non-sterol producing nematode Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18285–18290. doi: 10.1073/pnas.0907117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen JC, Wu ML, Huang KC, Lin WW. HMG-CoA reductase inhibitors activate the unfolded protein response and induce cytoprotective GRP78 expression. Cardiovasc Res. 2008;80:138–150. doi: 10.1093/cvr/cvn160. [DOI] [PubMed] [Google Scholar]
- 32.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 35.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, Adams PD, Anderson KI, Gottlieb E, Sansom OJ, Ryan KM. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 36.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Xia Y, Yeddula N, Leblanc M, Ke E, Zhang Y, Oldfield E, Shaw RJ, Verma IM. Reduced cell proliferation by IKK2 depletion in a mouse lung-cancer model. Nat Cell Biol. 2012;14:257–265. doi: 10.1038/ncb2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatzivassiliou G, Haling JR, Chen H, Song K, Price S, Heald R, Hewitt JF, Zak M, Peck A, Orr C, Merchant M, Hoeflich KP, Chan J, Luoh SM, Anderson DJ, Ludlam MJ, Wiesmann C, Ultsch M, Friedman LS, Malek S, Belvin M. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature. 2013;501:232–236. doi: 10.1038/nature12441. [DOI] [PubMed] [Google Scholar]
- 39.Moriceau G, Ory B, Mitrofan L, Riganti C, Blanchard F, Brion R, Charrier C, Battaglia S, Pilet P, Denis MG, Shultz LD, Monkkonen J, Redini F, Heymann D. Zoledronic acid potentiates mTOR inhibition and abolishes the resistance of osteosarcoma cells to RAD001 (Everolimus): pivotal role of the prenylation process. Cancer Res. 2010;70:10329–10339. doi: 10.1158/0008-5472.CAN-10-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gnant M, Mlineritsch B, Schippinger W, Luschin-Ebengreuth G, Postlberger S, Menzel C, Jakesz R, Seifert M, Hubalek M, Bjelic-Radisic V, Samonigg H, Tausch C, Eidtmann H, Steger G, Kwasny W, Dubsky P, Fridrik M, Fitzal F, Stierer M, Rucklinger E, Greil R, Marth C. Endocrine therapy plus zoledronic acid in premenopausal breast cancer. N Engl J Med. 2009;360:679–691. doi: 10.1056/NEJMoa0806285. [DOI] [PubMed] [Google Scholar]
- 41.Lin JH. Bisphosphonates: a review of their pharmacokinetic properties. Bone. 1996;18:75–85. doi: 10.1016/8756-3282(95)00445-9. [DOI] [PubMed] [Google Scholar]
- 42.Weiss HM, Pfaar U, Schweitzer A, Wiegand H, Skerjanec A, Schran H. Biodistribution and plasma protein binding of zoledronic acid. Drug Metab Dispos. 2008;36:2043–2049. doi: 10.1124/dmd.108.021071. [DOI] [PubMed] [Google Scholar]
- 43.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M, Jr, Yeaman C, Camonis JH, Zhao Y, White MA. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 44.Falsetti SC, Wang DA, Peng H, Carrico D, Cox AD, Der CJ, Hamilton AD, Sebti SM. Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol Cell Biol. 2007;27:8003–8014. doi: 10.1128/MCB.00057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kissil JL, Walmsley MJ, Hanlon L, Haigis KM, Bender Kim CF, Sweet-Cordero A, Eckman MS, Tuveson DA, Capobianco AJ, Tybulewicz VL, Jacks T. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res. 2007;67:8089–8094. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Zhu W, Liu YL, Wang H, Wang K, Li K, No JH, Ayong L, Gulati A, Pang R, Freitas-Junior L, Morita CT, Old-Field E. Chemo-Immunotherapeutic Anti-Malarials Targeting Isoprenoid Biosynthesis. ACS Med Chem Lett. 2013;4:423–427. doi: 10.1021/ml4000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta crystallographica Section D, Biological crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 49.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Structural diagram of bisphosphonates.
Fig. S2. FPPS/GGPPS activity and cell growth inhibited by bisphosphonates.
Fig. S3. ER stress induced by blocking protein prenylation.
Fig. S4. Comparison of drug combinations.
Fig. S5. Combination therapy in KRAS-shp53 lentiviral model.
Fig. S6. Working model of combination therapy.
Table S1. FPPS/GGPPS activity and cell growth inhibited by bisphosphonates.
Table S2. Data collection and refinement statistics.
Table S3. Summary of protein prenylation inhibited by various compounds.
Table S4. Summary of KRAS molecular weight (M.W.) measured by Mass spectrum.
Table S5. PCR primer sets used in the study.